Abstract
Granulosa cell (GC) is a critical somatic component of ovarian follicles to support oocyte development, while the regulatory role of long noncoding RNA (lncRNA) in GCs is largely unknown. Here, we identified a down-regulated lncRNA ZNF674-AS1 in GCs from patients with biochemical premature ovarian insufficiency (bPOI), and its expression correlates with serum levels of clinical ovarian reserve indicators. Functional experiments showed that ZNF674-AS1 is induced by energy stress, and regulates the proliferation and glycolysis of GCs, which possibly leads to follicular dysfunction. Mechanistically, low-expressed ZNF674-AS1 reduced the enzymatic activity of aldolase A (ALDOA), concomitant with promoting the association between ALDOA and v-ATPase to activate the lysosome localized AMP-activated protein kinase (AMPK). These findings identified a new lncRNA–ALDOA complex through which ZNF674-AS1 exerts its functions, expanding the understanding of epigenetic regulation of GCs function and POI pathogenesis.
Similar content being viewed by others
Introduction
In mammals, the ovarian follicle is the basic female reproductive unit which consists of oocytes and two types of somatic cells, granulosa cells (GCs) and theca cells1. GCs are supposed to provide physical and hormonal support to oocytes so that folliculogenesis can be successfully executed to fulfill female fertility2,3. Notably, GCs apoptosis initiates follicular atresia4. Follicular growth originates from primordial follicle formation to the final ovulation. Any elements that disrupt this process will diminish the ovarian reserve pool and/or accelerate ovarian follicle depletion, therefore resulting in premature ovarian insufficiency (POI)5, which is defined as partial or complete loss of ovarian function before the age of 40 years6. POI is a heterogeneous disorder affecting ~1% of women of reproductive age6. Clinically, three stages of POI have been described, including occult, biochemical, and eventually overt5,7,8. Patients with biochemical POI (bPOI) show raised concentrations of follicle-stimulating hormone (FSH), in whom the ovarian reserve has not yet been fully exhausted. Recognized causes of POI include genetic, autoimmune, and iatrogenic factors9, however, the majority remains unclear.
Long noncoding RNAs (lncRNAs) are transcripts longer than 200 nucleotides without protein-coding function10, which performs diverse regulatory roles in many biological processes, such as gene regulation, mRNA splicing, and protein stabilization11,12. Aberrant expression of lncRNAs has been observed in many pathological conditions, regulating tumor metastasis13, cell viability14, metabolism alteration15, and innate immunity16. However, the regulatory roles of lncRNAs in the GCs function are not completely characterized. Therefore, elucidating the role of lncRNAs underlying GCs function will expand the understanding of the molecular mechanisms of follicular development and benefit the diagnosis and treatment of POI.
In the present study, we identified a down-regulated lncRNA ZNF674-AS1 in GCs from patients with POI and demonstrated its essential regulatory role in the glycolysis and proliferation of GCs. ZNF674-AS1 functions through interactions with aldolase A (ALDOA), a key enzyme that modulates the glycolysis process17 and controls the activation of AMP-activated protein kinase (AMPK) at lysosome18,19,20. Moreover, our findings suggested that expression of ZNF674-AS1 in GCs correlates with serum levels of clinical ovary function indicators and that down-regulated ZNF674-AS1 might contribute to the development of POI.
Results
LncRNA ZNF674-AS1 is down-regulated in GCs from patients with bPOI and correlates with poor ovarian reserve
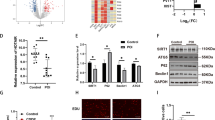
To uncover lncRNAs that are differentially expressed in GCs from patients with bPOI, we collected the GCs from eight patients with bPOI and nine age/BMI-matched healthy women for RNA-seq analysis (GEO: GSE158526). Results showed that 78 lncRNAs were differentially expressed in GCs from patients with bPOI (Supplementary Table 1). Subsequently, six lncRNAs (AC112721.1, ZNF674-AS1, RP4-545C24.1, GS1-358P8.4, SNAI3-AS1, and RP11-3D4.3) with highly conserved properties based on UCSC21 resources were singled out for qRT-PCR validation, and the down-regulated lncRNA ZNF674-AS1, which was validated in GC samples from 33 patients with bPOI and 41 age-matched controls (Fig. 1A), stood out by showing a significant correlation with serum levels of clinical ovary function indicators. In detail, the expression of ZNF674-AS1 showed a positive correlation with serum levels of basic estradiol in 33 patients with bPOI (Fig. 1B and Supplementary Fig. 1A). Furthermore, correlation analysis in all participants (bPOIs and Controls, n = 74) showed that the expression of ZNF674-AS1 was positively correlated with serum levels of basic estradiol and AMH, and negatively correlated with serum levels of basic FSH (Fig. 1C), suggesting the regulatory role of ZNF674-AS1 in ovary functions. Therefore, lncRNA ZNF674-AS1 was selected for further exploration. LncRNA ZNF674-AS1 is located at chromosome X with limited known functions (Fig. 1D). Coding Potential Assessment Tool showed the noncoding feature of lncRNA ZNF674-AS1 (Supplementary Fig. 1B). Collectively, lncRNA ZNF674-AS1 is down-regulated in patients with bPOI and ZNF674-AS1 expression is a promising indicator for ovarian reserve.
