Abstract
Base editor (BE), containing a cytidine deaminase and catalytically defective Cas9, has been widely used to perform base editing. However, the narrow editing window of BE limits its utility. Here, we developed a new editing technology named as base editor for programming larger C to U (T) scope (BE-PLUS) by fusing 10 copies of GCN4 peptide to nCas9(D10A) for recruiting scFv-APOBEC-UGI-GB1 to the target sites. The new system achieves base editing with a broadened window, resulting in an increased genome-targeting scope. Interestingly, the new system yielded much fewer unwanted indels and non-C-to-T conversions. We also demonstrated its potential use in gene disruption across the whole genome through induction of stop codons (iSTOP). Taken together, the BE-PLUS system offers a new editing tool with increased editing window and enhanced fidelity.
Similar content being viewed by others
Introduction
CRISPR technology is widely used in genome manipulation because of its efficiency and simplicity.1,2 After binding to the targeted sequence, Cas9 introduces DNA double-strand breaks (DSB) that are repaired by either non-homologous end joining (NHEJ) or homology-directed repair (HDR) pathways,3,4 which eventually results in the accumulation of insertions and deletions (indels) at the site of the DSB. To limit DNA damage during editing, CRISPR-based editing strategies that entail directly-targeted nucleotide conversion without DSB formation have been developed.5,6,7,8,9 BE1, the original version of base editor, consists of a catalytically inactive form of Cas9 (dCas9) and rAPOBEC1.6 BE2, the updated version, contains one uracil glycosylase inhibitor (UGI) at N-terminus of dCas9 for reducing uracil N-glycosylase (UNG)-mediated base excision repair (BER).6 BE2 was further optimized to BE3, with dCas9 replaced by the Cas9 nickase nCas9(D10A). BE3 enables direct and irreversible conversions of cytosine (C) to uracil (U) or thymine (T) at positions 4–8 (counting from the distal side of PAM) of the noncomplementary strand of DNA, and induces far fewer unwanted indels or translocations.6 This method has been used to introduce single-nucleotide polymorphisms of interest in cells, plants and animals,6,8,10,11,12,13,14,15,16,17,18,19,20 and enables the manipulation of disease-causing genes in human embryos.21,22,23 Recently, BE3 has also been used to disrupt genes by inducing stop codons (iSTOP).17,18,19
Although showing great potential, BE3 can introduce unwanted indels6,8 and unexpected non-C-to-T conversions.5,7,10,19,24 In addition, the narrow editing window limits the sites that are targetable by BE3. Several strategies have been successfully applied to enhance the fidelity of BE3. For example, using a high-fidelity nCas9 (HF-nCas9) significantly decreased the off-target mutagenesis by BE3.12 Co-expressing free uracil DNA glycosylase inhibitor (UGI) with BE3 enhanced the efficiency and fidelity of base editing.25 A new-generation base editor, BE4, with longer linkers between rAPOBEC1 and nCas9(D10A) and two copies of UGI for reducing uracil N-glycosylase (UNG)-mediated base excision repair (BER), displays much better efficiency in base editing. In addition, the DNA end-binding Gam protein26 has also been used to reduce the introduction of undesirable indels by BE4.14 Meanwhile, to broaden the genome-targeting scope, modified Cas9 variants with alternative PAM sequences have been used. Nevertheless, current base editing across the whole genome is limited by the 4th–8th targetable sites of sgRNA. New strategies are expected to increase the targeting scope and enhance the editing fidelity.
SunTag is a signal amplification system, containing multiple copies of a 19-amino-acid GCN4 peptide that is recognized by a single chain variable fragment (scFv) antibody.27 A small soluble tag GB1, which is a binding domain of protein G from group G Streptococcus,28 was fused to the C-terminus of scFv to eliminate protein aggregation. The SunTag system has been successfully applied in fluorescence imaging and targeted demethylation of specific DNA loci.27,29
We have exploited the SunTag system to create a new base editor, termed “the base editor for programming larger C to U (T) scope (BE-PLUS)”, which shows broader editing window and higher fidelity as compared with BE3. As an example of its applications, we demonstrated that BE-PLUS offers increased flexibility in inducing stop codons.
Results
A SunTag-based system for broadening the editing window
To enlarge the editing window of BE3, we engineered a new editing system (called “dCas9-GCN4”) consisting of dCas9 fused at the C-terminus to 10 copies of GCN4 peptide (SunTag), scFv-APOBEC-UGI-GB1 and sgRNA (Fig. 1a).27,29 To assess the efficiency of dCas9-GCN4, we used the episomal shuttle vector pSP189 as a reporter.25 In this method, the sgRNA is designed to target SupF tRNA gene on pSP189; C to U transitions at the tRNA would cause its inactivation, which is detected as a change in the blue colonies into white following transformation of the mutant pSP189 into lacZambE. coli25 (Supplementary information, Figure S1a). Briefly, we co-transfected plasmids expressing the three components of the dCas9-GCN4 system with pSP189 into HEK293FT, and 48 h later, we extracted the shuttle vector and transformed it into lacZambE. coli, before calculating the base conversion frequency, which is defined as the number of white colonies among the total number of colonies.
