Abstract
Cellular senescence is a fundamental cell fate playing a significant role throughout the natural aging process. However, the molecular determinants distinguishing senescence from other cell-cycle arrest states such as quiescence and post-mitotic state, and the specified mechanisms underlying cell-fate decisions towards senescence versus cell death in response to cellular stress stimuli remain less understood. Employing multi-omics approaches, we revealed that switching off the specific mitochondrial processing machinery involving the peptidase IMMP2L serves as the foundation of the senescence program, which was also observed during the mammalian aging process. Mechanistically, we demonstrate that IMMP2L processes and thus activates at least two substrates, mitochondrial metabolic enzyme glycerol-3-phosphate dehydrogenase (GPD2) and cell death regulator apoptosis-inducing factor (AIF). For cells destined to senesce, concerted shutdown of the IMMP2L-GPD2 and IMMP2L-AIF signaling axes collaboratively drives the senescent process by reprogramming mitochondria-associated redox status, phospholipid metabolism and signaling network, and simultaneously blocking cell death under oxidative stress conditions.
Similar content being viewed by others
Introduction
Cellular senescence is an irreversible cell cycle arrest state that can be triggered by multiple types of cellular stress including irreparable DNA damage, telomere attrition, oncogenic mutation, deregulated metabolism and oxidative stress.1, 2 Senescent cells share a combination of characteristics, including enlarged and flatten morphology, loss of proliferative capacity, increased senescence-associated ß-galactosidase (SA-ß-gal) activity, and pro-inflammatory senescence-associated secretory phenotype (SASP).3 Physiologically, cellular senescence plays complex roles in many biological processes such as development, wound healing, tumorigenesis and aging.4,5,6,7,8 During the natural aging process that takes place for decades, the senescent program is triggered in response to a series of multivariable stress challenges. Some of the senescent cells remain viable and accumulate in tissues and organs due to the potential aging-related decline in immune surveillance, and they would act as detrimental factors significantly accelerating the aging process via impairing tissue regeneration and reinforcing prolonged inflammatory response.9 Aging is the major risk factor for multiple types of geriatric diseases including cardiovascular disorder, stroke, diabetes, neurodegeneration and cancer,10 and emerging evidence suggests that selectively killing senescent cells could alleviate aging-related disorders in vivo.11,12,13,14 While extensive studies have been done to reveal the mechanisms underlying cellular senescence, it is critical to further elucidate the unique and specified program orchestrating the onset of this distinct cell fate because many currently defined characteristics implicated in senescence are also shared by other important cell fates and activities including cell cycle arrest, production of inflammatory factors, and activated stress responses.
In this study, we have systematically uncovered a signaling network orchestrated by a nuclear-encoded mitochondrial intermembrane peptidase IMMP2L for cell fate determination, senescence versus cell death, critical events associated with tissue homeostasis and aging process. Shutdown of IMMP2L signaling through either the inhibitory regulation of this peptidase by an irreversible, suicidal protease inhibitor SERPINB4 or its own downregulation is sufficient to initiate cellular senescence by reprogramming the mitochondrial functionality. Employing advanced proteomics, we have identified at least two mitochondrial target proteins processed by IMMP2L, including metabolic enzyme GPD2 and cell death regulator/electron transport chain complex I component AIF. In healthy cells, the IMMP2L-GPD2 axis catalyzes redox reactions to produce phospholipid precursor glycerol-3-phosphate, while under oxidative stress, IMMP2L cleaves AIF into its truncated pro-apoptotic form, leading to cell death initiation to remove cells with irreparable damage. For cells programmed to senesce, conversely, the IMMP2L-GPD2 axis is switched off to block phospholipid biosynthesis, leading to reduced availability of membrane building blocks for cell growth together with the disruption of mitochondrial localization of certain phospholipid-binding kinases, such as protein kinase C-δ (PKC-δ) and its downstream signaling. These alterations in mitochondria-associated metabolism and signaling network render the cells to enter a senescent state featuring high levels of reactive oxygen species (ROS). Simultaneously, as a vital collaborative mechanism, blockage of pro-apoptotic AIF generation due to the loss of IMMP2L ensures the viability of senescent cells under ROS-mediated oxidative stress. Taken together, we have mechanistically uncovered IMMP2L-mediated signaling as a key regulatory pathway in the control of fates of healthy, apoptotic and senescent cells. Furthermore, we propose that loss of IMMP2L signaling represents one of the fundamental events associated with cellular senescence as alteration of this network can be observed across a variety of tissue and cell types throughout the natural aging process.
Results
Exploration of the early cellular events associated with onset of senescence rather than quiescence
Previously, we found that epidermal growth factor (EGF) could act as a natural senescence-suppressor for normal human epithelial cells grown in culture, besides its traditional role as a mitogen to stimulate cell proliferation.15 The potent impact of EGFR signaling inhibition on inducing cell cycle arrest and senescence initiation in primary normal human bronchial epithelial (NHBE) cells prompted us to use this experimental condition as a tool to identify signaling pathways mediating the onset of cellular senescence program. Experimentally, short-term EGFR inhibitor Erlotinib treatment could induce a strong G1 cell cycle arrest in NHBE cells to lead them to senescence or quiescence, which can be distinguished by SA-ß-Gal staining (Supplementary information, Figure S1a and b). Therefore, comparing the gene expression profiles among normal proliferating NHBE cells versus senescent and quiescent cells collected from the same EGFR inhibition treatment condition allowed us to identify genes potentially playing a more specific role in senescence programming beyond regulating cell cycle arrest. As illustrated in Fig. 1a, after EGFR inhibitor treatment, we subsequently added to the culture a compound C12FDG that could be catalyzed into a green fluorescent product by the SA-ß-gal enzyme present at a high level in senescent cells.15, 16 Employing fluorescence-activated cell sorting (FACS), we were able to separately collect two different cell populations based on green fluorescence intensity. As a validation of our experimental approach, we found that the cell population exhibiting stronger fluorescence (senescent cell population) displayed multiple characteristics of cellular senescence in contrast to the cell population representing quiescent cells (Supplementary information, Figure S1b–e). Based on those observations, we then performed a transcriptomics analysis aimed to identify potential factors specifically implicated in senescence initiation.
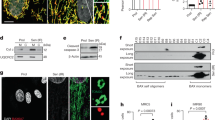
Study model to identify specific determinants of cellular senescence. a Schematic depiction of the strategy to separately collect the quiescent and senescent populations from the same group of treatment. b Principal component analysis (PCA) of transcriptional profiling reveals distinct signature of senescent cells. c Gene Ontology (GO) analysis of all genes specifically altered in the senescent population. See Supplementary information, Table S1 for list of senescence-associated genes. d Heatmap of all SERPIN genes involved in endopeptidase regulation. e Senescent NHBE cells are being detected by SA-ß-Gal assay. NHBE cells carrying a Dox-inducible (Tet-On) vector expressing target genes were grown with Dox (1 ug/mL) for 7days and SA-ß-Gal staining was then performed. Scale bar = 50 μm. f Quantification of senescent cells in indicated groups. Data are mean ± s.d.; Two-tailed, unpaired t-test was used. Data are representative of five independent areas (e)
Principal component analysis (PCA) of the transcriptomics data indicated that the senescent cells displayed a unique transcriptional signature compared with proliferating and quiescent cells (Fig. 1b), so we conducted gene ontology (GO) analysis for genes significantly and specifically altered in the senescent population (Fig. 1c; Supplementary information, Table S1). Among the top enriched pathways, a majority of them were associated with cell cycle regulation and DNA replication, reflecting the fact that the genes captured in the screen participate in the initiation and establishment of irreversible cell cycle arrest. Notably, since this experimental setting was aimed mainly to explore the early driving events of cellular senescence, we collected the cells at the earliest time point when those EGFR inhibitor-treated cells displayed significantly increased SA-ß-gal activity. Accordingly, at this very early stage, the senescent population has not fully developed the profound pro-inflammatory phenotype based on our transcriptional analysis, which was expected as notable upregulation of SASP factors occurs at the late stage of senescence program.17 Based on the GO analysis, our attention was drawn to the pathway of negative regulation of endopeptidase activity that contains members of the clade B serine protease inhibitor (SERPIN) superfamily that includes SERPINB1, B4, B7, B9 and B13, with their expression significantly upregulated in the senescence-positive NHBE cell population (Fig. 1d), suggesting its potential association with the establishment of senescent cell fate.
A negative regulator of endopeptidase SERPINB4 is sufficient and required in driving senescence initiation
Certain members of the SERPIN family have been previously implicated in senescence such as plasminogen activator inhibitor-1 (PAI-1), a secreted SERPINE family member involved in senescence induction.18, 19 Furthermore, transcription of the SERPINB3 and SERPINB4 loci has been found to be activated in oncogenic HRas-induced senescence despite that the mechanism underlying their function and significance during senescence remains largely unknown.20 To systematically explore the potential function of all these up-regulated SERPINs in the context of senescence initiation, we ectopically expressed each of the five SERPIN genes in NHBE cells and only SERPINB4 was able to exert a potent impact on senescence programming as determined by SA-ß-gal staining (Fig. 1e, f). Furthermore, the aforementioned SERPINB3 was not specifically upregulated in senescent population based on our transcriptome profiling, and ectopic expression of SERPINB3 also failed to induce senescence when compared with SERPINB4 (Supplementary information, Figure S2a, b). Since the level of ectopically expressed SERPINB4 was comparable to its endogenous state detected in the senescent population upon EGFR inhibition (Supplementary information, Figure S2c), we postulated SERPINB4 represents one of the most prominent factors identified in our screen involved in the program driving cellular senescence.
To explore this further, we also employed IMR-90 cells, a type of commonly used human diploid fibroblasts for senescence research throughout our subsequent studies. Consistent with the initial observation of a potent effect of SERPINB4 in inducing senescence in NHBE cells, we detected the appearance of several important hallmarks of cellular senescence that include enlarged cell size, strong SA-ß-gal staining (Fig. 2a, b), markedly decreased proliferation capacity (Fig. 2c) along with induction of the cyclin-dependent kinase inhibitor p16Ink4a (Fig. 2d). Importantly, blockage of SERPINB4 upregulation through shRNA knockdown (Fig. 2e) could suppress the induction of senescence in NHBE cells with EGFR inhibition (Fig. 2f, g), indicating the necessity of SERPINB4 induction in the initiation of senescence. Together with the observation of SERPINB4’s upregulation in lung-associated cells undergoing different types of senescence (Fig. 3c), these results suggest that induction of SERPINB4 represents a crucial event for the initiation of senescence programming.