A The expression level of ZNF674-AS1 was validated by qRT-PCR in GCs from patients with bPOI (n = 33) and controls (n = 41). Ct values were normalized to GAPDH. *p < 0.05, by two-tailed Student’s t test. B The correlation between the expression level of ZNF674-AS1 in GCs and the serum concentration of estradiol from patients with bPOI (n = 33) was analyzed by Pearson correlation analysis. C The correlation between the expression level of ZNF674-AS1 in GCs and the serum concentration of estradiol, AMH, FSH from all participants (n = 74) was analyzed by Pearson correlation analysis. D Schematic representation of localization of ZNF674-AS1 in chromosome X.
Silencing of lncRNA ZNF674-AS1 significantly inhibited the proliferation of GCs
To explore the roles of lncRNA ZNF674-AS1 on GC function, we silenced ZNF674-AS1 in KGN and COV434 cells via antisense LNA GapmeRs (Supplementary Fig. 1C). Decreased cell viability of KGN and COV434 cells was observed after ZNF674-AS1 knockdown (Fig. 2A, B). EdU assays showed that ZNF674-AS1 silence significantly inhibited the proliferation of KGN and COV434 cells (Fig. 2C–F). Furthermore, protein levels of proliferating cell nuclear antigen were decreased in ZNF674-AS1 silenced KGN and COV434 cells (Fig. 2G, H). Taken together, these results indicated that silencing lncRNA ZNF674-AS1 inhibited GC proliferation.
A, B The viability of KGN and COV434 cells after ZNF674-AS1 silencing was assessed by CCK8 assay. Results are expressed as the mean ± SD (n = 3). ***p < 0.001 by two-tailed Student’s t test. C, D The proliferation of KGN cells after ZNF674-AS1 silencing was evaluated by EdU staining assay. Results are expressed as the mean ± SD (n = 3). ***p < 0.001 by two-tailed Student’s t test. Scale bar: 200 μm. E, F The proliferation of COV434 cells after ZNF674-AS1 silencing was evaluated by EdU staining assay. Results are expressed as the mean ± SD (n = 3). **p < 0.01 by two-tailed Student’s t test. Scale bar: 50 μm. G, H PCNA protein levels in KGN and COV434 cells were analyzed by western blot after ZNF674-AS1 silencing. For gray value quantification of PCNA, data were normalized to the internal reference. Shown are mean ± SD. *p < 0.05, ***p < 0.001 by two-tailed Student’s t test.
We also detected the impact of ZNF674-AS1 knockdown on steroid hormone synthesis of KGN cells. However, results showed that levels of estradiol, as well as FSHR, CYP19A1 proteins, remain unchanged in ZNF674-AS1 silenced KGN cells (Supplementary Fig. 1D, E).
LncRNA ZNF674-AS1 interacts with ALDOA protein
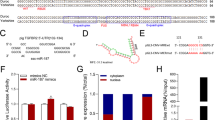
Subcellular fraction assay revealed that ZNF674-AS1 was predominantly distributed in the cytoplasm (Fig. 3A). To uncover the molecular mechanism of ZNF674-AS1 function, an RNA pulldown assay was performed to identify proteins that directly binds with ZNF674-AS1. After separated on SDS–PAGE and visualized by silver staining, a ~40 kDa band was specifically present in ZNF674-AS1 pulldown samples (Fig. 3B), which was cut out and subjected to mass spectrometry. Subsequently, ALDOA was identified as the protein binding with ZNF674-AS1 after analysis of mass spectrometry followed by western blot validation (Fig. 3C). To corroborate these findings, we used antibodies against ALDOA to immunoprecipitate endogenous RNAs from the total lysates of KGN cells. We observed a significant enrichment of ZNF674-AS1 in the anti-ALDOA immunoprecipitates compared with the IgG control (Fig. 3D). Therefore, we explored whether ZNF674-AS1 plays a role in the expression or subcellular localization of ALDOA protein. Western blot and immunofluorescence showed that silencing of ZNF674-AS1 had no affect the protein level and the subcellular localization of ALDOA (Fig. 3E and Supplementary Fig. 1F). Taken together, these results indicated that ZNF674-AS1 and ALDOA directly bind with each other.