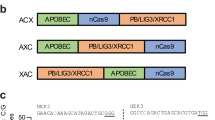
The generation of a new base editing system that broadens the base editing window. a Schematic overview of dCas9-GCN4. 10 × GCN4 is linked to the C-terminus of dCas9, while APOBEC and UGI are conjugated with scFv to form a fusion protein. When the two plasmids are co-transfected with sgRNA, dCas9-GCN4 is guided by sgRNA to the binding site. scFv-APOBEC-UGI is recruited around the binding site to induce C-to-T conversions. b Analysis of the base editing efficiency of dCas9-GCN4 and BE3 by blue/white colony screening. HEK293FT cells were transfected with the reporter vector pSP189 together with dCas9-GCN4 or BE3 as described in Materials and methods. The base editing efficiencies of dCas9-GCN4 and BE3 were tested using the white colony formation assay. The number of white colonies among the total number of colonies for sgRNA F1, F2, and F3 were calculated. The data was analysed using a two-tailed Student’s t-test. Error bars (±) indicate the standard deviations of 3 replicates. ns (not significant), P ≥ 0.05; **P < 0.01. c Sequencing analysis of the base editing window of dCas9-GCN4 and BE3 in the mutation reporter. White colonies were collected and subjected to Sanger sequencing. The average conversion rates of 12 positions of the protospacers (C1, C2, C5, C6, C9, C10, C11, C12, C14, C15, C18, and C19) targeted by 3 sgRNAs (sgRNA F1, F2, F3) were calculated. Error bars (±) indicate the standard deviations of 3 replicates
We initially screened for sgRNAs (L1, F1, F2, and F3) for their abilities to support BE2/BE3 catalysed base editing (all of these sgRNAs were effective for Cas9-mediated DNA cleavage based on the T7EN1 cleavage assay; Supplementary information, Figure S1b). We found that sgRNAs F1, F2, and F3 induced far more white colonies than sgRNA L1 (Supplementary information, Figure S1c), and so used the sgRNAs F1, F2, and F3 for subsequent experiments. The results showed that dCas9-GCN4 and BE3 induced comparable ratio of white colonies guided by sgRNA F1 (50.83 vs. 45.17%, P > 0.05), whereas dCas9-GCN4 induced higher ratio of white colonies guided by sgRNA F2 (76.33 vs. 62.73%, P < 0.01) and F3 (78.37 vs. 65.7%, P < 0.01) as compared with BE3 (Fig. 1b). These results indicate that dCas9-GCN4 is more efficient than BE3.
We then sequenced pSP189 recovered from the white colonies, and found that while BE3 edited Cs at positions 2–6 as reported,6 dCas9-GCN4 edited Cs at positions 1–14, demonstrating the successful enlargement of the editing window (Fig. 1c and Supplementary information, Figure S1d and e).
Optimizing the scFv-APOBEC system for higher efficiency and broader window
We next set out to optimize dCas9-GCN4. As nicking the DNA strand containing the unedited G would preferentially turn the U:G mismatch into the desired U:A/T:A pairing,6 we tried Cas9 nickase instead of dCas9. There are two kinds of Cas9 nickases, nCas9(D10A) and nCas9(H840A), which respectively produce cleavage of the complementary and non-complementary strand. Specifically, we fused GCN4 to N-terminus or C-terminus of nCas9(D10A) and nCas9(H840A), and these new systems were termed GCN4-D10A, GCN4-H840A, D10A-GCN4, and H840A-GCN4, respectively. We also fused GCN4 to N-terminus of dCas9 and termed the system GCN4-dCas9. dCas9-GCN4 was used as a control (Fig. 2a). We found that fusing GCN4 to dCas9 or nCas9(D10A), rather than nCas9(H840A), gave higher ratio of white colonies (Supplementary information, Figure S2a). Sequencing analysis indicates that different systems had similar editing window widths but GCN4-D10A resulted in the highest ratio of base editing (Supplementary information, Figure S2b). We then extended the analysis to 4 endogenous gene loci (DNMT3B, EMX1, FAP site 1, FANCF) in 293FT cells (Supplementary information, Table S3), where editing frequency was quantified by EditR.30 Consistent with the results in the reporter system, GCN4-D10A performed most effectively. Interestingly, GCN4-H840A and H840A-GCN4 showed much lower efficiency at endogenous loci than in reporter system (Fig. 2b), perhaps because pSP189 reporter system is derived from tRNA, which is dependent on secondary structure for activity and thus may be more sensitive to mutations.31,32,33 Overall, GCN4-D10A proves to be the optimal system and will be termed BE-PLUS hereafter.
