Abstract
Substitution of lysine 36 with methionine in histone H3.3 (H3.3K36M) is an oncogenic mutation that inhibits SETD2-mediated histone H3K36 tri-methylation in tumors. To investigate how the oncohistone mutation affects the function of SETD2 at the nucleosome level, we determined the cryo-EM structure of human SETD2 associated with an H3.3K36M nucleosome and cofactor S-adenosylmethionine (SAM), and revealed that SETD2 is attached to the N-terminal region of histone H3 and the nucleosome DNA at superhelix location 1, accompanied with the partial unwrapping of nucleosome DNA to expose the SETD2-binding site. These structural features were also observed in the previous cryo-EM structure of the fungal Set2–nucleosome complex. By contrast with the stable association of SETD2 with the H3.3K36M nucleosome, the EM densities of SETD2 could not be observed on the wild-type nucleosome surface, suggesting that the association of SETD2 with wild-type nucleosome might be transient. The linker histone H1, which stabilizes the wrapping of nucleosome DNA at the entry/exit sites, exhibits an inhibitory effect on the activities of SETD2 and displays inversely correlated genome distributions with that of the H3K36me3 marks. Cryo-EM analysis of yeast H3K36 methyltransferase Set2 complexed with nucleosomes further revealed evolutionarily conserved structural features for nucleosome recognition in eukaryotes, and provides insights into the mechanism of activity regulation. These findings have advanced our understanding of the structural basis for the tumorigenesis mechanism of the H3.3K36M mutation and highlight the effect of nucleosome conformation on the regulation of histone modification.
Similar content being viewed by others
Introduction
Histone modifications play pivotal roles in a multitude of cellular processes, including transcription, DNA replication, and DNA damage repair1. Tri-methylation of histone H3 on lysine 36 (H3K36me3), primarily deposited by histone methyltransferase (HMT) SETD2 in mammalian cells, occurs at gene bodies of active chromatin and serves as one of the essential histone marks associated with active transcription2,3. SETD2 directly associates with the hyperphosphorylated C-terminal domain (CTD) repeats of RNA polymerase II (pol II) through its Set2–Rpb1 interaction (SRI) domain to deposit the H3K36me3 marks co-transcriptionally4,5,6,7, and participates in the physiological regulation of chromatin condensation, histone exchange, pre-mRNA splicing, DNA damage repair, etc.8,9,10,11,12,13,14,15,16.
Dysfunction of the SETD2–H3K36me3 axis has been linked to a wide range of human malignancies. Frequent loss or mutations of the SETD2 gene have been observed in clear cell renal cell carcinoma, high-grade gliomas, esophageal squamous cell carcinoma, colorectal cancer, and acute leukemia17,18,19,20,21. Moreover, the substitution of lysine 36 with methionine in the histone H3 variant H3.3 (abbreviated as H3.3K36M) has been identified in chondroblastoma and head and neck squamous cell carcinomas22,23. This mutation results in loss of H3K36 methylation landscape and alteration of cancer-related gene expression24.
Structural and biochemical analysis of the catalytic domain of SETD2 toward an H3.3K36M peptide indicated that the oncohistone peptide occupies the substrate channel of SETD2 accompanied with the side chain of K36M inserted into the catalytic site, and inhibits the HMT activities of SETD2 against mononucleosomes25,26. Recent cryo-electron microscopy (cryo-EM) studies of several histone modifiers, such as human MLL and PRC2 complexes, bound to their natural nucleosome substrates revealed that the modifications of histone tails require specific recognition of nucleosome and even higher-order chromatin structures by the enzymes27,28,29. A recent cryo-EM structure of Chaetomium thermophilum Set2, a fungal homolog of SETD2, bound to a ubiquitinated nucleosome showed that the catalytic domain of Set2 makes extensive interactions with the N-terminus of histone H3 and the C-terminal tail of histone H2A, and stabilizes nucleosome DNA in the unwrapped conformation30. However, it remains poorly understood how the H3.3K36M mutation is recognized by human SETD2 at the nucleosome level and how it affects the physiological functions of human SETD2 in the context of chromatin structure.
SETD2 exclusively catalyzes H3K36 tri-methylation in mammalian cells, whereas mono- and di-methylations of H3K36 are implemented by the methyltransferases NSD1-3 and ASH1L2,31. By contrast, Set2, the yeast homolog of SETD2, is the sole enzyme responsible for the mono-, di-, and tri-methylations of H3K36 in yeast32. The functional domains of SETD2 are highly conserved in yeast Set233,34. It remains unclear what determines the different activities of human SETD2 and yeast Set2.
In this work, cryo-EM analysis of human SETD2 in complex with an H3.3K36M nucleosome or a wild-type nucleosome revealed that the catalytic domain of SETD2 is stably associated with the H3.3K36M mutant nucleosome in a similar manner as observed in the fungal Set2–nucleosome structure30. By contrast, the EM densities of SETD2 could not be observed on the wild-type nucleosome surface, suggesting that the association of SETD2 with wild-type nucleosome might be very dynamic. The detachment of nucleosome DNA at its entry/exit site is a prerequisite for the nucleosome-binding and catalytic activities of SETD2. Accordingly, the linker histone H1, which stabilizes the wrapping of nucleosome DNA at the entry/exit sites, causes an inhibitory effect on the in vitro HMT activities of SETD2 toward nucleosomes. ChIP-Seq analysis also indicates that the genomic distribution of histone H1 is inversely correlated with that of the H3K36me3 marks. In addition, structural characterization of yeast Set2 bound to the oncohistone or wild-type nucleosome reveals an evolutionarily conserved structural framework for nucleosome-binding and also provides structural insights into the mechanism of activity regulation in eukaryotes.
Results
Overall structure of human SETD2 in complex with an H3.3K36M nucleosome
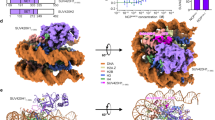
To investigate the effects of H3.3K36M oncogenic mutation on the functions of human SETD2 at the level of the nucleosome, we reconstituted human wild-type and H3.3K36M-mutant nucleosome core particles (hereafter, hNCPWT and hNCPH3.3K36M) in vitro and performed the cryo-EM analysis of SETD2 complexed with hNCPWT and hNCPH3.3K36M, respectively. A truncated SETD2 construct (residues 1382 to the C-terminus, with residues 1916–2467 replaced with a (Gly–Gly–Ser)3 linker) that contains both the catalytic domain and the SRI domain and exhibits optimal HMT activities toward nucleosome substrates was used for the structural and biochemical analysis (Fig. 1a; Supplementary Fig. S1a–c). Electrophoretic mobility shift assays (EMSAs) indicate that SETD2 binds hNCPWT or hNCPH3.3K36M with comparable affinities (Supplementary Fig. S1d). Cryo-EM single-particle analysis generated a global density map of the SETD2–hNCPH3.3K36M complex at a resolution of 3.1 Å (Supplementary Fig. S2 and Table S1). Masked classifications on the AWS (associated with SET) region and the active site of SETD2 further improved the local EM densities (Supplementary Fig. S2a). An atomic model of the SETD2–hNCPH3.3K36M complex was built by docking available crystal structures of SETD2 and nucleosome25 into the EM density map, followed by manual building (Fig. 1b, c).
