Abstract
Chromodomain helicase DNA binding protein (CHD) family plays critical roles in regulating gene transcription. The family is linked to cancer disease, but the family member’s role in tumorigenesis remains largely unknown. Here, we report that CHD6 is highly expressed in colorectal cancer (CRC). CHD6 knockdown inhibited cancer cell proliferation, migration, invasion, and tumorigenesis. Consistently, Villin-specific Chd6 knockout in mice attenuates cancer formation in AOM/DSS model. We found that aberrant EGF signals promoted the stability of CHD6 by diminishing ubiquitin-mediated degradation. EGF signal inhibits GSK3β activity, which in turn prevents phosphodegron formation of CHD6, thereby hindering E3 ligase FBXW7-mediated CHD6 ubiquitination and degradation. CHD6’s chromatin remodeler activity engages in binding Wnt signaling transcription factor TCF4 to facilitate the transcriptional expression of TMEM65, a mitochondrial inner membrane protein involved in ATP production and mitochondrial dynamics. In addition, Wnt signaling is also an upstream regulator of CHD6. CHD6 promoter contains TCF4 and β-catenin binding site, and CHD6 can be transcriptionally activated by Wnt ligand to facilitate TMEM65 transcription. Thus CHD6-TMEM65 axis can be regulated by both EGF and Wnt signaling pathways through two different mechanisms. We further illustrate that CHD6-TMEM65 axis is deregulated in cancer and that co-administration of Wnt inhibitor LGK974 and the anti-EGFR monoclonal antibody cetuximab largely restricted the growth of patient-derived xenografts of CRC. Targeting CHD6-TMEM65 axis may be effective for cancer intervention.
Similar content being viewed by others
Introduction
Colorectal cancer (CRC) is a genetic disease with accumulated genetic alterations during the progression and invasion1. CRC is highly aggressive and is the third leading malignance in the world population, causing near 900,000 death per year, and its incidence has been a health care challenge2. Despite treatment progress has been made, CRC continues to be one of the deadliest cancer types with different molecular phenotypes/strong resistance to therapies3 and very high mortality rate2. Thus, there is an urgent need to identify more molecular biomarkers for CRC. Further, it is critical to explore the molecular mechanisms underlying CRC progression, which may help provide new therapeutic targets.
Chromodomain helicase DNA-binding protein (CHD) family is composed of several members. This group of enzymes belongs to the SNF2 superfamily of ATP-dependent chromatin remodelers4. CHD proteins contain nine members and can be classified into three subfamilies on the basis of their functional domain similarity4. CHD activities are critical for a wide spectrum of cellular functions, including transcriptional regulation, cell growth, cell death, development, virus infection5, autophagy6, DNA damage response7, and genome integrity4. Deregulations of CHD in various human cancers are emerging, indicating that chromatin dynamics is critical during tumorigenesis. Subfamily III includes CHD6, CHD7, CHD8, and CHD9. These four enzymes are similar in their constituent domains, but they have non-redundant roles in the cell functions. The mechanisms behind their distinct and non-overlapping activities are unclear. Low CHD7 expression leads to enhanced survival of pancreatic cancer patients, suggesting that CHD7 promotes oncogenesis8. CHD8 is critical in recruiting E2F1 to the promoter of cyclin E2 for transcriptional activation9. The roles of CHD subfamily III proteins, including CHD6 and CHD9, in cancer remain to be characterized. CHD9 mutations have been found in gastric and colorectal cancers10. Functional analysis of the large CHD6 protein (2715 aa) has been hindered by its large size.
FBXW7 is an important component of the SCF (SKP1–CUL1–F-box protein) ubiquitin E3 ligase complex11. FBXW7 mutations are observed in cancers12,13,14,15,16. FBXW7 is a transcriptional target of tumor suppressor p5317,18. FBXW7 binds target proteins and facilitates their ubiquitination and degradation19. Most of the FBXW7 target proteins are oncoproteins, including c-MYC20,21, Cyclin E22, c-JUN23,24, mTOR25, Notch26, Aurora B27, FOXO4, and MCL-124,28,29,30,31,32. FBXW7 is a tumor suppressor19 as manifested in many types of cancers with FBXW7 mutation33,34. The functional activity of FBXW7 is important in hindering cancer growth. Nonetheless, many FBXW7 targets involved in tumorigenesis remain to be characterized.
Here, we show that CHD6 is overexpressed in CRC. We have characterized the upstream regulators of the CHD6 during tumorigenesis, including EGF, glycogen synthase kinase-3β (GSK3β), FBXW7α (hereafter abbreviated as FBXW7), and Wnt3a. Moreover, we identify CHD6 downstream target TMEM65, which is a mitochondrial protein overexpressed in many cancers and is involved in regulating mitochondrial dynamics. Our results shed light on the EGF-GSK3β axis in preventing FBXW7-mediated CHD6 degradation and the Wnt-β-catenin axis in promoting the CHD6 gene transcription during tumorigenesis, and illustrate a role of CHD6 in regulating downstream target gene TMEM65 through Wnt signaling and TCF4 transcriptional activation. CHD6 collaborates with TCF4 to positively regulate TMEM65 gene expression, thereby impacting mitochondrial homeostasis and promoting cancer growth and metastasis. Our understanding of the biological role of CHD6 in regulating TMEM65-mediated mitochondrial dynamics and metastasis of cancer reveals therapeutic strategies for cancer intervention and treatment.
Results
CHD6 is overexpressed in CRC
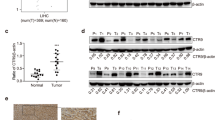
To identify the potential dysregulated genes in CRC, we searched for the TCGA database and found that CHD family members were all altered in CRC. CHD6 ranked first with a 35% alteration rate (Fig. 1a). From the TCGA pan-cancer data, we also demonstrated that CHD6 was highly amplified in CRC (Supplementary Fig. S1a). In addition, CHD6 mRNA (encoding 2715 aa) level was higher in CRC adenocarcinoma tissues than in normal tissues (Supplementary Fig. S1b). To further investigate this phenomenon, we also detected high CHD6 mRNA level in eighteen CRC cancer samples (Supplementary Fig. S1c). Furthermore, Kaplan–Meier analyses of the data from colon cancer dataset (GSE39582) revealed that high CHD6 level correlated with poor relapse-free survival (Supplementary Fig. S1d). Consistent with CHD6 mRNA upregulation, immunohistochemistry (IHC) staining of a panel of 104 CRC and 76 corresponding normal tissue specimens (tissue microarray (TMA)) showed that CHD6 protein expression is significantly higher in CRC than in adjacent normal tissue (66 of 76 paired samples (86.8%)) (Fig. 1b). Kaplan–Meier analysis curves showed that high CHD6 protein expression correlated with poor overall survival in CRC patients (Fig. 1c). Collectively, these data showed that CHD6 may play a critical role during CRC progression.
a Genomic alterations of CHD6 and other CHD6 family members in the TCGA Colorectal Adenocarcinoma database. b Representative images of IHC staining for CHD6 in human colon cancer and adjacent normal colon tissue (left). A plot showing the relative expression of CHD6 in 76 paired samples of CRC and adjacent normal colon tissue (right). Scale bars, 100 μm. Data are presented as means ± SD. ***P < 0.001. P values were calculated by two-tailed t-test. c Kaplan–Meier survival curves of overall survival duration based on CHD6 expression in the TMA containing 104 CRC cases. The receiver operating characteristic curve was used to define the cutoff, and log-rank analysis was used to test for significance. d Images of subcutaneous tumors derived from HCT116 cells expressing PLKO-Tet-on-shCHD6#1, with (shCHD6) or without doxycycline (shCTL) treatment (top). Growth curves of subcutaneous tumors (bottom). Data are presented as means ± SD (n = 6 per group). *P < 0.05. P value was calculated by two-way ANOVA. e Schematic depiction of generating Chd6 CKO mouse model. f Cartoon illustration of a cross between Chd6fl/fl and Villin-CreERT to breed Chd6fl/fl;Villin-CreERT (Chd6 CKO) mice. The Chd6 CKO mice were induced with tamoxifen (TAM) two weeks before AOM/DSS treatment. g Representative images of immunofluorescence staining (IF, left), H&E staining (middle), and TEM (right) of colon tissues obtained from the indicated unchallenged mice. Histology score analysis of H&E staining and quantification of mitochondrial length and cristae number of TEM were shown as bar graph. Data are presented as means ± SD. ns, no significance, **P < 0.01. P values were calculated by two-tailed t-test. h Representative images of colons from Chd6fl/fl + TAM (n = 6) and Chd6fl/fl;Villin-CreERT + TAM (n = 6) mice treated with AOM/DSS. Arrows indicate tumors. i Quantification of tumor incidence per colon (top) and colon length (bottom) of Chd6fl/fl + TAM (n = 6) or Chd6fl/fl;Villin-CreERT + TAM (n = 6) mice with AOM/DSS treatment. Data are presented as means ± SD. *P < 0.05, **P < 0.01. P values were calculated by two-tailed t-test. j Representative H&E staining and Ki67 IHC staining on the colon sections obtained from the indicated mice with AOM/DSS treatment (left). Histology score analysis of H&E staining and quantification of IHC staining were shown as bar graph (right). Data are presented as means ± SD. ***P < 0.001, **P < 0.01. P values were calculated by two-tailed t-test.
To identify the role of CHD6 in CRC, we introduced shRNA-mediated CHD6 knockdown (KD) in DLD1, HCT116, and SW620 cells and validated the KD efficiency by western blot (Supplementary Fig. S2a). CHD6 KD in these CRC cells inhibited/decreased cell proliferation, colony formation, cell migration, invasion, sphere formation, patient-derived organoid (PDO)35 growth, G1-S progression, cell survival, oxygen consumption rate (OCR), ATP production, and mitochondrial mass (FACS analysis of Mitotracker Red staining) (Supplementary Figs. S2b–e, S3a–g). Significantly, mouse subcutaneous CRC xenograft model showed that CHD6 KD (Dox-inducible CHD6 KD) showed a lower tumor burden (Fig. 1d; Supplementary Fig. S4a, b).
Villin-specific CHD6 knockout attenuates cancer formation in AOM/DSS model
To further investigate whether CHD6 in intestine is involved in tumorigenesis, we established Chd6fl/fl + TAM (tamoxifen) mice and inducible villin-specific conditional Chd6-knockout (Chd6fl/fl;Villin-CreERT + TAM) mice (Fig. 1e, f). Correct villin Cre-mediated excision of the Chd6 in intestine was confirmed by PCR (Supplementary Fig. S4c). We found that knockout of Chd6 did not show any histological difference (Fig. 1g) in the colon section when compared with the control group based on hematoxylin and eosin (H&E) staining, and that knockout of Chd6 did not show any difference in terms of goblet number as demonstrated by Alcian blue-periodic acid Schiff’s (AB-PAS) staining at three weeks after TAM injection (Supplementary Fig. S4d). However, transmission electron microscopy (TEM) analysis of mitochondria in mouse colon tissue demonstrated mitochondrial fragmentation phenotype (reduced mitochondrial length) and decreased number of cristae in Chd6-knockout colon cells (Fig. 1g). We then investigated the role of Chd6 in the colitis-associated CRC model. In this model, Chd6fl/fl + TAM mice (control wild type (WT)) and Chd6fl/fl;Villin-CreERT + TAM mice (Chd6 conditional knockout (CKO)) were injected with Azoxymethane (AOM), followed by Dextran Sodium Sulfate (DSS) treatment (Supplementary Fig. S4e). Notably, AOM/DSS models of Chd6 CKO mice seemed to have reduced number of tumors in the colon, a longer colon length (Fig. 1h, i), and better histology (Fig. 1j) on day 80 when compared with Chd6fl/fl mice. Chd6 CKO mice also tended to have less body weight loss during the DSS treatment (Supplementary Fig. S4f). Colorectal tissues from Chd6 CKO mice exhibited a significantly reduced Ki67 staining while tumors of control WT mice have abundant Ki67 staining (Fig. 1j). Immunofluorescence staining of CHD6 further validated the deletion of Chd6 in Chd6 CKO mice for AOM/DSS model (Supplementary Fig. S4g). Taken together, these data indicate that CHD6 has a pivotal role in colon cancer development.