Optimization of scFv-APOBEC system. a A scheme of 6 scFv-APOBEC systems. 10 × GCN4 is fused to the N-terminus or C-terminus of dCas9/nCas9(D10A or H840A), respectively. Every scFv-APOBEC system is labelled (left). b Analysis of the base editing window of 6 scFv-APOBEC systems by Sanger sequencing. The PCR products were collected and subjected to Sanger sequencing. The primers used for PCR amplification are listed (Supplementary information, Table S5). The average C-to-T conversion rates at 16 positions of protospacers (C3, C4, C5, C6, C7, C8, C9, C10, C11, C12, C13, C14, C17, C18, C19, and C20) targeted by 4 sgRNAs (DNMT3B, EMX1, FAP site 1, FANCF) were calculated. Error bars (±) indicate the standard deviations of 3 replicates. c Analysis of the base editing window of BE-PLUS and BE3 in endogenous genes by high-throughput deep sequencing. BE-PLUS was applied to edit seven different human genes distributed in different chromosomes. HEK293FT cells were co-transfected with different sgRNAs together with BE-PLUS or BE3. The PCR products of different loci were then subjected to deep sequencing. The average C-to-T conversion rate of every position (C1–C20) was calculated. Error bars (±) indicate the standard deviations of 3 replicates
We next characterized BE-PLUS in detail, using 7 sites at 7 different human genes located on different chromosomes (Supplementary information, Table S3). HEK293FT cells were co-transfected with plasmids expressing different sgRNAs together with different scFv-APOBEC systems (GCN4-D10A/dCas9) or BE3. Editing frequency was determined using the T7EN1 cleavage assay in conjunction with Sanger sequencing (Supplementary information, Figure S3). The results showed that BE-PLUS produced more C-to-T conversions inside of and outside of the 4th–8th base window (Supplementary information, Figure S3), confirming that BE-PLUS has a broadened work window and induces efficient base editing at endogenous loci.
To better understand the BE-PLUS system, we used deep-sequencing to compare the effects of BE-PLUS vs. BE3. A total of 69,100,000 reads were generated. The results showed that at PPEF1, FAP, IGF1, and IDO1 sites, BE-PLUS induced more C-to-T conversions at almost all positions. At the DNMT3B and EMX1 sites, BE-PLUS induced fewer C-to-T conversions at positions 4–8, but was more effective at positions 9–16 (Supplementary information, Figures S4 and 5). We then calculated the average C-to-T conversion rate in the protospacers of all 7 loci. As expected, BE-PLUS indeed induced far more C-to-T conversions at positions 9–16 as compared with BE3, although the efficiency at positions 4–8 was similar for the two editors (Fig. 2c).
BE-PLUS enhances the fidelity of base editing
BE3 usually causes indels and other undesirable base substitutions in addition to C-to-T conversions, including C-to-A and C-to-G conversions.6 Remarkably, BE-PLUS induced far fewer indels than BE3 at DNMT3B (2.11 vs. 17.63%, P < 0.01), EMX1 (1.83 vs. 5.28%, P < 0.05), FAP (0.3 vs. 1.62%, P < 0.05), IDO1 (0.42 vs. 0.51%, P > 0.05), MYOD1 (0.32 vs. 4.44%, P < 0.01), PPEF2 (0.34 vs. 0.97%, P < 0.000 1) and IGF1 (0.62 vs. 2.91%, P < 0.05) (Fig. 3a). Furthermore, the rate of undesirable substitutions (C-to-A and C-to-G) was also greatly reduced (0.73 vs. 2.63%, P < 0.001) (Fig. 3b, c). Thus, BE-PLUS dramatically enhances the fidelity of base editing.