a Schematic drawing of the domain organizations of human SETD2. The color scheme is the same as that of the SETD2–hNCPH3.3K36M structural model. b Cryo-EM density map of human SETD2–hNCPH3.3K36M complex at 3.1 Å shown from two orthogonal views. The cryo-EM map was segmented and colored according to the respective components of the SETD2–hNCPH3.3K36M complex. c Atomic model of human SETD2–hNCPH3.3K36M complex shown from two orthogonal views. The catalytic domain of SETD2 binds to the α-N of histone H3.3 as well as the nucleosome DNA at SHL1.
The SETD2–hNCPH3.3K36M complex structure resembles that of the fungal Set2–nucleosome complex30 (Fig. 1c; Supplementary Fig. S3a). The catalytic domain of SETD2 binds to the N-terminal α helix (α-N) of histone H3.3 as well as the nucleosome DNA at superhelix location 1 (SHL1), at which the histone H3.3 tail snuggly extends through the substrate-binding channel of SETD2 (Fig. 1c). Notably, only the catalytic domain of SETD2 could be unambiguously built in the cryo-EM map of the SETD2–hNCPH3.3K36M complex, which suggests that the rest of SETD2 regions might adopt flexible conformations on the mutant nucleosome (Fig. 1b, c). By contrast with the SETD2–hNCPH3.3K36M complex structure, cryo-EM analysis of the SETD2–hNCPWT complex indicates that the EM densities of the catalytic domain of SETD2 could not be observed on the surface of hNCPWT, despite that the SETD2 protein appeared to bind hNCPWT similarly to hNCPH3.3K36M in EMSA assays as well as in cryo-EM sample preparation (Supplementary Figs. S1d, S3b–e). This suggests that the interaction of SETD2 with hNCPWT might be very dynamic, resulting in rather flexible association of the enzyme–substrate complex as seen in our structural analysis. The H3.3K36M oncohistone mutation stabilizes the association of the catalytic domain of SETD2 with the mutant nucleosome, which leads to the visualization of clear EM densities of SETD2 on the H3.3K36M nucleosome.
Nucleosome recognition by the catalytic domain of SETD2
Similar to the structure of the fungal Set2–nucleosome complex30, a remarkable feature of the human SETD2–hNCPH3.3K36M complex structure is that the nucleosome DNA at SHL-6 and SHL-7 is detached from the core histones to expose the binding site of SETD2 on nucleosome (Fig. 2a; Supplementary Fig. S3a). Then, the AWS domain of SETD2 (SETD2AWS) is attached to the α-N helix of histone H3, which confers specific interactions between SETD2 and nucleosome (Fig. 2b). At the interface, the side chains of Tyr41, Arg49 and Arg52 from histone H3 stick out and form a hydrogen-bonding network with the main chain carbonyls of Gln1498, Leu1525, Lys1639 as well as the side chain of Asn1522 from SETD2AWS (Fig. 2b). The hydrophobic side chains of Met1497, Leu1521, Leu1525, Met1526 and Ile1527 from SETD2AWS further constrain the side-chain conformations of Tyr41, Arg49 and Arg52 of histone H3.3 (Fig. 2b).
a Nucleosome DNA at SHL-6 and SHL-7 is detached from the core histones in the SETD2–hNCPH3.3K36M complex structure. b Detailed view of the recognition interface between SETD2AWS and the α-N helix of histone H3.3. c Endpoint HMT assays carried out with wide-type or mutant hNCP, revealing that mutations of Arg49H3.3 and Arg52H3.3 to Glu drastically diminished the enzymatic activities of SETD2 towards hNCP. The HMT activities were reported for equal amounts of the SETD2 proteins. Error bars correspond to the SD of three replicate assays. Input of the HMT reactions is shown in Supplementary Fig. S4a. d Endpoint HMT assays carried out with wide-type or mutant hNCP, revealing that mutation of Arg49H3.3 and Arg52H3.3 to Glu severely impaired the enzymatic activities of NSD1 towards hNCP. The HMT activities were reported for equal amounts of the NSD1 proteins. Error bars correspond to the SD of three replicate assays. Input of the HMT reactions is shown in Supplementary Fig. S4c. e Path of the histone H3.3 tail along the SHL1 DNA, the active site and the α2 helix of SETD2. f Detailed view of the active site of SETD2 bound with the K36M mutation.
In vitro HMT assays of SETD2 showed that a nucleosome mutant with substitutions of Arg49H3.3 and Arg52H3.3 with glutamic acids (R49E&R52E) failed to be methylated by SETD2, which underscores the importance of these residues in SETD2-mediated H3K36 tri-methylation (Fig. 2c; Supplementary Fig. S4a). In addition, the AWS domain is commonly present N-terminally to the catalytic SET domain in multiple H3K36-specific methyltransferases, such as NSD1–3 and ASH1L (Supplementary Fig. S4b). The methyltransferase activity of NSD1 against the R49E&R52E mutant nucleosome is also dramatically diminished, suggesting that the specific recognition between AWS and the α-N helix of histone H3 might be a common mechanism involved in the methylation of the H3K36 site (Fig. 2d; Supplementary Fig. S4c).
Located N-terminally to the α-N helix of histone H3, the H3 tail region extends above the minor groove of SHL1 DNA, passes through the active site of the catalytic SET domain of SETD2, and then exits around the α2 helix of SETD2 (Fig. 2e; Supplementary Fig. S4d). The active center of SETD2–nucleosome structure resembles that observed in the crystal structures of SETD2 complexed an H3.3K36M peptide25 (Supplementary Fig. S4e). At the active site of SETD2, residues Val35, Met36 and Lys37 of the H3.3 tail, sandwiched within an intermolecular β-sheet structure, are stabilized through main-chain contacts with the Phe1606–Met1607–Ala1608 and Gln1667–Phe1668–Gln1669 strands of SETD2. Meanwhile, the side chain of Met36H3.3 is confined by the hydrophobic side chains of Tyr1579, Met1607, Phe1664 and Tyr1666 of SETD2, and its methylthio group approaches the methyl group of the cofactor S-adenosylmethionine (SAM) (Fig. 2f).
