Abstract
NF-κB, a family of transcription factors regulating diverse biological processes including immune responses, is activated by canonical and noncanonical pathways based on degradation of IκBα and processing of the IκB-like protein p100, respectively. Although p100 responds to noncanonical NF-κB stimuli for processing, it does not undergo degradation, but rather becomes accumulated, along with canonical NF-κB activation. We show here that the stability of p100 is tightly controlled by a deubiquitinase, Otub1. Otub1 deficiency not only promotes signal-induced p100 processing and noncanonical NF-κB activation but also causes steady-state p100 degradation, leading to aberrant NF-κB activation in the canonical pathway. B-cell-conditional deletion of Otub1 results in B-cell hyperplasia, antibody hyper-production, and lupus-like autoimmunity. Otub1-deficient B cells display aberrantly activated phenotypes and overproduce the cytokine IL-6, contributing to autoimmunity induction. Thus, maintenance of p100 stability by Otub1 serves as an unusual mechanism of NF-κB regulation that prevents autoimmunity.
Similar content being viewed by others
Introduction
Nuclear factor κB (NF-κB) is a family of transcription factors regulating different immune functions as well as cell growth and survival.1,2 Mammalian NF-κB includes five members, NF-κB1 p50, NF-κB2 p52, RelA, RelB, and c-Rel. These NF-κB proteins are normally sequestered in the cytoplasm as inactive complexes via interaction with inhibitory proteins, IκBs.1 Activation of NF-κB members is mediated by two major signaling pathways, the canonical and noncanonical pathways.3 Canonical pathway is based on degradation of the typical NF-κB inhibitor IκBα, causing nuclear translocation of p50, RelA, and c-Rel, which are often referred to as canonical NF-κB members. IκBα degradation is mediated through its phosphorylation by the IκB kinase (IKK) complex that is activated by numerous inducers, such as ligands of toll-like receptors (TLRs), proinflammatory cytokines, and antigens.4 The noncanonical NF-κB pathway, which mainly activates RelB and p52, is based on the proteolytic processing of the NF-κB2 precursor protein, p100.3 p100 contains a C-terminal portion that is structurally homologous to IκBα and functions as an IκB-like molecule. The processing of p100 is stimulated by a specific subset of TNF receptor (TNFR) superfamily members, such as lymphotoxin beta receptor (LTβR), B-cell-activating factor belonging to the TNF family (BAFF) receptor (BAFFR), and CD40. These receptors activate the NF-κB-inducing kinase (NIK) and its downstream kinase IKKα, which mediates phosphorylation of p100 to induce ubiquitin-dependent p100 processing (selective degradation of IκB-like portion), leading to generation of mature p52 and nuclear translocation of p52 and RelB.5,6,7
Although the primary role of p100 is to regulate noncanonical NF-κB activation, strong evidence suggests that p100 also contributes to the cytoplasmic sequestration of canonical NF-κB proteins. In addition to RelB and p52, several other NF-κB members are physically associated with p100 and sequestered in the cytoplasm by this atypical IκB.8,9,10 However, in contrast to IκBα, p100 does not undergo degradation or processing in cells stimulated with canonical NF-κB inducers, and the cytoplasmic p100/NF-κB complexes remain unchanged even when IκBα is completely degraded.8,10 Moreover, since the p100-encoding gene, Nfkb2, is a target of canonical NF-κB, p100 expression is strongly induced by canonical NF-κB stimuli.8,10,11 Thus, p100 is accumulated along canonical NF-κB activation and may play a role in preventing abnormal NF-κB activation.10 However, the molecular mechanism regulating the remarkable stability of p100 and the pathological consequences of deregulated p100 processing or degradation have not been defined.
In the present study, we performed an unbiased screening study to identify p100-binding proteins. We identified the deubiquitinase (DUB) Otub1 as a p100-interacting protein that inhibits the ubiquitination and degradation of p100. Otub1 deficiency causes spontaneous ubiquitination and degradation of p100 in both B cells and mouse embryonic fibroblasts (MEFs). Moreover, the Otub1 deficiency also prevented accumulation of p100 along with canonical NF-κB activation, leading to aberrant activation of both canonical and noncanonical NF-κB members. We provide genetic evidence that Otub1-mediated NF-κB regulation is crucial for maintaining normal B-cell homeostasis and function to prevent the development of autoimmunity.
Results
Otub1 is a p100-interacting protein
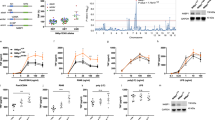
To characterize regulators of p100, we screened for p100-binding proteins using the proximity-dependent biotin identification (BioID) method.12 This approach involves expression of a bait protein fused with a promiscuous biotin ligase mutant (BirA R118R), which catalyzes biotinylation of proteins interacting with, or adjacent to, the bait when cells are provided with excess biotin. We constructed a retroviral vector encoding either the Myc-BioID control or Myc-BioID–p100 fusion protein and stably infected a B cell line, M12, frequently used for the study of noncanonical NF-κB signaling13,14 (Supplementary Fig. S1a, b). Streptavidin bead pull-down assays using biotin-treated cells revealed a large number of biotinylated protein bands in the Myc-BioID–p100-expressing cells in addition to several strong background bands present in both the control and Myc-BioID–p100-expressing cells (Supplementary Fig. S1b). Mass spectrometry analysis revealed a number of potential p100-binding proteins, including several NF-κB family members, NF-κB1, RelA, RelB, and c-Rel, known to interact with p100 (Supplementary Fig. S1c and Table S1). In addition, a DUB, Otub1, was identified as a major p100-interacting protein. Co-immunoprecipitation (coIP) assays confirmed the interaction between Otub1 and p100 (Fig. 1a). In contrast, Otub1 did not interact with IκBα and only weakly interacted with the NF-κB1 precursor protein p105, suggesting selectivity to p100 (Fig. 1a). When stably expressed in MEFs, Otub1 was also complexed with endogenous p100 (Fig. 1b). Furthermore, coIP assays also detected interaction of endogenous Otub1 and p100 in B cells (Fig. 1c).
Identification of Otub1 as a p100-binding protein. a Co-immunoprecipitation (coIP) analysis of p100/Otub1 binding (upper) and direct immunoblot analysis for monitoring protein expression (lower) using whole-cell lysates of HEK293 cells transfected with Flag-tagged Otub1 along with either an empty vector or expression vectors encoding the indicated IκB proteins. b Whole-cell lysates were prepared from Otub1–/– MEFs reconstituted with either an empty vector or Flag-Otub1 and subjected to coIP to detect Otub1 binding to endogenous p100 (upper) or direct immunoblot assays (lower). c CoIP analysis of endogenous Otub1/p100 interaction in whole-cell lysates of untreated wild-type splenic B cells. IgG pulldown was used as a negative control for p100 IP. d, e A schematic picture depicting the domains of Otub1 and Otub2 (d) and coIP analysis of p100/Otub1 interaction using whole-cell lysates of HEK293 cells transfected with the indicated expression vectors (e). f, g A schematic picture depicting the domains of p100 and its mutants (f) and coIP analysis of Otub1 interaction with p100 mutants using whole-cell lysates of HEK293 cells transfected with the indicated expression vectors (g, upper). Cell lysates were also subjected to direct immunoblotting to monitor expression of p100 mutants (p100 mut) and Otub1 (lower)
Otub1 contains an OTU catalytic domain as well as an N-terminal ubiquitin-association (UBA) domain15 (Fig. 1d). Deletion of the UBA domain of Otub1 abolished its interaction with p100 (Fig. 1e). Notably, this N-terminal domain is missing in the Otub1 homolog, Otub2 (Fig. 1d). Consistently, Otub2 failed to interact with p100 (Fig. 1e). Domain mapping studies of p100 revealed that a C-terminal truncation mutant of p100, 1–665, lacking a part of the ankyrin repeat domain, the death domain, and the phosphorylation site, retained the ability to bind Otub1 (Fig. 1f, g). However, further deletion of the rest part of the ankyrin repeat domain generated a mutant, 1–454, defective in Otub1 interaction (Fig. 1g). Consistently, internal deletion of ankyrin repeats 1–3 or ankyrin repeats 5–7 of p100 partially inhibited its interaction with Otub1, whereas a combined deletion of ankyrin repeats 1–3 and 5–7 of p100 completely abolished its ability to bind Otub1 (Fig. 1g). Thus, Otub1 physically interacts with p100 via the N-terminal UBA domain of Otub1 and the ankyrin repeat domain of p100.