EGF activation leads to CHD6 protein stabilization
Given the high expression level and the oncogenic role of CHD6 in CRC, we sought to uncover the upstream regulators of CHD6. We analyzed two publicly available datasets (GSE2109 and GSE14333) using Ingenuity Pathway Analysis (IPA) software (Ingenuity® Systems, www.ingenuity.com). The ten most significantly enriched cancer-related pathways commonly associated with high-CHD6 group in both datasets were selected and plotted (Fig. 2a). Among these pathways, we selected PI3K/AKT and EGF signaling pathways as potential upstream regulators of CHD6 since PI3K/AKT was prominently presented in both IPA (Fig. 2a) and gene set enrichment analysis (GSEA) (Fig. 2b), and EGFR signaling is highly activated in CRC. Therefore, we investigated the dynamics of EGFR activation and CHD6 regulation. Immunoblotting analysis demonstrated that EGF treatment increased the steady-state expression of CHD6 within 0.5 h without changing its mRNA level (Fig. 2c, d). As expected, PKB/AKT was activated in response to EGF treatment (Fig. 2c), suggesting that EGF-regulated CHD6 steady-state expression may involve PKB/AKT activation. Indeed, AKT inhibitor (MK2206) treatment significantly reduced CHD6 steady-state expression with or without EGF treatment (Fig. 2e, f). We further showed that EGF treatment reduced the turnover rate of CHD6 and decreased the ubiquitination level of CHD6 (Fig. 2g, h). These data showed that EGF signaling attenuates ubiquitin-mediated degradation of CHD6, thereby increasing CHD6 protein stability.
a Ten cancer-related pathways that were significantly associated with genes affected by CHD6 in CRC (GSE2109 and GSE14333). Hallmark pathways emerged following IPA ‘core analysis’. Enrichment scores were displayed as −log10 (P value) by Fisher’s exact test. b GSEA plot of AKT signaling pathway signature correlated with CHD6 highly related genes. Normalized enrichment score (NES) and nominal P value of correlation were shown. c Representative immunoblot analysis of the indicated proteins in HCT116 and DLD-1 cells treated with EGF (100 ng/mL) at the indicated time points. d CHD6 mRNA levels in the indicated cells treated with EGF (100 ng/mL) for the indicated hours. ns, not significant. e Representative immunoblot analysis of the indicated proteins in HCT116 cells treated with MK2206 (5 μM) at different time points. f Representative immunoblot analysis of the indicated proteins in HCT116 cells treated with or without MK2206, followed by EGF treatment. g Representative immunoblots showing CHD6 protein turnover rate in HCT116 cells treated with cycloheximide (CHX, 60 μg/mL), in the presence or absence of EGF (100 ng/mL) treatment (top). Quantification of g (bottom). IOD, integrated optical density. The relative density of CHD6 was normalized to Vinculin and then normalized to the t = 0 control. h Representative immunoblots showing ubiquitination of Flag-tagged CHD6, under EGF (100 ng/mL) treatment in 293T cells. Cells were treated with MG132 (10 μM) 6 h before harvest. The cell lysates were pulled down (PD) with nickel beads (Ni-NTA) and immunoblotted with the indicated antibodies. WCL whole cell lysate.
FBXW7 is involved in regulating CHD6 ubiquitination and degradation
To determine whether a specific E3 ligase is involved in CHD6 ubiquitination, we analyzed the CHD6 peptide sequence and found that FBXW7 binding motif (2125 LPTPXXT 2131) or degron is present in CHD6 (Fig. 3a). Indeed, FBXW7 overexpression decreased the steady-state expression of CHD6 while FBXW7 KD increased the CHD6 expression (Fig. 3b). Co-immunoprecipitation (co-IP) studies indicated the exogenous and endogenous interaction between CHD6 and FBXW7 (Fig. 3c, d). Notably, FBXW7 overexpression accelerated the turnover rate of CHD6 and increased the ubiquitination of CHD6 (Fig. 3e, f).
a Sequence alignment of the putative FBXW7-recognized degron on CHD6. [LIVMP] refers to a group of amino acids, Leu, Ile, Val, Met, Pro, are all allowed; {0,2}, 0, 1, and 2 positions of any kind of amino acids; (T), phosphorylated Thr;.., any kinds of amino acids. b Representative immunoblots showing CHD6 steady-state expression in HCT116 cells upon FBXW7 overexpression or KD. c The interaction between exogenous FBXW7 and CHD6 was determined by co-IP assay. 293T cells were transfected with Myc-tagged CHD6 and Flag-tagged FBXW7. The cell lysates were pulled down with Myc-tagged magnet beads and immunoblotted with the indicated antibodies. WCL: whole cell lysates. d The interaction between endogenous FBXW7 and CHD6 was determined by co-IP assay. HCT116 cell lysates were pulled down with an anti-CHD6 antibody and immunoblotted with FBXW7. e Representative immunoblots showing CHD6 protein turnover rate in HCT116 cells, with or without FBXW7 overexpression (left). Quantification of CHD6 turnover rate (right). Cells were transfected with the indicated plasmids in the presence of CHX (60 μg/mL) for the indicated times. CHD6 levels were normalized to Vinculin and then normalized to the t = 0 control. f 293T cells were transfected with the indicated plasmids and treated with MG132 (10 μM) 6 h before harvest. The cell lysates were pulled down with nickel beads and immunoblotted with an anti-Myc antibody. g, h Representative immunoblots showing the turnover rate of Flag-tagged CHD6 WT or Flag-tagged CHD6 T2127A/T2131A, with or without Myc-tagged FBXW7 overexpression in 293T cells (g). Quantification of CHD6 turnover rate (h). Transfected cells were treated with CHX (60 μg/mL) for the indicated times. Flag-tagged CHD6 levels were first normalized to Vinculin, and then to the t = 0 control. i 293T cells were transfected with the indicated plasmids and treated with MG132 (10 μM) 6 h before harvest. The cell lysates were pulled down with nickel beads and immunoblotted with an anti-Flag antibody. j 293T cells were transfected with the indicated plasmids. The cell lysates were immunoblotted with the indicated antibodies. k Representative immunoblots showing the turnover rate of Flag-tagged CHD6 WT or Flag-tagged CHD6 P2128L, with or without Myc-tagged FBXW7 overexpression in 293T cells (left). Transfected 293T cells were treated with CHX (60 μg/mL) for the indicated times. Quantification of CHD6 WT and P2128L mutant turnover rate (right). Flag-tagged CHD6 levels were first normalized to Vinculin, and then to the t = 0 control.
To confirm that FBXW7 targeted the specific binding motif (LPTPXXT) or phosphodegron for CHD6 degradation, we constructed the CHD6 T2127A/T2131A mutant within (2125 LPTPXXT 2131) motif. The results showed that FBXW7 accelerated the turnover rate of CHD6 WT but not the CHD6 T2127A/T2131A mutant (Fig. 3g, h). Again, CHD6 WT is vulnerable to FBXW7-mediated ubiquitination, while CHD6 T2127A/T2131A mutant is resistant to FBXW7’s impact (Fig. 3i). Based on the CRC genome sequence data, we identified a cancer-derived mutation (P2128L) in CHD6 within the binding motif. We then constructed the patient-derived CHD6 mutant (P2128L) and demonstrated that cancer-derived CHD6 P2128L is resistant to FBXW7-mediated degradation and turnover (Fig. 3j, k). Together, these results demonstrated that FBXW7-mediated downregulation of CHD6 requires the phosphorylation of FBXW7 motif on CHD6.
Tumor suppressor FBXW7 promotes CHD6 ubiquitination through binding phosphodegron of CHD6 in a GSK3β-dependent manner
Given that FBXW7 recognizes phosphorylated degron of target proteins for degradation, and the FBXW7 binding motif on CHD6 is a GSK3β-recognized motif (S/TXXXS/T) and phosphorylation site (Fig. 4a), we hypothesized that FBXW7-mediated CHD6 degradation depends on GSK3β-catalyzed phosphorylation. Indeed, CHD6 steady-state expression decreased with increasing amount of GSK3β (Fig. 4a), while GSK3 inhibitor (CHIR-99021) treatment seemed to increase the level of CHD6 (Fig. 4b). Moreover, FBXW7-mediated CHD6 downregulation was antagonized by CHIR-99021 treatment (Fig. 4c). The threonine phosphorylation level of CHD6 was increased in the presence of GSK3β, but was attenuated by CHIR-99021 treatment (Fig. 4d). Both endogenous and exogenous co-IP assays confirmed the interaction between CHD6 and GSK3β (Fig. 4e, f). More importantly, the presence of GSK3β can facilitate the ubiquitination of CHD6 (Fig. 4g). Congruently, CHIR-99021 treatment to inhibit GSK3β decreased the ubiquitination of CHD6 (Fig. 4h). To further confirm that GSK3β is involved in regulating CHD6 phosphorylation, we investigated the phosphorylation level of CHD6 (T2127A/T2131A) mutant in the presence of GSK3β. As expected, WT CHD6 threonine phosphorylation was enhanced by GSK3β. However, the CHD6 (T2127A/T2131A) mutant was resistant to GSK3β’s activity in terms of Thr phosphorylation (Fig. 4i), and thus was less vulnerable to GSK3β-mediated ubiquitination (Fig. 4j). Together, these results demonstrated that GSK3β-mediated downregulation of CHD6 requires the phosphorylation of CHD6 on phosphodegron motif (T2127/T2131).
a Consensus sequence of the putative GSK3β phosphorylation motif of CHD6 (T2127 and T2131) (top). Representative immunoblots showing CHD6 steady-state expression in HCT116 cells with increased GSK3β overexpression (bottom). b Representative immunoblots showing CHD6 steady-state expression in HCT116 and DLD-1 cells treated with GSK3 inhibitor CHIR-99021 (2 μM) at the indicated time points. c Representative immunoblots showing CHD6 steady-state expression in HCT116 cells transfected with the indicated plasmid in the presence of CHIR-99021 (2 μM). d 293T cells transfected with the indicated plasmids, in the presence or absence of CHIR-99021 (2 μM) treatment, were immunoprecipitated with an anti-Myc antibody and immunoblotted with the indicated antibodies. Threonine-phosphorylated CHD6 was shown. WCL: whole cell lysates. e HCT116 cell lysates were immunoprecipitated (IP) with an anti-CHD6 antibody and immunoblotted with anti-GSK3β antibody. f 293T cells transfected with the indicated plasmids were immunoprecipitated with an anti-Flag antibody and immunoblotted with the indicated antibodies. g 293T cells were transfected with the indicated plasmids and treated with MG132 (10 μM) 6 h before harvest. The cell lysates were pulled down with nickel beads and immunoblotted with the indicated antibodies. h 293T cells were transfected with the indicated plasmids, in the presence or absence of CHIR-99021 treatment. All plates were added with MG132 (10 μM) 6 h before harvest. The cell lysates were pulled down with nickel beads and immunoblotted with an anti-Flag antibody. i 293T cells were transfected with the indicated plasmids. The cell lysates were immunoprecipitated with an anti-Flag antibody and immunoblotted with the indicated antibodies. j 293T cells were transfected with the indicated plasmids and treated with MG132 (10 μM) 6 h before harvest. The cell lysates were pulled down with nickel beads and immunoblotted with the indicated antibodies.