Linker histone represses the deposition of H3K36me3 by SETD2
The attachment of SETD2 on the nucleosome requires partial unwrapping of the nucleosome DNA to expose the underlying SETD2-binding site. This particular conformation of the nucleosome often occurs during the passage of Pol II in transcription elongation35,36,37, which favors the co-transcriptional deposition of H3K36me3 by SETD2 in the gene bodies of active transcription regions. On the contrary, histone H1 interacts with the linker DNA at both the entry and exit sites of nucleosome38,39,40, which stabilizes the compact conformation of the nucleosome and might inhibit the association of SETD2 with nucleosome (Fig. 3a). To investigate the impact of linker histone on the implementation of H3K36 tri-methylation by SETD2, we assembled a 197-base pair (bp) nucleosome with the globular domain of histone H1.4 (residues 1–130) and examined the methyltransferase activities of SETD2 against the nucleosomes with or without the linker histone in vitro. As shown in Fig. 3b and Supplementary Fig. S5a, the binding of histone H1.4 resulted in an almost 90% decrease of the SETD2 activities toward the nucleosome, suggesting that the linker histone represses the SETD2-mediated H3K36 tri-methylation through tightening the DNA wrapping around core histones.
a Schematic drawing of the nucleosome bound with a linker histone H1. b HMT assays performed with SETD2 toward the 197-bp nucleosomes bound with histone H1.4. The input of the HMT reactions is shown in Supplementary Fig. S5a. c Integrative Genomics Viewer (IGV) tracks representing the distributions of H3K36me3, H1.4 and IgG ChIP-Seq results with two replicates. Rep1, replicate 1, Rep2, replicate 2. d Venn diagram illustration showing the overlap of H3K36me3, H1.4 and IgG peaks. The peaks in independent replicates were combined. e The percentages of H3K36me3, H1.4 and IgG peaks in different genomic elements. ChIP-Seq results in independent replicates were combined. The distributions were analyzed by HOMER with default parameters. f The enrichments of H3K36me3, H1.4 and IgG ChIP-Seq results at H1.4 peak regions. A 100-bp sliding window was used to scan 10-kb regions surrounding the peak center. Heatmaps illustrating ChIP-Seq read densities from 5 kb upstream to 5 kb downstream of the peak regions (in rows) were shown on a per-peak basis (in columns). g The distribution profiles of normalized H3K36me3, H1.4 and IgG read density from 5 kb upstream of the TSS to 5 kb downstream of the TES.
To further analyze whether the linker histone could inhibit the H3K36me3 in cells, we performed chromatin immunoprecipitation coupled with high-throughput sequencing (ChIP-Seq) by using H3K36me3 and H1.4 antibodies, respectively. The rabbit anti-mouse IgG was used as the negative control. Individual browser track views showed that the two replicates of each ChIP-Seq result were very similar (Fig. 3c). We calculated the read densities of each ChIP-Seq by a 10-bp window in the genome and then compared their correlations (Supplementary Fig. S5b). The two biological replicates of ChIP-Seq were highly consistent, further confirming that these ChIP-Seq results are repeatable and stable. Interestingly, we found that the correlations between H3K36me3 and H1.4 were very low, suggesting that H3K36me3 and H1.4 are distributed in different regions of the genome. To further compare the enrichments of H3K36me3 and H1.4, we called ChIP-Seq peaks and analyzed their overlap by combining the two replicates (Fig. 3d). The results showed that only around 1% of the called H3K36me3 peaks were overlapped with H1.4 peaks. More importantly, this low number of overlapped peaks was similar to that of the overlapped peaks between IgG control and other ChIP-Seq results. Supporting this, the H3K36me3 peaks were mainly distributed at exons and introns which were the gene body regions, and H1.4 peaks were enriched at intergenic regions and introns (Fig. 3e).
To gain insight into the chromatic incorporations of H3K36me3 and H1.4, we then compared the normalized density of ChIP-Seq reads at H3K36me3 and H1.4 peaks, respectively (Fig. 3f; Supplementary Fig. S5c). We found that H3K36me3 was barely detected at the H1.4 peaks where H1.4 was highly enriched. The reduction of the H1.4 signal was also detected at the H3K36me3 peaks, but with a lower level of reduction compared with that of H3K36me3 at H1.4 peaks. This may be because the H3K36me3 peaks were very broad and H1.4 could not be completely absent at these broad regions. We further compared the enrichment of H3K36me3 and H1.4 around the gene body regions where H3K36me3 was enriched (Fig. 3g). The H3K36me3 was enriched as a gradual increase from 5′ end to 3′ end of the gene body that is consistent with previous results24. H1.4 was enriched at the transcription starting site (TSS) where the H3K36me3 level was low. The results suggest that the distributions of H3K36me3 and H1.4 are mostly mutually exclusive in the chromatin. Still, future studies will be needed to determine whether this relationship is causal.
Structural comparison with yeast Set2–xNCPH3K36M complex
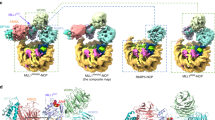
Set2 is the sole H3K36 methyltransferase that carries out mono-, di- and tri-methylation of H3K36 in yeast31. To understand the structural basis for nucleosome recognition by yeast Set2 and the underlying mechanism that determines the activity difference between yeast Set2 and human SETD2, we solved the cryo-EM structure of full-length yeast Set2 complexed with an H3K36M mutant Xenopus laevis nucleosome (hereafter, xNCPH3K36M) at an overall resolution of 3.3 Å (Fig. 4a, b; Supplementary Figs. S6, S7 and Table S1). In the yeast Set2–xNCPH3K36M complex, the catalytic domain of Set2 interacts with nucleosome in a similar manner as the human SETD2–hNCPH3.3K36M association and the fungal Set2–nucleosome interaction30 (Fig. 4b; Supplementary Fig. S3a). Nucleosome DNA unwrapping at SHL-6 and SHL-7 as well as the specific interaction between the α-N helix of histone H3 and the AWS domain of Set2 serve as the evolutionarily conserved structural features for the nucleosome binding by Set2 from yeast to human (Supplementary Fig. S8). In addition, cryo-EM analysis of yeast Set2 in complex with a Xenopus laevis wild-type nucleosome (hereafter, xNCPWT) also failed to reveal the EM density of Set2 on xNCPWT, consistent with our observations in human SETD2 studies (Supplementary Fig. S9).
a Schematic drawing of the domain organizations of yeast Set2. The color scheme is the same as that of the Set2–xNCPH3K36M structural model. b Cryo-EM density map and atomic model of yeast Set2–xNCPH3K36M complex shown from two orthogonal views. The cryo-EM map was segmented and colored according to the respective components of the Set2–xNCPH3K36M complex. c The detailed view of the active site in yeast Set2 and human SETD2. d HMT assays of wild-type and mutant Set2 (top) and SETD2 (bottom) proteins against nucleosome substrates. Mono-, di-, and tri-methylation levels of histone H3K36 are determined with antibodies of H3K36me1, H3K36me2, and H3K36me3. The input of the reactions is shown in Supplementary Fig. S10d. Each assay was repeated at least three times with similar results. e EMSAs of different truncations of Set2 (left) and SETD2 (right) with nucleosome at molar ratios of 1:1, 2:1, 4:1, and 8:1. The Native-PAGE gel is stained with ethidium bromide (EB) to show the gel shifting of NCP by Set2/SETD2. The input of the assays is shown in Supplementary Fig. S11d.
