Abstract
The interaction between cancer cells and molecular cues provided by tumor stromal cells plays a crucial role in cancer growth and progression. We have recently reported that the outcome of interaction between tumor cells and stromal cells is dependent on the gene expression signature of tumor cells. In the current study, we observed that several cancer cell lines, e.g., MCF7 breast cancer line, exhibited growth advantage when cultured in the presence of conditioned media (CM) derived from human bone marrow stromal stem cells (hBMSCs). Regarding the underlying molecular mechanism, we have identified CXCR7 as highly expressed by MCF7 cells and that it mediated the enhanced growth in response to hBMSC CM. Regarding the clinical relevance, we found an inverse correlation between the level of tumor gene expression of CXCR7 in bladder, breast, cervical, kidney, liver, lung, pancreatic, stomach, and uterine cancers, and patients’ overall survival. Interestingly, significant positive correlation between CXCR7 and CXCL12 gene expression (Pearson = 0.3, p = 2.0 × 10–16) was observed in breast cancer patients, suggesting a biological role for the CXCR7/CXCL12 genetic circuit in breast cancer biology. Our data provide insight into the molecular mechanisms by which stromal-derived microenvironmental cues mediate CXCR7 signaling and growth enhancement of breast cancer cells. Therapeutic targeting of this circuit might provide novel therapeutic opportunity for breast cancer.
Similar content being viewed by others
Introduction
Carcinogenesis is a complex process resulting from an interplay between malignant cells and microenvironmental cues, including extracellular matrix, endothelial cells, pericytes, immune infiltrating cells, and carcinoma-associated fibroblasts (CAFs)1. This interaction contributes to tumor growth, invasion, and metastasis. Among the tumor microenvironment components, the role of CAFs in cancer development is an area of intensive investigation. A number of studies have suggested that CAFs are derived from mesenchymal (stromal) stem cells (MSCs), which are multipotent stem cells present within the stroma of bone marrow and other organs2. The precise role of CAFs or MSCs in cancer development and progression remains controversial3,4. Our recent experimental studies suggested reciprocal interaction between cancer cells and MSCs5,6. In a co-culture system, we observed that the effects of human bone marrow-derived MSCs (hBMSCs) on cancer cells were largely dependent on the expression of IL1β and CDH1 by tumor cells6. However, the exact molecular factors secreted by hBMSCs and their cognate receptors that promote cancer growth remained largely unknown.
Chemokines and their receptors represent a family of signaling molecules that may play a role in breast cancer progression. Karnoub et al. reported a role for BMSC-derived C-C Motif Chemokine Ligand 5 in promoting breast cancer metastasis to the lung7. Similarly, secretion of stromal cell-derived factor 1 (SDF1, also known as C-X-C Motif Chemokine Ligand 12 (CXCL12)) by BMSCs has been implicated in bone metastases of breast cancer cells8. However, there are limited data on the effect of hBMSCs-derived factors on tumor growth. Herein, we performed a systematic approach to assess the role of secreted factors from hBMSCs in tumor growth in vitro and identified C-X-C Chemokine Receptor Type 7 (CXCR7) expressed by tumor cells as a key receptor driving cancer cell proliferation and survival.
Results
Effect of hBMSC-conditioned media (MCM) on in vitro cancer cell growth
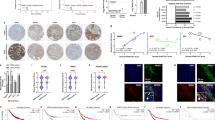
To assess the effect of MCM on tumor cell growth, tumor cell lines were cultured (day 0) in the presence of MCM control fresh medium (FM). Both MCM and FM were supplemented with 1% fetal bovine serum (FBS). We examined cell growth in cancer cells on day 2, day 4, and day 6 post exposure. Qualitative analysis revealed tumor cells cultured in MCM exhibited enhanced cell proliferation and reached confluency at an earlier time point compared to cells cultured in FM (Fig. 1). In particular, MCF7 exhibited the most pronounced response (Fig. 1b).