A The relative expression levels of ZNF674-AS1 in the cytoplasmic and nuclear fractions of KGN cells were detected by qRT-PCR. MALAT1 was the nuclear lncRNA control, Lamin B1 was the nuclear control, and GAPDH was the cytoplasmic control. B A specific band of ∼40 kDa (red box) were pulled down by ZNF674-AS1 and subsequently analyzed by mass spectrometry. C The specific association between ZNF674-AS1 and ALDOA was validated by western blot in the samples obtained from RNA pull-down. The antisense strand of ZNF674-AS1 was used as the negative control. D RIP assay was utilized to confirm the interaction between ZNF674-AS1 and ALDOA in KGN cells. Results are expressed as the mean ± SD (n = 3). **p < 0.01 by two-tailed Student’s t test. E ALDOA protein levels in KGN cells were analyzed by western blot after ZNF674-AS1 silencing. For gray value quantification of ALDOA, data were normalized to the internal reference. Shown are mean ± SD. n.s. no significance by two-tailed Student’s t test.
LncRNA ZNF674-AS1 is involved in the glycolysis and glucose metabolism of GCs
As reported, ALDOA plays a vital role in the glycolysis process, which converts fructose-1, 6-bisphosphate (F1,6BP) into glyceraldehyde-3-phosphate and dihydroxyacetone phosphate17. Therefore, we investigated whether ZNF674-AS1 regulates the enzymatic activity of ALDOA. As shown in Fig. 4A, B, the aldolase activity was obviously reduced in ZNF674-AS1 silenced KGN and COV434 cells. Moreover, we detected the concentrations of F1,6BP and lactic acid, middle and terminal products of glycolysis, to evaluate the influence of ZNF674-AS1 silence on the glycolysis of GCs. Results showed that the concentrations of F1,6BP and lactic acid were significantly decreased after ZNF674-AS1 knockdown (Fig. 4C, D), indicating the regulation of ZNF674-AS1 on glycolysis. Moreover, the ATP levels was reduced (Fig. 4E) and the ratio of ADP/ATP was significantly elevated in ZNF674-AS1 silenced KGN and COV434 cells (Fig. 4F), suggesting the abnormal process of glucose metabolism upon ZNF674-AS1 knockdown. Based on the results above, we hypothesized that lncRNA ZNF674-AS1 is necessary for the glycolysis and the glucose metabolism of GCs. To validate our hypothesis, we explored whether the energy status influences the expression of ZNF674-AS1. We detected the expression of ZNF674-AS1 in glucose-containing and glucose-free medium cultured cells. As shown in Fig. 4G, ZNF674-AS1 was significantly up-regulated in glucose-free medium cultured KGN and COV434 cells. Taken together, these data indicated that energy stress-induced ZNF674-AS1 is necessary for the glycolysis process and the glucose metabolism of GCs.
A, B The aldolase activities of KGN and COV434 cells after ZNF674-AS knockdown were examined by an Aldolase Activity Colorimetric Assay Kit and compared with control cells. Results are normalized to the total protein and expressed as the mean ± SD (n = 3), *p < 0.05, **p < 0.01, ***p < 0.001 by two-sided Student’s t test. C Levels of F1,6BP in KGN and COV434 cells after ZNF674-AS1 silencing were measured and normalized to the total protein. Results are expressed as the mean ± SD (n = 3), *p < 0.05, by the two-sided Student’s t test. D Lactic acid production in ZNF674-AS1 silenced and negative control KGN and COV434 cells was measured via Lactate Colorimetric Assay Kit. Results are expressed as the mean ± SD (n = 3), **p < 0.01, ***p < 0.001 by two-sided Student’s t test. E Levels of ATP in ZNF674-AS1 silenced and negative control KGN cells were measured and normalized to the total protein. Results are expressed as the mean ± SD (n = 3), **p < 0.01 by two-tailed Student’s t test. F The ratio of ADP/ATP in ZNF674-AS1 silenced and negative control KGN and COV434 cells were detected. Results are expressed as the mean ± SD (n = 3), **p < 0.01 by two-tailed Student’s t test. G The expression levels of ZNF674-AS1 were analyzed by qRT-PCR in glucose-containing and glucose-free medium cultured KGN and COV434 cells. Results were expressed as the mean ± SD (n = 3). **p < 0.01, ***p < 0.001 by two-tailed Student’s t test.