Structural comparison at the active sites of yeast Set2 and human SETD2 revealed that the side chain of the K36M mutation sticks into a hydrophobic pocket of Set2 (composed of residues Tyr149, Phe174, Met177, Phe234, and Tyr236) that is mostly conserved with that of human SETD2 (Fig. 4c). However, a remarkable difference in the side-chain conformations of Phe174Set2 and its counterpart Tyr1604SETD2 was observed between the two structures (Fig. 4c; Supplementary Fig. S10a–c). The side chain of Tyr1604SETD2 has pointed away from the hydrophobic H3.3M36-binding pocket, whereas the aromatic ring of Phe174 at the active site of Set2 is oriented toward the side chain of Met36H3, shaping the H3K36-binding pocket (Fig. 4c; Supplementary Fig. S10a, b). Mutagenesis analysis revealed that substitution of Phe174Set2 with Tyrosine (F174Y) severely impaired the mono-, di- and tri-methylation of H3K36 and a Phenylalanine-to-Alanine substitution (F174A) completely abolished the activities of Set2, suggesting an essential role of Phe174 in the recognition of H3K36 in yeast Set2 (Fig. 4d; Supplementary Fig. S10d). By contrast, similar mutations of Tyr1604 (Y1604F and Y1604A) of human SETD2 remarkably boosted the activities of SETD2 and resulted in the di-methylation and a slight mono-methylation in addition to the H3K36 tri-methylation (Fig. 4d; Supplementary Fig. S10d). In a previous study, the Y1604A mutant of SETD2 also showed elevated activities, indicating the important role of Y1604 in regulating the activity of SETD225. This observation implies that Tyr1604SETD2 might fine-tune the methyltransferase activity of SETD2 and also contribute to the determination of substrate specificity of SETD2. In addition, to investigate the effect of Phe1996 that corresponds to Tyr1604 in SETD2 on the methyltransferase activities of NSD1 (Supplementary Fig. S11a), an HMT activity assay was performed for further mutagenesis analysis. It was shown that the substitution of Phe1996NSD1 with Tyr (F1996Y) decreased the mono- and di-methylation of H3K36, and Phenylalanine-to-Alanine substitution (F1996A) more severely hampered the activities of NSD1 (Supplementary Fig. S11b). These results suggest an essential role of Tyr1604SETD2/Phe1996NSD1 in the methylation of H3K36.
Previous studies revealed that deletion of the SRI domain of Set2 (Set2SRI) abolished the interaction of Set2 with RNA pol II and impaired H3K36 methylation during transcription elongation in yeast6. The SRI domain was also reported to be required for the in vitro methyltransferase activity of Set25, which was observed in our analysis, as well (Supplementary Fig. 11c). In addition, EMSA assays demonstrated that lack of the SRI domain severely impaired the formation of the Set2–xNCPH3K36M complex, suggesting that Set2SRI is involved in nucleosome binding (Fig. 4e, left panel; Supplementary Fig. S11d, left panel). However, Set2SRI is not visible in the cryo-EM density map of the yeast Set2–xNCPH3K36M complex, which implies that Set2SRI might interact with nucleosome in a nonspecific way. In a similar analysis of the SRI domain of human SETD2 (SETD2SRI), although SETD2SRI could result in a three-fold increase in the activity of the catalytic domain of SETD2 against nucleosomes, it did not appear to participate in the binding of the nucleosome (Fig. 4e, right panel; Supplementary Figs. S1b and S11d, right panel). These results suggest that the SRI domain might regulate the activities of Set2 and SETD2 in different ways.
Discussion
In this work, the cryo-EM studies of human SETD2 methyltransferase complexed with an H3.3K36M mutant nucleosome or a wild-type nucleosome provide structural mechanisms for SETD2 catalysis as well as for inhibition of SETD2 by the oncogenic H3.3K36M mutation in the context of nucleosomes. We observed that the catalytic domain of SETD2 dynamically interacts with the wild-type nucleosome and the cofactor product S-adenosyl-l-homocysteine (SAH), suggestive of a transient binding mechanism for SETD2 and nucleosome. The ‘transient binding’ mechanism could allow rapid release of SETD2 from its nucleosome substrates, which might be suited to match the pol II-associated deposition of H3K36me3 marks with the transcription elongation rate (Fig. 5a). In contrast with the wild-type nucleosome, the H3.3K36M mutation stabilizes the complex of SETD2 with its nucleosome substrate and the cofactor SAM, resulting in the retention of SETD2 on nucleosome surface (Fig. 5a). Previous studies indicated that the H3.3K36M peptide binds to SETD2 in a more stable way than the wild-type H3.3K36 peptide does, and inhibits the activities of SETD224,26. Our in vitro HMT assays further demonstrated that the H3.3K36M nucleosome exhibited stronger inhibitory effects on the activities of SETD2, compared with that of the H3.3K36M peptide (Supplementary Fig. S12). These observations collectively suggest that the H3.3K36M mutant nucleosome could potentially reduce the amounts of SETD2 that is available for H3K36 tri-methylation co-transcriptionally, and consequently impede the establishment of H3K36 methylation landscape and lead to the alteration of gene transcription. Future studies are required to further explore the mechanism in vivo.
a In normal cells, SETD2/Set2 directly associates with RNA Pol II and implements H3K36 methylation on partially unwrapped nucleosomes, co-transcriptionally. After that, SETD2/Set2 is rapidly released from the nucleosomes, and the DNA wraps back to its canonical conformation. However, in cancer cells carrying the oncogenic H3.3K36M mutations, SETD2/Set2 is withheld on the H3.3K36M nucleosome, which impairs the normal functions of SETD2/Set2 and possibly affects the movement of RNA pol II during transcription elongation. b A schematic drawing shows that histone H1 stabilizes the compact conformation of the nucleosome and prohibits the binding of SETD2 on the nucleosome.