Negative endopeptidase regulator SERPINB4 is identified to be a crucial factor involved in cellular senescence. a Ectopic expression of SERPINB4 is sufficient to induce cellular senescence in IMR-90 cells as determined by SA-ß-Gal staining. IMR-90 cells carrying a Dox-inducible (Tet-On) vector expressing Luciferase and SERPINB4 were grown with or without Dox (1 ug/mL) for 7 days and SA-ß-Gal staining was then performed. Scale bar = 50 μm. b Quantification of senescent cells in each group. Data are mean ± s.d.; Two-tailed, unpaired t-test was used. Data are representative of five independent areas (a). c Pronounced cell cycle arrest has been observed upon SERPINB4 overexpression in both IMR-90 (Left) and NHBE (right) cells. 10 uM Edu has been added into medium 2 h prior to the detection of Alexa Fluor 647-labeled Edu using flow cytometry. Data are mean ± s.d.; Two-tailed, unpaired t-test was used. d Immunoblot for detecting upregulation of 16Ink4a in both NHBE and IMR-90 cells upon SERPINB4 ectopic expression. e SERPINB4 was stably knocked down by shRNAs as determined by qPCR in NHBE cells. Data are mean ± s.d.; Two-tailed, unpaired t-test was used. f SERPINB4 is required for Erlotinib-mediated senescence induction. Cells undergoing SERPINB4 shRNA knockdown have been treated with Erlotinib for 48 h, which was followed by SA-ß-Gal staining. Scale bar = 50 μm. g Quantification of SA-ß-Gal-positive cells indicates that SERPINB4 knockdown suppresses the senescence induction upon blockage of EGFR. Data are mean ± s.d.; Two-tailed, unpaired t-test was used. Data are representative of five independent areas (f)
SERPINB4-IMMP2L inhibitory signaling axis carries pivotal impacts on cellular senescence. a Detection of potential covalently bound complex formed by SERPINB4 and its target protease using SERPINB4 antibody. NHBE cells were treated with EGFR inhibitor Erlotinib (1 uM) for 48 h for senescence induction prior to cell lysate preparation and immunoblot. b Identification of IMMP2L to be the potential peptidase trapped and inhibited by SERPINB4. Left: A Flowchart depicting the strategy to identify the target peptidase inhibited by SERPINB4. Right: Immunoblot detection of IMMP2L within the SERPINB4-target peptidase complex using IMMP2L antibody. c Loss of IMMP2L is a fundamental event of senescence programming. NHBE cells undergoing replicative senescence and IMR-90 cells undergoing oncogenic Hras-induced senescence have been used. Expression of the oncogenic Hras (Tet-On) was induced by adding Dox (1 ug/mL) 7 days before detection. d IMMP2L knockdown is sufficient to induce cellular senescence as determined by SA-ß-Gal staining. Left: Representative images of SA-ß-Gal staining in both NHBE and IMR-90 cells. Right: Quantification of senescent cells in each group. Data are mean ± s.d.; Two-tailed, unpaired t-test was used. Data are representative of five independent areas. Scale bar = 50 μm. e Cell cycle arrest can be induced by IMMP2L knockdown. IMMP2L has been stably knocked down in IMR-90 cells and Edu (10 uM) has been added into medium 2 h prior to the detection of Alexa Fluor 647-labeled Edu using flow cytometry. Data are mean ± s.d.; Two-tailed, unpaired t-test was used. f Immunoblots revealing upregulation of 16Ink4a in both NHBE and IMR-90 cells upon IMMP2L knockdown. g IMMP2L knockdown in IMR-90 cells induced the production of SASP factors, represented by three cytokines, as determined by qPCR examination. Data are mean ± s.d.; Two-tailed, unpaired t-test was used. h Loss of IMMP2L is specifically associated with senescence, but not quiescence, as determined by immunoblot. Quiescence marker, p27; senescence marker, 16Ink4a. Growth to confluence has been used for quiescence induction. Replicative-senescent IMR-90 cells have been used as senescence context
SERPINB4 directly traps and inhibits nuclear-encoded mitochondrial peptidase IMMP2L to trigger senescence initiation
Considering SERPINB4 as a serine protease inhibitor, we proceeded to identify the direct protease/peptidase substrate(s) inhibited by SERPINB4. As mentioned, SERPINB4 belongs to the clade B of SERPIN superfamily, which is the only subfamily of SERPINs lacking a signal peptide and thus resides primarily within cells.21 Therefore, SERPINB4 could carry out its function by blocking the activity of an intracellular serine peptidase and/or protease(s). Previously, three different proteases were found inhibited by SERPINB4, including cathepsin G, human mast cell chymase and granzyme M.22, 23 However, most of those experiments were conducted in cell-free systems, and these proteases are also known to have a cell type-specific distribution, being mainly expressed in immune cells. More importantly, SERPINB1, which could inhibit these same proteases as SERPINB4,24 failed to induce senescence upon overexpression (Fig. 1e). Accordingly, we postulated that the primary intracellular target(s) of SERPINB4 for driving the cellular senescence process remained unknown.
Biochemically, SERPINs belong to the specific group of protease inhibitors employing a unique mechanism to inhibit protease activity. In each SERPIN’s C-terminal domain, a reactive center loop (RCL) acts as a “bait” which is attacked by the target protease, and the SERPIN would suicidally trap the protease to form a covalently bound complex to irreversibly block the target protease activity. Such a protein complex is assumed to be stable under SDS-PAGE and remains detectable upon Western blot analysis,23 with the presumed process illustrated in Supplementary information, Figure S2d.
To identify the potential protease target(s) of SERPINB4, we employed an SERPINB4 antibody that is capable of recognizing the covalent complex formed between SERPINB4 and its known target cathepsin G. Using this antibody, we were able to detect a protein band representing SERPINB4 and a specific band with a larger size than SERPINB4 in the SERPINB4-upregulated senescent cells (Fig. 3a) that presumably represents a covalently bound complex formed by SERPINB4 and its target protease/peptidase. We subsequently developed a strategy aiming to identify the target trapped in this complex as illustrated in Fig. 3b. In brief, the immunoblot suggested the molecular weight of this potential complex to be less than 65 KDa (Fig. 3a). Considering the assumed size of SERPINB4 fragment within the SERPIN-protease complex to be about 40 KDa,23 we estimated that the molecular weight of the protease/peptidase trapped by SERPINB4 should be less than 25 KDa, so this designated protein size was used as our primary criterion to explore the potential protease target(s) of SERPINB4. Based on two protease databases, MEROPS and Degradome,25, 26 over 600 human proteases and peptidases have been identified so far with only 19 of them smaller than 25 KDa (Supplementary information, Table S2). Moreover, the primary targets of SERPINs are thought to be serine proteases/peptidases that further reduce the finalists into four peptidases/proteases. Theoretically, the covalent complex should be detected by antibodies for both SERPINB4 and the target protease in immunoblotting. Among the four candidates on the short list, mitochondrial inner membrane protease subunit 2 (IMMP2L) and signal peptidase complex catalytic subunit 11A (SEC11A) were chosen for the test mainly considering their potential functional relevance.27, 28 As shown in Fig. 3b, we found that only the anti-IMMP2L antibody could detect the protein complex with the same pattern as the anti-SERPINB4 antibody, indicating one of the potential protease(s) trapped by SERPINB4 within the complex to be IMMP2L. Furthermore, the inactive covalently bound complex is often degraded via proteasome-mediated degradation to ensure irreversible inhibition of the protease.29, 30 Thereby, SERPINB4-mediated inhibition of its target endopeptidase eventually leads to the degradation of the enzyme, a pattern that was observed for IMMP2L during both replicative senescence and oncogene-induced senescence (Fig. 3c). Thus, loss of IMMP2L activity and/or expression represents a fundamental and universal event under a wide range of senescence contexts.
IMMP2L is a nuclear-encoded peptidase known to be involved in processing signaling peptides of mitochondrial intermembrane proteins.31 Hence, we reasoned that blockage of IMMP2L by SERPINB4 could negatively impact the functions of certain mitochondrial proteins due to the failed pro-peptide cleavage, consequently reprogramming the mitochondrial functionality underpinning senescence initiation. In agreement with this notion, mice harboring a non-coding intron mutation in the IMMP2L locus display early onsets of age-related disorders,28, 32 implicating a potential role for IMMP2L in regulating cellular senescence although the mechanism underlying the mouse phenotype was not adequately explored. To test whether suppressing the expression of IMMP2L, mimicking the inhibition of its activity by SERPINB4, could recapitulate the senescent features observed in our earlier studies, both NHBE and IMR90 cells were treated with two different shRNAs for IMMP2L knockdown. Consistent with our hypothesis, knockdown of IMMP2L in these cells led to display of multiple features of cellular senescence, including enhanced SA-β-gal staining (Fig. 3d), significantly decreased proliferation capacity (Fig. 3e) along with induction of p16Ink4a and SASPs (Fig. 3f, g). Furthermore, ectopic expression of IMMP2L could partially suppress the SERPINB4-induced senescent phenotype as shown by decreased production of several key components of SASP and reduced SA-β-gal staining (Supplementary information, Figure S2e and f). Importantly, expression of IMMP2L was not altered in quiescent cells that also feature a profound cell cycle arrest as determined by an increase in the level of cyclin-dependent kinase inhibitor p27 (Fig. 3h), suggesting that the prominent impact of IMMP2L on senescence induction occurs beyond solely regulating cell cycle. Taken together, our initial screen to identify signaling pathways playing a fundamental and universal role in mediation of the onset of cellular senescence led to the discovery of IMMP2L as a central component whose activity or expression is directly and uniquely associated with the senescent program.
Although our study identified SERPINB4 as a senescence modulator based on initial screen in NHBE cells, this peptidase inhibitor’s expression appears to be restricted in several specific types of cells and tissues such as lung-associated bronchial cells, whereas IMMP2L is universally expressed across different cell types and organs.33,34,35 Thus, the regulation of IMMP2L expression/activity can be possibly achieved in multiple ways at both transcriptional and post-transcriptional levels that largely depend on the contexts of cell types and senescence stimuli, and inhibition of IMMP2L by SERPINB4 represents only one of the regulatory pathways to switch off IMMP2L signaling in a specific context such as NHBE cells. Hence, with an aim to explore the more universal process associated with the control of senescence initiation, our subsequent effort has been focused on exploration of the mechanisms by which alterations of IMMP2L activity serve as the focal point for senescence initiation and/or maintenance in a wide-range of biological contexts.
Systematical identification of the critical mitochondrial substrates regulated by IMMP2L-mediated processing
IMMP2L is a nuclear-encoded mitochondrial peptidase that belongs to the mitochondrial protein processing machinery. For over a thousand proteins within the mammalian mitochondria, the majority of them are synthesized in the cytoplasm and then imported into mitochondria.31, 36 Mechanisms underlying mitochondrial protein import have been mainly studied in the model organism budding yeast Saccharomyces cerevisiae. Based on those studies, many of these nuclear-encoded mitochondrial proteins contain the N-terminal pre-sequences that can be recognized by surface receptors on the mitochondria, leading them to be imported into destined sites.37 Once imported, a group of peptidases including Icp55, Oct1, m-AAA protease, Pcp1, Imp1 and Imp2, acts to proteolytically regulate the maturation, stability and functional properties of those proteins.31 IMMP2L is the mammalian homologue for yeast Imp2, so we postulated that failure in N-terminal transit peptide processing of certain specific mitochondrial proteins due to loss of IMMP2L would disrupt their activities and trigger the onset of cellular senescence (Supplementary information, Figure S2g). Since the functionality and the downstream substrates of IMMP2L are largely unknown in the context of mammalian system, we employed and further customized a SILAC-associated proteomics technology named terminal amine isotopic labeling of substrates (TAILS)38, 39 to systematically identify the mitochondrial substrates of IMMP2L in a proteome-wide screen (a schematic description of our substrate identification strategy is shown in Fig. 4a). Theoretically, we expected that ectopically expressing extra IMMP2L would facilitate the processing of its target mitochondrial proteins more efficiently, and thus those substrates would predominantly exist in their processed mature form. On the other hand, the control group solely expressing endogenous IMMP2L may process the target proteins in a relatively less efficient manner resulting in comparatively lower production of processed substrates. In addition, since IMMP2L is assumed to function in protein processing instead of degradation, we expected that the total protein amount of target substrates counting both unprocessed and processed forms should remain at a similar level regardless of IMMP2L ectopic expression. Furthermore, we focused our attention on the around 100 proteins known to be localized inside the mitochondrial intermembrane space where IMMP2L presumably carries out its catalytic activity.40, 41
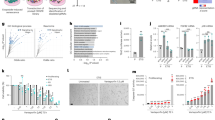
Systematic identification of mitochondrial protein(s) processed by peptidase IMMP2L using customized proteomics. a Schematic drawing of the TAILS experimental work flow. Exogenous IMMP2L was induced to express via adding doxycycline into the medium 48 h before detection. b Quantification of total GPD2 protein abundance between control and IMMP2L-overexpressing group. Left: The average relative abundance of all GPD2-associated peptides between two groups is similar. Right: MS1 spectrum of a representative GPD2-related peptide (LAFLNVQAAEEALPR) shows similar abundance between control and the induced group. c Specified comparison of processed GPD2 abundance between IMMP2L-overexpressing and control group. Left: the total extracted ion chromatogram (XIC) of TAILS-labeled N-terminal peptide (ADCISEPVNR) of processed GPD2 in two groups. Right: the relative higher abundance of N-terminal peptide of GPD2 observed in IMMP2L-overexpressing group indicates more GPD2 existing in processed form upon IMMP2L ectopic expression. d Quantification of total AIF protein abundance in control and IMMP2L-overexpressing group. Left: The average relative abundance of all AIF-associated peptides between two groups is similar. Right: MS1 spectrum of a representative AIF-related peptide (TGGLEIDSDFGGFR) shows similar abundance between control and the induced group. e Specified comparison of processed AIF abundance between IMMP2L-overexpressing and control group. Left: the total extracted ion chromatogram (XIC) of TAILS-labeled N-terminal peptide (SEGEEVPQDKAPSHVPFLLIGGGTAAFAAAR) of processed AIF in IMMP2L and control (non-detected) group. Right: TAILS-labeled peptide (SEGEEVPQDKAPSHVPFLLIGGGTAAFAAAR) representing truncated AIF can only be identified in the IMMP2L-overexpressing group indicating supplementary of extra IMMP2L could facilitate the conversion of mature AIF into truncated AIF. f Immunoblots were used to validate the findings from TAILS experiments. GPD2 and AIF antibodies, capable of detecting both unprocessed and processed forms, were employed for the experiments. Ectopic IMMP2L expression was induced using the same method as in TAILS in comparison with control cells. Left: total cell lysates from Hela cells. Right: total cell lysates from IMR-90 cells
Based on all the criteria described in Fig. 4a, two mitochondrial intermembrane space proteins, GPD2 and AIF, were identified as substrates of IMMP2L. Specifically, the whole mitochondrial proteomic profiling analysis indicated that the total protein levels of GPD2 and AIF remained similar in both control and IMMP2L-overexpressing samples (Fig. 4b, d). Regarding the abundance of TAILS-labeled N-terminus standing for the processed GPD2, we could detect those peptides in higher intensity in the IMMP2L-overexpressing group (Fig. 4c). Immunoblotting further validated the processing pattern of GPD2 that ectopic IMMP2L could enhance the conversion of GPD2 from pre-protein to its processed form (Fig. 4f). Regarding AIF, both the TAILS-MS and subsequent immunoblotting indicated that there was a significant increase of processed AIF upon IMMP2L ectopic expression whereas the processing was much less efficient under the endogenous condition (Fig. 4e, f). Taken together, these results strongly suggest that we have identified at least two substrates of IMMP2L including GPD2 and AIF that could potentially play roles in the IMMP2L signaling network.