Bright field (upper panels) images of FaDu, MCF7, MDA-MB-231, PC-3, HT-29, and MDA-MB-468 tumor cell lines cultured in MCM or FM for 2, 4, or 6 days (10×). Representative image of H and E-stained tumor cells cultured in MCM or FM (lower panels). Images were taken on day 6 (20× (left) and 60× (right) panel). MCM human bone marrow stromal stem cells conditioned media, FM fresh media
Secreted factors from hBMSC exhibited variable effects on different tumor cell lines
To rule out the possibility that the observed enhanced cell proliferation is mediated by general nonspecific factors released by mammalian cells, cancer cells were cultured under three different conditions: (i) in the presence of MCM derived from hBMSC, (ii) in the presence of own conditioned media (CM) (tumor conditioned medium) (TCM), or (iii) under FM. Cell viability was assessed using the alamarBlue assay on days 2, 4, and 6. As shown in Fig. 2, MDA-MB-231, PC-3, and HT-29 grew better when cultured in their own TCM (Fig. 2a–e). Although statistically significant, the effect of MCM on the growth of MDA-MB-231 and HT-29 on day 2 was modest. MDA-MB-468 grew equally well in their own TCM as compared to MCM (Fig. 2e). Interestingly, MCF7, and to a lesser extent FaDu on days 4 and 6, grew better in MCM compared to own TCM (Fig. 2a, b). Overall, all cell lines grew better in MCM compared to FM. In order to confirm these findings, we employed the co-culture trans-well assay experiment, where we cultured tumor cells in the lower chamber and hBMSC, tumor cells, or just media in the upper chamber. Tumor cells were included in the upper chamber in each experimental group to compensate for the total number of cells and to eliminate possible effects of general factors released by mammalian cells in a nonspecific manner. The two chambers are separated by a semipermeable membrane (pore size 0.4 µm) which allows only small molecules to pass through. MCF7 and MDA-MB-231 grew better under MCM condition (Fig. 3b, c), while PC-3 grew equally under their own TCM or MCM (Fig. 3d). No promoting effects of TCM or MCM on the growth of HT-29 and MDA-MB-468 were observed (Fig. 3e, f), while a modest growth-promoting effect of TCM and MCM was observed on FaDu cells, although it was not statistically significant (Fig. 3a). Taken together, our data revealed general promoting effects of MCM on the growth of MCF7, MDA-MB-231, PC-3, and FaDu cells.
FaDu, MCF7, MDA-MB-231, PC-3, HT-29, and MDA-MB-468 tumor cells were cultured in FM, TCM, or MCM for 2, 4, and 6 days. Cell viability on the indicated days was assessed using alamarBlue. Data are presented as mean ± S.E.M. from a minimum of three experiments, n ≥ 20. *P ≤ 0.05, **P ≤ 0.005, ***P ≤ 0.0005. p values were calculated using two-tailed Student test with equal variance. Black bars indicate compared experimental groups. MCM human bone marrow stromal stem cells conditioned media, TCM tumor-derived conditioned media, FM fresh media
Cell viability of the indicated tumor cell line cultured under different experimental conditions using the transwell system (0.4 µm). Tumor cells were cultured in the lower chamber, while the other treatment was in the upper chamber. Cell viability was assessed using alamarBlue assay on day 6. Data are presented as mean ± S.E.M. from a minimum of three experiments, n ≥ 40. *P ≤ 0.05, **P ≤0.005, ***P≤0.0005. p values were calculated using two-tailed Student test with equal variance. Black bars indicate compared experimental groups
CXCR7 plays an important role in mediating the promoting effects of hBMSCs on MCF7 cells
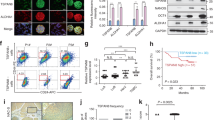
In order to identify potential surface receptors expressed on tumor cells that mediated the growth enhancement effects of MCM, we compared molecular signatures obtained from global gene expression analysis, between the tumor cell lines that were responsive to MCM (MCF7, FaDu, MDA-MB-231, and PC-3) and the nonresponsive cell lines (HT-29 and MDA-MB-468). Hierarchical clustering based on differentially expressed genes between the two groups is depicted in Fig. 4a. The top 100 upregulated genes in the responder group are shown in Supplementary Table 1. Interestingly, we observed that CXCR7 was upregulated >16.0 folds in the responder group compared to the nonresponders group. CXCR7, also known as ACKR3, is a chemokine receptor that binds to CXCL11 and CXCL12 (SDF1), while CXCR4 homodimer binds only to CXCL129. Expression of CXCR7, but not CXCR4, correlated with the cancer cell response to MCM (Fig. 4b).