BE-PLUS significantly enhances the fidelity of base editing. a Analysis of indels by deep sequencing. A total of 69,100,000 reads were generated and analysed. Reads containing at least 1 inserted or deleted nucleotides ±20 bp surrounding the protospacers were calculated as indel-containing reads. Indel frequency was calculated as the number of indel-containing reads among the total number of mapped reads. CTRL, negative control. Error bars (±) indicate the standard deviations of 3 replicates. ns (not significant), P ≥ 0.05; *P < 0.05; **P < 0.01; ****P < 0.000 1. b Analysis of unwanted base conversions. The average frequencies of C-to-A and C-to-G conversions of all mutant products induced by BE-PLUS and BE3 are presented. Error bars (±) indicate the standard deviations. ***P < 0.001. c Individual fractions of C-to-T, C-to-A and C-to-G conversions at 7 loci induced by BE-PLUS and BE3. The data shows a representative experiment from three independent experiments
To further define the specificity of BE-PLUS, 3 (DNMT3B, MYOD1 and EMX1) of the 7 loci were randomly selected for off-target analysis. Totally, 6 off-target sites (OTSs) for DNMT3B, 4 for MYOD1 and 3 for EMX1 were predicted by a web tool (http://crispr.mit.edu/). All these predicted off-target loci contain Cs within the editing window of BE-PLUS or BE3. All these sites were amplified by PCR and the PCR products were subjected to deep sequencing. The results showed that the mutation rates induced by BE-PLUS were comparable with those induced by BE3 (7.9 vs. 11.1%; P > 0.05) (Supplementary information, Figure S6a-d). Whereas for non-target mutations (C-to-T or G-to-A) beyond the protospacers, BE-PLUS causes fewer mutations within the ±100 bp regions flanking the 7 on-target and 13 off-target loci (0.013 vs. 0.033%, P < 0.001) (Supplementary information, Figure S6e and f). The results further confirmed the high fidelity of BE-PLUS.
BE-PLUS increases the genome-targeting scope of base editing and is a powerful functional genomic tool to disrupt genes
The enlargement of the base editing window (from position 4–8 to 4–16) predicts that BE-PLUS should target more sites across the whole genome than BE3. Indeed, bioinformatics analysis indicates that while BE3 targets only 20.4% of the Cs over the human genome, BE-PLUS targets 42.2% (Fig. 4a).
BE-PLUS is more efficient in inducing stop codons. a Cs editable by BE3 and BE-PLUS in the human genome were predicted based on the editing window of BE3 and BE-PLUS, respectively (all Cs and Gs in the chromosomes of the human reference genome hg38 were counted). Editable Cs are shown in green. b Scheme of GFP-iSTOP reporter system with 3 iSTOP codons (TGG) at positions 1–9 or 10–18 (from the distal side of the PAM). c Analysis of the GFP shutdown efficiency of BE-PLUS and BE3 using the GFP-iSTOP reporter system. The bar graph shows the GFP shutdown efficiency of BE-PLUS or BE3 when 1–9 or 10–18 TGG reporter vectors were used. GFP shutdown efficiency was calculated as the percentage of GFP knockout cells among the GFP-positive cells in the control group. Error bars (±) indicate the standard deviations of 3 replicates. ns, not significant, P ≥ 0.05; ***P < 0.001. d The plasmids expressing NME1 sgRNA (Supplementary information, Table S8) was co-transfected with either BE3 or BE-PLUS into HEK293FT cells. PCR products around the sgRNA binding site were subjected to Sanger sequencing (left), and T-A cloning and sequencing (right). e Statistical analysis of unique iSTOP sgRNAs or codons for BE3 and BE-PLUS in the entire human transcriptome. The unique iSTOP sgRNAs were identified over the genome dependent on the base editing window of BE-PLUS (from positions 4 to 16) or BE3 (from positions 4 to 8). The number of genes in the human genome that contain at least one unique iSTOP codon or are targeted by at least one iSTOP sgRNA for BE3 or BE-PLUS were calculated. f Statistical analysis of the iSTOP codons or sgRNAs for BE-PLUS or BE3. The bars represent the cumulative number of codons or sgRNAs for BE-PLUS or BE3. g The distribution of iSTOP codons over the targetable ORFs for BE-PLUS or BE3. The percentage of ORFs that can be disrupted and the relative positions of the iSTOP codons are shown
Base conversion in the ORFs can potentially introduce codon changes that can lead to amino acid substitutions or the introduction of early stop codons. Inspired by these insights, researchers have harnessed BE3 for gene disruption studies involving the induction of stop codons. By effectively converting C into T at 4 potentially inducible stop (iSTOP) codons, CAG (Gln), CGA (Arg), CAA (Gln) and TGG (Trp), BE3 can introduce TAG, TGA, or TAA stop codons to knock out genes.17,18 With its broader editing window, BE-PLUS should induce codon conversions more extensively. We utilized a GFP-iSTOP reporter system (unpublished data) to test this hypothesis. This system contains a target sequence fragment including three “TGG” codons at positions 1–9 or 10–18, immediately after the initiation codon “ATG” of the GFP coding sequence (referred to as the 1–9 TGG or 10–18 TGG target vector hereafter), and successful base editing is marked as decreases in GFP expression (Fig. 4b). BE-PLUS or BE3 and the indicated sgRNA were co-transfected with the reporter vector into HEK293FT cells. GFP was analysed by flow cytometry 72 h after transfection. While the GFP shutdown efficiencies for BE3-PLUS and BE3 were similar when the 1–9 TGG vector was transfected (84 vs. 83.33%, P > 0.05), the efficiency was 2-fold higher for BE-PLUS when the 10–18 TGG vector was applied (47.83 vs. 88.51%, P < 0.001) (Fig. 4c and Supplementary information, Figure S7). The Sanger sequencing confirmed that GFP knockout was caused by inducing stop codons in 1–9 TGG or 10–18 TGG (Supplementary information, Figure S8 and Table S7).