Alteration of IMMP2L-GPD2 signaling axis reprograms the metabolic state and signaling network associated with the initiation of cellular senescence
Consistent with our identification of GPD2 as one of the substrates of IMMP2L using human cells, a previous study reported that impaired processing of GPD2 was observed in IMMP2L mutant mice,28 indicating that processing GPD2 by IMMP2L is conserved in the mammalian system, although it was unclear whether IMMP2L-mediated processing was critical in regulating the activity of GPD2. More importantly, it remains largely unknown whether GPD2 represents the critical downstream target of IMMP2L which mediates its regulation of cell fate and the aging phenotype observed in mice.42 To address this question, we first examined different cell fates and found that downregulated IMMP2L-GPD2 axis was only observed during cellular senescence rather than quiescence (Supplementary information, Figure S3a), indicating that shutdown of the IMMP2L-GPD2 axis was specifically associated with the onset of senescence program instead of all non-dividing cell fates. GPD2 processing was also decreased upon IMMP2L knockdown in both NHBE and IMR-90 cells (Supplementary information, Figure S3b). Notably, in both types of cells with a longer term knockdown of IMMP2L or within senescence contexts, even the unprocessed GPD2 was diminished together with the loss of IMMP2L and processed GPD2, suggesting that a complete shutdown of the IMMP2L-GPD2 axis may require blockage of both expression and processing of components within this pathway. Moreover, knockdown of GPD2 with shRNAs could sufficiently induce cellular senescence recapitulating many features of IMMP2L knockdown cells (Fig. 5a–c; Supplementary information, Figure S3c and d). Together, these data suggest that the senescence program involving loss of IMMP2L is in part via suppression of its downstream pathway related to the metabolic enzyme GPD2.
IMMP2L-GPD2 axis programs metabolic and signaling activities associated with mitochondria for cell fate control. a–c GPD2 knockdown is sufficient to induce cellular senescence. a Immunoblot for detecting upregulation of p16Ink4a in IMR-90 cells with GPD2 knockdown by two different shRNAs. b, c SA-ß-Gal staining and its quantification of the same batches of IMR-90 cells. Data are representative of five independent areas in each group. Data are mean ± s.d., Two-tailed, unpaired t-test was used. Scale bar = 50 μm. d Schematic illustration of postulated IMMP2L-GPD2 axis on metabolic regulation. e The level of Glycerol 3-phosphate was increased in IMR-90 cells with IMMP2L overexpression. Left: Volcano plot for visualization of metabolites profiling comparing IMR-90 cells with or without IMMP2L overexpression. Log2 of the IMMP2L overexpression/control ratios of the quantifiable metabolites were used for X axis and -log10 of P value for each metabolite for Y axis. All metabolites showing over 1.3-fold difference (P < 0.05) in the IMMP2L overexpression group over the control are blue-labeled except glycerol 3-phosphate, which is labeled in red. The rest of the metabolites are marked by black dots. Right: Relative Abundance of Glycerol 3-phosphate in control and IMMP2L overexpression group. Data are mean ± s.d.; Two-tailed, unpaired t-test was used. f Principal component analysis (PCA) of lipidomics profiling focusing on the IMMP2L-GPD2 axis. IMMP2L (Tet-On) overexpression was achieved in IMR-90 cells by adding Dox (1 ug/mL) 48 h before detection. Normal proliferating (marked as control) and Hras-induced senescent IMR-90 cells were used in the same experiment. g Examine the impact of IMMP2L-GPD2 signaling on the lipidome. IMR-90 cells with IMMP2L ectopic expression, normal proliferating cells as the control, and cells with Hras-induced senescence are marked as the three groups of cells employed in the experiment. Each point represents one of the lipid molecules that were quantified, aggregated for ESI+and ESI−. Red dots represent the lipid molecules identified with abundance displaying a pattern of IMMP2L ≥ control > senescence among the three groups of cells. The rest of lipid molecules are labeled in blue. For all the red dots, the vast majority of them were phospholipids as shown in the chart on the right. h PKC-δ was identified as a phospholipid-binding candidate whose mitochondrial localization is regulated by IMMP2L-GPD2 signaling. Left, Scatter plot showing results of quantitative proteomic profiling of proteins collected from isolated mitochondria of control and IMMP2L-overexpressing HeLa cells. Log2 of the overexpression/control ratios of all the quantifiable proteins were used for X axis and log10 of summed peptide intensities (heavy plus light) for each protein for Y axis. All proteins showing over 1.5-fold increase in the IMMP2L overexpression group are red-labeled and the rest of the proteins are marked in black. Right, Relative abundance of PKC-δ between control and IMMP2L overexpression group based on the profiling. i Immunoblots to examine the abundance of IMMP2L-GPD2-PKC-δ associated with mitochondria. Mitochondria were isolated from IMR-90 cells with different contexts as indicated: IMMP2L overexpression, IMMP2L knockdown, undergone replicative senescence. COX IV was used as a loading control. j Excessive generation of ROS can be detected in IMR-90 cells with IMMP2L or GPD2 knockdown. The cells were stained with CellROX Deep Red Reagent (5 μM) by adding the probe to the medium 30 min before detection. The cells were then collected and analyzed using flow cytometry. k Examine the impact of IMMP2L-GPD2-PKC-δ axis on PDHE1α phosphorylation. Mitochondria were isolated from IMR-90 cells with Hras-induced senescence or HeLa cells with IMMP2L overexpression for immunoblots of indicated proteins
As illustrated in Fig. 5d, GPD2 is a critical component of the glycerol phosphate shuttle that connects cytoplasmic and mitochondrial metabolism, catalyzing the reduction of dihydroxyacetone phosphate (DHAP) to Glycerol 3-phosphate, which is coupled with oxidation of NADH.43 Considering our earlier finding that ectopic expression of IMMP2L could further facilitate the processing of GPD2 into its mature form, we probed its impact on cell metabolism regulation. From the metabolic profiling analysis, Glycerol 3-phosphate is one of the major metabolites displaying a significant increase in abundance among all molecules detected in the assay upon IMMP2L overexpression (Fig. 5e). Moreover, metabolite set enrichment analysis for all increased metabolites using MetaboAnalyst44 also indicated that metabolic pathways involving NAD+ synthesis and utilization of Glycerol 3-phosphate are the prevailing ones activated upon boosting the IMMP2L-medaited processing of GPD2 (Supplementary information, Figure S3e–g). These findings suggest that IMMP2L-mediated processing is essential to activate GPD2 enzymatically and GPD2 also represents the key substrate of IMMP2L from the aspect of metabolic control.
NAD+ and Glycerol 3-phosphate are the metabolites derived from activity of the mitochondrial IMMP2L-GPD2 signaling axis. Although NAD+ metabolism has been shown to be critically involved in aging and cellular senescence,45, 46 it may not fully account for the critical and unique roles of IMMP2L-GPD2 axis in regulating metabolism for cell fate regulation, considering the co-existence of multiple NAD+ biosynthetic pathways47 and our observation of only a mild increase of NAD+ level upon boosting the activity of the IMMP2L-GPD2 axis (Supplementary information, Figure S3h). To address this question, our focus was then centered on Glycerol 3-phosphate, which serves as a critical precursor for phospholipid biosynthesis.48 Since mitochondria are also regarded as one of the major cellular compartments responsible for phospholipid production,49 we then conducted a lipidomics profiling to systematically examine the impact of mitochondrial IMMP2L-GPD2 signaling axis on the status of lipid metabolism. IMR-90 cells with three different contexts were employed including cells ectopically expressing IMMP2L to boost the activity of IMMP2L-GPD2 axis towards overproduction of Glycerol 3-phosphate, normal proliferating cells, and senescent cells with intrinsically downregulated IMMP2L-GPD2 signaling. Each of these groups of samples displayed a distinct lipid signature (Fig. 5f). For over 1000 lipid compounds identified, we paid special attention to the lipids that were specifically decreased in the context of senescence, but were present at a similar or higher abundance in the context when the IMMP2L-GPD2 axis was further reinforced. Remarkably, over 90% of the lipid molecules fitting this specified criterion turned out to belong to different classes of phospholipids instead of other types of lipids (Fig. 5g; Supplementary information, Figure S3i). These data suggest that the mitochondrial IMMP2L-GPD2 axis is critically involved in phospholipid production by providing its precursor glycerol-3-phosphate, and inhibition of this pathway could lead to profound cell-cycle arrest due to a shortage of essential constituents for new membrane biogenesis. Moreover, phospholipids are known to serve as pathway regulators through their membrane-based recruitment of cytosolic proteins containing phospholipid-binding domains,50 which could thereby play a role to program mitochondrial signaling pathways for cell fate control. To probe this possibility, we isolated mitochondria for proteome-wide profiling with a specific interest in any cytosolic phospholipid-binding proteins that become more enriched in mitochondrial fractions upon boosting IMMP2L-GPD2 signaling. In this regard, we have identified PKC-δ as one of the top candidates fitting the criteria (Fig. 5h), which was subsequently validated by immunoblotting since either reinforcement or shutdown of IMMP2L-GPD2 signaling could alter the abundance of PKC-δ associated with mitochondria (Fig. 5i). We also isolated mitochondria from cells that have undergone cellular senescence and found the loss of IMMP2L-GPD2 signaling to be linked with a decrease in the level of mitochondria-associated PKC-δ (Fig. 5i).