CHD6 transcriptionally regulates the expression of TMEM65, a marker overexpressed in cancer
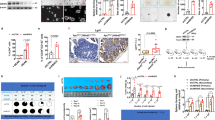
To identify the downstream signals of CHD6, we established Tet-on-inducible-shCHD6 construct for microarray analysis. The differentially expressed genes were shown in the volcano plot between the control (no Dox) and CHD6 KD cells (Dox) (Fig. 5a). The top five downregulated genes in CHD6 KD group were PRRG4, SPAST, TMEM65, PLEKHA8, and ZBTB18. To determine these potential downstream targets that may mediate the oncogenic effect of CHD6 in CRC, we performed correlation analysis with TCGA dataset and found that TMEM65 mRNA expression was significantly associated with CHD6 (Supplementary Fig. S5a); moreover, survival analyses showed that high transcriptional activation of TMEM65 (Supplementary Fig. S5b), but not other four genes, was associated with worse prognosis for overall survival in CRC patients. Consistent with the mRNA expression profiles, IHC analyses in our cohort of CRC patients also revealed that TMEM65 protein expression was significantly increased in CRC specimens compared with that in normal adjacent tissues (Fig. 5b). Further, high expression of TMEM65 protein was associated with poor prognosis for overall survival (Fig. 5c). These analyses indicate that TMEM65 may have an oncogenic effect during CRC development. The positive regulation of TMEM65 by CHD6 was further confirmed by RT-qPCR and western blot. Notably, CHD6 overexpression increased TMEM65 mRNA and protein levels, while CHD6 KD decreased TMEM65 mRNA and protein levels (Supplementary Fig. S5c, d). More importantly, CHD6 re-expression restored the expression of TMEM65 mRNA in CHD6 KD cells (Fig. 5d). Interestingly, TMEM65 can promote cancer cell growth (Supplementary Fig. S5e). To investigate whether the CHD6-TMEM65 axis plays a critical role in promoting tumorigenesis, we performed the following experiments. First, CHD6 KD led to inhibition of cell growth, but TMEM65 expression reversed, at least in part, this impact caused by CHD6 KD (Fig. 5e). Second, in the mouse xenograft cancer samples from Fig. 1d, TMEM65 mRNA levels were decreased in CHD6 KD tumors, as detected by RT-qPCR (Fig. 5f). Third, CHD6 KD tumors from Fig. 1d contained low levels of TMEM65 with concurrent lower Ki67 signals while control tumors showed relatively higher levels of TMEM65 and higher Ki67 signals based on IHC staining (Fig. 5g), suggesting that CHD6 and TMEM65 are positively correlated. Together, the correlation between CHD6 and TMEM65 could be recapitulated in mouse xenograft cancer model, and deregulation of TMEM65 level may play roles in CHD6-mediated tumorigenicity.
a Volcano plot generated from transcriptomic analyses of shCTL (no Dox) and shCHD6 (Dox) HCT116 cells. Dox, doxycycline. b Representative images of IHC staining for TMEM65 in human colon cancer and adjacent normal colon tissue (top). A plot showing the relative expression of TMEM65 in 76 paired samples of CRC and adjacent normal colon tissue (bottom). Scale bars, 50 μm. c Kaplan-Meier survival curves of overall survival duration based on TMEM65 protein expression in the 104 CRC patient tissues. The receiver operating characteristic curve was used to define the cutoff, and log-rank analysis was used to test for significance. d RT-qPCR analysis of TMEM65 in HCT116 cells expressing CHD6 shRNA with or without Flag-tagged CHD6 overexpression. e Growth curves of CHD6 KD HCT116 cells in the presence of TMEM65 overexpression (left); RT-qPCR analysis confirmed the efficiency of TMEM65 overexpression (right). f RT-qPCR analysis of CHD6 and TMEM65 in CHD6 KD HCT116 subcutaneous tumors from Fig. 1d. g Representative images of IHC staining for CHD6, TMEM65, Ki67, and PPOX in subcutaneous tumor sections from Fig. 1d (top) and quantification of IHC staining by ImageJ (bottom). h OCR of HCT116 cells with TMEM65 overexpression. i OCR of shCTL and shCHD6 HCT116 cells, with or without TMEM65 overexpression. j Quantification of cellular ATP in shCTL and shCHD6 HCT116 cells, with or without TMEM65 overexpression. k Immunoblot analysis of PPOX expression in shCTL and shCHD6 HCT116 cells. l Heme levels in control and CHD6 KD HCT116 cells, with or without TMEM65 overexpression. m Representative images of IHC staining for CHD6, TMEM65, PPOX, COXIV, OPA1, and p-Drp1 (Ser616) in the colon tissues obtained from the indicated mice with AOM/DSS treatment. Quantification of IHC staining shown at the bottom (samples from at least 3–6 mice for each genotype were analyzed). Assays were performed with three replicates. P values were calculated by two-tailed t-test (b, g, m), one-way ANOVA (d–f, j, l), or two-way ANOVA (e). Data are presented as means ± SD. ***P < 0.001, **P < 0.01, *P < 0.05, ns, not significant.
CHD6-TMEM65 axis regulates mitochondrial fusion process, OCR, and heme formation
TMEM65 is a mitochondrial inner membrane protein, and CHD6 KD inhibits OCR and ATP production (Supplementary Fig. S3e, f). We then sought to elucidate the function of CHD6-TMEM65 axis on mitochondrial homeostasis in CRC. TMEM65 overexpression led to increased OCR (Fig. 5h). Interestingly, TMEM65 expression rescued, at least in part, the reduced OCR and low ATP production caused by CHD6 KD (Fig. 5i, j). Further, CHD6 KD led to reduction of protoporphyrinogen oxidase (PPOX), a protein involved in heme synthesis (Fig. 5k, g). Heme regulates cytochrome c oxidase (COX) biogenesis, which plays essential roles in oxidative phosphorylation and ATP production. As a result, CHD6 KD led to a decrease of heme (measured after conversion to the autofluorescent precursor protoporphyrin IX (PPIX)) (Fig. 5l; Supplementary Fig. S5f). Again, TMEM65 increased heme level (Supplementary Fig. S5g), and its expression could also rescue, at least in part, the reduced heme production caused by CHD6 KD (Fig. 5l), suggesting that CHD6-TMEM65 axis increased oxidative metabolism. Furthermore, TMEM65 expression increased PPOX while TMEM65 KD decreased PPOX (Supplementary Fig. S5h). Co-IP result showed that TMEM65 can interact with PPOX (Supplementary Fig. S5i), suggesting that TMEM65 regulates PPOX through protein–protein interaction.
Given that mitochondrial protein TMEM65 impacts mitochondrial functions and interacts with PPOX, this observation raises an interesting issue regarding the association of TMEM65 with other mitochondrial proteins in the complex. Our gel filtration studies indicate that TMEM65 are present in complexes with different mitochondrial proteins, such as ATP5a, Mt-CO2, NDUFA8, ATP5b, and PPOX, reiterating its role in regulating mitochondrial homeostasis (Supplementary Fig. S5j). Together, CHD6 regulates mitochondrial functions through TMEM65. CHD6 KD leads to decreased TMEM65 (a PPOX interacting protein) and decreased oxidative phosphorylation and ATP production.
As mitochondrial morphology change was observed in Chd6-knockout mouse model (Fig. 1g), we then further examined mitochondrial dynamics and the expression of proteins involved in mitochondrial functions in CHD6 KD cells. Confocal microscopy demonstrated the irregular and shorter mitochondrial length observed in CHD6 KD cells (Supplementary Fig. S5k). Interestingly, expressing TMEM65 in CHD6 KD cells can reverse this phenomenon (Supplementary Fig. S6a); namely, cells with overexpression of TMEM65 showed longer mitochondria and increased mitochondrial mass even under CHD6 KD. Confocal microscopy showed the colocalization of TMEM65-GFP and mitochondria Mitotracker (Supplementary Fig. S6b), suggesting that TMEM65 may execute its rescue effect physically at the mitochondrial site. Consistent with the confocal picture, the morphology recorded by TEM demonstrated that CHD6 KD cells tended to have smaller mitochondria (possibly due to fission) (Supplementary Fig. S6c). Drp1 can be recruited to mitochondrial constriction site and assembled into higher-order oligomers to facilitate fission of mitochondria. We found that CHD6 KD led to increased Drp1 in the mitochondria, decreased COX-IV, and decreased mitochondrial transcription factor A (TFAM, an important protein interacting with mtDNA to be involved in transcription of mtDNA) (Supplementary Fig. S6d). This observation suggests that CHD6 KD has facilitated mitochondrial fission and perturbed the mitochondrial homeostasis.
Interestingly, TMEM65 expression led to longer fused mitochondria based on TEM images (Supplementary Fig. S6e). We found that TMEM65 expression led to the reduced p-Drp1 (Ser616), increased VDAC1, and reduced Parkin (an E3 ligase involved in mitophagy), indicating its role in mitochondrial fusion and mitophagy (Supplementary Fig. S6f). Further, TMEM65 expression led to more cristae per mitochondrion (Supplementary Fig. S6g). Consistent with the in vitro findings, we also detected that TMEM65, PPOX, and VDAC1 levels were reduced, whereas p-Drp1 level was increased in Chd6-knockout mice compared to control Chd6 fl/fl mice as demonstrated by immunoblotting (Supplementary Fig. S6h).
To examine whether the effect of CHD6-TMEM65 axis on mitochondrial function contributes to the development of colitis-associated neoplasia in the AOM/DSS model, we examined mitochondrial protein expression with IHC staining on colon tumor sections obtained from AOM/DSS-treated Chd6 fl/fl and Chd6-knockout mice. In line with observations of unchallenged Chd6-knockout mice, the mitochondrial deregulations again were also observed in AOM/DSS-treated Chd6-knockout mouse model (Fig. 5m), including reduced levels of TMEM65, PPOX, COX-IV, optic atrophy 1 (OPA1) and increased p-Drp1 level, suggesting that loss of impact of CHD6-TMEM65 axis on mitochondrial functions plays roles in alleviating tumor formation. In conclusion, the CHD6-TMEM65 axis is critical in the regulation of mitochondrial homeostasis during the AOM/DSS-induced carcinogenesis.
High CHD6 expression level in CRC promotes metastasis and tumorigenesis
Liver metastasis is commonly observed in CRC and is responsible for the high rate of mortality and morbidity in CRC patients. Many studies have shown that mitochondrial function is important for supporting cancer cell growth in the distant organs36,37,38. Indeed, in CRC patients with liver metastasis, metastatic liver cancer samples exhibited high levels of CHD6 and TMEM65 compared to CRC samples of primary site and its adjacent normal tissue sample (Fig. 6a). Thus, we hypothesized that disruption of CHD6-TMEM65 axis can impair the capacity of cancer cell to colonize the liver. To investigate our hypothesis, we established a CRC liver metastasis model by intra-splenic inoculation of Tet-on-inducible-shCHD6 HCT116 cells (1 × 106 cells per mouse) with or without ectopic expression of TMEM65 (Fig. 6b). The data showed that CHD6 KD led to reduced liver metastasis, which can be reversed, at least in part, by TMEM65 overexpression in terms of number of metastasis and tumor area (Fig. 6c, d), suggesting a role of CHD6-TMEM65 axis in facilitating metastasis. IHC staining on metastasized liver showed that CHD6 KD led to decreased metastatic cell proliferation (Ki67 staining), compromised expression of TMEM65, and reduced PPOX expression. Importantly, TMEM65 overexpression reversed, at least in part, these impacts from CHD6 KD (Fig. 6e). These results indicate that CHD6 and TMEM65 could be prognostic markers for CRC metastasis. We also validated the correlation of CHD6 and TMEM65 in 104 CRC patients by IHC of TMA and found that CHD6 and TMEM65 showed a significantly positive correlation (Fig. 6f, g). Kaplan–Meier analysis showed that the CHD6-high and TMEM65-high group tended to exhibit the poor overall survival compared to the CHD6-low and TMEM65-low group (Fig. 6h). Meanwhile, the analysis of clinical characteristics of CRC patients showed that high TMEM65 expression was positively correlated with pN status of CRC patients (Supplementary Table S1). Furthermore, multivariate Cox regression analysis revealed that TMEM65 expression is an independent prognostic factor for poor survival (Supplementary Table S2).
a Representative images of immunofluorescence staining for CHD6 and TMEM65 in CRC tissue of primary site, adjacent normal tissue, and liver metastasis tissue obtained from the same CRC patient (left). Red, CHD6; green, TMEM65; blue, DAPI. The staining intensity of CHD6 and TMEM65 was quantitated by ImageJ and presented as bar graphs (right). Data are presented as means ± SD. P values were calculated by one-way ANOVA. *P < 0.05, **P < 0.01. b Schematic diagram of the establishment of liver metastasis mouse model. Liver metastasis model was established by injection of HCT116 cells into the spleen of nude mice (no Dox, n = 6; Dox (shCHD6), n = 6; Dox+TMEM65 OE, n = 6). Dox, doxycycline. c Representative images of mouse liver tissue with metastatic tumors (left). Quantification of macroscopic liver metastases 4 weeks post inoculation (right). Data are presented as means ± SD. **P < 0.01; P values were calculated by one-way ANOVA. d Representative images of H&E staining on liver tissue sections (left) and quantification of metastatic tumor areas (right). The black dashed lines indicate the tumor borders; data are presented as means ± SD. *P < 0.05, **P < 0.01. P values were calculated by one-way ANOVA. e Representative images of IHC staining for CHD6, TMEM65, Ki67, and PPOX on liver metastasis sections (n = 6). Quantifications of IHC staining were shown as bar graphs. Data are presented as means ± SD. *P < 0.05, **P < 0.01, ***P < 0.001. P values were calculated by one-way ANOVA. f Representative images of IHC staining for CHD6 and TMEM65 in serial sections of patient TMAs that consist of 104 CRC cases. Case 1 is representative of a patient with CHD6 low-expressed colon cancer. Case 2 is representative of a patient with CHD6 high-expressed colon cancer. g χ2 analysis shows the correlation of CHD6 and TMEM65 expression in human CRC TMA specimens. h Kaplan–Meier survival curves of overall survival duration based on CHD6 and TMEM65 expression from TMA data. The receiver operating characteristic curve was used to define the cutoff, and log-rank analysis was used to test for significance.