Otub1 controls both basal and signal-induced p100 processing and noncanonical NF-κB activation
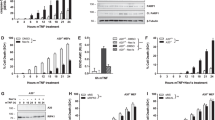
The processing of p100 is a central step in noncanonical NF-κB activation, which is stimulated by ligands of specific TNFR superfamily members, such as BAFF in B cells.3 To define the signaling function of Otub1, we generated Otub1 B-cell-conditional knockout (Otub1-BKO) mice (Supplementary Fig. S2) and examined the effect of Otub1 deficiency on p100 processing and NF-κB nuclear translocation in primary B cells. Consistent with their in vivo exposure to the noncanonical NF-κB inducer BAFF, freshly isolated wild-type B cells displayed constitutive processing of p100 for generation of p52, part of which was translocated to the nucleus (Fig. 2a). Remarkably, in the Otub1-deficient B cells, p100 level was drastically reduced, coupled with elevated nuclear p52 (Fig. 2a). Notably, the nuclear levels of other NF-κB members, particularly RelB and c-Rel, were also elevated in the Otub1-deficient B cells (Fig. 2a). This phenotype was in line with the association of p100 with not only RelB and p52 but also canonical NF-κB members.8,9,10 The enhanced p100 processing in Otub1-deficient cells was not due to reduced expression of TRAF2 and TRAF3, negative regulators of the noncanonical NF-κB pathway (Fig. 2a, b).
Otub1 regulates basal and signal-induced p100 processing and noncanonical NF-κB activation. a, b Immunoblot analysis of the indicated proteins using cytoplasmic (Cyt Ext) and nuclear (Nucl Ext) extracts of freshly isolated splenic B cells from wild-type (WT) and Otub1-BKO (BKO) mice (a) or Otub1+/+ (+/+) and Otub1–/– (–/–) primary MEFs (b). The p100 and p52 bands were quantified by densitometry based on three independent experiments and presented as the ratio of cytoplasmic p100 to nuclear p52 (a) or fold of cytoplasmic p100 and nuclear p52 with wild-type value being set to 1 (b). c Immunoblot analysis of p100 and p52 in whole-cell lysates of wild-type B cells or MEFs. The p100 and p52 bands were quantified by densitometry based on three independent experiments and presented as the ratio of p100 to p52 (right panel). d, e Immunoblot analysis of the indicated proteins using cytoplasmic or nuclear extracts of BAFF-stimulated splenic B cells (d) or anti-LTβR-stimulated primary MEFs (e). f Whole-cell lysates were prepared from MG132-treated (2 h) Otub1+/+ and Otub1–/– MEFs and subjected to p100 IP followed by detecting ubiquitinated p100 using antibodies detecting total ubiquitin chains (left) or K48-linked ubiquitin chains (right). The level of p100 and Actin in lysates was monitored by immunoblot. g p100 ubiquitination (upper panel) and direct immunoblot (lower panels) assays using whole-cell lysates of MG132-treated (for 2 h) HEK293 cells transfected with the indicated HA-tagged ubiquitin mutants (expressing only the indicated lysine with all other lysines mutated to arginines), either in the presence (+) or absence (–) of Flag-Otub1 and p100. h, i p100 ubiquitination (upper panel) and direct immunoblot (lower panels) assays using whole-cell lysates of MG132-treated (for 2 h) HEK293 cells transfected as indicated. D/C/H stands for Otub1 D88A/C91S/H265A mutant
Since B cells are exposed to noncanonical NF-κB inducers in vivo, we further examined the role of Otub1 in regulating the basal level of p100 processing using primary MEFs. As seen in B cells, Otub1 deficiency caused a profound reduction in the level of cytoplasmic p100 and an increase in the level of nuclear p52 and RelB at steady state (Fig. 2b). The reduced p100 protein level in Otub1-deficient cells was not due to suppression of Nfkb2 gene expression, as suggested by the comparable level of Nfkb2 mRNA in the wild-type and Otub1-KO MEFs (Supplementary Fig. S3a). Furthermore, the Otub1 deficiency also did not affect p100 translation, since similar levels of p100 were detected in wild-type and Otub1-BKO B cells when treated with a proteasome inhibitor, MG132 (Supplementary Fig. S3b). This result further suggested enhanced p100 degradation or processing in Otub1-deficient cells. In further support of this conclusion, p100 was more rapidly lost in Otub1-BKO B cells than in wild-type B cells upon incubation with a protein synthesis inhibitor, cycloheximide (Supplementary Fig. S3c).
To further examine whether Otub1 controlled the degradation or processing (generation of p52) of p100 under steady state, we performed immunoblot assays using whole-cell lysates. In splenic B cells, the Otub1 deficiency reduced the level of p100 and concomitantly increased the level of p52, suggesting upregulated processing (Fig. 2c). Otub1 deficiency in MEFs also markedly reduced the level of p100. Interestingly, however, the Otub1 deficiency did not cause accumulation of p52, although it significantly reduced the p100:p52 ratio (Fig. 2c). This result suggests that the basal loss of p100 in Otub1-KO MEFs was mainly via degradation (no generation of p52). Consistent with the role of p100 in p52/RelB regulation, the severe loss of p100 in Otub1-deficient MEFs was associated with nuclear translocation of p52 and RelB (Fig. 2b). The finding that Otub1 deficiency resulted in predominantly p100 degradation in MEFs, while p100 processing in B cells, indicated a cell type difference. However, it is important to note that B cells are constantly stimulated by BAFF in vivo, whereas the MEF culture lacks noncanonical NF-κB stimuli, which could also be a factor contributing to the differences in basal p100 processing versus degradation. Notwithstanding, these results demonstrate a crucial role for Otub1 in controlling the processing and degradation of p100.
To examine the role of Otub1 in regulating signal-induced noncanonical NF-κB activation, we stimulated the wild-type and Otub1-deficient B cells with BAFF, a well-defined noncanonical NF-κB inducer in B cells. As expected, stimulation of B cells with BAFF induced p100 processing and nuclear translocation of p52 and RelB in wild-type B cells (Fig. 2d). The Otub1-BKO B cells displayed enhanced p100 processing under both basal and BAFF-induced conditions, coupled with elevated nuclear expression of p52 and RelB (Fig. 2d). Moreover, the Otub1 deficiency strongly promoted anti-LTβR-induced p100 processing and p52/RelB nuclear expression in MEFs (Fig. 2e). These results were not due to alterations in upstream signaling events, since the Otub1 deficiency did not promote signal-induced degradation of TRAF3 and c-IAP1, phosphorylation of IKK and p100, or the accumulation of NIK (Supplementary Fig. S3d–g). Furthermore, the Otub1 deficiency also had no effect on the expression level of p105 and p50 (Supplementary Fig. S3d). Together, these results suggest that Otub1 specifically controls the processing and degradation of p100 under both steady-state and signal-induced conditions.
Otub1 inhibits K48 ubiquitination of p100
The processing of p100 is triggered by its ubiquitination.7,16 Since Otub1 is a DUB, we tested whether it controlled the fate of p100 through inhibiting p100 ubiquitination. Incubation of the Otub1-KO MEFs with a proteasome inhibitor, MG132, largely rescued the steady level of p100 (Fig. 2f). Furthermore, the MG132-mediated inhibition of p100 degradation in Otub1-KO MEFs resulted in marked accumulation of ubiquitinated p100 (Fig. 2f). Using a chain-specific ubiquitin antibody, we further demonstrated that p100 was conjugated with lysine (K) 48-linked polyubiquitin chains (Fig. 2f). Moreover, enhanced p100 ubiquitination was also detected in Otub1-deficient B cells under both untreated and BAFF-stimulated conditions (Supplementary Fig. S3h). These results suggested an important role for Otub1 in preventing p100 ubiquitination and degradation.
To further examine which type of p100 ubiquitin chains was regulated by Otub1, we employed a transfection model using ubiquitin mutants carrying a single K residue with the rest K residues being mutated to arginines. We found that p100 was conjugated with K11-, K33-, and K48-linked polyubiquitin chains, but not K29- and K63-linked polyubiquitin chain (Fig. 2g). Expression of Otub1 efficiently inhibited the K48 ubiquitination of p100 but did not affect the K11 or K33 type of p100 ubiquitination (Fig. 2g). Recent in vitro studies suggest that Otub1 could function both as a typical DUB to directly cleave ubiquitin chains from target proteins and as an atypical DUB that inhibits target protein ubiquitination through inhibiting the function of ubiquitin-conjugating enzymes (E2s). The latter function of Otub1 does not need its catalytic activity but requires its intact OTU domain.17,18 The DUB catalytic center of Otub1 has a conserved triad, composed of aspartate 88 (D88), cysteine 91 (C91), and histidine 265 (H265), and mutation of any of these residues abolishes the DUB catalytic activity in an in vitro assay.15 We found that mutation of neither C91 nor H265 interfered with the ability of Otub1 to inhibit p100 ubiquitination, although mutation of D88 or all of these key residues (D88A/C91S/H265A) abolished this function of Otub1 (Fig. 2h). Taken together with the previous in vitro study,15 this result indicates that the catalytic activity of Otub1 may not be essential for inhibiting p100 ubiquitination, implicating an atypical mechanism of Otub1 function, although further studies are required to confirm this possibility.