PKC-δ is a serine/threonine kinase51 and thus its decreased presence in mitochondria could reduce the phosphorylation level and hence alter the activities of its substrates within the organelle towards establishing a new metabolic and signaling landscape favoring cellular senescence. In this regard, we found the enhanced production of ROS as one of the primary consequences arising from loss-of-IMMP2L-mediated mitochondrial reprogramming (Fig. 5j), which is the condition known to act as a fundamental force driving the cell fate towards senescence.52 Moreover, we attempted to identify the specific protein(s) phosphorylated by PKC-δ that may serve as downstream effectors of this newly discovered pathway in the context of cellular senescence. PKC-δ has previously been shown to directly phosphorylate pyruvate dehydrogenase kinase (PDK), leading to phosphorylation-dependent inhibition of mitochondrial pyruvate dehydrogenase (PDH), a key regulator mediating pyruvate’s entrance into the tricarboxylic acid (TCA) cycle.53, 54 Notably, enhanced PDH activity due to its dephosphorylation is recognized as the causative event critically implicated in the execution of oncogene-induced senescence to suppress tumor initiation as well as cancer progression.55 Accordingly, we explored the possibility that IMMP2L signaling-mediated recruitment of PKC-δ to the mitochondria may serve as the upstream event responsible for modulating PDH phosphorylation in response to oncogenic insults and tumorigenesis. Mitochondria isolated from IMR-90 cells were probed and PDH dephosphorylation was found to be coupled with absence of PKC-δ on the mitochondria due to activation of the senescence program which switches off IMMP2L signaling against oncogenic transformation (Fig. 5k). Conversely, for the transformed HeLa cells, boosting the IMMP2L signaling to facilitate the recruitment of PKC-δ on the mitochondria can lead to profound increase of PDH phosphorylation (Fig. 5k). Collectively, starting from revealing GPD2 as a critical substrate of IMMP2L, we have systematically identified a new signaling hierarchy underlying the activity of IMMP2L-GPD2 axis for cell fate regulation.
Failure in the generation of pro-apoptotic AIF due to loss of IMMP2L confers resistance to oxidative stress and survival of senescent cells
Having illuminated the critical roles of IMMP2L-GPD2 axis in programing metabolism and signaling network for cell fate regulation, we turned our attention to the decision-making question of how cells can be specifically instructed into senescence but not cell death in response to high levels of ROS, which is a stress condition known to represent the driving force for both of these two distinct cell fates and derived from shutdown of the IMMP2L-GPD2 signaling axis. We reasoned that there must be a specified molecular machinery underlying the senescence program to guide the cells to avoid the fate of oxidative stress-induced cell death, which may also involve the regulation of IMMP2L along with another substrate of this peptidase identified in the aforementioned TAILS-MS proteomics screen, AIF, due to its functional relevance. Previous studies suggested that the mature AIF (mAIF) acts as a NADH-dependent oxidase and plays pivotal roles in regulating mitochondrial respiration in healthy cells such as terminally differentiated neurons.56 In response to certain cytotoxic insults, however, the N-terminus of mAIF is cleaved to generate the soluble truncated version of AIF (tAIF) possessing a pro-apoptotic activity, which can translocate into the nucleus to execute the lethal function.57 While the N-terminal proteolysis of mAIF into tAIF is critical for switching AIF from a pro-survival regulator of mitochondrial respiration into a pro-apoptotic factor, the specific peptidase involved in this processing remained largely unknown. In this regard, previous studies using cell-free systems have revealed that mitochondrial μ-calpain, which is a calcium-dependent protease, can process AIF into the pro-apoptotic form upon its activation.58,59,60 However, the involvement and necessity of μ-calpain for in vivo processing of AIF remain to be further scrutinized.61, 62
As shown below, we have now identified IMMP2L as a peptidase capable of catalyzing this in vivo pro-apoptotic conversion. To probe the cell death angle further, we also tested whether IMMP2L could act as a peptidase for the processing of other mitochondrial pro-apoptotic factors known to contain transit peptides,63 including SMAC, HtrA2 and Endonuclease G, and found that IMMP2L displayed a substrate-specificity in processing only mAIF (Fig. 6a). Notably, it was suggested that AIF, rather than other aforementioned mitochondrial death effectors, plays an essential role in oxidative stress-mediated cell death.64,65,66 Thereby, we reasoned that the IMMP2L-AIF axis likely represents the key regulatory pathway determining the susceptibility to oxidative stress-mediated cell death that needs to be switched off during the onset of cellular senescence. To better imitate the persistent presence of oxidative stress, glucose oxidase (GO) was employed as the stimulus, which can continuously generate H2O2 as illustrated in Fig. 6b. Cell viability assay indicated that cells with IMMP2L ectopic expression became more vulnerable under oxidative stress in comparison to control cells (Fig. 6c, d). Mechanistically, increased levels of IMMP2L could facilitate the conversion of mAIF to tAIF. Under the short-term GO treatment for cells with enforced IMMP2L expression, the oxidative stress was capable of disrupting mitochondrial outer membrane and enabling the release of a large portion of soluble pro-apoptotic tAIF from mitochondria to induce cell death (Fig. 6e).
The IMMP2L-AIF axis mediates oxidative stress-induced cell death and its shutdown is crucial to the survival of senescent cells under the stress condition. a Immunoblot to examine the impact of IMMP2L on the processing of mitochondrial-associated cell death effectors. Ectopic IMMP2L expression was induced by addition of Dox for 2 days. Antibodies capable of detecting specific proteins in both unprocessed and processed forms were used as indicated. b Schematic description of the approach for oxidative stress imitation. c IMR-90 cells with or without IMMP2L overexpression were grown for 48 h prior to replacement with fresh medium with or without addition of the oxidative stress-inducing agent glucose oxidase (50 mU/mL) for 2 h. Cells were then labeled with Annexin V and PI, and further analyzed by flow cytometry. Representative plots are shown. Frequency of Annexin V and/or PI-positive labeling was recoded as percentage of cell death. d Quantification of cell death rate based on flow cytometry analysis of data presented in panel c. Data are mean ± s.d.; Two-tailed, unpaired t-test was used. e The release of pro-apoptotic tAIF from mitochondria in control or cells with IMMP2L overexpression upon exposure to oxidative stress as described for panel c. Cells were fractionated into membrane and cytosolic fractions and immunoblots conducted for each fraction using indicated antibodies. Mitochondrial enrichment was seen with mitochondrial marker COX IV. f Prolonged oxidative stress facilitates the endogenous IMMP2L-mediated cleavage of endogenous AIF, which was suppressed due to loss of IMMP2L in cells with a senescent state. Early passage and replicative senescent IMR-90 cell were exposed to glucose oxidase (50 mU/mL) for 6 h as defined as the long-term oxidative stress condition before immunoblot analysis. g Senescent cells acquire resistance against oxidative stress. Early passage and senescent IMR-90 cells were treated with the same condition as in panel f. Cells were then labeled with Annexin V and PI, and further analyzed by flow cytometry. Representative plots are shown. Frequency of Annexin V- and PI-negative labeling was recoded as percentage of cell viability. h Quantification of cell viability based on flow cytometry analysis of panel g. Data are mean ± s.d.; Two-tailed, unpaired t-test was used
Under the senescence program, however, shutdown of IMMP2L signaling is thereby expected to ensure AIF remaining in its pro-survival form and thus drive cells into senescence but not cell death in the presence of high levels of ROS, which is one of the consequences of blockage of the IMMP2L-GPD2 axis as demonstrated earlier. Indeed, we found that oxidative stress in healthy cells could activate endogenous IMMP2L towards generating tAIF for cell death initiation (Fig. 6f). In contrast, with the same oxidative insult in cells undergoing either senescence or with IMMP2L knockdown, AIF remains in its pro-survival mature form without pro-apoptotic conversion normally catalyzed by its processing peptidase IMMP2L (Fig. 6f; Supplementary information, Figure S3j), leading to enhanced survival capacity of senescent cells (Fig. 6g, h). Taken together, these data demonstrate the critical role of IMMP2L for regulating AIF towards the execution of a cell death pathway in response to oxidative insult, and its shutdown under the senescence program can ensure a cell fate towards senescence instead of cell death upon high levels of ROS.
Declined IMMP2L signaling can be observed during natural mammalian aging process
For the natural aging process taking place throughout mammalian lifetime, cells within the body are confronted with a series of multivariable stress ranging from oncogenic activation to telomere shortening-induced replicative stress and DNA damage, leading to the generation of senescent cells which accumulate at least partly due to decline in immune clearance through chronological aging.9 Considering our in-depth mechanistic studies revealing IMMP2L’s critical roles in mediating cellular senescence regardless of its triggering origin, we investigated if decline of IMMP2L functionality is broadly implicated throughout the progressive function-declining process of natural aging. Our initial examination focused on mouse skeletal muscle mainly because symptom of declined skeletal muscle function is commonly associated with advanced aging67 and a recent study has uncovered that muscle stem cells switch their cell fate from quiescence into senescence at geriatric stage.68 We assessed the expression profile of components within the IMMP2L-assocaited pathways in wild-type mice of 2–3 (young) and 25–32 (geriatric) months of age. As shown in Fig. 7a, expression of the p16Ink4a, which is also a biomarker of mammalian aging besides being a senescence marker,69 is increased in the geriatric group as expected. Conversely, IMMP2L was markedly diminished at the geriatric stage. For GPD2, which needs to be processed by IMMP2L to become enzymatically active, we have observed that its total level was significantly decreased in aged animals, further supporting the notion that shutdown of the IMMP2L-GPD2 axis is universally implicated in cellular senescence both in vitro and in vivo. Considering that decline in the signaling of IMMP2L-GPD2 axis could predominantly suppress the biosynthesis of phospholipids based on our earlier in vitro studies, we examined aging-associated lipid metabolic alterations in skeletal muscle. Consistently, for all the lipid metabolites significantly decreased during the chronological aging process, the vast majority of them were various types of phospholipids (Fig. 7b). Intrigued by those results, we then extended the study into the context of human aging via using the whole blood samples from vigorous young and relatively fragile old adults. Importantly, significant elevation of p16Ink4a and downregulation of IMMP2L could be observed in samples from the aged cohort (Fig. 7c). Collectively, these pieces of physiological evidence suggest that loss of IMMP2L signaling is a fundamental event underlying cellular senescence during the mammalian natural aging process across tissues and species.
Loss of IMMP2L is associated with natural aging process. a Skeletal muscle tissues taken from young and geriatric C57BL/6 mice were used for immunoblot. b Lipid profiling for skeletal muscle tissue taken from young and geriatric C57BL/6 mice. All lipid molecules significantly declined in geriatric mice were shown (P value < 0.05). Data are mean ± s.d.; Two-tailed, unpaired t-test was used. c Expression of indicated genes were measured by RT-qPCR. White blood cell samples from indicated group are used for experiment. Data are mean ± s.d.; Two-tailed, unpaired t-test was used. d, e Deletions within intron 3 of IMMP2L genomic locus were detected by the CNVs assay in general populations compared with longevity group. d Schematic view of the IMMP2L genomic locus. qPCR probes corresponding to the intron region are marked with arrows. e Detection of intron deletions using CNV probe No.1; n = 10; Data are mean ± s.d.; Two-tailed, unpaired t-test was used
Healthy longevity of centenarians (CENs) represents a complex phenotype, with underlying contributions by environmental influence, genetic variations and environment-genome interactions.70,71,72,73 Previously, our study revealed that CENs possesses distinct genetic characteristics related to mitochondria,74 which prompted us to investigate if distinct patterns on the genomic locus of mitochondrial peptidase IMMP2L also exist in cohorts of CENs. Because copy number variations (CNVs) leading to intragenic deletions at the IMMP2L locus were observed in patients with neurological diseases,75, 76 we explored whether the disease-resistant CENs would display a different CNV profile from the general population at the IMMP2L locus. Accordingly, we employed the genomic samples of CENs, first-degree offspring (F1) of CENs and spouses of their offspring (F1SPs), and conducted CNV examinations on the IMMP2L locus with a focus on regions shown to contain prevalent deletions based on analysis of aforementioned diseases (Fig. 7d). Interestingly, we found that F1SPs seem to have more frequent deletions within intron 3 of IMMP2L when compared with CENs (Fig. 7e; Supplementary information, Figure S4). Considering that the offspring of centenarians inherit a potential advantage in the delay of developing age-related diseases,77 the incidence of IMMP2L intron deletions among these individuals appears to be slightly lower than their spouse group representing the general population. These results of genetic correlation analysis indicate that the centenarians display characteristics of better genomic integrity at the IMMP2L locus when compared with the general population which may be associated with their healthy longevity.