CHD6 collaborates with TCF4 to transcriptionally regulate TMEM65
Next, we tried to elucidate the underlying mechanisms by which CHD6 regulates TMEM65 expression. As a chromatin remodeler, CHD6 regulates gene transcription by increasing chromatin accessibility for transcription factors and the general transcription machinery39. We sought to identify the transcription factor that is involved in TMEM65 transcription. GSEA results showed that the expression of Wnt pathway genes was positively correlated with CHD6 and TMEM65 expression (Supplementary Fig. S7a, b). Our array data also showed that Wnt is the most downregulated hallmark pathway in CHD6 KD cells (Supplementary Fig. S7c), suggesting that Wnt pathway is critical for CHD6 signaling. Intriguingly, co-IP results showed that CHD6 interacted with both TCF4 and β-catenin (Fig. 7a). Moreover, gel filtration studies indicate that CHD6 are present in complexes (~2000 Kd) with TCF4 and β-catenin (Fig. 7b), suggesting that its chromatin remodeling role may be involved in regulating Wnt/ β-catenin signaling. By searching for the TCF4 consensus sequence, we found that TMEM65 promoter contains the TCF4 binding site located between −1327 and −1314 (Fig. 7c). We performed CHD6 chromatin immunoprecipitation (ChIP) analysis and found that CHD6 bound to this TCF4 binding site (−1327 to −1314) on TMEM65 promoter while CHD6 KD diminished the binding (Fig. 7d). We also performed TCF4 ChIP analysis to confirm TCF4 binding to this sequence of TMEM65 promoter (Fig. 7e). Importantly, CHD6 KD led to reduced binding of TCF4 to this sequence on TMEM65 promoter (Fig. 7e), suggesting that CHD6 is facilitating TCF4 binding to TMEM65 promoter.
a HCT116 cell lysates were immunoprecipitated with an anti-CHD6 antibody and were immunoblotted with the indicated antibodies. b Representative immunoblot analysis of fractions containing protein complex eluted from size exclusion chromatography. Cell lysate extracted from CHD6-transfected HEK293T cells was analyzed for CHD6-containing complexes using Superose 6 (GE) gel filtration column. Eluted fractions of protein complex were collected and revealed by immunoblotting with the indicated antibodies. CHD6, TCF4, and β-catenin were co-eluted at fractions 29–32. c TCF4 binding motif obtained from JASPAR (top). The potential binding site of TCF4 on the promoter of TMEM65 (bottom). d ChIP of CHD6 and IgG in control or CHD6 KD HCT116 cells, followed by qPCR for the loci identified by searching for binding sites on TMEM65 promoter. e ChIP of TCF4 and IgG in control and CHD6 KD HCT116 cells followed by qPCR for the loci of predicted binding sites on TMEM65 promoter. f RT-qPCR analysis of TMEM65 mRNA in HCT116 cells treated with Wnt pathway inhibitors IWR-1-endo (20 μM), XAV-939 (1 μM), or vehicle. g RT-qPCR analysis of TMEM65 mRNA in HCT116 cells cultured with L-Wnt3a-expressing cell CM. h RT-qPCR analysis of TMEM65 mRNA in HCT116 cells cultured with L-Wnt3a-expressing cell CM, in the presence or absence of Wnt pathway inhibitor IWR-1-endo (20 μM). i TMEM65-reporter luciferase activity in 293T cells with or without CHD6 overexpression. j TMEM65-reporter luciferase activity in 293T cells with overexpression of β-catenin and TCF4 alone, or in combination. k TCF4 binding motif obtained from JASPAR (top). The potential binding site of TCF4 on the promoter of CHD6 (bottom). l ChIP of TCF4 and IgG in control or CHD6 KD HCT116 cells, followed by qPCR for the loci on CHD6 promoter. m Immunoblot and RT-qPCR analysis of CHD6 in HCT116 cells treated with Wnt pathway inhibitors LGK974. n Immunoblot and RT-qPCR analysis of CHD6 in HCT116 cells cultured with L-Wnt3a-expressing cell CM. o Immunoblot and RT-qPCR analysis of CHD6 in HCT116 cells cultured with L-Wnt3a-expressing cell CM, in the presence or absence of Wnt pathway inhibitors LGK974 (10 μM). p ChIP of β-catenin and IgG in HCT116 cells, followed by qPCR for the loci on CHD6 promoter. Assays were performed with three replicates. P values were calculated by two-tailed t-test (d, e, i, l, p) or one-way ANOVA (f–h, j, m–o). Data are presented as means ± SD. ***P < 0.001, **P < 0.01, *P < 0.05, ns, not significant.
To further demonstrate the impact of Wnt signaling on regulating TMEM65 gene expression, we employed Wnt pathway inhibitors (IWR-1-endo and XAV-939) and showed that they can downregulate mRNA expression of TMEM65 (Fig. 7f). On the contrary, conditioned medium (CM) containing L-Wnt3a led to the elevation of TMEM65 gene expression (Fig. 7g). Congruently, L-Wnt3a-mediated gene elevation of TMEM65 can be attenuated by the presence of Wnt pathway inhibitor IWR-1-endo (Fig. 7h). Together, Wnt signaling may enhance the binding between CHD6 and TCF4 to facilitate TCF4 binding to TMEM65 promoter, thereby strengthening transcriptional expression of TMEM65. Further, we have performed a TMEM65 luciferase reporter gene assay to show that CHD6 can transcriptionally activate TMEM65 promoter (Fig. 7i). Consistently, β-catenin and TCF4 collaborate to transcriptionally activate TMEM65 promoter efficiently (Fig. 7j) based on a TMEM65 luciferase reporter gene assay. Domain mapping assay demonstrated that ATPase/Helicase domain of CHD6 is required for CHD6 interaction with TCF4, illustrating the physical interaction of these two proteins (Supplementary Fig. S7d, e). Detailed studies indicated that ATPase domain of CHD6 is critical for binding TCF4 (Supplementary Fig. S7f, g). Thus, CHD6 ATPase domain physically associates with TCF4 to transcriptionally regulate TMEM65.
Interestingly, we also found that CHD6 promoter contains the TCF4 binding site located between −1458 and −1445 (Fig. 7k). TCF4 ChIP analysis demonstrated that TCF4 bound to this binding site (−1458 to −1445) on CHD6 promoter while CHD6 KD diminished the TCF4 binding (Fig. 7l). Surprisingly, we also found that Wnt pathway inhibitors (LGK974, IWR-1-endo, and XAV939) downregulated CHD6 expression (Fig. 7m; Supplementary Fig. S7h) while CM containing L-Wnt3a elevated CHD6 expression (Fig. 7n). Consistently, the positive impact of CM on CHD6 gene expression can be antagonized by Wnt inhibitors including LGK974, IWR-1-endo, and XAV939 (Fig. 7o; Supplementary Fig. S7i). β-catenin ChIP analysis indicated that β-catenin also bound to CHD6 promoter (Fig. 7p). These results indicate that Wnt signaling may enhance the transcription of CHD6 through TCF4/β-catenin’s transcriptional activity. This could explain, at least in part, the upregulation of CHD6 mRNA level detected in CRC patient samples since Wnt signaling is highly deregulated in CRC. Together, CHD6 regulates TMEM65 transcriptional expression through enhancing TCF4/β-catenin’s activity via Wnt signaling. CHD6 gene itself is also a transcriptional target of TCF4/β-catenin in response to Wnt activity.
Combination treatment of Cetuximab and Wnt inhibitor mitigated tumor growth in CHD6-high human patient-derived xenograft (PDX) CRC model with better efficacy through hindering CHD6-TMEM65 signaling axis
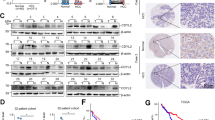
To validate the relevance of our findings to human CRC and to further examine whether hindering CHD6-TMEM65 signaling axis can control the capacity of tumor formation in human CRC, we established PDXs40 by implanting primary tumor samples resected from CRC patients into the immunocompromised mice. Two CRC PDX sets (all have WT RAS gene) were established, and the expression levels of CHD6 were characterized. Two PDXs contain high CHD6, while the other two PDXs contain low CHD6 (Fig. 8a). As EGF signaling positively regulates CHD6 expression, we rationalized that anti-EGFR monoclonal antibody Cetuximab can be exploited for treatment of CHD6-overexpressing CRC (Fig. 8b). Significantly, administration of the Cetuximab in the established CHD6-high PDX tumors can mitigate tumor progression effectively as revealed by the reduced tumor volume and Ki67 staining (Fig. 8c, d). By contrast, Cetuximab had a minimal impact on reducing tumor volume and Ki67 staining of CHD6-low PDX tumors (Fig. 8c, d) even though they all have WT RAS gene. The administration of Cetuximab in CHD6-high group dramatically diminished the expression of TMEM65, COXIV while Cetuximab’s impact on CHD6-low group is less effective as demonstrated by immunoblotting (Fig. 8e).
a Immunoblot images showing CHD6 expression in four cases of PDX tumors. b Schematic diagram of the cetuximab treatment after the establishment of PDX tumors in immunocompromised mice. c Growth of the indicated PDX tumors that were treated with either Cetuximab or PBS as control. n = 4 biological replicates. Data are presented as means ± SD. ***P < 0.001, ns, not significant. P values were calculated by two-way ANOVA. d Representative images of IHC staining for Ki67 in PDX tumor sections with the indicated treatments (left) and quantification of Ki67-positive areas (right). n = 4, data are presented as means ± SD. **P < 0.01, ns, not significant. P values were calculated by two-tailed t-test. e Immunoblot images showing the indicated protein levels analyzed from the PDX tumors that were treated with either Cetuximab or PBS. f Schematic diagram of the Cetuximab and LGK974 combination treatment after the establishment of PDX tumors in immunocompromised mice. g Representative images (left) of the PDX tumors that were harvested at the end of the experiment. Growth curves showing the proliferation of PDX tumors in each indicated treatment group (right). n = 4 biological replicates. Data are presented as means ± SD. ***P < 0.001. P values were calculated by two-way ANOVA. h Representative images (left) and quantifications (right) of IHC staining for CHD6, TMEM65, cleaved caspase-3, and Ki67 in PDX tumors from the indicated treatment groups. Data are presented as means ± SD. ***P < 0.001, **P < 0.01, *P < 0.05. P values were calculated by one-way ANOVA.