The noncanonical NIK induces p100 phosphorylation, which in turn promotes its ubiquitination by the SCFβTrCP for processing.7,16 In addition, the E3 ligase SCFFbw7 has been shown to mediate GSK3-induced ubiquitin-dependent degradation of p100 in cancer cells.19,20,21 As seen with its function in regulating the basal ubiquitination of p100, Otub1 efficiently inhibited the ubiquitination induced by both NIK and GSK3/Fbw7 (Fig. 2i). Furthermore, this function of Otub1 was abolished by its D88A mutation (Fig. 2i). These results indicate that Otub1 may control the ubiquitination of p100 stimulated by both NIK/βTrCP and other regulators, such as GSK3/Fbw7.
Otub1 deficiency causes aberrant NF-κB activation by canonical inducers
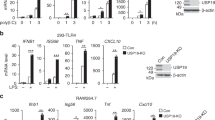
Although p100 predominantly regulates the nuclear translocation of p52 and RelB in response to noncanonical NF-κB stimuli, p100 also binds and sequesters canonical NF-κB members.8,9,10 Unlike IκBα, p100 does not undergo degradation in response to canonical NF-κB inducers.8 In fact, since Nfkb2 is a target gene of NF-κB, p100 is often accumulated along with canonical NF-κB activation.8,10,11 To date, it is unknown how the remarkable stability of p100 is regulated. Our identification of Otub1 as a DUB of p100 prompted us to examine whether Otub1 also controlled the stability of p100 along with canonical NF-κB activation. Stimulation of wild-type B cells with several TLR ligands, Pam3CSK4, LPS, and R848, resulted in marked accumulation of p100 (Fig. 3a). Interestingly, Otub1 was also induced along with the induction of p100, at both the protein and mRNA levels (Fig. 3a, b). CoIP assays revealed that the accumulated p100 and Otub1 were physically associated (Fig. 3c). In contrast to the canonical NF-κB inducers, the noncanonical NF-κB inducer BAFF did not induce Otub1 expression or influence Otub1/p100 interaction (Supplementary Fig. S4). These findings suggested a role for Otub1 in maintaining the stability of the IκB-like protein p100 during canonical NF-κB activation. Indeed, Otub1 deletion largely prevented the accumulation of p100 protein in B cells stimulated with the TLR ligand Pam3CSK4 and in MEFs stimulated with TNF-α (Fig. 3d, e). This phenotype was not due to inhibition of Nfkb2 gene expression, since the wild-type and Otub1-BKO cells had comparable levels of Nfkb2 mRNA upon stimulation with Pam3CSK4 (Fig. 3f). Consistent with the role of Otub1 in preventing ubiquitin-dependent p100 degradation, inhibition of proteasome in Otub1-deficient B cells rescued the p100 level and resulted in accumulation of ubiquitinated p100 (Fig. 3g). Therefore, Otub1-mediated p100 deubiquitination plays a crucial role in mediating p100 stability and accumulation along with canonical NF-κB activation.
Otub1 maintains p100 stability and prevents abnormal canonical NF-κB activation. a Immunoblot analysis of the indicated proteins using whole-cell lysates of wild-type splenic B cells stimulated with the indicated TLR ligands. b qRT-PCR analysis of Otub1 mRNA in Pam3CSK4-stimulated wild-type splenic B cells. c Whole-cell lysates of Pam3CSK4-stimulated (24 h) wild-type splenic B cells were subjected to IP using either a control IgG or anti-p100 followed by detecting the co-precipitated Otub1 and p100 by immunoblot (upper). Otub1 and p100 expression level was also analyzed by immunoblot (lower). d, e Immunoblot analysis of the indicated proteins using cytoplasmic or nuclear extracts of wild-type (WT) and Otub1-BKO splenic B cells (d) or Otub1+/+ and Otub1–/– MEFs (e) stimulated as indicated. f qRT-PCR analysis of Nfkb2 mRNA in wild-type splenic B cells stimulated with Pam3CSK4 for the indicated time periods. The value given for the amount of PCR product present in wild-type at time 0 was set to 1. g p100 ubiquitination (upper) and immunoblot (lower) analysis using whole-cell lysates prepared from wild-type (WT) and Otub1-BKO splenic B cells that were stimulated with Pam3CSK4 for 8 h and then incubated with MG132 for 2 h. h–j Immunoblot analysis of the indicated proteins using cytoplasmic or nuclear extracts prepared from wild-type or Otub1-BKO splenic B cells stimulated as indicated
The findings described above raised the question of whether Otub1 played a role in regulating NF-κB activation stimulated by canonical inducers. While Pam3CSK4 only weakly induced the nuclear translocation of p52 and RelB in wild-type B cells, induction of these noncanonical NF-κB members was profoundly enhanced in Otub1-BKO B cells (Fig. 3d). In addition, the nuclear translocation of the canonical NF-κB members, RelA and c-Rel, was also enhanced in the Otub1-deficient B cells, particularly during the early times of stimulation (Fig. 3d). Similarly, the canonical NF-κB inducer TNF-α barely activated p52 in wild-type MEFs, but the Otub1 deficiency enabled potent p52 activation by TNF-α (Fig. 3e). The Otub1 deficiency also promoted TNF-α-stimulated activation of other NF-κB members (Fig. 3e). The effect of Otub1 deficiency on NF-κB activation was not due to enhanced IκBα degradation (Fig. 3d and data not shown), emphasizing the role of Otub1 in regulating the ‘super-IκB’ function of p100.
To further examine the role of Otub1 in regulating NF-κB activation, we stimulated the B cells for shorter time periods with Pam3CSK4 and two other canonical NF-κB inducers, including the BCR stimulator anti-IgM and the TLR7/8 ligand R848. Under such conditions, it was once again clear that Otub1 deficiency caused drastic loss of p100 without altering the pattern of IκBα degradation (Fig. 3h–j). Otub1 deletion also did not influence the fate of another IκB-like molecule, p105 (Fig. 3h–j). Importantly, the Otub1 deficiency greatly promoted the nuclear translocation of both canonical and noncanonical NF-κB members (Fig. 3h–j). Similar results were obtained with TNF-α-stimulated Otub1-deficient MEFs (Supplementary Fig. S5a). Furthermore, the Otub1 deficiency also promoted TNF-α-stimulated expression of a number of genes known to be regulated by NF-κB (Supplementary Fig. S5b). Thus, Otub1 deficiency causes aberrant NF-κB activation by canonical stimuli.