The silence of ZNF674-AS1 promotes the activation of AMPK
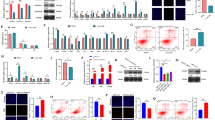
AMPK is a sensor monitoring availability of nutrients and energy and regulates cell growth22. Therefore, we investigated whether ZNF674-AS1 regulates the activation of AMPK. Interestingly, the phosphorylation levels of AMPK Thr172 and its substrates ACC were elevated after ZNF674-AS1 silencing (Fig. 5A, B). Furthermore, activation of AMPK by its activator Acadesine (AICAR) repressed the proliferation of KGN cells (Fig. 5C–F), which mimics the impact after ZNF674-AS1 knockdown. These results suggested that ZNF674-AS1 may execute its function by activating AMPK.
A, B Western blot showing the levels of phosphorylated proteins of the AMPK signaling pathway in ZNF674-AS1 silenced and negative control KGN and COV434 cells. For gray value quantification of proteins, data were normalized to the internal reference. Shown are mean ± SD. n.s. no significance, *p < 0.05, **p < 0.01 by two-tailed Student’s t test. C Western blot showing the phosphorylation of AMPK protein in AICAR or DMSO treated KGN cells. For gray value quantification of proteins, data were normalized to the internal reference. Shown are mean ± SD. n.s. no significance, *p < 0.05 by two-tailed Student’s t test. D The CCK8 assay showed the cell viability of ZNF674-AS1 silenced, AICAR treated and negative control KGN cells. Results are expressed as the mean ± SD (n = 3). **p < 0.01, ***p < 0.001 by two-tailed Student’s t test. E, F EdU staining assay showing the proliferation ability of ZNF674-AS1 silenced, AICAR treated, and negative control KGN cells. Results are expressed as the mean ± SD (n = 3). **p < 0.01, ***p < 0.001 by two-tailed Student’s t test. Scale bar: 100 μm.
ZNF674-AS1 regulates the proliferation of GCs through ALDOA/v-ATPase-dependent AMPK activation
AMPK activation is a hierarchical process including AMP-dependent and AMP-independent mechanisms23. In AMP-independent mechanisms, ALDOA binding to v-ATPase is the major premise for the activation of AMPK that located at the lysosome18,20. Therefore, we explored whether ZNF674-AS1 activates AMPK in an ALDOA/v-ATPase-dependent manner. We found that silence of ZNF674-AS1 promoted ALDOA binds to ATP6V1B2 (Fig. 6A), a non-catalytic subunit of v-ATPase. Furthermore, knockdown of ATP6V1B2 via siRNAs had no obvious impact on AMPK activation, however, abolished the activation effect of ZNF674-AS1 silence on AMPK of KGN and COV434 cells (Supplementary Fig. 1G and Fig. 6B, C). These results revealed that ZNF674-AS1 activates AMPK through the ALDOA/v-ATPase-dependent pathway. In addition, CCK8 and EdU assays showed that the inhibitory effect of ZNF674-AS1 silence on KGN and COV434 proliferation was reversed by co-transfected ATP6V1B2 siRNAs (Fig. 6D–I). Collectively, these data indicated that ALDOA/v-ATPase-dependent AMPK activation is responsible for the regulation of ZNF674-AS1 on the proliferation of GCs.
A Western blot showing the protein level of ATP6V1B2 immunoprecipitated by anti-ALDOA antibodies in ZNF674-AS1 silenced and negative control KGN cells. For gray value quantification of ATP6V1B2, data were normalized to ALDOA. Shown are mean ± SD. **p < 0.01 by two-tailed Student’s t test. B, C Western blot showing the levels of phosphorylated proteins involved in the AMPK signaling pathway in ZNF674-AS1 silenced, ATP6V1B2 silenced, co-silenced and negative control KGN and COV434 cells. For gray value quantification of proteins, data were normalized to the internal reference. Shown are mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001 by two-sided Student’s t test. D, E The CCK8 assay showed the cell viability of ZNF674-AS1 silencing, ATP6V1B2 silenced, co-silenced and negative control KGN and COV434 cells. Results are expressed as the mean ± SD (n = 3). ***p < 0.001 by two-tailed Student’s t test. F–I EdU staining assay showing the proliferation ability of ZNF674-AS1 silenced, ATP6V1B2 silenced, co-silenced and negative control KGN and COV434 cells. Results are expressed as the mean ± SD (n = 3). **p < 0.01, ***p < 0.001 by two-tailed Student’s t test. Scale bars: 100 μm (F) and 50 μm (H).
Discussion
In this study, we reported a down-regulated lncRNA ZNF674-AS1 in GCs from patients with bPOI, which was essential for the glycolysis and proliferation of GCs (Fig. 7). ZNF674-AS1 was an energy stress-responsive lncRNA that functioned through binding with ALDOA in GCs. Decreased expression of ZNF674-AS1 resulted in reduced enzymatic activity of ALDOA and promoted the activation of AMPK as well, therefore inhibiting the proliferation of GCs.