Importantly, these data show that the role of CHD6 in activating TMEM65 pathway and promoting tumorigenesis can be recapitulated in PDX CRC clinical samples. We additionally illustrated that the regulation network of EGF-AKT signaling in stabilizing CHD6, and consequent accumulation of TMEM65 during cancer formation can be blocked by Cetuximab treatment. Cetuximab is frequently used to treat CRC patients with WT Ras/Raf. However, among these patients, only half respond well, suggesting that other gene status needs to be considered. Our PDX experiments demonstrate the effectiveness of Cetuximab in suppressing tumor growth in CHD6-high CRC and also point out that in addition to WT Ras/Raf status, expression level of CHD6 is critical for determining the treatment efficacy of Cetuximab.
Our data that in addition to EGF, CHD6 is also upregulated by Wnt signaling support the exploration of Cetuximab in combination with Wnt inhibitor in treating CRC exhibiting high CHD6 expression. Next, we set up a CRC PDX model for drug combination efficacy assays and found that the combination of Cetuximab and Wnt inhibitor (LGK-974) was more efficient in hindering tumor growth than Cetuximab or the LGK-974 inhibitor alone in high CHD6-expressing PDX (case no: 241808) as revealed by the reduced tumor volume, diminished TMEM65 staining, increased cleaved caspase-3 staining, and decreased Ki67 staining (Fig. 8f–h). Together, Cetuximab as monotherapy and Cetuximab plus LGK-974 inhibitor as combination therapy may be considered for therapeutic design for CHD6-high, Ras/Raf WT CRC patients.
Discussion
CHD family members, including CHD6, are involved in gene regulation through chromatin remodeling in an ATP-dependent manner. They use chromodomains to bind histone tails and employ the SWI2/SNF2-like ATPase/helicase domain to remodel chromatin by moving histones, but our knowledge about upstream regulators and downstream targets of CHD6 has not been characterized. EGF-PKB/AKT signaling is highly activated in CRC, and the signal targets are not completely known. Here we characterize that CHD6 is overexpressed in CRC and is a critical regulator of Wnt-TCF4 signaling involved in cell proliferation and promoting tumorigenesis. We show that EGF, GSK3β, FBXW7, and Wnt have activities in regulating CHD6 expression. FBXW7 is identified as an E3 ubiquitin ligase that binds and destabilizes CHD6. Notably, CHD6 deregulates mitochondrial homeostasis by positively regulating TMEM65 gene expression through Wnt and TCF4 signaling. Our data shed light on CHD6 upstream regulatory circuit and reveal how EGF/Wnt oncogenic signal promotes CHD6’s downstream activity toward TMEM65 pathway to cause deregulation of mitochondrial dynamics and subsequently promote cancer metastasis and tumorigenesis (Fig. 9).
Model depicts that CHD6 regulates mitochondrial functions by promoting transcription of TMEM65 in response to EGF and Wnt stimulation. Upon activation of the EGFR-GSK3β axis, CHD6 is not vulnerable to FBXW7-mediated ubiquitination and is thus stabilized. Under the activation of Wnt signaling, β-catenin/TCF4 directly bind to the promoter of CHD6 to enhance its transcription. CHD6 in turn facilitates transcriptional activity of β-catenin/TCF4 (Wnt signaling) to enhance the expression of TMEM65, thereby facilitating tumorigenesis via regulating mitochondrial dynamics, ATP production, and metastasis.
CHD6 is a cancer-associated marker
CHD6 has been mapped to a region of amplification in CRC41. It is mutated in both CRC and bladder cancer42,43. However, these defects were not further characterized and biological activities in cancer were not unveiled. Our data fill this knowledge gap by identifying for the first time that CHD6 is highly expressed in CRC, and by characterizing its oncogenic activities including impacts on cell proliferation, migration, invasion, mitochondrial homeostasis, and tumorigenesis. CHD6 interacts with several proteins, such as MLL, CDK19, BRD7, and CTCF, but the significance of these interactions remains largely unknown44. It remains to be characterized whether the association of these proteins is critical for CHD6’s oncogenic role. It is important to point out that despite the high degree of sequence identity (~50%) among the group III subfamily members (CHD6–9), so far these members share non-redundant functional roles in the cell. Other than CHD6, roles of other members in CRC development required further investigation. Importantly, as indicated in our analysis (Fig. 1a), CHD6 is mutated in CRC. Indeed, we identified a cancer-specific missense mutation (P2128L) of CHD6 on FBXW7 substrate binding site from CRC patients. However, the significance and functional impact of this mutant have not been elucidated. We demonstrated that this cancer-derived CHD6 mutant is resistant to FBXW7-mediated degradation, suggesting that its continual stability/activity is critical in mediating tumorigenesis.
Establishing intestine tissue-specific CHD6-knockout mice
CHD6 Δexon12 (to delete the ATPase domain) mice have been characterized45. The deletion leads to cerebellar defects and ataxia. So far, no tissue-specific Chd6-knockout mouse is employed to investigate its role in cancer. CHD6’s impact on human disease has been documented in a chromosomal translocation t (18;20) (q21.1; q11.2) event between CHD6 (located at 20q12) and the basic helix-loop-helix transcription factor 4 gene (located at chromosome 18), which resulted in mental retardation syndrome46. However, CHD6’s biological role remains an enigma. We set up a Chd6 CKO mice by using the CRISPR-Cas9 genome-editing system to understand the genetic contribution of Chd6 during the development of tumorigenesis. Villin-specific knockout of Chd6 using Cre-loxP system led to slower formation of cancer upon the treatment of AOM/DSS, suggesting the critical role of Chd6 in promoting the growth of colon cancer. Our CRISPR-mediated knockout of Chd6 can be utilized in other types of cancer studies if using proper Cre expressing system, such as liver or prostate cancer. It would be a useful tool to understand the roles of CHD6 in other cancers.
EGF signaling in decelerating CHD6 degradation
EGF/AKT signaling is highly active in many types of cancers, and it becomes a critical therapeutic target for anti-cancer drugs. Although AKT activation is frequently observed in advanced clinical stages in many types of cancers, the consequence or target of aberrant activation of AKT remains not fully understood. GSK3β is inhibited through AKT-dependent phosphorylation. Our studies demonstrate that GSK3β mediates CHD6 phosphorylation, which destabilizes CHD6 through enhancing FBXW7-mediated CHD6 ubiquitination. Therefore, EGF/AKT activation can increase the stability of CHD6 to drive the activity of downstream signals. The fact that FBXW7 mediates CHD6 degradation is reminiscent of the regulatory mechanism of many FBXW7 targets. For examples, FBXW7 binds to SREBP phosphorylated by GSK3β. The binding leads to SREBP degradation47. FBXW7 is a tumor suppressor48 protein through recognizing and degrading several oncoproteins, including FOXO4, cyclin E, c-JUN, Aurora B, and MYC20,24,31,32. It is reasonable to have a tumor suppressor E3 ligase FBXW7, which is frequently mutated in colon cancer, to degrade a potential oncogenic protein CHD6. Our study adds a new member to the target list of FBXW7 E3 ligase.
Role of CHD6-TMEM65 axis in regulating mitochondrial dynamics
TMEM65 is localized at the mitochondrial inner membrane49,50, but little is known about its regulation or other biological functions. Noticeably, TMEM65 is overexpressed in cancer, which is consistent with the role of its upstream regulator CHD6 as CHD6 is also highly expressed in CRC.
Mitochondria are very dynamic organelles. They constantly undergo fission and fusion to impact the shape, size, number of mitochondria, subcellular transport, mitochondrial quality control (mitophagy), and programmed cell death (apoptosis) in the cell. The balance of fission and fusion events manifested mitochondrial dynamics, which is regulated by a number of mitochondria-shaping proteins such as Drp1. Drp1 (DNM1L) has a role in mitochondrial fission via mediating membrane fission through its oligomerization and interacting with membrane-associated tubular structures. We showed that TMEM65 can downregulate p-Drp1, suggesting that TMEM65 may have a role in antagonizing mitochondrial fission. Activated fission and/or inhibited fusion results in fragmented mitochondria, whereas active fusion and/or inhibited fission leads to mitochondrial elongation. Indeed, CHD6 KD leads to upregulation of p-Drp1, which in turn causes fragmented mitochondria (fission), while TMEM65 expression can reverse this impact caused by CHD6 KD. That is, TMEM65 can downregulate p-Drp1 to manifest mitochondrial elongation (fusion). Determining how CHD6/TMEM65 can downregulate p-Drp1 warrants further investigation. Fused mitochondria seem to promote increased oxidative metabolism, ATP production, and reduced ROS. These observations mediated by CHD6/TMEM65 expression are consistent with the role of regulated mitochondrial dynamics in cancer metastasis and tumorigenesis51,52. It is obvious that CHD6/TMEM65 expression has a role of increasing ATP production in mitochondria (bioenergetics) and another role of resisting apoptosis. It is then important to point out that both aspects are regulated by mitochondria53 and are critical in cancer development.
In addition, mitochondrial deregulation results in a variety of diseases54. It is conceivable that CHD6/TMEM65 deregulation may have a pathological impact on these diseases. For example, defects in TMEM65 lead to a mitochondrial disorder manifested as a complex encephalomyopathic phenotype. Further, TMEM65 interacts with connexin 43 (Cx43)55, and its KD affects the protein level of Cx4355 to regulate heart function. It remains to be determined whether CHD6 affects the expression of Cx43.
We observed that TMEM65 overexpression leads to more cristae in mitochondria compared to controls. This increase of cristae number could result in higher levels of respiratory chain proteins and the ATP synthase56, thereby boosting oxidative phosphorylation activity. This phenomenon is consistent with the role of CHD6/TMEM65 in increasing OCR and ATP production.
CHD6 interacts with TCF4 to regulate Wnt signaling
CHD6 interacts with TCF4 and β-catenin, implying its role in Wnt signaling. Its interaction with TCF4 is reminiscent of the observation that CHD6 functions as an epigenetic modulator, coordinating chromatin structure for CFTR expression44. We showed that CHD6 engages in the Wnt signaling pathway and modulates TMEM65 expression through its binding to TMEM65 promoter in a TCF4-collaborating manner. Interestingly, CHD6 is positively regulated by Wnt signaling as its expression is upregulated by TCF4 activity and Wnt ligand. Given that EGF can also stabilize CHD6, it is then conceivable that CHD6 participates in both EGF and Wnt signaling pathways, which have major impacts in promoting the development of CRC (Fig. 9).
Cetuximab treatment in CHD6-high CRC
Cetuximab is frequently used to treat CRC patients based on the WT Ras gene status. Nevertheless, among these patients, only half can respond well, suggesting that more detailed studies are required to understand the discrepancy. We found that Cetuximab has different treatment efficacies in suppressing tumor growth based on the expression level of CHD6 in two sets of WT Ras PDX models. Treating CHD6-high PDX tumors with Cetuximab can mitigate tumor progression effectively. In contrast, Cetuximab had a less impact on the growth of CHD6-low PDX tumors even though these tumors have WT Ras gene status. Our results may help explain why not all WT Ras CRC tumors respond to Cetuximab well. We interpret these results as an indication that the status of CHD6 needs to be considered before the administration of Cetuximab. Given that both EGF and Wnt signaling pathways can positively regulate CHD6 activity, it lends credence to the possibility that targeting EGFR-ERK activation (Cetuximab) plus Wnt signaling (Wnt inhibitor) might have a better synergistic effect in treating CHD6-high CRC. Indeed, Cetuximab plus Wnt inhibitor LGK-974 as a combination treatment strategy for CHD6-high, Ras/Raf WT CRC PDX studies demonstrated the feasibility (Fig. 8f–h). Our findings bear important prognostic and therapeutic implications for the treatment of CRC.
Taken together, our data uncover a link of EGF signaling, GSK3β/FBXW7 regulation, CHD6 stability, TMEM65 expression, mitochondrial dynamics, Wnt signaling, and tumorigenicity. The role of EGF/GSK3β in regulating CHD6 stability through FBXW7 and the activity of Wnt/TCF4/β-catenin signaling in regulating CHD6 transcription highlight important layers of regulation on the activation of CHD6 during tumorigenicity. Further development of compounds that promote FBXW7-mediated CHD6 degradation and inhibit Wnt signaling can be a rational cancer therapy for CHD6-overexpressing cancers.