B-cell-specific Otub1 deletion causes B-cell hyperplasia and aberrant antibody production
The peripheral survival and maturation of B cells rely on coordinated action of canonical and noncanonical NF-κB pathways stimulated by BCR and BAFFR signals, respectively.22 Because of the unique function of Otub1 in regulating both canonical and noncanonical NF-κB activation, we examined the effect of B-cell-conditional deletion of Otub1 on B-cell homeostasis and antibody production. Compared with wild-type control mice, the Otub1-BKO mice had a significant increase in B-cell frequencies and decrease in T-cell frequencies in the spleen, inguinal lymph nodes, and peritoneal cavity (Fig. 4a, b). This phenotype was due to increased absolute numbers of B cells (Fig. 4b). The Otub1-BKO mice had even more striking increase in the absolute B-cell numbers in the spleen and inguinal lymph nodes (iLNs) due to higher total cell numbers (Fig. 4b). Within the peritoneal cavity, there are two types of B cells, B1 and B2.23 Interestingly, the Otub1 deficiency increased the frequency of B2 cells and decreased the frequency of B1 cells (Fig. 4c, d), which was apparently due to a drastic increase in the absolute number of B2 cells (Fig. 4d). This finding was consistent with the crucial role of noncanonical NF-κB in mediating the development of B2 but not B1 cells.24
Otub1-BKO mice display B-cell hyperplasia and aberrant antibody production. a, b Flow cytometric analyses of B220+ or CD19+ B cells and TCRβ+ T cells in the spleen (SP), inguinal lymph nodes (iLN), and peritoneal cavity (PerC) of wild-type (WT) and Otub1-BKO mice (6–8 months old). Data are presented as a representative plot (a) and summary graphs of mean ± SD values based on multiple mice (b, each circle represents a mouse). c, d Flow cytometric analyses of B1 (CD19+CD23–) and B2 (CD19+CD23+) cells in the peritoneal cavity of wild-type (WT) and Otub1-BKO mice (6–8 months old). e, f Flow cytometric analyses of immature (Imm, B220+CD93+) and mature (B220+CD93−) B cells as well as follicular (FO, B220+CD21intCD23+) and marginal zone (MZ, B220+CD21hiCD23−) B cells in the spleen of wild-type and Otub1-BKO mice (6–8 months old). g A representative image of spleen from wild-type and Otub1-BKO mice (6–8 months old). h, i ELISA of the indicated serum antibody isotypes (A) or fecal IgA based on multiple wild-type and Otub1-BKO mice (6–8 months old; each circle represents a mouse). j, k Flow cytometric analysis of IgA+ B cells in the spleen (SP) and Peyer’s patches (PP) of wild-type and Otub1-BKO mice (6–8 months old). Data are representative of three independent experiments. P values are determined by two-tailed unpaired t-test. *P < 0.05, **P < 0.01, ***P < 0.001
Compared to the wild-type control mice, the Otub1-BKO mice had a significant increase in the frequency and absolute number of mature B cells and concomitantly reduced immature B cells (Fig. 4e, f). Within the mature B-cell population, marginal zone (MZ) B cells were profoundly increased in the Otub1-BKO mice (Fig. 4e, f). Although Otub1-BKO mice had a reduced frequency of follicular B cells, this phenotype was apparently due to the expansion of MZ cell population (Fig. 4e, f). Indeed, the absolute number of follicular B cells was not reduced, but even higher, in the Otub1-BKO mice than in the wild-type control mice (Fig. 4f). These results were in line with the important role of canonical and noncanonical NF-κB members in promoting B-cell maturation and MZ B-cell differentiation.22
Consistent with their B-cell hyperplasia, the Otub1-BKO mice had splenomegaly (Fig. 4g). Furthermore, compared to the wild-type control mice, the Otub1-BKO mice also had significantly increased concentration of serum antibodies, including IgA, IgM, IgG2a, and IgG2b, as well as elevated fecal IgA concentration (Fig. 4h, i). These mutant animals displayed increased frequencies of IgA+ B cells in both the spleen and the Peyer’s patches (Fig. 4j, k). These results demonstrated a central role for Otub1 in controlling B-cell homeostasis and antibody production under steady conditions.
Otub1 regulates NF-κB activation and B-cell functions via both NIK-dependent and -independent mechanisms
NIK is a kinase that is required for noncanonical NF-κB activation and B-cell survival, and NIK deficiency is associated with severe reduction in the number of peripheral B cells, particularly mature B cells and MZ B cells.25,26,27 Because of the role of Otub1 in controlling noncanonical NF-κB activation, we tested whether the B-cell abnormalities in Otub1-BKO mice were dependent on NIK. For these studies, we generated mutant mice carrying B-cell-specific deficiencies in both NIK and Otub1 (NIK/Otub1-dBKO). Consistent with prior studies,26,27 NIK B-cell-conditional KO (NIK-BKO) mice had reduced peripheral B-cell numbers compared to wild-type mice (Fig. 5a, b). Interestingly, the NIK/Otub1-dBKO mice displayed a rescue of the B-cell defect (Fig. 5a, b). The Otub1 deletion in NIK-BKO mice also greatly enhanced the frequency and absolute number of peritoneal B2 cells (Fig. 5c, d), a B-cell subset known to be regulated by NIK.24 Similarly, the Otub1 deletion rescued the defect of NIK-BKO mice in B-cell maturation, as shown by the significantly increased splenic mature B cells, particularly the MZ B cells, in the NIK/Otub1-dBKO mice (Fig. 5e, f). These results suggest that Otub1 deletion could largely rescue the defect of NIK-BKO mice in B-cell development and maturation.
Otub1 regulates both NIK-dependent and NIK-independent NF-κB activation and B-cell development. a, b Flow cytometric analyses of B220+ B cells and TCRβ+ T cells in the spleen (SP), inguinal lymph nodes (iLN), and peritoneal cavity (PerC) of WT, NIK-BKO, or NIK/Otub1 double BKO (dBKO) mice (8–10 weeks old). Data are presented as a representative plot (a) and summary graphs of mean ± SD values based on multiple mice (b, each circle represents a mouse). c, d Flow cytometric analyses of B1 (CD19+CD23−) and B2 (CD19+CD23+) cells in the peritoneal cavity of WT, NIK-BKO, or NIK/Otub1-dBKO mice (8–10 weeks old), presented as a representative plot (c) and summary graphs (d). e, f Flow cytometric analyses of immature (Imm, B220+CD93+) and mature (B220+CD93−) B cells as well as follicular (FO, B220+CD21intCD23+) and marginal zone (MZ, B220+CD21hiCD23−) B cells in the spleen of WT, NIK-BKO, or NIK/Otub1-dBKO mice (8–10 weeks old), presented as a representative plot (e) and summary graphs (f). g–i Immunoblot analysis of the indicated proteins using cytoplasmic or nuclear extracts of BAFF- or anti-IgM-stimulated splenic B cells (g, i) or anti-LTβR-stimulated MEFs (h) prepared from the indicated mice. Data are representative of three independent experiments. P values are determined by two-tailed unpaired t-test. *P < 0.05, **P < 0.01, ***P < 0.001
NIK-induced p100 processing involves phosphorylation of p100 at two C-terminal residues, S866 and S870.7 A mutant mouse strain, Nfkb2Lym1, carries a point mutation in the Nfkb2 gene and produces a p100 mutant, Lym1, lacking the C-terminal phosphorylation site and defective in NIK-induced processing.28 Like NIK-deficient mice, the heterozygous Nfkb2Lym1/+ mice display severe defect in peripheral B cells.28 We found that Otub1 deletion in B cells could significantly increase the total peripheral B cells, mature B cells, and follicular and MZ B cells in the Nfkb2Lym1/+ mice (Supplementary Fig. S6a–d). These results emphasize the role of Otub1 in regulating B cells via both NIK-dependent and NIK-independent mechanisms.
Because of the findings described above, we examined whether Otub1 could control NF-κB activation via both NIK-dependent and NIK-independent mechanisms. Compared to wild-type B cells, the Otub1-BKO B cells displayed both p100 basal processing and enhanced BAFF-induced p100 processing, coupled with p52/RelB nuclear translocation (Fig. 5g). Furthermore, consistent with the essential role of NIK in noncanonical NF-κB activation,7 B cells derived from NIK-BKO mice were defective in BAFF-induced p100 processing and p52/RelB nuclear expression (Fig. 5g). Remarkably, compared with NIK-BKO B cells, the NIK/Otub1-dBKO B cells still displayed p100 basal reduction, although these dBKO cells were defective in BAFF-induced p100 processing and noncanonical NF-κB activation (Fig. 5g). Similar to the results obtained with B cells, Otub1 deficiency in MEFs caused p100 basal processing and noncanonical NF-κB activation both in the presence (Otub1–/– cells) and absence (NIK–/–Otub1–/– cells) of NIK (Fig. 5h). To examine whether Otub1 also regulated canonical NF-κB activation in an NIK-independent manner, we stimulated the NIK-BKO and NIK/Otub1-dBKO B cells with the canonical NF-κB inducer anti-IgM. Remarkably, even under the NIK-deficient conditions, Otub1 deletion in B cells strongly promoted the activation of NF-κB members by anti-IgM (Fig. 5i). Similar results were obtained with B cells derived from the Otub1-BKO/Nfkb2lym1/+ mice (Supplementary Fig. S6e, f). Thus, in addition to regulating NIK-dependent noncanonical NF-κB signaling, Otub1 prevents NIK-independent p100 degradation and NF-κB activation.
Otub1 deletion in B cells promotes autoimmunity
B cells play an important role in the pathogenesis of systemic autoimmunity, particularly systemic lupus erythematosus (SLE).29 Because of the B-cell hyperplasia and antibody hyper-production phenotypes of Otub1-BKO mice, we examined whether the Otub1 deficiency in B cells promoted autoimmunity. Interestingly, at the age of 10 months, the Otub1-BKO mice had significantly higher concentration of serum autoantibodies against double-stranded DNA (dsDNA) and nuclear antigen (Fig. 6a), a typical feature of SLE.30 The 10-month-old mice, although not younger mice (8 weeks of age), also had antibody deposition to kidney glomeruli (Fig. 6b), another phenotype often associated with SLE (Fig. 6a). These results suggest that Otub1 deficiency in B cells causes spontaneous development of autoimmunity with features of SLE.