Discussion
Cellular senescence is one of the fundamental cell fates that plays important roles during the natural aging process. However, the distinct signaling program controlling this cell fate has remained less understood as many associated characteristics of cellular senescence, including cell cycle arrest, pro-inflammatory factor secretion and activated response to stress, are also shared by other important cell fates including quiescence, terminal differentiation and cell death. As described in this report, we have uncovered a signaling hierarchy that appears to be the switch controlling the cell fate decisions of healthy, senescent and apoptotic cells, which involves the collaboration between IMMP2L-GPD2 and IMMP2L-AIF signaling pathways centered on programming mitochondrial functionality as illustrated in Fig. 8.
Model of IMMP2L-mediated signaling on cell fate regulation. In healthy cells growing in normal condition, IMMP2L could regulate metabolic enzyme GPD2 towards achieving metabolic and signaling fitness associated with mitochondria. Upon exposure to oxidative stress, IMMP2L could then process AIF into its truncated form with pro-apoptotic activity, leading to clearance of irreparably damaged cells. Conversely, shutdown of the IMMP2L-GPD2 axis could establish the metabolic and signaling foundation for the cell fate favoring senescence with high levels of ROS, while simultaneous blockage of IMMP2L-AIF signaling could secure the survival of senescent cells in such stress condition
In the aftermath of encountering various types of stress stimuli, senescent cells are still viable and metabolically active, and this long-lasting state needs to be initiated and maintained by a specific metabolic program.55 In healthy cells, IMMP2L-mediated pre-sequence cleavage empowers the processed GPD2 to become metabolic active which enables a redox reaction to produce phospholipid precursor Glycerol 3-phosphate to maintain metabolic fitness of the cells. During the initiation of cellular senescence, shutdown of the IMMP2L-GPD2 axis would lead to reducing the supplies of phospholipid needed for both cell membrane synthesis and regulation of certain phospholipid-binding kinase pathways such as PKC-δ-associated signaling. These alterations could act in concert to maintain cells in a long-term stable cell-cycle arrest state featuring high levels of ROS. Currently, metabolic interventions and supplementations have been tested aiming to increase both healthy span and life span of human population.78 For example, therapeutic strategies to restore NAD+ levels have been shown to ameliorate some aging-related disorders.46 Considering that blockage of the IMMP2L-GPD2 axis is critically involved in the initiation of cellular senescence and GPD2 is highly expressed in muscle compared with other tissues,79 thorough investigations are needed to examine whether a combination of nutrition supplementations, which focus on achieving GPD2-related metabolic fitness regarding levels of both NAD+ and phospholipids, and specialized physical exercise/training modes, that aim at improving GPD2-assocaited mitochondrial functionality within skeletal muscle, could systematically protect cells against stress and thus may be potentially developed into geriatric medicine benefiting elderly populations.
In addition to elucidating IMMP2L’s critical role in metabolic programming underpinning the senescent state featuring high levels of ROS, our study has also uncovered that IMMP2L is concurrently involved in the critical pathway regulating cell death signaling to ensure the survival of senescent cells under oxidative stress. When normal cells are exposed to oxidative stress, IMMP2L could process AIF into its pro-apoptotic form to initiate cell death, which may help to remove cells with irreparable damage to maintain both cell and tissue homeostasis. In contrast, under oxidative stress stimuli, loss of IMMP2L switches off the path towards cell death due to the deficiency in the generation of pro-apoptotic AIF, consequently ensuring the damaged cells to be programmed into the senescent state and avoiding the fate of cell death. Considering that loss of IMMP2L can be physiologically observed across tissues and species, the decline in the rates of IMMP2L-AIF-dependent cell death in favor of cellular senescence may represent an additional mechanism accounting for the accumulation of viable senescent cells in tissues throughout the aging process, in addition to the acknowledged progressive incompetency of immune clearance.
Furthermore, while cooperative blockage of IMMP2L-GPD2 and IMMP2L-AIF signaling pathways forms the mechanistic basis for the establishment of a senescent state, activation of these two signaling branches under the control of the master regulator IMMP2L may need to be mutually exclusive in a physiological context, since activated IMMP2L-GPD2 axis represents the pro-survival arm that maintains the cellular fitness of mitochondria-associated metabolism and signaling network; whereas the other pro-apoptotic arm involving IMMP2L-mediated truncation of AIF is aimed at removing cells with irreparable damage upon oxidative stress exposure. Indeed, our attempts to probe the functional effect of enhancing the expression of IMMP2L on cellular senescence have yielded results with limited impact, although they are consistent with a critical role of this pathway in the determination of this cell fate versus cell death (data not shown). Future study focused on illuminating the mechanisms underlying the dynamic regulation of IMMP2L’s catalytic activity towards processing either GPD2 or AIF respectively will further advance the understanding of how IMMP2L, sitting in the crossroads, plays pivotal roles in the fate determination of healthy, senescent and apoptotic cells.
Given that the aging population is rapidly increasing worldwide,80 it is crucial to advance the development of geriatric medicine to extend healthy lifespan and ultimately support active and healthy aging.81 Previous studies suggested that selective killing of senescent cells in vivo would be a beneficial strategy to prevent the onsets of aging-associated disorders and facilitate the rejuvenation of individuals.11,12,13,14 Because cell fate decision towards senescence is a complex process involving the collaborations of multiple signaling pathways, which includes both senescence-unique and non-dividing cell fates-shared mechanisms, further in-depth understanding of the distinct mechanisms specifically underpinning the senescence program is critical to develop targeted therapeutics harnessing cellular senescence with minimum side effects on normal proliferating, quiescent and post-mitotic cells. In this regard, we have demonstrated that shutdown of IMMP2L signaling is uniquely associated with senescence along with illustrating its crucial implication on protecting senescent cells against the fate of cell death, which could potentially inspire the development of drugs targeting senescent cells.
Furthermore, a major challenge in the aging field remains in the lack of a definite and universal functional biomarker to identify cellular senescence in vivo besides p16Ink4a.82, 83 In this study, we have found that alterations of IMMP2L-mediated signaling are observed during cellular senescence associated with the natural mammalian aging process, suggesting that loss of IMMP2L may be a potential candidate of biomarker for senescence in vivo and also an indicator of aging at the molecular level. Finally, understanding the genetic basis associated with human longevity and healthy aging could provide significant biological insights into identification of protective factors and developing rejuvenated therapeutics for age-related disorders. In this regard, we have found that integrity of the genomic locus of IMMP2L is associated with healthy longevity. It will be of interest to conduct detailed investigations to understand if genomic integrity is the underlying mechanism to regulate IMMP2L properly to maintain tissue and organ homeostasis and integrity throughout the aging process, ultimately contributing to healthy longevity. Taken together, our findings that bridge mechanistic studies to genetic analysis ranging from cellular organelle to human populations have uncovered a new signaling hierarchy orchestrated by IMMP2L for cell fate determination.
Materials and methods
Cell culture
IMR-90 fetal lung fibroblasts, HeLa and HEK293T cells were obtained from ATCC. Normal Human Bronchial Epithelial Cells (NHBE) derived from healthy donors were obtained from LONZA. IMR-90 and HeLa cells were cultured in Eagle’s Minimum Essential Medium (EMEM) supplemented with 10% FBS and 1 × Penicillin Streptomycin. NHBE cells were kept in BEGM Basal Epithelial Growth Medium. HEK293T cells were maintained in Dulbecco’s modified eagle medium (DMEM) supplemented with 10% FBS. For cells undergoing replicative senescence, IMR-90 cells and NHBE cells were kept passaging for over 90 days and 40 days respectively when senescence was confirmed by cell cycle arrest, SA-β-gal activity and changes in morphology. For oncogenic Hras-induced senescence, IMR-90 cells carrying a Dox-inducible (Tet-One) vector expressing HrasV12 were grown with Dox (1 ug/mL) for 9 days.
For SILAC labeling, constructed inducible IMMP2L-expressing HeLa cells were divided into two parts and cultured in light DMEM medium containing 12C614N2‐Lys (K0) and 12C614N4‐Arg (R0) or heavy DMEM medium containing 13C614N2‐Lys (K6) and 13C615N4‐Arg (R10), respectively. Cells were cultured and passaged until the labeling efficiency reached 98%. Then exogenous IMMP2L was induced to express in light medium via adding Dox (1 ug/mL) 48 h before detection.
Human whole blood samples
Individuals from longevity families were recruited from Hainan province, China for the study. The research protocol was approved by the Ethics Committee at Kunming Institute of Zoology, Chinese Academy of Sciences. Informed consent was obtained from all participants prior to study and all individuals were generally healthy. Peripheral blood samples were obtained from venipuncture.
Mouse experiments
Wild type C57BL/6 were used at different ages. Mice with 2–3 months old were considered as young ones and 25–32 months old mice were regarded as geriatric. All experiments were approved by the Duke Institutional Animal Care and Use Committee.
Fluorescence activated cell sorting for collecting senescent cells
NHBE cells were cultured with DMSO or EGFR inhibitor Erlotinib (1 μM) for 20 h. After the treatment, cells continued to be incubated with C12FDG (33 μM) for 1 h. Cell were then suspending in ice-cold FACS buffer (2% FBS, 2 mM EDTA in PBS). FSC versus SSC set helped exclude the dead cells and then the green fluorescence gating based on relative β-gal activity is used to separately sort out quiescent (β-gal negative) and senescent (β-gal positive) population derived from Erlotinib-treated group. Cells treated with DMSO only were also collected using the gating criteria for quiescent cell population. Cell sorting was carried out using the Beckman MoFlo Astrios EQ sorter.
Transcriptional profiling and quantitative PCR
RNAs from control, quiescence, senescence groups were extracted by QIAGEN RNeasy Mini Kit. RNA processing and microarray analysis were done by Duke Sequencing and Genomic Technologies Core facility using GeneChip Human Genome U133 Plus 2.0 Array (Affymetrix). Based on data analysis using Partek Genomics Suite, genes with p value less than 0.05 and fold change more than 1.5 comparing senescence group with non-senescence group (both control and quiescence groups) were considered as genes specifically enriched for senescence program, which were then being used for gene ontology analysis using MetaCore.
For RNA quantitative PCR for cultured cells, primers DNA oligos were synthesized from IDT that have been listed in Supplementary information, Table S3, total RNAs from each sample were reversely transcribed by advanced iScript cDNA synthesis kit (Bio-Rad), and further mixed with primers and SYBR green mix (Affymetrix) followed by qPCR analysis using Mastercycler Realplex system (Eppendorf). For RNA quantitative PCR using human peripheral blood samples. Blood samples of 33 subjects containing Geriatric group containing 18 centenarians (CEN), young group containing 15s-degree offspring (F2) were treated with red blood cell lysis buffer (Tiangen) and then centrifuged at 4000 rpm for 10 min to isolate white blood cells. Total RNA from white blood cells was extracted using Trizol reagent (Invitrogen). The synthesized cDNA was then used for qPCR analysis using the CFX Connect Real-Time System (Bio-Rad) with GoTaq qPCR Master Mix (Promega).
Copy number variation assay
Blood samples of 30 subjects containing 10 centenarians (CEN), 10 their first-degree offspring (F1), 10 spouses of their offspring (F1SP) were randomly chosen for copy number assay. Genomic DNA (gDNA) was extracted using the AxyPrep Blood Genomic DNA Miniprep Kit (Axygen). gDNA with an A260/A280 ratio of greater than 1.7 was qualified and then diluted to 5 ng/μL using nuclease-free water to make 5 × stock solution. Reaction was performed on a 384-well plate using a copy number assay kit (Applied Biosystems). Four replicates of each sample were used for statistical analysis. Copy number was analyzed using the CopyCaller™ Software (Applied Biosystems). Probes for different locations of the target gene IMMP2L were from Applied Biosystems and listed in the Supplementary information, Table S3.