Materials and methods
Animal studies
All animal experimental procedures were approved by the Animal Ethical and Welfare Committee of Sun Yat-sen University.
Mice and genotyping
Chd6flox/flox mice were established via CRISPR/Cas9 system. Chd6flox/flox mice were intercrossed with Villin-CreERT mice to obtain Chd6flox/flox;Villin-CreERT mice. All mouse lines were maintained on a C57Bl/6 genetic background. For genotyping, tail or colon tissues were cut into 2-mm pieces and digested with 50 μL tail buffer at 95 °C for 5 min, followed by incubation at room temperature until the samples were completely digested. Samples were then incubated with 1 μL proteinase K at 55 °C overnight, followed by heat-inactivation at 95 °C for 5 min. Ethanol was used to wash and precipitate genome DNA. 1 μL of purified genomic DNA was used in the PCR genotyping reaction. The amplified products were detected by agarose gel electrophoresis.
AOM/DSS model
Two groups of mice were confirmed by genotyping: Chd6flox/flox (6 mice) and Chd6flox/flox;Villin-CreERT (6 mice). For tumor induction, 6-week-old mice were injected with 10 mg/kg AOM (Sigma) intraperitoneally (i.p.) one time at the beginning of the experiment (day 0). On day 5, 2% DSS (MP Biologicals) was given in drinking water for 7 days followed by regular drinking water for 2 weeks. This cycle (7 days DSS plus 14 days water) was repeated twice with 1.5% DSS. Mice were sacrificed at 80 days after AOM injection. To induce Chd6 knockout, mice were administered with 50 mg/kg tamoxifen (Sigma, 10 mg/mL in 90% corn oil) for 5 days 2 weeks before AOM injection.
Xenograft model
Xenograft cancer models were established as previously described57,58. Briefly, 5-week-old female BALB/c nude mice were purchased from GemPharmatech Co., Ltd. 1 × 106 HCT116 cells, containing stably tetracycline-inducible shCHD6 construct, were subcutaneously injected into the right flank. When tumor volumes reached 50–80 mm3, all the tumor-bearing mice were randomly divided into two groups of 6 animals per group. Animals were either i.p. injected with doxycycline (50 mg/kg, Selleck) or vehicle every three days. Tumor widths and lengths were measured twice per week using a caliper. Tumor volumes were calculated by the formula: V (mm3) = (width2 × length)/2.
Liver metastasis model
1 × 106 HCT116 cells, containing stably tetracycline-inducible shCHD6 with or without TMEM65 overexpression, were injected into the spleen of BALB/c nude mice under general anesthesia. The mice in CHD6 KD group were i.p. injected with 50 mg/kg doxycycline every three days. On day 30, mice were sacrificed by CO2 inhalation.
The PDX model
PDX experiments were performed as previously described59. Basically, four KRAS/BRAF WT PDX lines were selected. Patient-derived tumor tissues were cut into small fragments (3 mm3) and subcutaneously implanted into immunocompromised mice (GemPharmatech Co., Ltd.). For single drug treatment, when tumor volume reached ~50 mm3, mice were randomly allocated to two groups and were i.p. injected with either cetuximab (40 mg/kg, Selleck) or vehicle once a week. For combination treatment, when tumor volume reached ~50 mm3, mice were randomly allocated to the following treatment groups: (1) vehicle control; (2) cetuximab (40 mg/kg, Selleck); (3) LGK974 (3 mg/kg, Apexbio), (4) cetuximab and LGK974 using doses described above. Administration of cetuximab was the same as addressed above. LGK974 was administered to mice via i.p. injection every day. Mouse weights and tumor volumes were measured every three days during the experiments.
TEM
HCT116 cells and mouse colon tissues were fixed with fixation solution (Servicebio, G1102). After 24 h, samples were washed 3 times with 0.1 M cacodylate buffer and post-fixed with 1% osmium tetroxide (Ted Pella Inc) for 2 h at room temperature. Then the samples were dehydrated in a graded ethanol series and embedded into SPI-Pon 812 (Structure Probe, Inc.). The embedding models were polymerized at 60 °C for 2 days. Ultrathin sections (60–80 nm) were cut with a Leica Ultracut microtome (Leica UC7) and stained with 2% uranyl acetate and 2.6% lead citrate. TEM imaging was performed on a Hitachi TEM system (HT7800). The lengths of mitochondria in TEM images were measured using ImageJ.
Organoid culture
Human CRC organoid was cultured as described previously60,61. In brief, fresh CRC tumor tissues were washed with cold PBS containing penicillin-streptomycin, and cut into 3–5 mm fragments. Pieces were digested with EDTA (5 mM) on ice for 60 min with mixing. After being digested into clumps of cells, the sample was mixed with Matrigel and seeded into a 24-well plate. After Matrigel polymerization (10 min at 37 °C), 500 μL/well advanced DMEM/F12 medium containing 10 mM HEPES, 100 U/mL penicillin/streptomycin, 2 mM GlutaMAX, 1× B27, 1× N2 (Life Technologies), 10 nM gastrin I (Biogems), 500 ng/mL R-spondin1 (Peprotech), 10 μM SB202190 (Sigma), 10 μM Y-27632 (Abmole), 50 ng/mL recombinant EGF, 500 nM A83-01 (Biogems), 100 ng/mL recombinant Noggin (Peprotech), 10 mM nicotinamide (Sigma), 1 mM N-acetylcysteine (Sigma) was added to each well containing organoids. On the second day, the organoids were transduced with shCTL or shCHD6 lentivirus using polybrene (10 μg/mL) (Millipore, TR-1003-G).
Cell culture and transfection
All colorectal cell lines and HEK293T cells used in this study were obtained from ATCC, confirmed to be mycoplasma-free, and incubated in humidified incubator at 37 °C with 5% (vol/vol) CO2. HCT116, SW620, and HEK293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FBS. DLD-1 cells were cultured in RPMI 1640 medium (RPMI) with 10% FBS. LipofectamineTM 2000 (Invitrogen) and polyethylenimine (Polysciences, 24765) were used for cell transfection in this study following the manufacturer’s standard protocol.
Plasmids, cloning, and lentivirus production
The cDNAs for CHD6, TMEM65, TCF4, and FBXW7 were amplified by PCR. These cDNAs were cloned into a pCMV5 vector to generate fusion proteins with N-terminal Flag or Myc tag. CHD6 cDNA was also subcloned into pEGFP-N1 vector (Addgene) to produce a fusion protein with a GFP tag at the C-terminus of CHD6. TMEM65-Flag was also re-cloned into a lentiviral vector pWPI. CHD6 mutant was generated by using a Fast Mutagenesis Kit V2 (Vazyme) according to the manufacturer’s instructions. The resulting plasmids were verified by sequencing.
For CHD6 KD, lentiviral shRNA constructs were purchased from (GeneCopoeia). We screened 6 hairpins targeting human CHD6 transcripts and found two independent sequences that reduced protein levels by > 80% (CHD6 shRNA #31: 5′-GCACAGAAGATCAAGCGATTT-3′; CHD6 shRNA #32: 5′-GCGAGTATAAGAACAGTAACC-3′). For the doxycycline-inducible CHD6 KD, the shRNAs targeting CHD6 were inserted into Tet-pLKO-puro vector (Addgene). Tet-pLKO-shCHD6-1 (CHD6 shRNA #1: 5′-CATTCCAGCAATCATAGTTAA-3′) was designed to target 3′UTR of CHD6. The shRNA sequence in Tet-pLKO-shCHD6-2 is the same as shCHD6-31 (5′-GCACAGAAGATCAAGCGATTT-3′). For the doxycycline-inducible FBXW7 KD, the shRNA sequence (5′-TGATACATCAATCCGTGTTTG-3′) was inserted into Tet-pLKO-puro vector. For TMEM65 KD, the sequence (5′-TCCAGGTTAGGCCTGTCAATT-3′) was inserted into pLKO.1 puro vector.
To prepare lentivirus, HEK293T cells were seeded into a 10 cm dish at a density of 1 × 107, and were co-transfected with 10 μg shRNA lentiviral plasmid, 5 μg psPAX2 and 5 μg pMD2.G using polyethylenimine (Polysciences, 24765). The supernatant containing lentivirus was collected at 48 and 72 h after transfection and were filtered through Millex-GP Filter Unit (0.45 μm pore size, Millipore). HCT116, DLD-1, and SW620 cells were infected twice with filtered viral supernatant containing 10 μg/mL polybrene (Millipore, TR-1003-G).
Western blot analysis
Cells were washed twice with cold PBS and lysed in RIPA buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 0.1% SDS, 1% Triton X-100, 1% sodium deoxycholate, phosphatase and protease inhibitors). Sample proteins were separated by SDS-PAGE and transferred onto PVDF membranes. For protein turnover assay, cells were treated with 60 μg/mL cycloheximide (MDBio, Inc.) for the indicated times before collection. The following antibodies were used in the immunoblotting and immunoprecipitation experiments: CHD6 (1:500, Abcam, ab114095), CHD6 (1:1000, Santa Cruz, sc-393445), TMEM65 (1:500, Sigma, HPA025020), p-AKT (Ser473) (1:1000, Cell Signaling, 4060S), AKT (1:2000, Cell Signaling, 2920S), Vinculin (1:4000, Cell Signaling, 4650S), FBXW7 (1:5000, Abcam, ab109617), COX IV (1:5000, Cell Signaling, 4850), PPOX (1:2000, Santa Cruz, sc-271768), VDAC1 (1:2000, Santa Cruz, sc-390996), mtTFA (1:1000, Santa Cruz, sc-166965), p-Drp1 (Ser616) (1:800, Cell Signaling, 4494S), Drp1 (1:2000, Proteintech, 12957-1-AP), Parkin (1:1000, Proteintech, 14060-1-AP), GAPDH (1:4000, Proteintech, 60004-1-Ig), Flag-tag (1:5000, Sigma, F1804), HA-tag (1:5000, Cell Signaling, 3724S), Myc-tag (1:5000, Cell Signaling, 2276S), β-Catenin (1:4000, BD, 610153), TCF4 (1:1000, Santa Cruz, sc-166699), GSK3β (1:4000, Cell Signaling, 9832S).
Immunoprecipitation
Cells were lysed in IP lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.1% Triton X-100, 0.1% NP-40, 1 mM EDTA, phosphatase and protease inhibitors). Lysates were incubated either with CHD6 antibody or IgG at 4 °C overnight. Protein A/G agarose beads (Santa Cruz, sc-2003) were added into each sample and then incubated at 4 °C for 3 h. Samples were washed 4 times using cold IP lysis buffer and boiled with 2× SDS loading buffer. For Flag-tag immunoprecipitation, cell lysate supernatants were incubated with Flag-M2 agarose beads at 4 °C overnight.
Immunofluorescence
Frozen sections were blocked in 2% bovine serum albumin for 1 h at room temperature. After blocking, sections were incubated with CHD6 (1:10, Santa Cruz, sc-393445) and TMEM65 (1:100, Sigma, HPA025020) antibodies overnight at 4 °C. The samples were subsequently incubated with Alexa Fluor 488- and Alexa Fluor 594-conjugated secondary antibodies. Nuclei were stained with DAPI (Invitrogen). Fluorescence signals were imaged using fluorescence microscope (Leica).
Ubiquitination assay
For CHD6 ubiquitination assay, cells were transfected with the indicated plasmids for 48 h. MG132 (10 μmol/mL, Selleck) was added to the culture media for 6 h. Then cells were collected and lysed in Buffer A (6 M guanidine-HCl, pH 8.0, 10 mM imidazole, 0.1 M Na2HPO4/NaH2PO4). Cell lysates were incubated with Ni-NTA agarose (Invitrogen) at room temperature for 4 h and washed with Wash Buffer (25 mM Tris-HCl, pH 6.0, 20 mM imidazole). Eluted proteins were analyzed by SDS-PAGE and immunoblotting with the indicated antibodies.