a Hierarchical clustering based on differentially expressed genes between tumor cell lines that exhibited growth advantage (MCF7, FaDu, MDA-MB-231, and PC-3) compared to those that did not exhibit growth advantage (HT-29 and MDA-MB-468). b Bar chart depicting the expression of CXCR7 and CXCR4 on the indicated tumor cell lines. c Effect of inhibition of CXCR4 (using WZ811) or inhibition of CXCR7 on tumor cell growth in the presence of recombinant CXCL12 (SDF1) or hBMSC-derived CM. Data are presented as mean ± S.E.M. from three experiments
Previous studies have suggested a role for SDF1/CXCL12 and its receptor CXCR4 in regulating cell migration and survival10, and a role for CXCR7 in mediating cancer tumor survival, and development11. Thus, we investigated the role of CXCR7 signaling in promoting tumor cell survival. Since MCF7 expressed the highest levels of CXCR7 (Fig. 4b), it was employed in the subsequent experiments. Incubating MCF7 with exogenous CXCL12 (SDF1) promoted cell growth and these effects were partially abolished by cotreatment with CXCR4 (WZ811) small-molecule inhibitor (Fig. 4c). Interestingly, MCM promoted MCF7 proliferation, which was not affected by CXCR4 inhibition (Fig. 4b). siRNA-mediated inhibition of CXCR7 expression diminished the growth enhancement effect of MCM, suggesting that signaling via CXCR7 is a regulatory mechanism promoting MCF7 growth in response to secreted factors present within MCM. To determine the clinical relevance of our observations, interrogation of the expression of CXCR7 in bladder, breast, cervical, kidney, liver, lung, pancreatic, stomach, and uterine cancers revealed significant poor overall survival in patients with tumors exhibiting elevated gene expression levels of CXCR7 (Fig. 5). Network analysis on the cancer genome atlas (TCGA) breast cancer dataset revealed interaction between CXCL12 and CXCR7 (ACKR3), and a number of G-protein family members (GNG5, GNB4, GNB2, GNG12, GNG7, GNGT1, and GNAI3, Fig. 6a). Significant correlation between CXCR7 and CXCL12 was also observed in the same patient cohort, suggesting a regulatory role for CXCR7 and CXCL12 in breast cancer biology (Fig. 6b). Schema depicting the role of hBMSCs in promoting tumor cells via CXCR7 signaling is illustrated in Fig. 6c.
a Scatter plot depicting the correlation between CXCR7 and CXCL12 expression in breast cancer. Pearson and Spearman correlations and associated p values are indicated. b Network analysis illustrating the interaction between CXCL12, CXCR7, and various members of the G-protein family. c Schema illustrating CXCL12 released by stromal cell to promote tumor cell survival and proliferation via binding to CXCR7 on breast cancer cells
Discussion
Identification of factors regulating the interaction between cancer cells and microenvironment is an area of intensive investigation as it provides understanding of cancer development as well as targets for therapy. In this paper, we have demonstrated that factors secreted by stromal cells regulate cell proliferation of cancer cells and identified signaling through chemokine receptor CXCR7 as a possible mechanism.
Malignant tumors are composed of a multitude of cell types in addition to the malignant cells, and that include stromal cells, also known cancer-associated fibroblasts. Conflicting data exist in the literature regarding the precise role of stromal cells in cancer development and progression7,12. We have previously proposed that the discrepancy observed in the published results may be explained by differences in cancer type, as we observed that the interaction between tumor cells and stromal cells is dependent on the gene expression signature of tumor cells. In addition, we have identified IL1β and CDH1 expression by tumor cells as predictive markers for this interaction5,6.
We observed that cancer cell lines vary in the degree of responsiveness in terms of enhanced cell proliferation when exposed to CM derived from stromal cells. Interestingly, MCF7 breast cancer cell line exhibited the most pronounced response. These data corroborate our previous observations demonstrating increased cell growth of MCF7 cells when cocultured with hBMSCs in vitro6. Our data suggest that cancer cell–stromal cell interaction is cell type specific and dependent not only on the factors secreted by stromal cells, but also on the biological characteristics of cancer cells.
In order to identify the possible mechanism for differences in responsiveness to conditioned medium of stromal cells between different cancer cell types, we examined the molecular signature of cancer cells by using global gene expression analysis. We identified CXCR7 as highly expressed in MCF7 cells and as a possible mediator of the enhanced cell growth of MCF7 in response to stromal cell CM. Previous studies have examined the role of chemokine receptors in breast cancer biology. MCF7 and HT-29 (a colon cancer cell line) expressed similar levels of CXCR4, but only MCF7 expressed higher levels of CXCR7, which correlated with enhanced cell growth advantage in response to stromal cell CM. Also, signaling via CXCR7 has been reported to promote cancer cell growth11. At a molecular level, CXCR4 binds to CXCL12/SDF-1, whereas CXCR7 homodimer and CXCR7/CXCR4 heterodimer bind to both CXCL11 and CXCL12/SDF-1. We have previously reported that stromal cells express CXCL12, but not CXCL1113. Our loss-of-function studies showed that inhibition of CXCR7, but not CXCR4, was able to abolish the observed increase in cell proliferation in MCF7 cells, suggesting that CXCR7/CXCL12 genetic circuit regulates the growth responses in MCF7 cells.