Otub1-BKO mice develop lupus-like autoimmunity. a ELISA of basal concentrations of autoantibodies reacting against double-stranded DNA (anti-dsDNA) or nuclear antigen (ANA) in the serum of unimmunized wild-type or Otub1-BKO mice (10 months old). b Immunofluorescent microscopy of IgG deposits in kidney sections from young (8 weeks) or older (10 months) unimmunized wild-type and Otub1-BKO mice. Sections were stained with DAPI (blue) and Alexa Fluor 488 anti-mouse IgG (green). Scale bar, 100 μm. Quantification of the intensity of signal was performed by ImageJ software. Data are presented as a representative plot (left panel) and summary graph (right panel). c A representative spleen image (left) and a summary graph of spleen weight (right, each circle represents a mouse) of wild-type and Otub1-BKO mice that were either not treated (NT) or injected (i.p.) with CD4 T cells from BM12 mice. d, e Flow cytometric analysis of Fas+GL-7+ germinal center (GC) B cells and B220−CD138+ plasma cells in wild-type and Otub1-BKO mice that were either not treated (NT) or injected (i.p.) with CD4 T cells from BM12 mice for the indicated time periods. Data are presented as a representative plot (left) and summary graph based on multiple recipient mice (right). f ELISA of autoantibodies reacting against double-stranded DNA (anti-dsDNA) and nuclear antigen (ANA) in serum from wild-type and Otub1-BKO mice injected (i.p.) with CD4 T cells from BM12 mice for the indicated time periods. g Immunofluorescent microscopy of IgG deposits in kidney sections from wild-type and Otub1-BKO mice injected (i.p.) with CD4 T cells from BM12 mice for the indicated time periods. Sections were stained with DAPI (blue) and FITC anti-mouse IgG (green). Scale bar, 100 μm. Quantification of the fluorescent signal was performed by ImageJ software. Data are presented as a representative plot (left panel) and summary graph (right panel). Data are representative of three independent experiments. P values are determined by two-tailed unpaired t-test. *P < 0.05, **P < 0.01, ***P < 0.001
To further examine the role of B-cell-specific Otub1 in regulating autoimmunity, we employed an experimental model of SLE based on adoptive transfer of T cells from BM12 mice to C57BL/6 mice.31 The BM12 mice are identical to C57BL/6 mice except for three amino acid substitutions in MHCII, which cause alloactivation of donor CD4 T cells by antigen-presenting cells of the C57BL/6 recipient mice. The activated donor CD4 T cells function as helper T cells for activation of self-reactive recipient B cells, resulting in production of autoantibodies and development of SLE-like autoimmune symptoms.31 The transfer of BM12 T cells to wild-type recipient mice resulted in enlargement of the spleen, indicative of autoimmune responses (Fig. 6c). This phenotype was much more striking for the Otub1-BKO recipient mice (Fig. 6c). The Otub1-BKO recipient mice transferred with BM12 CD4 T cells also had a significantly higher frequency of germinal center B cells and plasma cells than the wild-type recipient mice (Fig. 6d, e). Moreover, compared with the wild-type recipient mice, the Otub1-BKO recipient mice displayed profoundly higher titer of serum autoantibodies reacting against dsDNA and nuclear antigen at different time points of BM12 T-cell transfer (Fig. 6f). Furthermore, the Otub1-BKO recipient mice had considerably higher levels of kidney glomerular deposition with IgG (Fig. 6g). Collectively, these results suggest that Otub1 functions as a B-cell-specific regulator of autoimmune responses in mice. Currently, there have been no reports that link Otub1 to human autoimmune diseases. However, genome-wide association studies indicate the association of Otub1 polymorphism with IgA glomerulonephritis (GWAS Central).
Otub1-deficient B cells display activated phenotype and aberrantly produce IL-6
The development of lupus-like autoimmune disease in Otub1-BKO mice could be partially contributed by the aberrant B-cell maturation and hyperplasia. However, since the Otub1-deficient B cells had deregulated activation of both canonical and noncanonical NF-κBs, we surmised that the Otub1-deficient B cells might also undergo abnormal activation, contributing to the overt autoimmunity phenotype. Indeed, the Otub1-deficient B cells had markedly increased the expression of several surface molecules, MHCII, CD86, CD23, CD21, and ICOSL (Fig. 7a), known to be associated with B-cell activation.32,33,34 Furthermore, Otub1-deficient B cells also displayed hyper-expression of CD23 and CD86 in the BM12 SLE model (Fig. 7b).
Otub1 deficiency causes aberrant B-cell activation. a Flow cytometric analysis of the indicated activation markers on gated B220+ B cells from unimmunized wild-type or Otub1-BKO mice (8–10 weeks old), presented as representative histograms (upper) and summary graph of median fluorescence intensity (MFI) (lower). b Flow cytometric analysis of CD23 and CD86 on gated B220+ B cells from wild-type or Otub1-BKO mice injected (i.p.) with BM12 CD4 T cells for the indicated time periods. Data are presented as representative histograms (left) and summary graph of median fluorescence intensity (MFI) (right). c ELISA of IL-6 in serum from 10-month-old untreated wild-type and Otub1-BKO mice. d ELISA of IL-6 in the serum of 8-week-old wild-type and Otub1-BKO mice injected with BM12 CD4 T cells for the indicated time periods. e, f ELISA (e) and qRT-PCR (f) analysis of IL-6 expression in wild-type or Otub1-BKO splenic B cells that were either not treated (–) or stimulated (+) with the indicated inducers. g ELISA of autoantibodies reacting against nuclear antigen (ANA) or double-stranded DNA (anti-dsDNA) in the serum of wild-type and Otub1-BKO mice treated for the indicated time periods with BM12 CD4 T cells along with repeated injection (every 3 days) with either anti-IL-6 or a rat IgG1 isotype control. h, i Flow cytometric analysis of the frequency of Fas+GL-7+ germinal center (GC) B cells (h) and B220−CD138+ plasma cells (i) in wild-type and Otub1-BKO mice treated for 3 weeks with BM12 CD4 T cells along with repeated injection (every 3 days) with either anti-IL-6 or a rat IgG1 isotype control. Data are representative of two independent experiments. P values are determined by two-tailed unpaired t-test. *P < 0.05, **P < 0.01, ***P < 0.001
To further examine the effect of Otub1 deficiency on B-cell activation, we measured serum cytokines in wild-type and Otub1-BKO mice. Interestingly, the Otub1-BKO mice had markedly higher concentration of serum IL-6 than wild-type control mice at the age of 10 months, when spontaneous autoimmunity occurred (Fig. 7c). Increased serum IL-6 concentration was also detected in Otub1-BKO mice along with autoimmunity induction in BM12 lupus model (Fig. 7d). In line with these findings, the Otub1-BKO B cells displayed IL-6 hyper-expression upon in vitro activation by several canonical NF-κB inducers, including Pam3CSK4, CpG, and R848 (Fig. 7e). The IL-6 expression was further elevated upon costimulation with the noncanonical NF-κB inducer CD40L (Fig. 7e, f). Although Il-6 is known as a target gene of canonical NF-κB, the role of noncanonical NF-κB in IL-6 induction has been elusive. To address this issue, we employed B cells isolated from the Nfkb2Lym1/+ mice expressing a processing-defective p100 mutant.28 Stimulation of wild-type B cells with the canonical NF-κB inducer CpG together with the noncanonical NF-κB inducer BAFF or CD40L led to synergistic induction of Il-6 mRNA and protein (Supplementary Fig. S7a, b). However, the Nfkb2Lym1/+ B cells were defective in such synergistic induction of IL-6 expression (Supplementary Fig. S7a, b). In line with these results, induction of IL-6 in wild-type B cells was potently inhibited by a selective inhibitor of NIK (Supplementary Fig. S7c, d). These results suggest that Otub1 controls IL-6 induction through regulating canonical and noncanonical NF-κB activation.
B-cell-derived cytokines, particularly IL-6, have been implicated in the pathogenesis of autoimmune diseases, including SLE.35,36 To directly examine the role of IL-6 in regulating autoimmunity in the Otub1-BKO mice, we performed IL-6-blocking studies using an IL-6-neutralizing antibody. As expected, the Otub1-BKO mice produced much higher amounts of ANA and anti-dsDNA autoantibodies than the wild-type control mice upon treatment with BM12 CD4 T cells (Fig. 7g). Importantly, injection with an IL-6-neutralizing antibody efficiently inhibited the autoantibody production, erasing the differences between the Otub1-BKO and wild-type control recipient mice (Fig. 7g). Consistently, injection of anti-IL-6 also greatly reduced the frequency of germinal center B cells and plasma cells and eliminated the hyper-production of these B cells in Otub1-BKO recipient mice (Fig. 7h, i). Collectively, these results suggest that Otub1 controls the homeostasis and activation of B cells, thereby preventing the development of autoimmune disorders.