Lentivirus production and transduction
The lentiviral inducible expression vector pCDH-TetOne-MCS-EF1-Puro84 carrying SERPINB4, IMMP2L, Luciferase, SERPINB1, SERPINB3, SERPINB7, SERPINB9, SERPINB13 ORFs were individually cotransfected with the packaging plasmids psPAX2, pCI-VSVG using Lipofectamine LTX into HEK293T cells. For NHBE cells, lentiviral supernatant were concentrated using Lenti-X concentrator (Clontech) and then applied for 8 h with 2 μg/mL polybrene. For viral transduction in IMR-90 cells, cells were infected with viruses for 24 h with 4 μg/mL polybrene. 48 h post virus transduction; both NHBE and IMR-90 cells were cultured in selection medium containing 1 ug/mL puromycin for two days. After selection, 1 ug/mL Dox was added into medium to induce gene expression and cells were then collected for subsequent experiment. For control and the shRNA constructs (Supplementary information, Table S3) targeting SERPINB4, IMMP2L and GPD2, all the virus production and transduction procedures remain the same as that for ectopic expression.
SA-β-gal assay
SA-β-gal assay was conducted using Senescence β-Galactosidase Staining Kit (Cell Signaling). In general, indicated groups of cells were washed twice in PBS and then fixed using 2% formaldehyde and 0.2% glutaraldehyde in PBS for 5 min at room temperature. After further rinse in PBS again, cells will be stained with β-Galactosidase Staining Solution at 37 °C ranging from 8 h to overnight. Microscopic analyses were performed using an Olympus CK40 microscope with DP20 camera.
Flow cytometry analysis
Detection of SA-β-gal activity using flow cytometry. Erlotinib-induced senescent cells has been treated with C12FDG prior to detection of SA-β-gal positive cells using flow cytometry. For measuring cell proliferation, indicated groups of cells were incubated with 10 uM EdU for 2 h and the subsequent processing and labeling were performed using Click-iT EdU flow cytometry assay kit (Thermo Fisher). For measuring the levels of ROS, the cells were stained with 5 μM of CellROX Deep Red Reagent (Thermo Fisher) by cultured with the probe for 30 min. The cells were then collected and analyzed using flow cytometry. Regarding the cell death measurement, indicated groups of cells were harvested and immediately stained using Dead Cell Apoptosis Kit (Thermo Fisher). All flow cytometry analyses were performed on BD FACSCanto IIs. Data were analyzed using the Flow Jo software.
Western blot analyses
For immunoblot assay, cell culture related protein samples were prepared using lysis buffer (20 mM Tris-Cl, 100 mM NaCl, 0.5 mM EDTA) containing protease and phosphatase inhibitors with sonication. For protein samples from tissue, samples were mixed with T-PER buffer (Thermo Scientific) and further homogenized using TissueRuptor (QIAGEN). All samples were collected and boiled in loading buffer for 10 min. Protein concentrations were determined with the BCA Protein Assay (Pierce). Cell lysates were loaded on NuPAGE Bis-Tris protein gel (Thermo Scientific) and transferred to PVDF membrane (Millipore). Membranes were blocked with 5% BSA and visualized with Supersignal West Pico Chemiluminescent Substrate Kit (Thermo Fisher).
The following antibodies were used for immunoblots. Beta-Actin (Sigma, A5316, 1:5000), p16 (BD, 551154, 1:1000), SERPINB4 (Aviva, ARP42085_P050, 1:1000), IMMP2L (Abcam, ab103326, 1:1000), SEC11A (Proteintech, 14753-1-AP, 1:1000), IMMP2L (Aviva, ARP60563_P050, 1:1000), AIF (Santa Cruz, sc-13116, 1:1000), GPD2 (Santa Cruz, sc-393620, 1:1000), Phospho-Rb (Cell Signaling, 8516 S, 1:1000), Rb (BD, 554136, 1:1000), IL-6 (Cell Signaling, 12153, 1:1000), SMAC (Cell Signaling, 15108, 1:1000), HtrA2 (Santa Cruz, sc-271528, 1:1000), ENDOG (Santa Cruz, sc-365359, 1:1000), COX IV (Cell Signaling, 4850 P, 1:1000), GAPDH (Cell Signaling, 5174, 1:5000), Mouse HRP (Thermo Fisher, G-21040, 1:5000), Rabbit HRP (Thermo Fisher, G-21234, 1:5000), PKC-δ (Cell Signaling, 9616 S, 1:1000), PKC-δ (LSBio, C98923, 1:1000), Phospho-PDHE1a (Millipore Sigma, AP1064, 1: 5000), PDHE1a (Cell Signaling, 3205 S, 1:1000).
Mitochondria-based proteome profiling and TAILS experiment
Isolated mitochondrial pellets were resuspended in lysis buffer (100 mM HEPES (pH 8.0), 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS and 1 mM PMSF) containing protease inhibitor cocktail (1 tablet per 10 mL buffer) and sonicated on ice for 3 min. The lysates were clarified by centrifugation at 21,130 × g for 10 min at 4 °C, and then the supernatant was collected. The amount of mitochondrial proteins was quantified using standard BCA protein assay kit. For quantitative analysis, 200 µg proteins of both samples were combined prior to subsequent procedures. Guanidine hydrochloride were added to the lysate to a final concentration of 4 M for mitochondrial protein denature. Then the proteins were reduced with 5 mM dithiothreitol (DTT) at 56 °C for 30 min and alkylated with 15 mM iodoacetamide (IAA) in the dark at room temperature for 30 min. The alkylation was quenched by DTT at the final concentration of 20 mM at room temperature.
For SILAC-labeled proteome profiling experiment, 100 µg protein mixture were transferred to the 30 KDa molecular weight cut-off filter, spun down and washed twice with 100 mM HEPES buffer (pH 8.0). Then the proteins were resuspended in 100 μL 50 mM HEPES (pH 8.0) in the filter and digested by trypsin at a protein to enzyme ratio of 50:1 (w/w) (overnight, 37 °C). Then the digested peptides were filtered, desalted, and fractionated into six fractions using home-made C18 Stage-Tip.85 Peptide samples were vacuum dried for HPLC−MS/MS analysis.
For the TAILS enrichment experiment, 300 µg protein mixture were extracted. Then the primary amines were blocked at the protein level with 40 mM formaldehyde and 20 mM sodium cyanoborohydride (overnight, pH 6.5, 37 °C). The dimethyl labeling reaction was quenched with 100 mM Tris-HCl (pH 6.8) at 37 °C for 4 h. The mixture was digested with trypsin. The tryptic peptides were incubated with hyperbranched polyglycerol-aldehydes polymer at a polymer:peptide ratio of 5:1 (w/w) and with 20 mM sodium cyanoborohydride (pH 7, overnight, 37 °C). The reaction was quenched with 100 mM Tris-HCl (pH 6.8) at 37 °C for 30 min and the peptides with demethylation-blocked N-terminal were recovered by ultrafiltration. The filtered peptides were desalted and fractionated into 3 fractions by home-made C18 Stage-Tip, and vacuum dried for HPLC−MS/MS analysis.
For the HPLC-MS/MS analysis, the peptides were analyzed on an Orbitrap Fusion mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA) equipped with an EASY-nLC 1000 ultra-high-pressure liquid chromatography(HPLC) system (Thermo Fisher Scientific, Waltham, MA, USA). The peptides were dissolved in mobile phase buffer A (0.1% formic acid (FA), 2% acetonitrile in water) and loaded onto an in-home made capillary column (75 μm ID × 15 cm) with C18 resin (3 μm, 100 Å, Dikma, China) by an autosampler. Then the peptides were separated and eluted with a linear gradient of 8%–32% of mobile phase buffer B (0.1% FA in 90% acetonitrile) for 50 min, 32%–48% buffer B for 5 min and 48%–80% buffer B for 5 min at the flow rate of 300 nL/min on HPLC system. The eluted peptides were ionized under high voltage (2.2 KV) and detected by Orbitrap Fusion using nano-spray ion source (NSI). For MS1 scan, peptides with 350–1300 m/z were detected in the Orbitrap at the resolution 70,000 at 200 m/z. The automatic gain control (AGC) target was set to 5 × 105, and maximum ion injection time was set to 50 ms. The ions with intensity above 5 × 103 were subjected to fragmentation via high energy collision induced dissociation (HCD) with 32% normalized collision energy (NCE). The fragmented ions were analyzed in LTQ. The AGC target in the ion trap was set to 7 × 103, and the isolation width was 2 m/z.
Regarding the database search, for the SILAC labeled proteome profiling, the raw data were searched with MaxQuant software (version 1.4.1.2) based on Andromeda search engine against a composite target/decoy database to estimate false discovery rate (FDR). The target Human database (version 20170507, 88817 protein sequences) was downloaded from UniProt database (http://www.uniprot.org/), and the decoy database was derived from pseudoreversed sequences of the target proteins. Lys0 and Arg0 were set as “light label,” Lys6 and Arg10 were set as “Heavy labels.” Enzyme specificity with trypsin was used, and maximum missing cleavages was set as two. The mass error tolerance for precursor ions was 20 ppm and fragment ions was 0.5 Da. The fixed modification was set as Carbamidomethyl (C) and variable modifications were oxidation (M), acetylation (Protein N-term). False discovery rate (FDR) thresholds for protein and peptide were set at 0.01.
For the TAILS analysis, the raw data were searched with Proteome Discoverer software (version 1.4; Thermo Fisher Scientific Inc.) based on Mascot search engine (version 2.3.01; Matrix Science, London, UK) with percolator. The strategy of target/decoy database search was used to estimate false discovery rate (FDR). The target Human database was downloaded from UniProt database, and the decoy database was derived from pseudoreversed sequences of the target proteins. Enzyme specificity with trypsin was used, and maximum missing cleavages was set as two. The mass error tolerance for precursor ions was 20 ppm and fragment ions was 0.5 Da. The fixed modification was set as Carbamidomethyl (C) and variable modifications were oxidation (M), acetylation (Protein N-term), dimethylation (K), dimethylation (N-term), stable isotope labeled Lys (+6.02013 Da), stable isotope labeled Arg (+8.02013 Da). At peptide level, the FDR was set as 1%. At the protein level, peptide-spectrum match (PSM) with at least high confidence and a strict maximum parsimony principle were applied. The peptides with Mascot score lower than 20 were removed. For quantitative analysis, the area of extracted ion currents (XICs) of the corresponding peptides were calculated and compared between control and IMMP2L overexpression samples. The Neo-N-termini peptides with H/L ratio lower than 0.67, or only identified in IMMP2L overexpression samples were picked out, which indicated that these peptides were generated by IMMP2L cleavage. Also the mature protein N-terminal peptides with H/L ratio higher than 1.5, or only identified in the control sample were picked out, which indicated the corresponding proteins may be the substrate of IMMP2L. Quantitative result of each candidate was manually checked and recalculated.
Mitochondrial isolation and cell fractionation
For proteome profiling and TAILS experiment, mitochondria have been isolated using the QProteome mitochondria isolation kit (QIAGEN). For all the immunoblot analysis, mitochondria isolation kit (Miltenyibiotec) was used for the isolation. For experiment involving the usage of subcellular fractions, cells were fractionated using the Subcellular Protein Fractionation Kit (Thermo Fisher) according to the manufacture’s instruction.
NAD/NADH quantification assay
The cytosolic fractions obtained from subcellular fraction assay were used for quantifying NAD/NADH using an NAD/NADH Quantification Kit (Promega), as described by the manufacturer.
Polar metabolite profiling
IMR-90 cell pellets were dissolved in 1 mL pre-cooled 80% methanol/water and incubate for 15 min at −80 °C, the supernatants collected post centrifuge were then split and transferred to two new Eppendorf tubes (one tube is for back up) and dried in vacuum concentrator at room temperature. The dry pellets were reconstituted into 30 uL sample solvent (water:methanol:acetonitrile, 2:1:1, v/v) and 3 uL was further analyzed by liquid chromatography-mass spectrometry (LC-MS). Ultimate 3000 UHPLC (Dionex) is coupled to Q Exactive Plus-Mass spectrometer (QE-MS, Thermo Scientific) for metabolite profiling. A hydrophilic interaction chromatography method (HILIC) employing an Xbridge amide column (100 × 2.1 mm i.d., 3.5 um; Waters) is used for polar metabolite separation.