By convention, POI derives from the insufficient primordial follicle and accelerated follicle loss. Within follicles, GCs are the somatic cells to support and maintain the development of oocyte and follicle2,3. At the initiation of oocyte growth, flattened pregranulosa cells enclosed the oocytes to protect them from cell death24. Thereafter, GCs proliferate mitotically and undergo shape change so that they could provide sufficient materials, such as metabolic substrate, estradiol, to drive folliculogenesis and prevents follicular atresia3. In the present study, we showed that down-regulated lncRNA ZNF674-AS1 significantly inhibited the proliferation of GCs. Similarly, lncRNA GCAT1 has been shown to regulate GCs proliferation in patients with bPOI25. Our study provided new regulatory evidence conducted by lncRNA that mediates GCs proliferation.
We identified a novel interaction between lncRNA ZNF674-AS1 and ALDOA, and showed that this lncRNA–protein complex was essential for the aldolase activity and glycolysis process in GCs. Glucose is the major substrate for ATP synthesis in GCs26. Of note, in order to minimize oxidative damage, glycolysis is the predominant energy metabolism pathway in oogonia and primordial follicle27. With regard to the course of oocyte maturation, oocyte up-regulates the expression of glycolytic genes in GCs to promote the production of pyruvate and lactate, which are readily utilized by the oocyte28,29. In the present study, ZNF674-AS1 was inducible under energy stress and insufficient synthesis of lactate and ATP was observed in ZNF674-AS1 silenced KGN cells. Therefore, follicular growth might be restrained in a state of low expression of ZNF674-AS1.
ALDOA is a central enzyme in the glycolysis process which catalyzes F1,6BP to glyceraldehyde 3-phosphate and dihydroxyacetone phosphate17. Several lncRNAs have been reported to mediate the expression of certain glycolytic enzymes30,31, suggesting the epigenetic modulation of glycolysis. However, we revealed a distinct mechanism that ZNF674-AS1 regulates glycolysis via direct binding to ALDOA and modulating its aldolase activity without affecting its expression and subcellular distribution, which provided a better understanding of the mechanisms by which lncRNAs conduct their regulatory functions. Similarly, lncRNA HULC promotes glycolysis by modulating enzymatic activities of glycolytic enzymes in hepatocellular carcinoma cell lines15. To our knowledge, ZNF674-AS1 is the first lncRNA involved in the aldolase activity of ALDOA that is involved in the metabolism homeostasis of GCs.
We demonstrated that aberrant AMPK activation is the downstream pathway of ZNF674-AS1 regulating GCs proliferation. AMPK is a sensor of cellular energy status and a regulator of cell growth23,32. It is well established that AMPK inhibits the proliferation of different cell types including GCs33,34. Our present data re-emphasized the inhibitory impact of AMPK on cell growth. AMPK activation depends on its subcellular localization and levels of cellular AMP19,20. Under the early stage of energy deficiency, AMPK localized at lysosome is exclusively activated through the association of ALDOA with v-ATPase20. Our results showed that down-regulated ZNF674-AS1 promoted ALDOA binding to v-ATPase, thereby activating AMPK. Furthermore, silencing ATP6V1B2, the non-catalytic subunit of v-ATPase, reversed the ZNF674-AS1 silencing-induced activation of AMPK and inhibition of GCs proliferation. Collectively, our findings suggest that ZNF674-AS1 exerts its function at least in part through regulating AMPK activation at lysosome.
In conclusion, we identified a down-regulated lncRNA ZNF674-AS1 in GCs from patients with POI and demonstrated its regulatory function in the proliferation, glycolysis, and AMPK activation of GCs through interaction with ALDOA. Our study revealed a novel lncRNA–aldolase complex through which the lncRNA exerts its functions, providing new evidence of epigenetic regulation of GCs function and the development of POI.
Materials and methods
Clinical samples
33 women with bPOI and 41 age and BMI-matched control women undergoing in-vitro fertilization or intracytoplasmic sperm injection and embryo transfer treatment due to tubal obstruction or male factors in the Reproductive Hospital affiliated to Shandong University were selected in this study. bPOI is defined as follows: (i) <40 years of age; (ii) 10 IU/l < basal serum FSH < 25 IU/l; (iii) AMH < 1.1 ng/ml; and (iv) regular menstruation (every 23–35 days). Patients with chromosomal abnormalities or with a history of ovarian surgery, chemotherapy, or radiotherapy were excluded. The clinical characteristics of all participants are shown in Table 1.