Discussion
The nfkb2 gene product p100 functions as the precursor protein of p52 as well as an IκB-like NF-κB inhibitor. Under normal conditions, p100 is remarkably stable and does not respond to canonical NF-κB stimuli for degradation or undergo constitutive processing. In response to selective members of the TNFR superfamily, p100 undergoes processing in a NIK-dependent manner to generate p52 and mediate nuclear translocation of sequestered NF-κB members, particularly p52 and RelB. How the stability of p100 is regulated has been enigmatic. Results presented in the present study identified Otub1 as an essential factor that maintains the stability of p100. Our data suggest that Otub1-mediated p100 stabilization is crucial for controlling the activation of both canonical and noncanonical NF-κBs and preventing the development of autoimmune symptoms.
Using the BioID protein–protein interaction screening approach, we identified Otub1 as a DUB physically interacting with p100. The Otub1/p100 interaction required the ankyrin repeats of p100 and the N-terminal UBA domain of Otub1. This molecular interaction appeared to be highly specific, since Otub1 displayed weak or no activity to interact with two other IκB molecules, p105 and IκBα, which also contain ankyrin repeats. The requirement of UBA domain of Otub1 for p100 binding suggests the possibility that p100 ubiquitin chains may facilitate the Otub1/p100 interaction. This possibility was also suggested by the constitutive ubiquitination of p100 occurring prominently in Otub1-deficient cells. It is likely that p100 undergoes dynamic ubiquitination under steady conditions, but the ubiquitinated p100 is rapidly bound by Otub1 and deubiquitinated to prevent its constitutive processing or degradation. We found that Otub1 could efficiently inhibit p100 conjugation with K48-linked ubiquitin chains. Consistently, Otub1 deficiency not only promoted NIK-induced p100 processing but also caused spontaneous p100 degradation under steady conditions. The E3 ligase that mediates steady-state ubiquitination and degradation of p100 is unclear. NIK-stimulated p100 processing involves recruitment of the SCFbTrCP E3 ligase to p100.16 However, since Otub1 deficiency also caused substantial loss of p100 under NIK-deficient conditions, the spontaneous p100 ubiquitination in Otub1-deficient cells may involve additional E3s. Another E3, SCFFbw7, has been shown to mediate p100 degradation in cancer cells in a GSK3b-dependent manner.19,20,21 We found that Otub1 inhibits K48 ubiquitination of p100 induced by both NIK and GSK3b/Fbw7, although it remains to be examined whether the basal degradation of p100 in Otub1-deficient cells is dependent on Fbw7. Future studies will also map the ubiquitination sites of p100 regulated by Otub1.
Our data suggest that Otub1 not only regulates p100 processing and noncanonical NF-κB activation but also plays a role in controlling canonical NF-κB activation. In addition to its function in regulating noncanonical NF-κB signaling, p100 interacts with and inhibits canonical NF-κB members.8,28 Since p100 does not respond to canonical NF-κB inducers for degradation, a large proportion of NF-κB proteins remain sequestered in the cytoplasm by p100 even when IκBα is completely degraded.8 In fact, canonical NF-κB activation is often associated with accumulation of p100 due to the induction of Nfkb2 gene expression. We found that Otub1 deficiency in both B cells and MEFs caused drastic reduction in the steady level of p100, which was apparently due to p100 degradation since the p100 loss could be blocked upon incubation with a proteasome inhibitor. The Otub1 deficiency also abolished signal-induced p100 accumulation along with canonical NF-κB activation, which was coupled with aberrant activation of various NF-κB members. Thus, the Otub1-mediated p100 stabilization plays a crucial role in preventing abnormal activation of both canonical and noncanonical NF-κB members.
Previous in vitro studies suggest that Otub1 and Otub2 regulate virus-induced type I interferon expression through inhibiting the ubiquitination of TRAF3 and TRAF6 in the VISA (or MAVS) signaling complex.37 Otub1 knockdown does not alter the expression level of TRAF3 and TRAF6, suggesting that Otub1 may control the nondegradative ubiquitination of these TRAF members in the interferon pathway.37 We also found in our present study that Otub1 deficiency in B cells or MEFs had no obvious effect on the fate of TRAF3 under both steady-state and BAFF-induced conditions. Our data further suggest that Otub1 does not regulate upstream signaling events of the noncanonical NF-κB pathway but functions as a direct regulator of p100. A previous study suggests that Otub1 deubiquitinates c-IAP1 within the TREAK receptor signaling complex.38 Otub1 knockdown inhibits TREAK-stimulated activation of canonical NF-κB and MAPK pathways and promotes TREAK-stimulated cell death.38 However, this function of Otub1 appears to be specific for the TREAK signaling pathway.38 Indeed, we found that Otub1 deficiency in B cells or MEFs greatly promoted, rather than inhibited, NF-κB activation by different canonical NF-κB inducers, including TLR ligands and TNFα. This phenotype was not due to hyper-activation of canonical NF-κB signaling events, such as IKK activation and IκBα degradation, but rather caused by reduction of the IκB-like molecule p100. As such, our study uncovered a novel signaling function of Otub1.
We obtained genetic evidence that Otub1 was essential for maintaining normal B-cell homeostasis and preventing aberrant B-cell activation. The Otub1-BKO mice had B-cell hyperplasia with a particular expansion of mature B cells and MZ B cells, and the Otub1-deficient B cells displayed a hyper-activated phenotype characterized by upregulated expression of MHCII, CD86, CD23 and several other surface markers associated with B-cell activation. The deregulated B-cell expansion and activation may play an important role in the aberrant production of antibodies and autoimmune phenotypes of Otub1-BKO mice. In addition, we found that the cytokine IL-6 was aberrantly induced in Otub1-deficient B cells and accumulated in the serum of Otub1-BKO mice. B cell-derived IL-6 has been implicated in the pathogenesis of autoimmune diseases, including SLE, although the mechanism underlying Il-6 gene expression in B cells has been poorly defined.35,36 Indeed, our antibody neutralization studies revealed that deregulated IL-6 production contributed to different autoimmune symptoms in the Otub1-BKO mice, including upregulation of germinal center B cells and plasma cells and overproduction of autoantibodies. Although Il-6 is known as a target gene of canonical NF-κB, the role of noncanonical NF-κB in Il-6 gene regulation has been elusive. Our present study demonstrated a crucial role of noncanonical NF-κB in Il-6 gene induction. We found that canonical and noncanonical NF-κB inducers synergistically stimulated the expression of IL-6 in B cells, which was severely attenuated by genetic disruption or pharmacological inhibition of noncanonical NF-κB activation. Thus, the hyper-production of IL-6 in Otub1-BKO mice was in line with the aberrant activation of canonical and noncanonical NF-κB in Otub1-deficient B cells.
Materials and methods
Mice
Otub1-targeted mice (Otub1tm1a(EUCOMM)Hmgu; KO first allele) were generated by the International Mouse Phenotyping Consortium (IMPC) by targeting exons 2 and 3 of the Otub1 gene using a FRT-LoxP vector (Supplementary Fig. S2a). Embryos of the Otub1-targeted mice were obtained from the European Mutant Mouse Archive (EMMA) for producing live mice in the Genetically Engineered Mouse Facility of MD Anderson Cancer Center. The Otub1-targeted mice were crossed with an FLP deleter mice (Rosa26-FLPe; Jackson Laboratory) to generate Otub1-flox mice, which were further crossed with Cd19-Cre and Rosa26-Cre-ER mice to generate Otub1 B-cell-conditional KO (BKO; Otub1f/fCd19-Cre) and tamoxifen-inducible Otub1f/f CreER mice, respectively. In some experiments, Otub1-BKO and Otub1f/f CreER mice were further crossed with NIK-flox (provided by Genentech Inc.) or Nfkb2lym1 (provided by R. Starr, Walter and Eliza Hall Institute of Medical Research) mice for generating double mutant mice. Nfkb2lym1/+ heterozygous mice were used in experiments since they display a strong phenotype in impaired noncanonical NF-κB activation and function.28 BM12 mice were purchased from Jackson Laboratory. Nfkb2lym1/+ mice were on 129 genetic background, and all the other mouse strains were on C57BL/6 background. Genotyping was performed as indicated in Supplementary Table S2. Mice were maintained in specific pathogen-free facility, and all animal experiments were conducted in accordance with protocols approved by the Institutional Animal Care and Use Committee of the University of Texas MD Anderson Cancer Center. Outcomes of animal experiments were collected blindly and recorded based on ear-tag numbers of the experimental mice.
Antibodies and reagents
Antibodies and reagents used in this study are listed in Supplementary Table S3.