Histopathological scoring
Histopathological conditions were scored in a blinded fashion using a method previously published with minor modifications62. Briefly, the condition of ulcerative colitis and dysplasia of each mouse colon were graded ranging from scales 0 to 5 according to the following criteria: 0, no changes; 1, minimal inflammatory cell infiltration, with or without minimal epithelial dysplasia; 2, mild to diffuse inflammatory cell infiltration with occasional spreading to the submucosa with erosions, minimal to mild mucin depletion and epithelial dysplasia; 3, mild to moderate inflammatory cell infiltration that were sometimes transmural with ulceration, moderate dysplasia, and mucin depletion; 4, marked inflammatory cell infiltration that were often transmural and associated with ulceration, marked dysplasia and mucin depletion; 5, marked transmural inflammation with severe ulceration, gland loss and dysplasia.
Immunohistochemistry
Immunohistochemistry was performed following a standard protocol. Paraffin-embedded sections were dewaxed in xylene and rehydrated in gradient ethanol. Antigen retrieval was performed using 0.01 M sodium citrate in a high-pressure cooker. After cooling down, sections were incubated in 3% H2O2 for 10 min and blocked with goat serum for 1 h, followed by primary antibody incubation at 4 °C overnight. On the second day, the sections were incubated with secondary antibodies for 15 min at room temperature and developed with diaminobenzidine. Ki67 (Cell Signaling, 9449), CHD6 (Santa Cruz, sc-393445), TMEM65 (Sigma, HPA025020), COX IV (Cell Signaling, 4850), PPOX (Santa Cruz, sc-271768), p-Drp1(Ser616) (Cell Signaling, 4494S), OPA1 (Santa Cruz, sc-393296) and cleaved Caspase-3 (Asp175) (Cell Signaling, 9664) antibodies were used.
RNA isolation and RT-qPCR
Total RNA was extracted using TRIzol Reagent (Invitrogen, #15596026) according to the manufacturer’s protocol, and reverse-transcribed into cDNA using ReverTra Ace® qPCR RT Master Mix (TOYOBO). The expression of target genes was detected using the 2× SYBR Green qPCR Master Mix (Biotool, #B21203) by a LightCycler® 480 II (Roche).
OCR
Prior to assay, cells were seeded on XF24 microplate (5 × 104 cells per well) and cultured at 37 °C and 5% CO2 overnight. Sensor cartridge was hydrated in calibrant and incubated in a 37 °C, non-CO2 incubator. For the XF Cell Mito Stress Test Kit (Agilent, 103015-100), assay medium was prepared by supplementing XF Base Medium (Agilent, 102353-100) with 10 mM glucose (Sigma, G7528), 1 mM pyruvate, and 2 mM glutamine. Cells were washed with assay medium twice, and incubated at 37 °C, non-CO2 incubator for 45 min. Oligomycin, FCCP and Rotenone/antimycin A were each resuspended in assay medium and added to the microplate. The OCR was determined by a Seahorse XF24 analyzer (Agilent Technologies Co., Ltd.).
ATP assay
Total ATP production
The total ATP was detected using ATP Bioluminescence Assay Kit CLS II (Roche, #11699695001). 100 μL of cell suspension (1 × 106 cells/mL) was diluted to 9 volumes of boiling TE buffer (100 mM Tris, 4 mM EDTA, pH 7.75) and was incubated at 100 °C for 2 min. After incubation, samples were centrifuged at 1000× g for 1 min, and 50 μL supernatant of each sample was transferred to a well of 96-well microplate. ATP level was detected by adding 50 μL luciferase to each sample, and the bioluminescent signal was measured with a Microplate Reader (BioTek Instruments, Inc.).
Glycolytic and mitochondrial ATP production
For the determination of glycolytic ATP production rate (glycoATP Production Rate) and mitochondrial ATP production rate (mitoATP Production Rate), Agilent Seahorse XF Real-Time ATP Rate Assay Kit (103592-100) was used according to the manufacturer’s protocol. The glycoATP production rate is equivalent to Glycolytic Proton Efflux Rate (Glucose + 2 ADP + 2 Pi = 2 Lactate + 2 ATP + 2 H2O + 2 H+). ECAR data indicates the total proton Efflux Rate, and the mitochondrial Proton Efflux Rate can be calculated according to the ECAR that is changed by adding rotenone and antimycin A. The mitoATP Production Rate is calculated according to Equation: mitoATP Production Rate (pmol ATP/min) = OCRATP (pmol O2/min) × 2 (pmol O/pmol O2) × P/O (pmol ATP/pmol O). OCRATP can be calculated as the OCR that is inhibited by adding oligomycin (ATP synthase inhibitor). Assay medium was prepared by supplementing XF Medium-phenol free, pH 7.4 (Agilent, 103575-100) with 10 mM glucose (Sigma, G7528), 1 mM pyruvate (Gibco®, 11360070), 2 mM glutamine (Gibco®, 25030081). Oligomycin, Rotenone, and antimycin A were prepared prior to assay.
Heme content (PPIX fluorescence)
Heme content measurement assay was performed as described previously63. Briefly, cells were collected and resuspended in 500 μL of 20 mM oxalic acid (Sangon, A610400) and stored in a closed box at 4 °C overnight. On the second day, 500 μL of 2 M oxalic acid (heat for dissolution) was added and mixed with the stored samples. 500 μL of the mixture was heated at 98 °C for 30 min, and the remaining samples were kept at room temperature. All prepared mixtures were centrifuged at 16,000× g for 2 min. 200 μL of mixture from each sample was transferred to a black microtiter plate and the fluorescence (Excitation: 400 nm; Emission: 620 nm) was measured in a fluorescence microplate reader (BioTek Instruments, Inc.). The fluorescence of corresponding unboiled samples was used to correct for background fluorescence.
L-Wnt3a CM
L-Wnt3a-expressing cells were seeded into 10 cm dish, and cultured in DMEM supplemented with 10% FBS. After 4 days, cell culture supernatant was collected, filtered, and kept at 4 °C. The cells were cultured for another 3 days for a second supernatant collection. The second supernatant was combined with the first collection and stored at −80 °C.
ChIP-qPCR
ChIP was performed as previously described58,64. Briefly, control and CHD6 KD HCT116 cells were cross-linked using 1% formaldehyde for 20 min at room temperature. Reactions were quenched by treating with 0.125 M glycine for 5 min at room temperature. Then cells were washed with cold PBS and collected in 1 mL ChIP lysis buffer (5 mM HEPES, pH 8.0, 85 mM KCl, 0.5% NP-40, protease inhibitors). Chromatin fragmentation was performed using a Diagenode BioruptorPico sonicator (30 s on, 30 s off for 10 cycles). Clear chromatin extracts were incubated with 20 μL Magna ChIP™ Protein A + G Magnetic Beads (Millipore, 16–663) and 2 µg antibody (CHD6, Santa Cruz, sc-393445; β-Catenin, BD, 610153; TCF4, Santa Cruz, sc-166699) or IgG overnight at 4 °C. Immunoprecipitated chromatin was washed with low-salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.1, 150 mM NaCl) twice, washed with high-salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.1, 500 mM NaCl) twice, and then washed twice with LiCl buffer (10 mM Tris-HCl, pH 8.0, 250 mM LiCl, 1% NP-40, 1% deoxycholic acid and 1 mM EDTA) and once with TE buffer (pH 8.0). Sample beads were resuspended in 50 μL elution buffer (10 mM Tris-Cl, pH 8.0, 5 mM EDTA, 300 mM NaCl, 0.5% SDS) with 5 μL proteinase K (20 mg/mL) at 65 °C overnight. DNA was purified using PCR purification Kit (Omega, D2500-02). Eluted DNA was used for qPCR.
Cell lysate fractionation analysis
Fractionation of cell lysates by size-exclusion chromatography was performed as previously described with minor modification58. Briefly, CHD6-transfected HEK293T cells were lysed in lysis buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 100 mM EDTA, 0.5% NP-40, 0.1% Triton X-100). The cleared supernatants of each cell lysates were run through a size-exclusion column Superose 6 (GE), and proteins were eluted by PBS (0.01 M phosphate, 0.15 M NaCl, pH 7.4) in GE AKTA avant150 chromatography system at a flow rate of 0.4 mL/min. A total of 40 fractions (0.3 mL/fraction) were collected. Collected protein fractions were resolved with SDS-PAGE, followed by immunoblotting with the indicated antibodies.
Bioinformatics analysis
RNA was extracted from HCT116 cells with or without CHD6 KD (dox-inducible CHD6 KD). HG-U133 Plus 2.0 GeneChips (Affymetrix) was used to examine the gene expression profiles according to the manufacturer’s instructions. Analysis of microarray data was performed in R programming environment (http://cran.r-project.org), and the Bioconductor (http://bioconductor.org) R packages simpleaffy (http://bioinformatics.picr.man.ac.uk/simpleaffy) and EnhancedVolcano (https://github.com/kevinblighe/EnhancedVolcano) were used for analyzing Affymetrix data and generating volcano plot. TCGA expression and mutation data were downloaded from cBioportal65,66. The CRC datasets (GSE31595, GSE2109, among others) were downloaded from the Gene Expression Omnibus (GEO), the Oncomine, The Cancer Genome Atlas and The Human Protein Atlas databases. For the CRC study, tumors were stratified with median CHD6 expression. The datasets of GSE31595 and GSE2109 were annotated by GSEA (Broad Institute) on KEGG and Cancer Hallmark Pathways databases. The GEO2R web application (https://www.ncbi.nlm.nih.gov/geo/geo2r/) was used to computerize the differentially expressed genes (adjusted P value < 0.01) by comparing the gene expression profiles of CHD6-high and -low CRC (GES2109 and GES14333). The resulting gene list was submitted for pathway analysis using the ‘Core Analysis’ function in the IPA (Qiagen Silicon Valley).
Patient tissue samples and TMA analysis
All cancer patient samples were collected with the patients’ written informed consent and approval from the Institutional Review Board of the Sixth Affiliated Hospital of Sun Yat-sen University. For CHD6 expression level analysis, 18 paired CRC and adjacent normal colon tissues were collected from the Department of Surgery at the Sixth Affiliated Hospital of Sun Yat-sen University. Total RNA was isolated, and CHD6 mRNA level was detected by RT-qPCR. The human TMAs were purchased from Outdo Biotech (Shanghai Outdo Biotech Co., Ltd.). The TMAs contained 180 human colon tissue cores, including 76 paired specimens of CRC and corresponding normal tissues and 28 identified CRC specimens. The immunostained slides were scanned using Slide Scanning System SQS-1000 (TEKSQRAY). TMA images were analyzed with HALO image analysis software (Indica Labs).
Statistical analysis
The differences between two groups were measured by Student’s t-test (GraphPad Prism 6). Multiple group comparisons were performed by one-way ANOVA followed by Tukey test (comparing all pairs of columns) or Dunnett test (comparing all columns vs control column). The statistical significances about cell proliferation and tumor growth in different groups were calculated by two-way ANOVA. Survival analysis was performed using SPSS 25 by Kaplan–Meier survival curve and verified by log-rank test. The Pearson correlation was calculated using a package written in the R language (http://cran.r-project.org). χ2 test was used to examine the relationship between groups. Cox proportional hazards regression for univariate and multivariate analyses were employed to evaluate which clinicopathologic factors had prognostic values. The P values and hazard ratios are shown and P < 0.05 was defined as statistically significant.
Data availability
The array data has been deposited in the GEO database with accession code GSE163124.
References
Fearon, E. R. & Vogelstein, B. A genetic model for colorectal tumorigenesis. Cell 61, 759–767 (1990).
Siegel, R. L., Miller, K. D. & Jemal, A. Cancer statistics, 2019. CA Cancer J. Clin. 69, 7–34 (2019).
Kocarnik, J. M., Shiovitz, S. & Phipps, A. I. Molecular phenotypes of colorectal cancer and potential clinical applications. Gastroenterol. Rep. 3, 269–276 (2015).
Mills, A. A. The chromodomain helicase DNA-binding chromatin remodelers: Family traits that protect from and promote cancer. Cold Spring Harb. Perspect. Med. 7, a026450 (2017).
He, Y. et al. IFN-κ suppresses the replication of influenza A viruses through the IFNAR-MAPK-Fos-CHD6 axis. Sci. Signal. 13, eaaz3381 (2020).