RNA isolation and qRT-PCR analysis
GCs were collected and purified as previously described35. Total RNAs were isolated from GCs or KGN and COV434 cells using TRIzol reagent (Invitrogen, USA). The PrimeScript™ RT reagent Kit (TaKaRa, China) was applied to synthesize complementary DNA according to the manufacturer’s protocol. Quantitative RT-PCR was performed using SYBR Green Master Mix (TaKaRa), and GAPDH was used as a reference gene. Primer sequences are listed in Supplementary Table 2.
Cell culture
The human GC-like tumor cell line KGN was obtained from the RIKEN BioResource Center (Japan)36. The human granulosa-like tumor cell line COV434 was obtained from Prof. Ying Xu of Nanjing University in China. Cells were cultured in DMEM (HyClone, USA) or DMEM/high glucose (COV434) supplemented with 10% FBS (Biological Industries, ISR) and 1% penicillin–streptomycin (Invitrogen). Cells were cultured at 37 °C in a humidified atmosphere containing 5% CO2.
Cell transfection
Antisense LNA GapmeRs were obtained from QIAGEN. The siRNA duplexes were synthesized and purchased from GenePharma (China). KGN and COV434 cells were transfected with Antisense LNA GapmeRs and/or siRNAs using Lipofectamine 3000 Reagent (Invitrogen) according to the manufacturer’s protocols. The sequences of siRNA and Antisense LNA GapmeRs are provided in Supplementary Table 3.
Cell viability assay
The viability of KGN and COV434 cells was evaluated using Cell Counting Kit-8 (CCK8, Beyotime, China) assays. 4000 cells were seeded and transfected in 96-well plates. Then, CCK-8 solution was added at the different time periods and the cells were incubated for 2 h at 37 °C. Finally, the absorbance was measured at 450 nm.
EdU proliferation assay
For the cell proliferation assay, KGN or COV434 cells were seeded and transfected into 96-well plates. 48 h after transfection, the proliferation was detected through the EdU Cell Proliferation Assay Kit (EdU, Ribobio, China) according to the manufacturer’s instructions.
Western blot
Cells were lysed in SDS buffer supplemented with proteinase inhibitor cocktail (Cell Signaling Technology, USA). The protein concentrations were determined with a BCA assay kit (Invitrogen). Approximately 20 μg cellular proteins were resolved by SDS–PAGE and transferred to polyvinylidene fluoride membranes (Millipore, USA). Specific primary antibodies were incubated with the membranes at 4 °C overnight followed by secondary antibodies (Proteintech, China). Then, membranes were incubated with the ECL chemiluminescence kit (Millipore) and imaged using ChemiDoc MP Imaging System (BIO-RAD, USA). The details of the antibodies are given in Supplementary Table 4.
Cytoplasm/nucleus fractionation
Cytoplasmic and nuclear fractions were prepared using a PARISTM Kit (Invitrogen) according to the manufacturer’s instructions, and the RNAs were then subjected to qRT-PCR analysis.
Biotin-labeled RNA pulldown assay
RNA was transcribed in vitro using the MEGAscrip T7 Transcription Kit (Invitrogen) and biotinylated using the Pierce RNA 3′ End Desthiobiotinylation Kit (Invitrogen) according to the manufacturers’ instructions. Then, RNA pull-down assays were performed using the Pierce Magnetic RNA–Protein PullDown Kit according to the manufacturer’s instructions. Co-precipitated proteins were subjected to MS analysis or western blot.
RNA immunoprecipitation (RIP)
RIP assay was performed using the EZ-Magna RIP kit (Millipore) according to the manufacturer’s instructions. Cellular proteins from KGN cells were lysed in RIP lysis buffer with RNase inhibitor and protease inhibitor cocktail. Then, cell lysates were incubated with specific primary antibodies at 4 °C overnight. Simultaneously, the same amount of cellular proteins were incubated with A homologous IgG. The co-precipitated RNA was extracted using TRIzol reagent and quantified by qRT-PCR as described above. The details of the antibodies are given in Supplementary Table 4.
Immunofluorescence staining
KGN cells were fixed in 4% paraformaldehyde at room temperature for 20 min. Cells were then permeabilized and blocked in 0.3% Triton X-100/10% BSA/PBS for 30 min. Incubation for primary antibody was done at 4 °C overnight followed by incubation with Alexa Fluor secondary antibody for 1 h at room temperature in the dark. The details of the antibodies are given in Supplementary Table 4.