Plasmid
Myc-BirA(R118G) cDNA was amplified by PCR from the pcDNA3.1 myc-BioID (Addgene) and inserted into the pCLXSN(GFP) retroviral vector to generate pCLXSN(GFP)-myc-BioID vector. Human NFKB2 cDNA insert was transferred to the pCLXSN(GFP)-myc-BioID vector to generate the pCLXSN(GFP)-Myc-BioID-p100 vector. Expression vectors encoding HA-IκBα, p105, HA-GSK3β, Flag-Fbw7, Flag-HA-Otub1, Flag-HA-Otub2, HA-ubiquitin, and HA-tagged ubiquitin mutants (K11, K29, K33, K48, K63) were purchased from Addgene. p105 constructs were subcloned into pCMV4-HA expression verctor. Flag-Otub1 and Flag-Otub1(C91S) were provided by Shinichiro Nakada (Institute for Advanced Co-Creation Studies, Osaka University). Expression vectors encoding Flag-Otub1(ΔN), Flag-Otub1(D88A), Flag-HA-Otub1(D88A), Flag-Otub1(H265A), and Flag-Otub1(D88A/C91S/H265A) were created by site-directed mutagenesis. Primers for site-directed mutagenesis are listed in Supplementary Table S4. The pCMV4 expression vectors encoding human p100 and its truncation mutants (1–859, 1–753, 1–665, 1–454, ΔAnk1–3, ΔAnk5–7 and ΔAnk1–3/5–7) were as described.7,39 All DNA constructs used were verified by DNA sequencing.
Cell culture and stimulation
The human embryonic kidney 293 cells were cultured in DMEM containing 10% FBS. The cells were seeded in six-well plates and were transfected by the calcium phosphate method. Mouse M12 B cells (M12.4.1) were cultured in RIPM medium containing 10% FBS. To create M12 cells stably expressing Myc-BioID-p100 and Myc-BioID, the M12 B cells were infected with pCLXSN(GFP)-Myc-BioID-p100 or the control pCLXSN(GFP)-Myc-BioID retroviral vectors as previously described.40 To minimize non-specific interactions in the BioID assays, cells expressing low levels of Myc-BioID or Myc-BioID-p100 proteins were isolated by flow cytometric cell sorting based on GFPlow marker. Primary MEFs were cultured in DMEM containing 10% FBS and used within the first five passages for experiments. To produce Otub1+/+ and Otub1–/– MEFs, heterozygous Otub1f/+ CreER mice were bred to obtain Otub1+/+CreER and Otub1f/f CreER embryos from the same pregnant female, and the prepared MEFs were treated for 24 h with 1 μM 4-hydroxytamoxifen (Sigma) and cultured for 5 additional days to induce Otub1 deletion for generating Otub1+/+ and Otub1–/– MEFs. Other mutant and control MEFs were prepared using the same approach. For Otub1 reconstitution, Otub1–/– MEFs were infected with MSCV retroviruses encoding Otub1 or Otub1(D88A) mutant and enriched by puromycin selection. Splenocytes were prepared with nylon mesh followed by lysis of red blood cells. B cells and T cells were purified from the splenocyte samples with anti-B220- or anti-CD4-conjugated magnetic beads (Miltenyl Biotec), respectively.
For gene induction and signaling studies, B cells were stimulated with Pam3CSK4 (1 g/mL), CpG (25 mM), R848 (1 g/mL), LPS (500 ng/mL), anti-IgM (5 g/mL), CD40L (250 ng/mL), or BAFF (50–200 ng/mL), and MEFs were stimulated with anti-LTβR (125–500 ng/mL) or TNF-α (20 ng/mL). For ELISA and qRT-PCR analyses, B cells were stimulated with the indicated agents at a density of 2 × 105 cells per well in 96-well plates (for ELISA) or 2 × 106 cells per well in 24-well plates (for qRT-PCR).
Proximity biotinylation (BioID) and affinity capture of p100 interactors
BioID was performed essentially as described41 with some modifications. Approximately 4 × 107 M12 cells expressing Myc-BioID-p100 or the control protein Myc-BioID were cultured in medium supplemented with 50 μM biotin for 12 h. The cells were lysed in 1 mL of lysis buffer (50 mM Tris pH 7.5, 150 mM NaCl, 5 mM EDTA, 1 mM DTT, 0.5% TX-100, 0.1% SDS, and protease inhibitor), and the cell lysates were applied to an Amicon Ultra-4 10K (Millipore) column and spun for 30–60 min (4,000×g, 4 °C). The collected samples were resuspended in solubilization buffer (50 mM Tris, pH 7.4, 250 mM NaCl, 0.1% SDS, 1% Triton X-100, pH 7.4, 1 mM DTT, and protease inhibitors), sonicated, and centrifuged. The supernatant was incubated with 500 μL of streptavidin-conjugated magnetic beads (MyOne Streptavadin C1; Thermo Fisher Scientific). After several washes, the beads were collected for mass spectrometry analysis of the isolated biotinylated proteins at the Protein Proteomics Facility of Baylor College of Medicine. One-tenth of the sample was reserved for immunoblot analyses.
Immunoblot analysis, immunoprecipitation, and ubiquitination assays
Whole-cell lysates or subcellular extracts were prepared and subjected to immunoblot analysis, immunoprecipitation, and ubiquitination assays as described.42
Lupus-like autoimmune disease induction and analyses
A mouse model of lupus-like disease was induced by adoptive transfer of BM12 CD4 T cells as described.43 Briefly, 7.5 × 106 CD4 T cells, purified from BM12 mice, were intraperitoneally (i.p.) injected into the indicated recipient mice (8–10 weeks of age). Sera were collected at the indicated time points for measuring the titer of autoantibodies reacting against dsDNA (anti-dsDNA) and nuclear antigen (ANA) using an ELISA kit (Alpha Diagnostic). Formalin-fixed frozen mouse kidney sections were stained with Alexa Fluor 488-conjugated goat anti-mouse IgG (Thermo Fisher Scientific) for analysis of antibody deposition using a fluorescent microscope (Zeiss). Fluorescence intensity was quantified by ImageJ. Basal serum levels of Ig isotypes and fecal IgA in untreated mice were also analyzed by ELISA (Southern Biotechnology Associates).
IL-6 neutralization
Mice were administered i.p. with 0.5 mg of an anti-mouse IL-6-neutralizing antibody (Bio X cell) or a rat IgG1 isotype control antibody (Bio X cell), and the administration was subsequently repeated every 3 days at a lower dose (0.25 mg). For the BM12 lupus model, the first administration was given on day 1 of BM12 CD4 T-cell challenge.
Flow cytometry
Cell suspensions were subjected to flow cytometry analyses using an LSRII flow cytometer (BD Biosciences), as described previously.44 The data were analyzed using FlowJo software.
qRT-PCR
RNA preparation and qRT-PCR assays were as described44 using gene-specific primers listed in Supplementary Table S5.
Statistical analysis
Two-tailed unpaired t-tests were performed using Prism software. P values <0.05 were considered significant, and the level of significance was indicated as *P < 0.05, **P < 0.01, and ***P < 0.001.
References
Hayden, M. S. & Ghosh, S. Shared principles in NF-kappaB signaling. Cell 132, 344–362 (2008).
Vallabhapurapu, S. & Karin, M. Regulation and function of NF-kappaB transcription factors in the immune system. Annu. Rev. Immunol. 27, 693–733 (2009).
Sun, S. C. The non-canonical NF-kappaB pathway in immunity and inflammation. Nat. Rev. Immunol. 17, 545–558 (2017).
Hayden, M. S. & Ghosh, S. NF-kappaB, the first quarter-century: remarkable progress and outstanding questions. Genes Dev. 26, 203–234 (2012).
Senftleben, U. et al. Activation of IKKa of a second, evolutionary conserved, NF-kB signaling pathway. Science 293, 1495–1499 (2001).
Sun, S. C. Non-canonical NF-κB signaling pathway. Cell Res. 21, 71–85 (2011).
Xiao, G., Harhaj, E. W. & Sun, S. C. NF-kappaB-inducing kinase regulates the processing of NF-kappaB2 p100. Mol. Cell 7, 401–409 (2001).
Sun, S.-C., Ganchi, P. A., Beraud, C., Ballard, D. W. & Greene, W. C. Autoregulation of the NF-kB transactivator Rel A (p65) by multiple cytoplasmic inhibitors containing ankyrin motifs. Proc. Natl Acad. Sci. USA 91, 1346–1350 (1994).
Basak, S. et al. A fourth IkappaB protein within the NF-kappaB signaling module. Cell 128, 369–381 (2007).