Metabolomics data analysis-LC-MS peak extraction and integration were performed using commercial available software Sieve 2.2 (Thermo Scientific). The peak area was used to represent the relative abundance of each metabolite in different samples.
Lipidomics profiling in IMR-90 Cells
Flash-frozen cell pellets including control and IMMP2L-overexpressing IMR-90 cells together with senescent IMR-90 cells (oncogenic Hras-induced senescence) were homogenized in a minimum volume (100 uL) 50 mM ammonium bicarbonate (AmBic) with probe sonication, and concentration normalized based on a Bradford assay. To 115 uL of each sample lysate, 200 uL MeOH containing 1:100 dilution of the SPLASH LipidoMix (Avanti Polar Lipids) deuterated internal standards was added, and samples were mixed at 25 °C on Thermomixer for 5 min. An additional 600 uL of MTBE was added to each well, and samples were mixed on a thermomixer at room temperature for 60 min. Samples were centrifuged to perform phase separation, 2000 × g for 10 min at room temperature. 800 uL of the upper organic layer was transferred to glass LC vials and dried under N2 evaporation. Samples were resuspended in 50 uL of 4/3/1 v/v/v 2-propanol/MeCN/water, with shaking for 10 min at 37 °C. The samples were then immediately placed in the autosampler for analysis. The samples were analyzed using Ultraperformance Liquid Chromatography/Electrospray Ionization/Tandem Mass Spectrometry (UPLC/ESI/MS/MS) using a method allowing chromatographic resolution of many of the lipids by class and chain length/unsaturation. 5 uL injections were used in ESI+mode and 10 uL in ESI- mode, and UPLC separation of the lipids was performed on a Waters Acquity UPLC (Milford, MA) using a Waters Acquity 2.1 mm × 10 mm 1.7 μm CSH C18 column. Mobile phase A was 10 mM ammonium formate and 0.1% formic acid in 40/60 v/v water/MeCN. Mobile phase B was 0.1% formic acid in 10/90 v/v MeCN/2-propanol. Samples were introduced into a Synapt G2 HDMS mass spectrometer (Waters) and analyzed in acquisition Sensitivity mode (~15,000 Rs), with 0.4 s MS full scan followed by one 0.4 s data-dependent MS/MS scan. Dynamic exclusion was enabled within 1.2 Da for 30 s.
Raw data was imported into Progenesis QI (Nonlinear Dynamics) for peak-picking and data alignment between samples. This approach allows the same lipid “features” (as determined based on an accurate mass and retention time coordinate) to be quantified across all samples, resulting in minimal missing data for any sample. Raw data was aligned using default parameters, with the exception that lockmass correction was enabled using Leucine-Ekephalin (556.2771 ESI+, 554.2615 ESI-). ESI+ion adducts allowed were M+H, M+NH4, and M+Na. For ESI-, ion adducts allowed were M-H, M+Cl, M-2H, and M-H20-H. Raw peak intensities (without intensity scaling) as well as the robust mean scaled intensities for each analysis are reported for all lipid “compounds” detected. All lipid “compounds,” which consist of an accurate mass, retention time, and MS/MS spectra as available, were subjected to metabolite database searching against the LipidMAPS (http://www.lipidmaps.org/). A series of curation steps were undertaken in order to provide the set of lipid metabolites which were robustly quantified and subsequently an estimation of those which were differentially expressed; This curation process suggests that there are approximately 1000 robustly quantified lipid species in the ESI+data, and ~500 in the ESI data.
Lipid profiling in mice skeletal muscle
Weighed about 7 mg of skeletal muscle tissue in a new 2-mL Eppendorf tube on ice. Added 200 uL of ice-cold 80% methanol (HPLC grade) and stirred using TissueRuptor (QIAGEN) until tissue was well mixed with solvent. Added another 200-uL ice-cold 80% methanol, vortex for 1 min. Added 960 uL of ice-cold MTBE and vortex for 1 min. Added 240 uL of ice-cold water, vortex for 2 min. The final extraction solvent composition is MeOH/MTBE/H2O 1:3:1 (v/v/v, all HPLC grade). Centrifuge at 3500 × g 4°C for 10 min, two layers formed. Top layer contained lipids, while bottom layer contained polar metabolites. Transferred the top layer supernatant that contains lipids into two tubes so that each tube had 2.5-3 mg tissue. Dried the top layer supernatant in vacuum concentrator at room temperature. The dry pellets were reconstituted into 50 uL isopropanol (IPA) (per 3 mg tissue). Vortexed for 1 min and centrifuged at 20,000 × g at 5 °C. 3 uL supernatant was injected and further analyzed by liquid chromatography-mass spectrometry (LC-MS). Ultimate 3000 UHPLC (Dionex) was coupled to Q Exactive Plus-Mass spectrometer (QE-MS, Thermo Scientific) for lipids profiling. A reversed phase liquid chromatography method (RPLC) employing an Xbridge BEH C18 column (100 × 2.1 mm i.d., 2.5 um; Waters) at 40 °C was used for lipid separation. The mobile phase A is 60:40 (v/v) acetonitrile:water with 0.1% formic acid and 10 mM ammonium formate. The mobile phase B is 90:10 (v/v) isopropanol:acetonitrile with 0.1% formic acid and 10 mM ammonium formate. The linear gradient used is as follows: 0 min, 40% B; 1.5 min, 40% B, 5 min, 85% B; 12 min, 97% B, 16 min, 97% B, 16.5 min, 40% B, 20.5 min, 40% B. The flow rate was 0.2 mL/min. LC-MS peak extraction and integration were performed using commercial available software Sieve 2.2 (Thermo Scientific). The peak area was used to represent the relative abundance of each metabolite in different samples.
Data availability
The RNA transcriptional profiling data involved in this study has been uploaded to NCBI GEO datasets (GSE100014). The proteomics raw data have been uploaded to the ProteomeXchange Consortium86 with the data set identifier PXD006602 (ID: reviewer44025@ebi.ac.uk; password: E26iAcnV).
Statistical information
Statistical Analyses were performed using GraphPad Prism software. Significance was tested by unpaired two-tailed student t-test. A p value less than 0.05 was considered significant. *p < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
References
Collado, M., Blasco, M. A. & Serrano, M. Cellular senescence in cancer and aging. Cell 130, 223–233 (2007).
Campisi, J. Aging cellular senescence, and cancer. Annu. Rev. Physiol. 75, 685–705 (2013).
Rodier, F. & Campisi, J. Four faces of cellular senescence. J. Cell Biol. 192, 547–556 (2011).
Munoz-Espin, D. et al. Programmed cell senescence during mammalian embryonic development. Cell 155, 1104–1118 (2013).
Storer, M. et al. Senescence is a developmental mechanism that contributes to embryonic growth and patterning. Cell 155, 1119–1130 (2013).
Jun, J. I. & Lau, L. F. The matricellular protein CCN1 induces fibroblast senescence and restricts fibrosis in cutaneous wound healing. Nat. Cell Biol. 12, 676–685 (2010).
Xue, W. et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature 445, 656–660 (2007).
Herbig, U., Ferreira, M., Condel, L., Carey, D. & Sedivy, J. M. Cellular senescence in aging primates. Science 311, 1257 (2006).
van Deursen, J. M. The role of senescent cells in ageing. Nature 509, 439–446 (2014).
Lopez-Otin, C., Blasco, M. A., Partridge, L., Serrano, M. & Kroemer, G. The hallmarks of aging. Cell 153, 1194–1217 (2013).
Chang, J. et al. Clearance of senescent cells by ABT263 rejuvenates aged hematopoietic stem cells in mice. Nat. Med. 22, 78–83 (2016).
Baker, D. J. et al. Naturally occurringp16(Ink4a)-positive cells shorten healthy lifespan. Nature 530, 184–189 (2016).
Baar, M. P. et al. Targeted apoptosis of senescent cells restores tissue homeostasis in response to chemotoxicity and aging. Cell 169, 132–147 e116 (2017).
Jeon, O. H. et al. Local clearance of senescent cells attenuates the development of post-traumatic osteoarthritis and creates a pro-regenerative environment. Nat. Med. 23, 775–781 (2017).
Alexander, P. B. et al. EGF promotes mammalian cell growth by suppressing cellular senescence. Cell Res. 25, 135–138 (2015).
Debacq-Chainiaux, F., Erusalimsky, J. D., Campisi, J. & Toussaint, O. Protocols to detect senescence-associated beta-galactosidase (SA-betagal) activity, a biomarker of senescent cells in culture and in vivo. Nat. Protoc. 4, 1798–1806 (2009).
Young, A. R. et al. Autophagy mediates the mitotic senescence transition. Genes Dev. 23, 798–803 (2009).
Elzi, D. J. et al. Plasminogen activator inhibitor 1--insulin-like growth factor binding protein 3 cascade regulates stress-induced senescence. Proc. Natl Acad. Sci. USA 109, 12052–12057 (2012).
Kortlever, R. M., Higgins, P. J. & Bernards, R. Plasminogen activator inhibitor-1 is a critical downstream target of p53 in the induction of replicative senescence. Nat. Cell Biol. 8, 877–884 (2006).
Catanzaro, J. M. et al. Oncogenic Ras induces inflammatory cytokine production by upregulating the squamous cell carcinoma antigens SerpinB3/B4. Nat. Commun. 5, 3729 (2014).
Silverman, G. A. et al. Human clade B serpins (ov-serpins) belong to a cohort of evolutionarily dispersed intracellular proteinase inhibitor clades that protect cells from promiscuous proteolysis. Cell. Mol. life Sci. (CMLS) 61, 301–325 (2004).
Schick, C. et al. Squamous cell carcinoma antigen 2 is a novel serpin that inhibits the chymotrypsin-like proteinases cathepsin G and mast cell chymase. J. Biol. Chem. 272, 1849–1855 (1997).
de Koning, P. J. et al. Intracellular serine protease inhibitor SERPINB4 inhibits granzyme M-induced cell death. PLoS ONE 6, e22645 (2011).
Cooley, J., Takayama, T. K., Shapiro, S. D., Schechter, N. M. & Remold-O’Donnell, E. The serpin MNEI inhibits elastase-like and chymotrypsin-like serine proteases through efficient reactions at two active sites. Biochemistry 40, 15762–15770 (2001).
Rawlings, N. D., Barrett, A. J. & Bateman, A. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 40, D343–350 (2012).
Quesada, V., Ordonez, G. R., Sanchez, L. M., Puente, X. S. & Lopez-Otin, C. The Degradome database: mammalian proteases and diseases of proteolysis. Nucleic Acids Res. 37, D239–243 (2009).
Oue, N. et al. Signal peptidase complex 18, encoded by SEC11A, contributes to progression via TGF-alpha secretion in gastric cancer. Oncogene 33, 3918–3926 (2014).
Lu, B. et al. A mutation in the inner mitochondrial membrane peptidase 2-like gene (Immp2l) affects mitochondrial function and impairs fertility in mice. Biol. Reprod. 78, 601–610 (2008).
Hirst, C. E. et al. The intracellular granzyme B inhibitor, proteinase inhibitor 9, is up-regulated during accessory cell maturation and effector cell degranulation, and its overexpression enhances CTL potency. J. Immunol. 170, 805–815 (2003).
Bhatia, S. R. et al. Using C. elegans to identify the protease targets of serpins in vivo. Methods Enzymol. 499, 283–299 (2011).
Schmidt, O., Pfanner, N. & Meisinger, C. Mitochondrial protein import: from proteomics to functional mechanisms. Nat. Rev. Mol. Cell Biol. 11, 655–667 (2010).
George, S. K., Jiao, Y., Bishop, C. E. & Lu, B. Mitochondrial peptidase IMMP2L mutation causes early onset of age-associated disorders and impairs adult stem cell self-renewal. Aging Cell 10, 584–594 (2011).
Uhlen, M. et al. Proteomics. Tissue-based map of the human proteome. Science 347, 1260419 (2015).
Calabrese, F. et al. Serpin B4 isoform overexpression is associated with aberrant epithelial proliferation and lung cancer in idiopathic pulmonary fibrosis. Pathology 44, 192–198 (2012).