Measurements of aldolase activity, F1, 6BP, lactate, ATP, and ADP levels
The aldolase activity was determined by Aldolase Activity Colorimetric Assay Kit (BioVision, USA) according to the manufacturer’s protocol. The Lactate Colorimetric Assay Kit II (BioVision) and the PicoProbeTM Fructose-1,6-Bisphosphate Assay Kit (BioVision) were applied to detect the levels of lactate and F1,6BP. The intercellular ATP levels were detected by ATP detection kit (Beyotime) and ADP/ATP ratio was determined using ApoSENSOR™ ADP/ATP Ratio Bioluminescent Assay Kit (BioVision) according to manufacturer’s instructions.
AMPK activation assay
AICAR was used as a pharmacological activator of AMPK, which was purchased from APExBIO (USA). KGN cells were treated with AICAR at the final concentration of 0.1 mM for 24 h to evaluate the effect of AMPK activation.
Co-immunoprecipitation assay
KGN cells were lysed in Pierce IP lysis buffer. The lysates were incubated overnight at 4 °C with specific primary antibodies followed by binding to PureProteom Protein A/G Magnetic Beads (Millipore) at room temperature for 1 h. The immunoprecipitates were then washed and applied to western blot. The details of the antibodies are given in Supplementary Table 4.
Statistics
Statistical analysis was performed using the SPSS software and GraphPad Prism 8 (GraphPad Software, USA). The statistical significance was analyzed by the two-tailed Student’s t test, and the Mann–Whitney U test. Pearson correlation and linear regression analyses were used to determine the correlation between ZNF674-AS1 and clinical characteristics in the bPOI group (n = 33) or all participants (bPOIs and Controls, n = 74).
References
Fortune, J. E. Ovarian follicular growth and development in mammals. Biol. Reprod. 50, 225–232 (1994).
Jaffe, L. A. & Egbert, J. R. Regulation of mammalian oocyte meiosis by intercellular communication within the ovarian follicle. Annu. Rev. Physiol. 79, 237–260 (2017).
El-Hayek, S. & Clarke, H. J. Control of oocyte growth and development by intercellular communication within the follicular niche. Results Probl. Cell Differ. 58, 191–224 (2016).
Matsuda, F., Inoue, N., Manabe, N. & Ohkura, S. Follicular growth and atresia in mammalian ovaries: regulation by survival and death of granulosa cells. J. Reprod. Dev. 58, 44–50 (2012).
Tucker, E. J., Grover, S. R., Bachelot, A., Touraine, P. & Sinclair, A. H. Premature ovarian insufficiency: new perspectives on genetic cause and phenotypic spectrum. Endocr. Rev. 37, 609–635 (2016).
European Society for Human R, Embryology Guideline Group on POI, Webber, L. et al. ESHRE Guideline: management of women with premature ovarian insufficiency. Hum. Reprod. 31, 926–937 (2016).
Soules, M. R. et al. Executive summary: Stages of Reproductive Aging Workshop (STRAW). Fertil. Steril. 76, 874–878 (2001).
Welt, C. K. Primary ovarian insufficiency: a more accurate term for premature ovarian failure. Clin. Endocrinol. 68, 499–509 (2008).
De Vos, M., Devroey, P. & Fauser, B. C. Primary ovarian insufficiency. Lancet 376, 911–921 (2010).
Batista, P. J. & Chang, H. Y. Long noncoding RNAs: cellular address codes in development and disease. Cell 152, 1298–1307 (2013).
Kopp, F. & Mendell, J. T. Functional classification and experimental dissection of long noncoding RNAs. Cell 172, 393–407 (2018).
Rinn, J. L. & Chang, H. Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 81, 145–166 (2012).
Wu, N. et al. LINC00941 promotes CRC metastasis through preventing SMAD4 protein degradation and activating the TGF-beta/SMAD2/3 signaling pathway. Cell Death Differ. 28, 219–232 (2021).
Frank, F. et al. The lncRNA growth arrest specific 5 regulates cell survival via distinct structural modules with independent functions. Cell Rep. 32, 107933 (2020).
Wang, C. et al. Interactome analysis reveals that lncRNA HULC promotes aerobic glycolysis through LDHA and PKM2. Nat. Commun. 11, 3162 (2020).
Zhang, P., Cao, L., Zhou, R., Yang, X. & Wu, M. The lncRNA Neat1 promotes activation of inflammasomes in macrophages. Nat. Commun. 10, 1495 (2019).
Chang, Y. C., Yang, Y. C., Tien, C. P., Yang, C. J. & Hsiao, M. Roles of aldolase family genes in human cancers and diseases. Trends Endocrinol. Metab. 29, 549–559 (2018).
Zhang, C.-S. et al. Fructose-1,6-bisphosphate and aldolase mediate glucose sensing by AMPK. Nature 548, 112–116 (2017).