Legarda-Addison, D. & Ting, A. T. Negative regulation of TCR signaling by NF-{kappa}B2/p100. J. Immunol. 178, 7767–7778 (2007).
Liptay, S., Schmid, R. M., Nabel, E. G. & Nabel, G. J. Transcriptional regulation of NF-kappa B2: evidence for kappa B-mediated positive and negative autoregulation. Mol. Cell. Biol. 14, 7693–7703 (1994).
Roux, K. J., Kim, D. I., Raida, M. & Burke, B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell. Biol. 196, 801–810 (2012).
Liao, G., Zhang, M., Harhaj, E. W. & Sun, S. C. Regulation of the NF-kappaB-inducing kinase by tumor necrosis factor receptor-associated factor 3-induced degradation. J. Biol. Chem. 279, 26243–26250 (2004).
Morrison, M. D., Reiley, W., Zhang, M. & SC., S. An atypical tumor necrosis factor (TNF) receptor-associated factor-binding motif of B cell-activating factor belonging to the TNF family (BAFF) receptor mediates induction of the noncanonical NF-kappaB signaling pathway. J. Biol. Chem. 280, 10018–10024 (2005).
Balakirev, M. Y., Tcherniuk, S. O., Jaquinod, M. & Chroboczek, J. Otubains: a new family of cysteine proteases in the ubiquitin pathway. EMBO Rep. 4, 517–522 (2003).
Fong, A. & Sun, S.-C. Genetic evidence for the essential role of beta-transducin repeat-containing protein in the inducible processing of NF-kB2/p100. J. Biol. Chem. 277, 22111–22114 (2002).
Nakada, S. et al. Non-canonical inhibition of DNA damage-dependent ubiquitination by OTUB1. Nature 466, 941–946 (2010).
Wiener, R., Zhang, X., Wang, T. & Wolberger, C. The mechanism of OTUB1-mediated inhibition of ubiquitination. Nature 483, 618–622 (2012).
Arabi, A. et al. Proteomic screen reveals Fbw7 as a modulator of the NF-kappaB pathway. Nat. Commun. 3, 976 (2012).
Busino, L. et al. Fbxw7alpha- and GSK3-mediated degradation of p100 is a pro-survival mechanism in multiple myeloma. Nat. Cell Biol. 14, 375–385 (2012).
Fukushima, H. et al. SCF(Fbw7) modulates the NFkB signaling pathway by targeting NFkB2 for ubiquitination and destruction. Cell Rep. 1, 434–443 (2012).
Kaileh, M. & Sen, R. NF-kappaB function in B lymphocytes. Immunol. Rev. 246, 254–271 (2012).
Hardy, R. R. & Hayakawa, K. Perspectives on fetal derived CD5+ B1 B cells. Eur. J. Immunol. 45, 2978–2984 (2015).
Shinkura, R. et al. Alymphoplasia is caused by a point mutation in the mouse gene encoding Nf-kappa b-inducing kinase. Nat. Genet. 22, 74–77 (1999).
Dejardin, E. The alternative NF-kappaB pathway from biochemistry to biology: pitfalls and promises for future drug development. Biochem. Pharmacol. 72, 1161–1179 (2006).
Brightbill, H. D. et al. Conditional deletion of NF-kappaB-inducing kinase (NIK) in adult mice disrupts mature B cell survival and activation. J. Immunol. 195, 953–964 (2015).
Hahn, M., Macht, A., Waisman, A. & Hovelmeyer, N. NF-kappaB-inducing kinase is essential for B-cell maintenance in mice. Eur. J. Immunol. 46, 732–741 (2016).
Tucker, E. et al. A novel mutation in the Nfkb2 gene generates an NF-kappa B2 “super repressor”. J. Immunol. 179, 7514–7522 (2007).
Liossis, S. C. & Staveri, C. B cell-based treatments in SLE: past experience and current directions. Curr. Rheumatol. Rep. 19, 78 (2017).
Kaul, A. et al. Systemic lupus erythematosus. Nat. Rev. Dis. Primers 2, 16039 (2016).
Eisenberg, R. The chronic graft-versus-host model of systemic autoimmunity. Curr. Dir. Autoimmun. 6, 228–244 (2003).
Bonnefoy, J. Y., Lecoanet-Henchoz, S., Aubry, J. P., Gauchat, J. F. & Graber, P. CD23 and B-cell activation. Curr. Opin. Immunol. 7, 355–359 (1995).
Rickert, R. C. Regulation of B lymphocyte activation by complement C3 and the B cell coreceptor complex. Curr. Opin. Immunol. 17, 237–243 (2005).
Chang, M., Jin, W. & Sun, S. C. Peli1 facilitates TRIF-dependent Toll-like receptor signaling and proinflammatory cytokine production. Nat. Immunol. 10, 1089–1095 (2009).
Barr, T. A. et al. B cell depletion therapy ameliorates autoimmune disease through ablation of IL-6-producing B cells. J. Exp. Med. 209, 1001–1010 (2012).
Arkatkar, T. et al. B cell-derived IL-6 initiates spontaneous germinal center formation during systemic autoimmunity. J. Exp. Med. 214, 3207–3217 (2017).
Li, S. et al. Regulation of virus-triggered signaling by OTUB1- and OTUB2-mediated deubiquitination of TRAF3 and TRAF6. J. Biol. Chem. 285, 4291–4297 (2010).
Goncharov, T. et al. OTUB1 modulates c-IAP1 stability to regulate signalling pathways. EMBO J. 32, 1103–1114 (2013).
Liao, G. & Sun, S. C. Regulation of NF-kappaB2/p100 processing by its nuclear shuttling. Oncogene 22, 4868–4874 (2003).
Rivera-Walsh, I., Cvijic, M. E., Xiao, G. & Sun, S. C. The NF-kappa B signaling pathway is not required for Fas ligand gene induction but mediates protection from activation-induced cell death. J. Biol. Chem. 275, 25222–25230 (2000).
Roux, K. J., Kim, D. I. & Burke, B. BioID: a screen for protein–protein interactions. Curr. Protoc. Protein Sci. 74, Unit 19.23 (2013).
Jin, J. et al. Epigenetic regulation of the expression of Il12 and Il23 and autoimmune inflammation by the deubiquitinase Trabid. Nat. Immunol. 17, 259–268 (2016).
Klarquist, J. & Janssen, E. M. The bm12 inducible model of systemic lupus erythematosus (SLE) in C57BL/6 mice. J. Vis. Exp. 105, e53319 (2015).
Li, Y. et al. Cell intrinsic role of NF-kappaB-inducing kinase in regulating T cell-mediated immune and autoimmune responses. Sci. Rep. 6, 22115 (2016).
Acknowledgements
We thank Genentech Inc. for NIK-flox mice, Walter and Eliza Hall Institute of Medical Research for Nfkb2lym1 mice, and S. Nakada for Otub1 expression vectors. We thank the personnel from the MS Proteomics facility at Baylor College of Medicine for mass spectrometry analysis of the BioID samples. We also thank the personnel from the flow cytometry, DNA analysis, genetically engineered mouse facility, and animal facilities at The MD Anderson Cancer Center for technical assistance. This study was supported by grants from the National Institutes of Health (GM84459, AI64639, and AI057555).
Author information
Authors and Affiliations
Contributions
Y.L. designed and performed the research, prepared the figures, and wrote part of the manuscript; J.-Y.Y., X.X., Z.J., L.Z., J.S., D.L., M.G., X.Z., H.S.L. and X.C. contributed experiments; A.J. performed MS, which was analyzed by S.Y.J. and J.Q.; S.S.W. was involved in the supervision of H.S.L.; and S.-C.S. supervised the work and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Li, Y., Yang, JY., Xie, X. et al. Preventing abnormal NF-κB activation and autoimmunity by Otub1-mediated p100 stabilization. Cell Res 29, 474–485 (2019). https://doi.org/10.1038/s41422-019-0174-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41422-019-0174-3
This article is cited by
-
Deubiquitylating Enzyme OTUB1 Facilitates Neuronal Survival After Intracerebral Hemorrhage Via Inhibiting NF-κB-triggered Apoptotic Cascades
Molecular Neurobiology (2024)
-
Phosphorylation of OTUB1 at Tyr 26 stabilizes the mTORC1 component, Raptor
Cell Death & Differentiation (2023)
-
OTUB1 promotes osteoblastic bone formation through stabilizing FGFR2
Signal Transduction and Targeted Therapy (2023)
-
Expression of TRAF6 in peripheral blood B cells of patients with myasthenia gravis
BMC Neurology (2022)
-
Deubiquitination of MYC by OTUB1 contributes to HK2 mediated glycolysis and breast tumorigenesis
Cell Death & Differentiation (2022)