The association of SETD2 with nucleosome requires the unwrapping of nucleosome DNA at its entry/exit site to expose the N-terminal region of histone H3 (Fig. 2a). Rapid DNA unwrapping and rewrapping, also called ‘DNA breathing’, results from the intrinsic structural dynamics of nucleosome41. It frequently occurs during the passage of RNA pol II, where SETD2 implements H3K36 tri-methylation co-transcriptionally. We also found that the linker histone H1 represses the methyltransferase activities of SETD2 toward nucleosomes and that it displays distinct distributions from the H3K36me3 marks in the genome (Fig. 3). Histone H1 binds the nucleosome linker DNA at the entry and exit sites, which stabilizes the compact conformation of the nucleosome and blocks the binding site of SETD2 on the nucleosome surface (Fig. 5b). Previous studies revealed that eviction of histone H1 from chromatin enhances gene transcription activation42,43,44. In addition to its effect on chromatin decondensation, the histone H1 displacement could also restore the dynamics of nucleosome conformation and favors H3K36 tri-methylation and other histone modifications accompanied with active transcription. Previous findings that H3.3 maintained a decondensed chromatin state and antagonizes H1 incorporation in the early development of mice also suggest mutually exclusive functions of histone H3.3 and H1 on chromatin regulation45.
The cryo-EM structures of human SETD2 and its yeast homolog Set2 complexed with the H3K36M nucleosome also provide an evolutionarily conserved structural framework for the recognition of nucleosome by the histone H3K36 methyltransferases. The AWS domains of SETD2 and Set2 specifically interact with the α-N helix of histone H3, where the side chains of Arg49 and Arg52 of H3 are inserted into a conserved concaved surface of AWS (Fig. 2b; Supplementary Fig. S8b). The R49E&R52E mutant nucleosome failed to be methylated by SETD2, Set2, and the H3K36 di-methyltransferase NSD1, highlighting the importance of this specific recognition in H3K36 methylation (Fig. 2c, d; Supplementary Fig. S8c). A similar interaction interface was also observed in the fungal Set2–nucleosome structure30, suggesting that it provides a common structural basis for H3K36 methylation. In addition, the recognition interfaces of the K36M mutation with the active site of SETD2 and Set2 are mostly similar except for the side-chain conformations of the Phe174Set2/Tyr1604SETD2 pair (Fig. 4c). The biochemical analysis further indicated that the two residues play opposite roles in the activity regulation of Set2 and SETD2, and Tyr1604SETD2 also participates in the determination of substrate specificity in SETD2 (Fig. 4d). A ‘Phe/Tyr switch’ mechanism has been previously proposed for controlling the methylation states of several SET-domain-containing HMTs46,47. Residues Y149 and F234 of Set2 were proposed to serve as the ‘Phe/Tyr switch’ in yeast48. It was shown that yeast Set2 bearing a Y149F mutation predominately catalyzes H3K36 tri-methylation, and an F234Y substitution results in the mono- and di-methylation of H3K3648. As human SETD2 and yeast Set2 have the identical residues (Tyr149Set2/Tyr1579SETD2, Phe234Set2/Phe1664SETD2) at the two positions (Supplementary Fig. S10c), the Phe174Set2/Tyr1604SETD2 pair identified in this study might serve as another type of ‘Phe/Tyr switch’ that accounts for the different methyltransferase activities between human SETD2 and yeast Set2.
In summary, our studies provide structural insights into the oncogenic effects of the H3.3K36M mutation on the molecular functions of the H3K36 methyltransferase SETD2 at the nucleosome level, and demonstrate the regulatory effects of nucleosome conformation dynamics on SETD2-mediated H3K36 tri-methylation. Comparative structural analysis of human SETD2 and yeast Set2 complexed with nucleosomes further revealed conserved structural features for the recognition of nucleosome and identified structural elements that account for the difference of the enzyme activities. The key residues and interaction interfaces revealed in this work may be targeted for further functional investigation of SETD2 and also for the mechanistic elucidation of SETD2-related tumorigenesis.
Materials and methods
Protein expression and purification
Human SETD2, yeast Set2, and their truncations or mutants were cloned into a modified pET28b vector with a 6× His-SUMO tag fused at the N-terminus. The proteins were individually expressed in E. coli Transetta (DE3) and were purified with Ni-NTA agarose beads (QIAGEN) in lysis buffer (50 mM Tris–HCl, pH 7.5, 500 mM NaCl, 10% glycerol, 5 mM 2-mercaptoethanol, 1 mM PMSF, 5 mM benzamidine, 1 µg/mL leupeptin, and 1 µg/mL pepstatin). The 6× His-SUMO tag was removed through Ulp1 protease digestion and the proteins were further purified sequentially through Mono S 10/100 GL cation exchange chromatography (GE Healthcare) in Column Buffer A (25 mM MES, pH 6.0) and column buffer B (25 mM MES, pH 6.0 and 1 M NaCl) and HiLoad Superdex 200 gel filtration chromatography (GE Healthcare) in column buffer C (25 mM Tris–HCl, pH 7.5 and 150 mM NaCl). The purified proteins were concentrated to 5 mg/mL and stored at –80 °C in small aliquots.
Preparation of nucleosomes
Wild-type or H3/H3.3K36M-mutant nucleosomes were in vitro reconstituted from four core histones (H3/H3.3 or H3/H3.3K36M, H4, H2A, and H2B) and the 147-bp or 197-bp Widom 601 DNA as previously described49. The human or Xenopus laevis histones and their mutants were expressed in E. coli BL21 (DE3) as inclusion bodies and were purified through Q Sepharose HP chromatography (GE Healthcare) and SP Sepharose HP chromatography (GE Healthcare) in denaturing buffer (20 mM Tris–HCl, pH 7.5, 8 M urea, 0–0.5 M NaCl, 1 mM EDTA, and 5 mM 2-mercaptoethanol), sequentially. After dialysis and lyophilization, the histone proteins were assembled into histone octamers and were purified on a HiLoad Superdex 200 gel filtration column in refolding buffer (20 mM Tris–HCl, pH 7.5, 2 M NaCl, 1 mM EDTA, and 5 mM 2-mercaptoethanol). Then, the histone octamers were mixed with the Widom 601 DNA at a molar ratio of 0.9:1.0, and the mixture was dialyzed against reconstitution buffer (10 mM Tris–HCl, pH 7.5, 1 mM EDTA, 1 mM DTT, and 0.15–2 M KCl). The reconstituted nucleosomes were further purified through HiLoad Superdex 200 gel filtration chromatography and were then stored in small aliquots at –80 °C.
H1 deposition mediated by NAP-1
Human NAP-1 and H1.4_1–130aa truncation were cloned into a modified pETDuet vector with or without a 6× His tag fused at the N-terminus, respectively. The proteins were individually expressed in E. coli BL21 (DE3) cells as previously described50. His-NAP-1 was purified from the supernatant of the bacterial lysate using Ni-NTA agarose beads, followed by Mono Q 10/100 GL anion exchange chromatography (GE Healthcare). H1.4_1–130aa was purified through Mono Q 10/100 GL anion exchange chromatography (GE Healthcare) and Mono S 10/100 GL cation exchange chromatography, sequentially.