Sivaprasad, U. et al. A nonredundant role for mouse Serpinb3a in the induction of mucus production in asthma. J. Allergy Clin. Immunol. 127, 254–261 (2011). 261 e251-256.
Mossmann, D., Meisinger, C. & Vogtle, F. N. Processing of mitochondrial presequences. Biochim Biophys. Acta 1819, 1098–1106 (2012).
Chacinska, A., Koehler, C. M., Milenkovic, D., Lithgow, T. & Pfanner, N. Importing mitochondrial proteins: machineries and mechanisms. Cell 138, 628–644 (2009).
Kleifeld, O. et al. Isotopic labeling of terminal amines in complex samples identifies protein N-termini and protease cleavage products. Nat. Biotechnol. 28, 281–288 (2010).
Kleifeld, O. et al. Identifying and quantifying proteolytic events and the natural N terminome by terminal amine isotopic labeling of substrates. Nat. Protoc. 6, 1578–1611 (2011).
Ieva, R. et al. Mitochondrial inner membrane protease promotes assembly of presequence translocase by removing a carboxy-terminal targeting sequence. Nat. Commun. 4, 2853 (2013).
Hung, V. et al. Proteomic mapping of the human mitochondrial intermembrane space in live cells via ratiometric APEX tagging. Mol. Cell 55, 332–341 (2014).
Bharadwaj, M. S., Zhou, Y., Molina, A. J., Criswell, T. & Lu, B. Examination of bioenergetic function in the inner mitochondrial membrane peptidase 2-like (Immp2l) mutant mice. Redox Biol. 2C, 1008–1015 (2014).
Lee, Y. J., Jeschke, G. R., Roelants, F. M., Thorner, J. & Turk, B. E. Reciprocal phosphorylation of yeast glycerol-3-phosphate dehydrogenases in adaptation to distinct types of stress. Mol. Cell Biol. 32, 4705–4717 (2012).
Xia, J., Sinelnikov, I. V., Han, B. & Wishart, D. S. MetaboAnalyst 3.0—making metabolomics more meaningful. Nucleic Acids Res. 43, W251–257 (2015).
Wiley, C. D. et al. Mitochondrial dysfunction induces senescence with a distinct secretory phenotype. Cell Metab. 23, 303–314 (2016).
Zhang, H. et al. NAD(+) repletion improves mitochondrial and stem cell function and enhances life span in mice. Science 352, 1436–1443 (2016).
Verdin, E. NAD(+) in aging, metabolism, and neurodegeneration. Science 350, 1208–1213 (2015).
Vance, J. E. Phospholipid synthesis and transport in mammalian cells. Traffic 16, 1–18 (2015).
Tamura, Y., Sesaki, H. & Endo, T. Phospholipid transport via mitochondria. Traffic 15, 933–945 (2014).
Lemmon, M. A. Membrane recognition by phospholipid-binding domains. Nat. Rev. Mol. Cell Biol. 9, 99–111 (2008).
Rosse, C. et al. PKC and the control of localized signal dynamics. Nat. Rev. Mol. Cell Biol. 11, 103–112 (2010).
Jiang, P., Du, W., Mancuso, A., Wellen, K. E. & Yang, X. Reciprocal regulation of p53 and malic enzymes modulates metabolism and senescence. Nature 493, 689–693 (2013).
Churchill, E. N., Murriel, C. L., Chen, C. H., Mochly-Rosen, D. & Szweda, L. I. Reperfusion-induced translocation of deltaPKC to cardiac mitochondria prevents pyruvate dehydrogenase reactivation. Circ. Res. 97, 78–85 (2005).
Qvit, N., Disatnik, M. H., Sho, E. & Mochly-Rosen, D. Selective phosphorylation inhibitor of delta protein kinase c-pyruvate dehydrogenase kinase protein-protein interactions: application for myocardial injury in vivo. J. Am. Chem. Soc. 138, 7626–7635 (2016).
Kaplon, J. et al. A key role for mitochondrial gatekeeper pyruvate dehydrogenase in oncogene-induced senescence. Nature 498, 109–112 (2013).
Hangen, E., Blomgren, K., Benit, P., Kroemer, G. & Modjtahedi, N. Life with or without AIF. Trends Biochem Sci. 35, 278–287 (2010).
Otera, H., Ohsakaya, S., Nagaura, Z., Ishihara, N. & Mihara, K. Export of mitochondrial AIF in response to proapoptotic stimuli depends on processing at the intermembrane space. EMBO J. 24, 1375–1386 (2005).
Polster, B. M., Basanez, G., Etxebarria, A., Hardwick, J. M. & Nicholls, D. G. Calpain I induces cleavage and release of apoptosis-inducing factor from isolated mitochondria. J. Biol. Chem. 280, 6447–6454 (2005).
Mizukoshi, S. et al. Activation of mitochondrial calpain and release of apoptosis-inducing factor from mitochondria in RCS rat retinal degeneration. Exp. Eye Res. 91, 353–361 (2010).
Norberg, E. et al. An increase in intracellular Ca2+is required for the activation of mitochondrial calpain to release AIF during cell death. Cell Death Differ. 15, 1857–1864 (2008).
Joshi, A., Bondada, V. & Geddes, J. W. Mitochondrial micro-calpain is not involved in the processing of apoptosis-inducing factor. Exp. Neurol. 218, 221–227 (2009).
Wang, Y. et al. Calpain activation is not required for AIF translocation in PARP-1-dependent cell death (parthanatos). J. Neurochem. 110, 687–696 (2009).
Kroemer, G., Galluzzi, L. & Brenner, C. Mitochondrial membrane permeabilization in cell death. Physiol. Rev. 87, 99–163 (2007).
Barbouti, A., Amorgianiotis, C., Kolettas, E., Kanavaros, P. & Galaris, D. Hydrogen peroxide inhibits caspase-dependent apoptosis by inactivating procaspase-9 in an iron-dependent manner. Free Radic. Biol. Med. 43, 1377–1387 (2007).
Son, Y. O. et al. Apoptosis-inducing factor plays a critical role in caspase-independent, pyknotic cell death in hydrogen peroxide-exposed cells. Apoptosis 14, 796–808 (2009).
Yu, J. Y. et al. Continuous presence of H(2)O(2) induces mitochondrial-mediated, MAPK- and caspase-independent growth inhibition and cytotoxicity in human gingival fibroblasts. Toxicol. Vitr. 26, 561–570 (2012).
Doherty, T. J. Invited review: aging and sarcopenia. J. Appl. Physiol. 95, 1717–1727 (2003).
Sousa-Victor, P. et al. Geriatric muscle stem cells switch reversible quiescence into senescence. Nature 506, 316–321 (2014).
Liu, Y. et al. Expression ofp16(INK4a) in peripheral blood T-cells is a biomarker of human aging. Aging Cell 8, 439–448 (2009).
Erikson, G. A. et al. Whole-genome sequencing of a healthy aging cohort. Cell 165, 1002–1011 (2016).
Christensen, K., Johnson, T. E. & Vaupel, J. W. The quest for genetic determinants of human longevity: challenges and insights. Nat. Rev. Genet. 7, 436–448 (2006).
Murabito, J. M., Yuan, R. & Lunetta, K. L. The search for longevity and healthy aging genes: insights from epidemiological studies and samples of long-lived individuals. J. Gerontol. A Biol. Sci. Med. Sci. 67, 470–479 (2012).
Brooks-Wilson, A. R. Genetics of healthy aging and longevity. Hum. Genet. 132, 1323–1338 (2013).
He, Y. H. et al. Mitochondrial DNA content contributes to healthy aging in Chinese: a study from nonagenarians and centenarians. Neurobiol. Aging 35, 1779 e1771–1774 (2014).
Gimelli, S. et al. Interstitial 7q31.1 copy number variations disrupting IMMP2L gene are associated with a wide spectrum of neurodevelopmental disorders. Mol. Cytogenet. 7, 54 (2014).
Bertelsen, B. et al. Intragenic deletions affecting two alternative transcripts of the IMMP2L gene in patients with Tourette syndrome. Eur. J. Hum. Genet. 22, 1283–1289 (2014).
Terry, D. F. et al. Lower all-cause, cardiovascular, and cancer mortality in centenarians’ offspring. J. Am. Geriatr. Soc. 52, 2074–2076 (2004).
Lopez-Otin, C., Galluzzi, L., Freije, J. M., Madeo, F. & Kroemer, G. Metabolic control of longevity. Cell 166, 802–821 (2016).
Koza, R. A. et al. Sequence and tissue-dependent RNA expression of mouse FAD-linked glycerol-3-phosphate dehydrogenase. Arch. Biochem. Biophys. 336, 97–104 (1996).
Beard, J. R. & Bloom, D. E. Towards a comprehensive public health response to population ageing. Lancet 385, 658–661 (2015).
Kaeberlein, M., Rabinovitch, P. S. & Martin, G. M. Healthy aging: the ultimate preventative medicine. Science 350, 1191–1193 (2015).
He, S. & Sharpless, N. E. Senescence in health and disease. Cell 169, 1000–1011 (2017).
Sharpless, N. E. & Sherr, C. J. Forging a signature of in vivo senescence. Nat. Rev. Cancer 15, 397–408 (2015).
Hu, J. et al. MiR-215 is induced post-transcriptionally via HIF-drosha complex and mediates glioma-initiating cell adaptation to hypoxia by targeting KDM1B. Cancer Cell 29, 49–60 (2016).
Wisniewski, J. R., Zougman, A. & Mann, M. Combination of FASP and stagetip-based fractionation allows in-depth analysis of the hippocampal membrane proteome. J. Proteome Res. 8, 5674–5678 (2009).
Vizcaino, J. A. et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44, 11033 (2016).
Acknowledgements
We thank the Duke Flow Cytometry Shared Resource (FCSR) for assistance with flow cytometry, Duke Sequencing and Genomic Technologies for assistance with transcriptional analysis, laboratory of Dr. Jason Locasale for polar metabolite analysis (CCSG grant: P30CA01423), and Duke Proteomics and Metabolomics Shared Resource for lipidomics profiling. This work was supported by Chinese Academy of Sciences (QYZDBSSW-SMC020 and 152453KYSB20160006 to Q.K.), National Basic Research Program of China (2014CBA02004 to M.T.) and Chinese Academy of Sciences (XDA12020314 to M.T.), National Institutes of Health (R01-CA154586 to X.F.W.); CTSA grant (UL1TR001117 to X.F.W.).
Author information
Authors and Affiliations
Contributions
Conceptualization, L.Y. and X.F.W.; Methodology, L.Y. and M.T.; Formal Analysis, L.Y., L.Z., L.Q., X.L., J.L., and M.T.; Investigation, L.Y., L.Z., L.Q., Y.D., D.H., X.L., J.L., J. W.T., H.Y., X.C., and J.H.; Resources, Q.K. and M.T.; Data Curation, L.Y. and M.T.; Writing-Original Draft, L.Y.; Writing-Review & Editing, L.Y. and X.F.W.; Visualization, L.Y.; Supervision, Q.K., M.T. and X.F.W., Funding Acquisition, Q.K., M.T., and X.F.W.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Rights and permissions
About this article
Cite this article
Yuan, L., Zhai, L., Qian, L. et al. Switching off IMMP2L signaling drives senescence via simultaneous metabolic alteration and blockage of cell death. Cell Res 28, 625–643 (2018). https://doi.org/10.1038/s41422-018-0043-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41422-018-0043-5
This article is cited by
-
Antagonizing the irreversible thrombomodulin-initiated proteolytic signaling alleviates age-related liver fibrosis via senescent cell killing
Cell Research (2023)
-
Mitochondrial ClpP serine protease-biological function and emerging target for cancer therapy
Cell Death & Disease (2020)
-
Cellular senescence: from anti-cancer weapon to anti-aging target
Science China Life Sciences (2020)
-
Profile of Dr. Xiao-Fan Wang
Science China Life Sciences (2020)
-
Sanofi-Cell Research outstanding paper award of 2018
Cell Research (2019)