We extended the analysis to endogenous genes, using 7 sites at 6 different human genes (Supplementary information, Table S8). NME1 is one of the genes that cannot be disrupted by BE3 as there is no suitable sgRNA. We designed a sgRNA targeting one iSTOP codon with C at position 14. As expected, the C was converted to T by BE-PLUS, rather than BE3, to induce an early stop codon (Fig. 4d). Then, more sgRNAs were designed to test BE-PLUS. We used 2 sgRNAs that are effective for BE-PLUS only (Cs located at position 11 for RAD21, at 10–11 and 13–14 for KCNS1), and 2 sgRNAs that are effective for both systems (Cs located at positions 3–4 for RAC1, and 8–9 for KCNV2) (Supplementary information, Figure S9a and b). We also designed 2 sgRNAs targeting CDK10. For CDK10 sg1, the iSTOP codon locates beyond BE3, but within BE-PLUS editing window (C at position 13), while for CDK10 sg2, the same iSTOP codon locates within both BE3 and BE-PLUS editing window (C at position 5) (Supplementary information, Figure S9c and d). The editing was detected by Sanger sequencing of PCR products and further determined by T-A cloning and sequencing (Supplementary information, Table S9). The results showed that both BE3 and BE-PLUS induced stop codon at predicted positions (Supplementary information, Figure S9a–d), confirming the versatility of BE-PLUS in inducing stop codon.
To compare the versatility of base editors in gene disruption, we used bioinformatics to enumerate the human genes amenable to the iSTOP strategy,17 on the condition that the editing window for BE3 and BE-PLUS are positions 4–8 and 4–16, respectively. Although the two editors can target at least one unique iSTOP codon at comparable numbers of genes (18,067 vs. 18,582 for BE3 and BE-PLUS, respectively), BE-PLUS is able to target multiple unique iSTOP codons at many more genes as compared with BE3 (e.g., 6947 genes bearing 20 unique codons for BE-PLUS vs. 2794 genes for BE3). Similarly, with the same number of available sgRNAs, BE-PLUS system can target more genes (e.g., 10,690 genes targeted by 20 unique sgRNAs for BE-PLUS vs. 4380 genes for BE3) (Fig. 4e). Overall, the numbers of targetable iSTOP codons and available sgRNAs for BE-PLUS almost double those for BE3 over the entire ORF (388,080 vs. 210,415) (Fig. 4f). Finally, as inducing stop codons near N-terminus of ORF would be more efficient for disrupting genes,34 we analysed the distribution of iSTOP codons on ORF. BE-PLUS can introduce stop codons to the first 30% portion of the ORF at ~90% of genes, compared 60% by BE3 (Fig. 4g). Thus, BE-PLUS offers more flexibility for gene targeting via the iSTOP strategy.
Discussion
Base editors such as BE3 enable a wide variety of research and medical applications. One of the main limitations is the narrow width of the editing window, which renders sites beyond the window difficult to edit.16 In addition, indel levels were 4–12% by BE3 in some cases6,8 and unwanted base conversions, such as C-to-A or C-to-G conversions, were reported to occur at comparable level to C-to-T conversions.16 Therefore, base editors with improved editing scope and enhanced fidelity are desirable.
In this study, we designed a novel base-editing tool, BE-PLUS, which features broader editing window, and far fewer indels and non-C-to-T conversions.
To demonstrate its potential, we applied our new system to gene disruption by iSTOP. Comprehensive analysis revealed that, with broader window, there were far more sgRNAs available for BE-PLUS to induce stop codons across the whole genome. Consistently, BE-PLUS performed much more efficient iSTOP in a GFP-iSTOP reporter system as well as endogenous genes. Besides, BE-PLUS may also be used in other fields, such as directed evolution, regulator element screening, etc., which would further increase the application of BE-PLUS.