Li, M. et al. Transient receptor potential V channels are essential for glucose sensing by aldolase and AMPK. Cell Metab. 30, 508–524 (2019). e512.
Zong, Y. et al. Hierarchical activation of compartmentalized pools of AMPK depends on severity of nutrient or energy stress. Cell Res. 29, 460–473 (2019).
Kent, W. J. et al. The human genome browser at UCSC. Genome Res. 12, 996–1006 (2002).
Garcia, D. & Shaw, R. J. AMPK: mechanisms of cellular energy sensing and restoration of metabolic balance. Mol. Cell 66, 789–800 (2017).
Lin, S. C. & Hardie, D. G. AMPK: sensing glucose as well as cellular energy status. Cell Metab. 27, 299–313 (2018).
Pepling, M. E. & Spradling, A. C. Mouse ovarian germ cell cysts undergo programmed breakdown to form primordial follicles. Dev. Biol. 234, 339–351 (2001).
Li, D. et al. lncRNA GCAT1 is involved in premature ovarian insufficiency by regulating p27 translation in GCs via competitive binding to PTBP1. Mol. Ther.—Nucleic Acids 23, 132–141 (2021).
Collado-Fernandez, E., Picton, H. M. & Dumollard, R. Metabolism throughout follicle and oocyte development in mammals. Int. J. Dev. Biol. 56, 799–808 (2012).
Bermejo-Alvarez, P., Lonergan, P., Rizos, D. & Gutierrez-Adan, A. Low oxygen tension during IVM improves bovine oocyte competence and enhances anaerobic glycolysis. Reprod. Biomed. Online 20, 341–349 (2010).
Sutton-McDowall, M. L., Gilchrist, R. B. & Thompson, J. G. The pivotal role of glucose metabolism in determining oocyte developmental competence. Reproduction 139, 685–695 (2010).
Sugiura, K., Pendola, F. L. & Eppig, J. J. Oocyte control of metabolic cooperativity between oocytes and companion granulosa cells: energy metabolism. Dev. Biol. 279, 20–30 (2005).
Fan, C. et al. Role of long non-coding RNAs in glucose metabolism in cancer. Mol. Cancer 16, 130 (2017).
Chen, J. et al. Long non-coding RNA PVT1 promotes tumor progression by regulating the miR-143/HK2 axis in gallbladder cancer. Mol. Cancer 18, 33 (2019).
Herzig, S. & Shaw, R. J. AMPK: guardian of metabolism and mitochondrial homeostasis. Nat. Rev. Mol. Cell Biol. 19, 121–135 (2018).
Kayampilly, P. P. & Menon, K. M. AMPK activation by dihydrotestosterone reduces FSH-stimulated cell proliferation in rat granulosa cells by inhibiting ERK signaling pathway. Endocrinology 153, 2831–2838 (2012).
Kayampilly, P. P. & Menon, K. M. Follicle-stimulating hormone inhibits adenosine 5’-monophosphate-activated protein kinase activation and promotes cell proliferation of primary granulosa cells in culture through an Akt-dependent pathway. Endocrinology 150, 929–935 (2009).
Xu, X. et al. Impaired telomere length and telomerase activity in peripheral blood leukocytes and granulosa cells in patients with biochemical primary ovarian insufficiency. Hum. Reprod. 32, 201–207 (2017).
Nishi, Y. et al. Establishment and characterization of a steroidogenic human granulosa-like tumor cell line, KGN, that expresses functional follicle-stimulating hormone receptor. Endocrinology 142, 437–445 (2001).
Acknowledgements
This work was supported by the National Key Research & Developmental Program of China (2017YFC1001100); the National Natural Science Foundation of China (82071609); the Taishan Scholars Program for Young Experts of Shandong Province (tsqn20161069); Shandong Science Fund for Distinguished Young Scholars (JQ201720). The authors thank all participants.
Author information
Authors and Affiliations
Contributions
Y.Q. and S.Z. performed study concept and design; D.L. conducted the experiment; X.W. and Y.D. contributed to the acquisition of results; D.L. and X.W. wrote the manuscript; G.L. provided technical and material support. All authors read and approved the final paper.
Corresponding author
Ethics declarations
Funding
The authors received no specific funding for this work.
Conflict of interest
The authors declare no competing interests.
Ethics statement
The study was authorized and approved by the Institutional Review Board of Reproductive Medicine of Shandong University, and written informed consent was obtained from all participants.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by I. Amelio
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, D., Wang, X., Li, G. et al. LncRNA ZNF674-AS1 regulates granulosa cell glycolysis and proliferation by interacting with ALDOA. Cell Death Discov. 7, 107 (2021). https://doi.org/10.1038/s41420-021-00493-1
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41420-021-00493-1