Kargapolova, Y. et al. Overarching control of autophagy and DNA damage response by CHD6 revealed by modeling a rare human pathology. Nat. Commun. 12, 3014 (2021).
Moore, S. et al. The CHD6 chromatin remodeler is an oxidative DNA damage response factor. Nat. Commun. 10, 241 (2019).
Colbert, L. E. et al. CHD7 expression predicts survival outcomes in patients with resected pancreatic cancer. Cancer Res. 74, 2677–2687 (2014).
Caldon, C. E. et al. Estrogen regulation of cyclin E2 requires cyclin D1 but not c-Myc. Mol. Cell. Biol. 29, 4623–4639 (2009).
Kim, M. S., Chung, N. G., Kang, M. R., Yoo, N. J. & Lee, S. H. Genetic and expressional alterations of CHD genes in gastric and colorectal cancers. Histopathology 58, 660–668 (2011).
Cardozo, T. & Pagano, M. The SCF ubiquitin ligase: Insights into a molecular machine. Nat. Rev. Mol. Cell Biol. 5, 739–751 (2004).
Calhoun, E. S. et al. BRAF and FBXW7 (CDC4, FBW7, AGO, SEL10) mutations in distinct subsets of pancreatic cancer: Potential therapeutic targets. Am. J. Pathol. 163, 1255–1260 (2003).
Kemp, Z. et al. CDC4 mutations occur in a subset of colorectal cancers but are not predicted to cause loss of function and are not associated with chromosomal instability. Cancer Res. 65, 11361–11366 (2005).
Song, J. H., Schnittke, N., Zaat, A., Walsh, C. S. & Miller, C. W. FBXW7 mutation in adult T-cell and B-cell acute lymphocytic leukemias. Leuk. Res. 32, 1751–1755 (2008).
Sancho, R. et al. F-box and WD repeat domain-containing 7 regulates intestinal cell lineage commitment and is a haploinsufficient tumor suppressor. Gastroenterology 139, 929–941 (2010).
Davis, R. J., Welcker, M. & Clurman, B. E. Tumor suppression by the Fbw7 ubiquitin ligase: Mechanisms and opportunities. Cancer Cell 26, 455–464 (2014).
Perez-Losada, J., Mao, J. H. & Balmain, A. Control of genomic instability and epithelial tumor development by the p53-Fbxw7/Cdc4 pathway. Cancer Res. 65, 6488–6492 (2005).
Mao, J. H. et al. Fbxw7/Cdc4 is a p53-dependent, haploinsufficient tumour suppressor gene. Nature 432, 775–779 (2004).
Welcker, M. & Clurman, B. E. FBW7 ubiquitin ligase: A tumour suppressor at the crossroads of cell division, growth, and differentiation. Nat. Rev. Cancer 8, 83–93 (2008).
Welcker, M. et al. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc. Natl. Acad. Sci. USA 101, 9085–9090 (2004).
Grim, J. E. et al. Isoform- and cell cycle-dependent substrate degradation by the Fbw7 ubiquitin ligase. J. Cell Biol. 181, 913–920 (2008).
Klotz, K. et al. SCF(Fbxw7/hCdc4) targets cyclin E2 for ubiquitin-dependent proteolysis. Exp. Cell Res. 315, 1832–1839 (2009).
Wei, W., Jin, J., Schlisio, S., Harper, J. W. & Kaelin, W. G. Jr The v-Jun point mutation allows c-Jun to escape GSK3-dependent recognition and destruction by the Fbw7 ubiquitin ligase. Cancer Cell 8, 25–33 (2005).
Nateri, A. S., Riera-Sans, L., Da Costa, C. & Behrens, A. The ubiquitin ligase SCFFbw7 antagonizes apoptotic JNK signaling. Science 303, 1374–1378 (2004).
Mao, J. H. et al. FBXW7 targets mTOR for degradation and cooperates with PTEN in tumor suppression. Science 321, 1499–1502 (2008).
O’Neil, J. et al. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to gamma-secretase inhibitors. J. Exp. Med. 204, 1813–1824 (2007).
Teng, C. L. et al. FBXW7 is involved in Aurora B degradation. Cell Cycle 11, 4059–4068 (2012).
Yada, M. et al. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J. 23, 2116–2125 (2004).
Koepp, D. M. et al. Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science 294, 173–177 (2001).
Tsunematsu, R. et al. Mouse Fbw7/Sel-10/Cdc4 is required for notch degradation during vascular development. J. Biol. Chem. 279, 9417–9423 (2004).
Inuzuka, H. et al. SCF(FBW7) regulates cellular apoptosis by targeting MCL1 for ubiquitylation and destruction. Nature 471, 104–109 (2011).
Chen, J. et al. CSN6 drives carcinogenesis by positively regulating Myc stability. Nat. Commun. 5, 5384 (2014).
Ojesina, A. I. et al. Landscape of genomic alterations in cervical carcinomas. Nature 506, 371–375 (2014).
Kourtis, N. et al. FBXW7 modulates cellular stress response and metastatic potential through HSF1 post-translational modification. Nat. Cell Biol. 17, 322–332 (2015).
Bolhaqueiro, A. C. F. et al. Ongoing chromosomal instability and karyotype evolution in human colorectal cancer organoids. Nat. Genet. 51, 824–834 (2019).
Scheid, A. D., Beadnell, T. C. & Welch, D. R. Roles of mitochondria in the hallmarks of metastasis. Br. J. Cancer 124, 124–135 (2021).
Andrzejewski, S. et al. PGC-1alpha promotes breast cancer metastasis and confers bioenergetic flexibility against metabolic drugs. Cell Metab. 26, 778–787.e5 (2017).
LeBleu, V. S. et al. PGC-1alpha mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nat. Cell Biol. 16, 992–1003 (2014).
de Dieuleveult, M. et al. Genome-wide nucleosome specificity and function of chromatin remodellers in ES cells. Nature 530, 113–116 (2016).
Byrne, A. T. et al. Interrogating open issues in cancer precision medicine with patient-derived xenografts. Nat. Rev. Cancer 17, 254–268 (2017).
Ali Hassan, N. Z. et al. Integrated analysis of copy number variation and genome-wide expression profiling in colorectal cancer tissues. PLoS One 9, e92553 (2014).
March, H. N. et al. Insertional mutagenesis identifies multiple networks of cooperating genes driving intestinal tumorigenesis. Nat. Genet. 43, 1202–1209 (2011).
Mouradov, D. et al. Colorectal cancer cell lines are representative models of the main molecular subtypes of primary cancer. Cancer Res. 74, 3238–3247 (2014).
Sancho, A. et al. CHD6 regulates the topological arrangement of the CFTR locus. Hum. Mol. Genet. 24, 2724–2732 (2015).
Lathrop, M. J. et al. Deletion of the Chd6 exon 12 affects motor coordination. Mamm. Genome 21, 130–142 (2010).
Kalscheuer, V. M. et al. Disruption of the TCF4 gene in a girl with mental retardation but without the classical Pitt-Hopkins syndrome. Am. J. Med. Genet. A 146A, 2053–2059 (2008).
Sundqvist, A. et al. Control of lipid metabolism by phosphorylation-dependent degradation of the SREBP family of transcription factors by SCF(Fbw7). Cell Metab. 1, 379–391 (2005).
Fuchs, S. Y. Tumor suppressor activities of the Fbw7 E3 ubiquitin ligase receptor. Cancer Biol. Ther. 4, 506–508 (2005).
Nazli, A. et al. A mutation in the TMEM65 gene results in mitochondrial myopathy with severe neurological manifestations. Eur. J. Hum. Genet. 25, 744–751 (2017).
Nishimura, N., Gotoh, T., Oike, Y. & Yano, M. TMEM65 is a mitochondrial inner-membrane protein. PeerJ 2, e349 (2014).
Srinivasan, S., Guha, M., Kashina, A. & Avadhani, N. G. Mitochondrial dysfunction and mitochondrial dynamics—The cancer connection. Biochim. Biophys. Acta Bioenerg. 1858, 602–614 (2017).
Altieri, D. C. Mitochondrial dynamics and metastasis. Cell. Mol. Life Sci. 76, 827–835 (2019).
Burke, P. J. Mitochondria, bioenergetics and apoptosis in cancer. Trends Cancer 3, 857–870 (2017).
Schapira, A. H. Mitochondrial diseases. Lancet 379, 1825–1834 (2012).
Sharma, P. et al. Evolutionarily conserved intercalated disc protein Tmem65 regulates cardiac conduction and connexin 43 function. Nat. Commun. 6, 8391 (2015).
Frey, T. G. & Mannella, C. A. The internal structure of mitochondria. Trends Biochem. Sci. 25, 319–324 (2000).
Fang, L. et al. ERK2-dependent phosphorylation of CSN6 is critical in colorectal cancer development. Cancer Cell 28, 183–197 (2015).
Choi, H. H. et al. EGF relays signals to COP1 and facilitates FOXO4 degradation to promote tumorigenesis. Adv. Sci. 7, 2000681 (2020).
Li, K. et al. ILF3 is a substrate of SPOP for regulating serine biosynthesis in colorectal cancer. Cell Res. 30, 163–178 (2020).
Roper, J. et al. In vivo genome editing and organoid transplantation models of colorectal cancer and metastasis. Nat. Biotechnol. 35, 569–576 (2017).
Qin, B. et al. CSN6-TRIM21 axis instigates cancer stemness during tumorigenesis. Br. J. Cancer 122, 1673–1685 (2020).
Meira, L. B. et al. DNA damage induced by chronic inflammation contributes to colon carcinogenesis in mice. J. Clin. Invest. 118, 2516–2525 (2008).
Vandekeere, S. et al. Serine synthesis via PHGDH is essential for heme production in endothelial cells. Cell Metab. 28, 573–587.e13 (2018).
Zhao, D. et al. Synthetic essentiality of chromatin remodelling factor CHD1 in PTEN-deficient cancer. Nature 542, 484–488 (2017).
Cerami, E. et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012).
Gao, J. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1 (2013).
Acknowledgements
This research was supported by the National Key R&D Program of China (2020YFA0803300), the National Natural Science Foundation of China (81630072), Shenzhen Municipal Government (KQTD20170810160226082), the Natural Science Foundation of Guangdong Province (2021A1515012081), the Basic and Applied Basic Research Program of Guangzhou City (202102020084), the Guangzhou Science and Technology Program Project (202206010167). We thank L.K.F., P.Z., Y.Z.H., J.Y.F., Q.T., and L.S.X. for providing technical help.
Author information
Authors and Affiliations
Contributions
M.-H.L. and Q.L. conceived the project. B.Z. and Q.L. designed and participated in many of the experiments. B.Z. performed data analysis. Q.L. performed the bioinformatics analysis, IHC staining, and ChIP experiments. B.Z., Q.L., H.G., B.Q., W.X.W., W.J.W., X.M., K.L., and H.J. performed in vivo experiments. B.Z., F.Y., Q.P., and H.L. performed ubiquitination assay. B.Z., Q.L., A.T., and H.G. cloned plasmids. W.J.W. established the PDX model. M.-H.L. directed the project and wrote the manuscript. All authors reviewed and approved the manuscript for publication.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhang, B., Liu, Q., Wen, W. et al. The chromatin remodeler CHD6 promotes colorectal cancer development by regulating TMEM65-mediated mitochondrial dynamics via EGF and Wnt signaling. Cell Discov 8, 130 (2022). https://doi.org/10.1038/s41421-022-00478-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41421-022-00478-z
This article is cited by
-
Mitochondrial dynamics and colorectal cancer biology: mechanisms and potential targets
Cell Communication and Signaling (2024)
-
The EIF3H-HAX1 axis increases RAF-MEK-ERK signaling activity to promote colorectal cancer progression
Nature Communications (2024)
-
The involvement of E3 ubiquitin ligases in the development and progression of colorectal cancer
Cell Death Discovery (2023)
-
FBXW7β loss-of-function enhances FASN-mediated lipogenesis and promotes colorectal cancer growth
Signal Transduction and Targeted Therapy (2023)