The mechanism underlying how BE-PLUS enhances editing fidelity is unknown. The fidelity might benefit from increased UGI molecules delivered to the target site in the BE-PLUS system (as such increase dramatically reduced UNG-mediated BER14,35), and/or from the tandem uracil intermediates converted from multiple target Cs within the same DNA strand.14
In conclusion, we have created a new base editor, BE-PLUS, featuring increased editing window and enhanced fidelity of base editing, which adds a powerful weapon to the base editing toolkit.
Materials and methods
Plasmid construction
BE3 has been previously reported.6 pST1374-scFv-APOBEC-UGI-GB1 was generated by fusing the scFv fragment to the N-terminus and UGI-GB1 to the C-terminus of the APOBEC fragment. Then, scFv-APOBEC-UGI-GB1 was inserted into the pST1374 vector. The scFv and GB1 fragments were amplified from Addgene plasmid 60904 by PCR amplification. The APOBEC and UGI fragments were amplified from Addgene plasmid 73021 by PCR amplification. The plasmid pST1374-N-NLS-flag-linker-D10A (Addgene, 51130) and pST1374-N-NLS-flag-linker-H840A (Addgene, 51129) were used as templates to acquire nCas9(D10A/H840A) sequences. Two mutations (D839A and N863A) were introduced into Cas9-H840A to eliminate the function of the HNH nuclease domain.36 The amino acid sequence of the GCN4s used was EELLSKNYHLENEVARLKK. The amino acid sequence of the linker between each GCN4 peptide unit was GSGSGGSGSGGSGSGSGGSGSGGSGSG. 10 × GCN4 was fused to either the N- or C-terminus of nCas9(D10A), nCas9(H840A), and dCas9. The shuttle vector pSP189 was a gift from Dr. Jia Chen’s laboratory. Oligos (Supplementary information, Table S1) used to generate sgRNA expression plasmids were annealed and cloned into the BsaI sites of pGL3-U6-sgRNA-PGK-Puro (Addgene, 51133). All plasmid sequences were verified by Sanger sequencing.
Cell culture and transfection
HEK293FT cells (from ATCC) have been tested to exclude mycoplasma contamination, and were maintained in Dulbecco’s modified Eagle’s medium (HyClone, SH30243.01) supplemented with 10% foetal bovine serum (HyClone, SV30087) at 37 °C under 5% CO2. Cells were seeded in 12-well plates at a density of 2 × 105 per well. The BE-PLUS treatment cells were transfected with 200 μl Opti-MEM containing 5 μl LipofectamineTM 2000 (Thermo Fisher Scientific, 11668019), 1 μg pST1374-GCN4-dCas9 (pST1374-GCN4-D10A or pST1374-GCN4-H840A), 1 μg scFv-APOBEC-UGI-GB1, and 0.5 μg sgRNA-expressing plasmid. The BE3 treatment cells were transfected with 100 μl Opti-MEM containing 5 μl LipofectamineTM 2000, 2 μg BE3 plasmid and 0.5 μg sgRNA-expressing plasmid. Twenty-four hours after transfection, blasticidin (InvivoGen, ant-bl-1) at a final concentration of 10 μg/ml and puromycin (InvivoGen, nt-pr-1) at a final concentration of 2 μg/ml were added to the media. Forty-eight hours after the transfection, genomic DNA was extracted from the cells using the phenol-chloroform method.
T7EN1 cleavage assay
The sequence around the target site was amplified using Phanta® Max Super-Fidelity DNA Polymerase (Vazyme, P505-d3). Primers used for PCR amplification are listed (Supplementary information, Tables S2 and 5). The PCR products were purified using a PCR cleanup kit (Axygen, AP-PCR-500) according to the manufacturer’s instructions. In total, 200 ng purified PCR products were annealed and digested with T7EN1 (NEB, M0302L) for 30 min and separated using a 2.5% agarose gel.
Selection of off-target sites
Off-target sites of DNMT3B, EMX1 and MYOD1 were predicted by a web tool (http://crispr.mit.edu/). These off-target sites and primers are listed in supplementary information (Supplementary information, Tables S4 and 6).
High-throughput DNA sequencing of on-target and off-target sites
PCR primers were designed at ±100 bp surrounding the on-target sites and are listed (Supplementary information, Table S5). Non-specific sequences in the PCR products were eliminated by gel electrophoresis. PCR products with different barcodes were pooled together for deep sequencing using the Illumina Nextseq 500 (2 × 150) platform at CAS-MPG Partner Institute for Computational Biology Omics Core, Shanghai, China.
Base conversion calculation at on-target and off-target sites
The adapter pair of the pair-end reads were removed using AdapterRemoval version 2.2.2, and pair-end read alignments of 11 bp or more bases were combined into a single consensus read. All process reads were then mapped to the target sequences using the BWA-MEM algorithm (BWA v0.7.16a). For each site, the mutation rate was calculated using bam-readcount with parameters -q 20 -b 30.