For H1 deposition, the NAP-1 and H1.4 proteins were mixed at a molar ratio of 2:1 in the buffer of 20 mM Tris–HCl, pH 7.5, 0.5 mM EDTA, 100 mM NaCl, 1 mM DTT, 10% glycerol, and 0.1 mM PMSF, and were incubated at 30 °C for 15 min. Then, an equimolar ratio of nucleosomes and linker histone/NAP-1 complexes were mixed and further incubated at 30 °C for 30 min. His-tagged NAP-1 was removed by passing through Ni-NTA beads, and the H1–nucleosomes were concentrated to 5 mg/mL and stored at –80 °C in small aliquots.
Sample preparation and cryo-EM data collection
The SETD2 or Set2 proteins were mixed with the wild-type or K36M-mutant NCPs and the cofactor SAH or SAM at a molar ratio of 8:1:10 and were incubated at 4 °C for 30 min. Then, the SETD2–hNCPH3.3K36M, Set2–xNCPH3K36M, SETD2–hNCPWT, or Set2–xNCPWT complexes were purified using the method of GraFix51. In brief, a continuous 10%–30% gradient of glycerol with a 0–0.1% gradient of glutaraldehyde was generated on a BioComp gradient master. Then, the SETD2–hNCPH3.3K36M, Set2–xNCPH3K36M, SETD2–hNCPWT or Set2–xNCPWT mixture was loaded on top of the glycerol gradient and the ultracentrifugation was performed at the speed of 35,000 rpm at 4 °C for 15 h, using an SW41Ti or SW60Ti rotor (Beckman). The GraFix fractions containing the SETD2–hNCPH3.3K36M, Set2–xNCPH3K36M, SETD2–hNCPWT, or Set2–xNCPWT complex were collected and dialyzed against 25 mM Tris–HCl, pH 7.5, and 150 mM NaCl, and were concentrated to 1 mg/mL. 2 µL aliquots of the samples were applied to glow-discharged holey carbon grids (Quantifoil R1.2/1.3, 300 mesh), and the grids were blotted for 2.5 s and were then plunged into liquid ethane cooled by liquid nitrogen, using a Vitrobot Mark IV (FEI).
The samples were observed under a Titan Krios transmission electron microscope (FEI) operated at 300 kV. The images were collected on a K3 direct electron detector (Gatan) with a pixel size of 1.09 or 1.10 Å. The SETD2–hNCPH3.3K36M, Set2–xNCPH3K36M, and Set2–xNCPWT datasets have a defocus range of –0.7 to –2.8, –0.8 to –3.3, and –0.8 to –2.6 µm, respectively; and each micrograph was dose-fractioned to 32 frames with 0.1 s exposure time for each frame. The total accumulated dose of each micrograph is 50.0 e−/Å2. The dataset of the SETD2–hNCPWT complex collected on a Falcon 3EC detector has a defocus range of –1.0 to –2.0 µm, and each micrograph was dose-fractioned to 38 frames with 1.3 s exposure time for each frame. The total accumulated dose of each micrograph is 40.0 e−/Å2. The imaging conditions were also listed in Supplementary Table S1.
Image processing and model building
A total of 3931 cryo-EM images of the SETD2–hNCPH3.3K36M complex were collected on a K3 detector, and motion correction was performed on the dose-fractioned image stacks using MotionCor2 with dose weighting52,53. The CTF parameters of each image were determined with Gctf54. Particle picking, 2D classification, 3D initial model, 3D classification, 3D auto-refine, CTF refinement, and Bayesian polishing were performed with RELION-355. An overview of the data processing procedure was shown in Supplementary Fig. S2a. After two rounds of 2D classification and two rounds of 3D classification with exhaustive angular searches, a total of 339,177 particles that belong to the SETD2–hNCPH3.3K36M complex were processed with 3D auto-refine and solvent-masked post-processing. To improve the map density of SETD2, the particles were further processed through masked 3D classifications with partial signal subtraction56, and a cryo-EM map of the SETD2–hNCPH3.3K36M complex was finally calculated from 154,984 particles at an overall resolution of 3.1 Å. The resolution estimation was based on the gold-standard Fourier shell correlation (FSC) 0.143 criterion and the local resolution was estimated with ResMap57,58. The cryo-EM datasets of the Set2–xNCPH3K36M, SETD2–hNCPWT, and Set2–xNCPWT complexes were processed similarly to that of the SETD2–hNCPH3.3K36M complex.
Model building was carried out by fitting the available structures of NCP and human SETD2 (PDB codes: 5X7X, 6J99, and 5JJY) in the EM density maps of the SETD2–hNCPH3.3K36M and Set2–xNCPH3K36M complexes using UCSF Chimera59. The model was then manually built in Coot and real-space refined with secondary structure restraints in Phenix60,61.
HMT assay
For an 8-µL HMT reaction, 0.2 µM wild-type or mutant SETD2 and Set2 proteins, 20 µM SAM, and 2 µM NCP were mixed in the buffer of 50 mM Tris–HCl, pH 8.0, 2 mM DTT, 5% glycerol, and 0.4 mg/mL BSA, and were incubated at 30 °C for 2 and 3 h, respectively. The reaction was stopped by adding 2 μL of 0.5% trifluoroacetic acid (TFA) and the HMT activity was evaluated using an MTase-GloTM Methyltransferases Assay Kit (Promega). The luminescent signal that corresponds to the production of SAH was measured using the EnSpire Alpha Multimode plate reader (PerkinElmer) in a white 384-well plate. Each reaction was run in triplicate and was reported as the means ± standard deviation (SD).
For evaluation of the activity specificity of SETD2 and Set2, the HMT reaction mixtures were separated on a 15% SDS–PAGE gel, and the mono-, di- and tri-methylation of histone H3K36 was detected by western blotting using the corresponding antibodies (H3K36me1 antibody: #A2364, ABclonal; H3K36me2 antibody: #A2365, ABclonal; H3K36me3 antibody: #A2366, ABclonal).
EMSA
SETD2 or Set2 was mixed with 1 µM NCPWT or NCPH3K36M at a molar ratio of 1:1, 2:1, 4:1, or 8:1 in a total volume of 20 µL. After incubation on ice for 1 h, 10 µL of each mixture was run on a 6% Native-PAGE gel and the gel was stained with EB to show the shift of NCPWT or NCPH3K36M. The same amount of each mixture was loaded on a 13.5% SDS–PAGE gel and detected by Coomassie blue staining to show the input of the EMSA assays.