Non-target mutation analysis
The C-to-T and G-to-A mutations within ± 100 bp regions surrounding the 7 on-target and 13 off-target loci were calculated using approximately 94,000,000 sequence reads.
Indel frequency calculation
Indels were calculated based on reads containing at least 1 inserted or deleted nucleotide spanning from 20 nucleotides upstream of to 20 nucleotides downstream of the protospacers. Indel frequency was calculated as the number of indel-containing reads/total mapped reads.
Analysis of iSTOP by BE3 or BE-PLUS
A genome-wide iSTOP was analysed as follows: First, all editable sites in the chromosomes of the human reference genome hg38 as well as available sgRNAs were predicted. The editing window was considered as positions 4–16 for BE-PLUS and 4–8 for BE3. Second, Snpeff was used to annotate sites where stop codons can be generated. The well-curated RefSeq database was used for coding genes, and RefSeq IDs beginning with “NM” were selected. In addition, any sgRNAs matching with other sites in the genome were discarded. Finally, iSTOP codons and unique sgRNAs were identified.
Flow cytometry
293FT cells were seeded on a 24-well plate. To test BE3 system, cells were transfected with BE3 (600 ng), GFP-iSTOP reporter vector (20 ng) and sgRNA-expressing plasmid (200 ng). To test BE-PLUS system, cells were transfected with pST1374-GCN4-dCas9/pST1374-GCN4-D10A (400 ng), pST1374-scFv-APOBEC-UGI-GB1 (400 ng), GFP-iSTOP reporter vector (20 ng) and sgRNA expressing plasmid (200 ng). Three days after transfection, the cells were harvested, and the GFP fluorescence was analysed by BD LSRFortessaTM flow cytometry. Scatter plots were generated using FlowJo software.
Data availability
Deep sequencing data can be accessed at the “National Omics Data Encyclopedia (NODE) of CAS-MPG Partner Institute for Computational Biology(PICB), Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences” (accession No: NODEP00371730)
References
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013).
Mali, P. et al. RNA-guided human genome engineering via Cas9. Science 339, 823–826 (2013).
Hsu, P. D., Lander, E. S. & Zhang, F. Development and applications of CRISPR-Cas9 for genome engineering. Cell 157, 1262–1278 (2014).
Jasin, M. & Haber, J. E. The democratization of gene editing: insights from site-specific cleavage and double-strand break repair. DNA Repair. 44, 6–16 (2016).
Hess, G. T. et al. Directed evolution using dCas9-targeted somatic hypermutation in mammalian cells. Nat. Methods 13, 1036–1042 (2016).
Komor, A. C., Kim, Y. B., Packer, M. S., Zuris, J. A. & Liu, D. R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016).
Ma, Y. et al. Targeted AID-mediated mutagenesis (TAM) enables efficient genomic diversification in mammalian cells. Nat. Methods 13, 1029–1035 (2016).
Nishida, K. et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 353, pii: aaf8729 (2016).
Yang, L. et al. Engineering and optimising deaminase fusions for genome editing. Nat. Commun. 7, 13330 (2016).
Lu, Y. & Zhu, J. K. Precise editing of a target base in the rice genome using a modified CRISPR/Cas9 system. Mol. Plant 10, 523–525 (2017).
Zong, Y. et al. Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat. Biotechnol. 35, 438–440 (2017).
Rees, H. A. et al. Improving the DNA specificity and applicability of base editing through protein engineering and protein delivery. Nat. Commun. 8, 15790 (2017).
Zhang, Y. et al. Programmable base editing of zebrafish genome using a modified CRISPR-Cas9 system. Nat. Commun. 8, 118 (2017).
Komor, A. C. et al. Improved base excision repair inhibition and bacteriophage Mu Gam protein yields C:G-to-T: Abase editors with higher efficiency and product purity. Sci. Adv. 3, eaao4774 (2017).
Komor, A. C., Badran, A. H. & Liu, D. R. Editing the genome without double-stranded DNA breaks. ACS Chem. Biol. 13, 383–388 (2017).
Kim, Y. B. et al. Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nat. Biotechnol. 35, 371–376 (2017).
Billon, P. et al. CRISPR-mediated base editing enables efficient disruption of eukaryotic genes through induction of STOP codons. Mol. Cell 67, 1068–1079 e1064 (2017).
Kuscu, C. et al. CRISPR-STOP: gene silencing through base-editing-induced nonsense mutations. Nat. Methods 14, 710–712 (2017).
Kim, K. et al. Highly efficient RNA-guided base editing in mouse embryos. Nat. Biotechnol. 35, 435–437 (2017).
Chadwick, A. C., Wang, X. & Musunuru, K. In vivo base editing of PCSK9 (proprotein convertase Subtilisin/Kexin Type 9) as a therapeutic alternative to genome editing. Arterioscler. Thromb. Vasc. Biol. 37, 1741–1747 (2017).
Li, G. et al. Highly efficient and precise base editing in discarded human tripronuclear embryos. Protein & Cell 8, 776–779 (2017).
Liang, P. et al. Correction of beta-thalassemia mutant by base editor in human embryos. Protein Cell 8, 811–822 (2017).
Ma, H. et al. Correction of a pathogenic gene mutation in human embryos. Nature 548, 413–419 (2017).
Komor, A. C., Badran, A. H. & Liu, D. R. CRISPR-based technologies for the manipulation of eukaryotic genomes. Cell 169, 559 (2017).
Chen, J., Miller, B. F. & Furano, A. V. Repair of naturally occurring mismatches can induce mutations in flanking DNA. eLife 3, e02001 (2014).
d’Adda di Fagagna, F., Weller, G. R., Doherty, A. J. & Jackson, S. P. The Gam protein of bacteriophage Mu is an orthologue of eukaryotic Ku. EMBO Rep. 4, 47–52 (2003).
Tanenbaum, M. E., Gilbert, L. A., Qi, L. S., Weissman, J. S. & Vale, R. D. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell 159, 635–646 (2014).
Gronenborn, A. M. et al. A novel, highly stable fold of the immunoglobulin binding domain of streptococcal protein G. Science 253, 657–661 (1991).
Morita, S. et al. Targeted DNA demethylation in vivo using dCas9-peptide repeat and scFv-TET1 catalytic domain fusions. Nat. Biotechnol. 34, 1060–1065 (2016).
Kluesner, M. G. et al. EditR: A novel base editing quantification software using Sanger sequencing. bioRxiv 213496, doi: https://doi.org/10.1101/213496 (2017).
Johnston, J. M., Yu, M. T. & Carroll, W. L. A shuttle vector system for the investigation of immunoglobulin gene hypermutation: absence of enhanced mutability in intermediate B cell lines. Mol. Immunol. 29, 1005–1011 (1992).
Ashman, C. R. & Davidson, R. L. High spontaneous mutation frequency in shuttle vector sequences recovered from mammalian cellular DNA. Mol. Cell. Biol. 4, 2266–2272 (1984).
Hauser, J., Levine, A. S. & Dixon, K. Unique pattern of point mutations arising after gene transfer into mammalian cells. EMBO J. 6, 63–67 (1987).
Krivega, I. & Dean, A. LDB1-mediated enhancer looping can be established independent of mediator and cohesin. Nucleic Acids Res. 45, 8255–8268 (2017).
Wang, L. et al. Enhanced base editing by co-expression of free uracil DNA glycosylase inhibitor. Cell Res. 27, 1289–1292 (2017).
Mali, P. et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 31, 833–838 (2013).
Acknowledgements
We thank members of Huang and Chen laboratories for helpful discussions. We are grateful to Dr. Tian Chi from ShanghaiTech University for excellent language editing. This work was supported by the National Key R&D Program (2017YFC1001903, 2016YFC0905901), the National Science Foundation of China (31471400, 39870046, 81270605, 30971066, 81470324) and Major technological innovation plan of hospital (SWH2016ZDCX1003, SWH2016ZDCX1010).
Author information
Authors and Affiliations
Contributions
X.H., J.C., S.F., and W.J. designed the study. W.J., S.F., W.Y., G.L., G.Y., Y.L., Y.Z., L.Z., and Y.H. performed the experiments. W.J., S.F., and S.H. analysed the data. W.J. and S.F. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Jiang, W., Feng, S., Huang, S. et al. BE-PLUS: a new base editing tool with broadened editing window and enhanced fidelity. Cell Res 28, 855–861 (2018). https://doi.org/10.1038/s41422-018-0052-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41422-018-0052-4
This article is cited by
-
CRISPR technologies for genome, epigenome and transcriptome editing
Nature Reviews Molecular Cell Biology (2024)
-
Recent advances in therapeutic CRISPR-Cas9 genome editing: mechanisms and applications
Molecular Biomedicine (2023)
-
Development of a versatile nuclease prime editor with upgraded precision
Nature Communications (2023)
-
Base Editing and Prime Editing: Potential Therapeutic Options for Rare and Common Diseases
BioDrugs (2023)
-
A split prime editor with untethered reverse transcriptase and circular RNA template
Nature Biotechnology (2022)