ChIP-Seq and data analysis
ChIP-Seq was conducted as described before with modifications described below62. Cells were cross-linked with 1% formaldehyde and quenched in 125 mM glycine. Nuclei were extracted by lysis buffer (10 mM Tris–HCl, pH 7.5, 10 mM NaCl, 0.5% NP-40), and then digested in MNase digestion buffer (20 mM Tris–HCl, pH 7.5, 15 mM NaCl, 60 mM KCl, 1 mM CaCl2) with MNase at 37 °C for 20 min. After mixing with equal amounts of 2× STOP buffer (100 mM Tris–HCl, pH 8.1, 20 mM EDTA, 200 mM NaCl, 2% Triton X-100, 0.2% sodium deoxycholate), the digested nuclei were sonicated for 5 min with biorupter (30 s on/30 s off). The extracted chromatin was incubated with 2 μg of anti-H1.4 (Cell Signaling Technology, Cat. #41328), anti-H3K36me3 (Active Motif, Cat. #61101) antibodies, and rabbit anti-mouse IgG (Abcam, Cat. #ab6709) at 4 °C overnight, respectively. The antibody-bound chromatin was then incubated with 30 μL of protein G-magnetic beads (Life Technologies) for 2 h. The beads were extensively washed before the release of enriched DNAs. DNA libraries were prepared by TruPrep DNA library prep kit (Vazyme) and subsequently sequenced on the Illumina HiSeq 4000 platform.
ChIP-Seq reads were cleaned by trim-galore and then aligned to the human genome, hg19, by bowtie263. The duplicated PCR reads were further cleaned by SAMtools64. The H3K36me3 and H1.4 ChIP-Seq peaks were identified by MACS265 with the parameters of the broad peak for H3K36me3, and the narrow peak calling for H1.4. BEDTools66 and deeptools67 were used to calculate the read density at different regions.
Data availability
The EM density maps have been deposited in the Electron Microscopy Data Bank with accession codes EMD-31039, EMD-31040, EMD-31041, and EMD-31042. The final models have been submitted to the RCSB Protein Data Bank under the accession codes 7EA5 and 7EA8. ChIP-Seq data have been submitted to the GEO database with GEO number GSE148235. All other data are available from the corresponding author upon reasonable request.
References
Kouzarides, T. Chromatin modifications and their function. Cell 128, 693–705 (2007).
Edmunds, J. W., Mahadevan, L. C. & Clayton, A. L. Dynamic histone H3 methylation during gene induction: HYPB/Setd2 mediates all H3K36 trimethylation. EMBO J. 27, 406–420 (2008).
Allis, C. D. et al. New nomenclature for chromatin-modifying enzymes. Cell 131, 633–636 (2007).
Li, M. et al. Solution structure of the Set2–Rpb1 interacting domain of human Set2 and its interaction with the hyperphosphorylated C-terminal domain of Rpb1. Proc. Natl Acad. Sci. USA 102, 17636–17641 (2005).
Wang, Y., Niu, Y. & Li, B. Balancing acts of SRI and an auto-inhibitory domain specify Set2 function at transcribed chromatin. Nucleic Acids Res. 43, 4881–4892 (2015).
Kizer, K. O. et al. A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Mol. Cell. Biol. 25, 3305–3316 (2005).
Rebehmed, J., Revy, P., Faure, G., de Villartay, J. P. & Callebaut, I. Expanding the SRI domain family: a common scaffold for binding the phosphorylated C-terminal domain of RNA polymerase II. FEBS Lett. 588, 4431–4437 (2014).
Matsuda, A. et al. Highly condensed chromatins are formed adjacent to subtelomeric and decondensed silent chromatin in fission yeast. Nat. Commun. 6, 7753 (2015).
Venkatesh, S. et al. Set2 methylation of histone H3 lysine 36 suppresses histone exchange on transcribed genes. Nature 489, 452–455 (2012).
Jones, B. Chromatin: Lys36 sets limits for histone exchange. Nat. Rev. Mol. Cell Biol. 13, 602–603 (2012).
Pai, C. C. et al. A histone H3K36 chromatin switch coordinates DNA double-strand break repair pathway choice. Nat. Commun. 5, 40911 (2014).
Carvalho, S. et al. SETD2 is required for DNA double-strand break repair and activation of the p53-mediated checkpoint. Elife 3, e02482 (2014).
Jha, D. K. & Strahl, B. D. An RNA polymerase II-coupled function for histone H3K36 methylation in checkpoint activation and DSB repair. Nat. Commun. 5, 3965 (2014).
Guo, R. et al. BS69/ZMYND11 reads and connects histone H3.3 lysine 36 trimethylation-decorated chromatin to regulated pre-mRNA processing. Mol. Cell 56, 298–310 (2014).
Luco, R. F. et al. Regulation of alternative splicing by histone modifications. Science 327, 996–1000 (2010).
Zhu, X. et al. Identification of functional cooperative mutations of SETD2 in human acute leukemia. Nat. Genet. 46, 287–293 (2014).
Dalgliesh, G. L. et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature 463, 360–363 (2010).
Yuan, H. et al. Histone methyltransferase SETD2 modulates alternative splicing to inhibit intestinal tumorigenesis. J. Clin. Invest. 127, 3375–3391 (2017).
Salem, M. E. et al. Comparative molecular analyses of esophageal squamous cell carcinoma, esophageal adenocarcinoma, and gastric adenocarcinoma. Oncologist 23, 1319–1327 (2018).
Fontebasso, A. M., Liu, X. Y., Sturm, D. & Jabado, N. Chromatin remodeling defects in pediatric and young adult glioblastoma: a tale of a variant histone 3 tail. Brain Pathol. 23, 210–216 (2013).
Mar, B. G. et al. Mutations in epigenetic regulators including SETD2 are gained during relapse in paediatric acute lymphoblastic leukaemia. Nat. Commun. 5, 3469 (2014).
Behjati, S. et al. Distinct H3F3A and H3F3B driver mutations define chondroblastoma and giant cell tumor of bone. Nat. Genet. 45, 1479–1482 (2013).
Papillon-Cavanagh, S. et al. Impaired H3K36 methylation defines a subset of head and neck squamous cell carcinomas. Nat. Genet. 49, 180–185 (2017).
Fang, D. et al. The histone H3.3K36M mutation reprograms the epigenome of chondroblastomas. Science 352, 1344–1348 (2016).
Yang, S. et al. Molecular basis for oncohistone H3 recognition by SETD2 methyltransferase. Genes Dev. 30, 1611–1616 (2016).
Zhang, Y. et al. Molecular basis for the role of oncogenic histone mutations in modulating H3K36 methylation. Sci. Rep. 7, 43906 (2017).
Park, S. H. et al. Cryo-EM structure of the human MLL1 core complex bound to the nucleosome. Nat. Commun. 10, 5540 (2019).
Xue, H. et al. Structural basis of nucleosome recognition and modification by MLL methyltransferases. Nature 573, 445–449 (2019).
Poepsel, S., Kasinath, V. & Nogales, E. Cryo-EM structures of PRC2 simultaneously engaged with two functionally distinct nucleosomes. Nat. Struct. Mol. Biol. 25, 154–162 (2018).
Bilokapic, S. & Halic, M. Nucleosome and ubiquitin position Set2 to methylate H3K36. Nat. Commun. 10, 3795 (2019).
Wagner, E. J. & Carpenter, P. B. Understanding the language of Lys36 methylation at histone H3. Nat. Rev. Mol. Cell Biol. 13, 115–126 (2012).
Strahl, B. D. et al. Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol. Cell. Biol. 22, 1298–1306 (2002).
Hacker, K. E. et al. Structure/function analysis of recurrent mutations in SETD2 protein reveals a critical and conserved role for a SET domain residue in maintaining protein stability and histone H3 Lys-36 trimethylation. J. Biol. Chem. 291, 21283–21295 (2016).
McDaniel, S. L. & Strahl, B. D. Shaping the cellular landscape with Set2/SETD2 methylation. Cell. Mol. Life Sci. 74, 3317–3334 (2017).
Kujirai, T. et al. Structural basis of the nucleosome transition during RNA polymerase II passage. Science 362, 595–598 (2018).
Farnung, L., Vos, S. M. & Cramer, P. Structure of transcribing RNA polymerase II-nucleosome complex. Nat. Commun. 9, 5432 (2018).
Ehara, H. et al. Structural insight into nucleosome transcription by RNA polymerase II with elongation factors. Science 363, 744–747 (2019).
Bednar, J. et al. Structure and dynamics of a 197 bp nucleosome in complex with linker histone H1. Mol. Cell 66, 384–397 (2017).
Zhou, B. R. et al. Structural insights into the histone H1-nucleosome complex. Proc. Natl Acad. Sci. USA 110, 19390–19395 (2013).
Zhou, B. R. et al. Structural mechanisms of nucleosome recognition by linker histones. Mol. Cell 59, 628–638 (2015).
Zhou, K., Gaullier, G. & Luger, K. Nucleosome structure and dynamics are coming of age. Nat. Struct. Mol. Biol. 26, 3–13 (2019).
Vicent, G. P. et al. Four enzymes cooperate to displace histone H1 during the first minute of hormonal gene activation. Genes Dev. 25, 845–862 (2011).
Zhang, Q., Giebler, H. A., Isaacson, M. K. & Nyborg, J. K. Eviction of linker histone H1 by NAP-family histone chaperones enhances activated transcription. Epigenet. Chromatin 8, 30 (2015).
Shimada, M. et al. Gene-specific H1 eviction through a transcriptional activator→p300→NAP1→H1 pathway. Mol. Cell 74, 268–283 (2019).
Lin, C. J., Conti, M. & Ramalho-Santos, M. Histone variant H3.3 maintains a decondensed chromatin state essential for mouse preimplantation development. Development 140, 3624–3634 (2013).
Collins, R. E. et al. In vitro and in vivo analyses of a Phe/Tyr switch controlling product specificity of histone lysine methyltransferases. J. Biol. Chem. 280, 5563–5570 (2005).
Couture, J. F., Dirk, L. M., Brunzelle, J. S., Houtz, R. L. & Trievel, R. C. Structural origins for the product specificity of SET domain protein methyltransferases. Proc. Natl Acad. Sci. USA 105, 20659–20664 (2008).
DiFiore, J. V. et al. Unique and shared roles for histone H3K36 methylation states in transcription regulation functions. Cell Rep. 31, 107751 (2020).
Dyer, P. N. et al. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 375, 23–44 (2004).
Syed, S. H. et al. Single-base resolution mapping of H1-nucleosome interactions and 3D organization of the nucleosome. Proc. Natl Acad. Sci. USA 107, 9620–9625 (2010).
Kastner, B. et al. GraFix: sample preparation for single-particle electron cryomicroscopy. Nat. Methods 5, 53–55 (2008).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Grant, T. & Grigorieff, N. Measuring the optimal exposure for single particle cryo-EM using a 2.6 Å reconstruction of rotavirus VP6. Elife 4, e06980 (2015).
Zhang, K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7, e42166 (2018).
Bai, X. C., Rajendra, E., Yang, G., Shi, Y. & Scheres, S. H. Sampling the conformational space of the catalytic subunit of human gamma-secretase. Elife 4, e11182 (2015).
Scheres, S. H. & Chen, S. Prevention of overfitting in cryo-EM structure determination. Nat. Methods 9, 853–854 (2012).
Kucukelbir, A., Sigworth, F. J. & Tagare, H. D. Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2014).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta. Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta. Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Fang, D. et al. H3.3K27M mutant proteins reprogram epigenome by sequestering the PRC2 complex to poised enhancers. Elife 7, e36696 (2018).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Feng, J., Liu, T., Qin, B., Zhang, Y. & Liu, X. S. Identifying ChIP-seq enrichment using MACS. Nat. Protoc. 7, 1728–1740 (2012).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Ramirez, F. et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016).
Acknowledgements
We thank staff of the Electron Microscopy System, the Mass Spectrometry System, and the Protein Preparation System at Shanghai Institute of Precision Medicine for instrument support and technical assistance. This work was supported by the National Key R&D Program of China (2016YFA0501803 and 2017YFA0504504 to J.H.), the National Natural Science Foundation of China (32022036, 31570766, U1632130 to J.H.), China Postdoctoral Science Foundation (2018M642037 to Y.L.), the SHIPM-sigma fund (BJ1-7009-18-1303 and JY201805 to J.H.), and fund from State Key Laboratory of Esophageal Cancer Prevention and Treatment (K2020-0013 to J.H.). J.H. is a member of the innovative research team of high-level local universities in Shanghai.
Author information
Authors and Affiliations
Contributions
Y.L. performed sample preparation, EM data collection, and biochemical analyses; H.X., M.C., and G.B. helped with EM data collection and analyses; D.F. and Y.Z. performed ChIP-Seq analysis; Z.M., Y.Y., and S.S. helped with biochemical analysis; J.H. designed and supervised the research; J.H. and D.F. wrote the manuscript with help from all other authors.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Liu, Y., Zhang, Y., Xue, H. et al. Cryo-EM structure of SETD2/Set2 methyltransferase bound to a nucleosome containing oncohistone mutations. Cell Discov 7, 32 (2021). https://doi.org/10.1038/s41421-021-00261-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41421-021-00261-6
This article is cited by
-
Structural and functional specificity of H3K36 methylation
Epigenetics & Chromatin (2022)
-
Mechanistic basis of the increased methylation activity of the SETD2 protein lysine methyltransferase towards a designed super-substrate peptide
Communications Chemistry (2022)
-
SMYD5 catalyzes histone H3 lysine 36 trimethylation at promoters
Nature Communications (2022)
-
Computational Study of Methionine Methylation Process Catalyzed by SETD3
Interdisciplinary Sciences: Computational Life Sciences (2022)
-
SETD2: from chromatin modifier to multipronged regulator of the genome and beyond
Cellular and Molecular Life Sciences (2022)