Our findings have clinical relevance, as shown by our survival analysis of a cohort of cancer patients with different types of cancers. Levels of gene expression of CXCR7 were positively correlated to increased overall mortality across different cancer types. Similar to our findings, a recent meta-analysis demonstrated that high expression of CXCR7 is associated with higher risk of lymph node metastasis (LNM), higher tumor grade, poorer overall survival (OS), and shorter recurrence-free survival, across multiple cancer types14. These findings suggest that CXCR7 expression can be employed as a prognostic marker for multiple cancer types. Future longitudinal clinical studies are needed to corroborate the prognostic value of CXCR7 expression and to provide a quantitative estimate of its predictive value. Also, pharmacological targeting of CXCR7 signaling represents a possible approach for breast cancer therapy.
Conclusion
Our data provide an insight into stromal-derived microenvironmental cues that interact with cancer cells, and identify CXCR7 signaling as an important cancer cell growth-promoting factor and that tumor tissue expression level is an unfavorable prognostic marker in breast and several other cancer types.
Materials and methods
Cell lines and culture
Cell lines used in the current study covered a broad spectrum of cancer types, including breast (MDA-MB-231, MDA-MB-468, MCF7), colon (HT-29), head and neck (FaDu), and prostate (PC-3). Tumor cell lines were purchased from CLS Cell Lines Service GmbH (Eppelheim, Germany) or American Type Culture Collection (ATCC; Manassas, VA). The well-characterized telomerized hBMSC cell line (hBMSC-TERT) was used as a model for primary hBMSCs15. All cell lines were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 4500 mg/l d-glucose, 4 mM l-glutamine, 110 mg/l sodium pyruvate, 10% FBS, 1% penicillin–streptomycin, and nonessential amino acids (NEAA).
Preparation of conditioned media
Tumor-derived conditioned media (TCM) and hBMSC CM (MCM) were prepared using same protocol. Tumor cells (MDA-MB-231, MDA-MB-468, MCF7; HT-29; FaDu; and PC-3) and hBMSCs were seeded individually in six-well plates at 1 × 106/well (4 ml total) in DMEM supplemented with 10% FBS, 1% NEAA, and 1% penicillin/streptomycin, and incubated at 37 °C and 5% CO2. At 70–80% confluence, the cells were washed with serum-free DMEM, and the media were then replaced with fresh DMEM media supplemented with 1% FBS, 1% NEAA, and 1% penicillin/ streptomycin, and incubated at 37 °C and 5% CO2. After 72 h, the media were collected and centrifuged at 300 × g for 10 min to remove any cellular content and debris, and were then stored at −80 °C until used.
Light microscopy
Tumor cells were cultured on chamber slides (Lab-Tek chamber slides; Nunc, Naperville, IL) at a density of 0.06 × 106/ml in MCM, TCM, or FM, and then incubated at 37 °C under 5% CO2. On day 6, cells were fixed in 4% paraformaldehyde followed by hematoxylin and eosin (H&E) staining. Stained slides were mounted, covered, and scanned using ScanScope (×20 or ×60 magnification). The Nikon® ECLIPSE Ti-U inverted microscope was used to image the unstained tumor cells' confluency (×10 magnification).
Measurement of cell viability
Tumor cells were seeded in 96-well plates at a density of 6 × 103/100 μl/well. Each cell line was seeded on day 0 in fresh media (FM), TCM, or MCM, and was incubated at 37 °C and 5% CO2 (each experiment was done at least three times). Cell viability was assessed on day 2, 4, and 6 using the alamarBlue assay. At the indicated time points, 10 μl of alamarBlue reagent (BUF012B; AbD Serotec, Kidlington, UK) was added to each well and was incubated for 3 h at 37 °C. Subsequently, plates were read using BioTek Synergy 2 Multi-Mode plate reader (BioTek, Winooski, VT).
Transwell experiments
Tumor cells were seeded in the lower compartments of a transwell system (0.4 µm pore size, BD Biosciences) at a density of 0.06 × 106/1 ml, while the upper inserts contained hBMSCs, same tumor cell line or FM. Cells under various experimental conditions were incubated at 37 °C and 5% CO2. On day 6, cell viability was assessed using alamarBlue as described above.
Transfection and small-molecule inhibitor experiments
The validated siRNA targeting human CXCR7 (assay ID: 109229) and FAM-labeled scrambled control siRNA (cat no. AM4620) were purchased from Applied Biosystems (Invitrogen, Carlsbad, CA, USA). Transfection was performed using a reverse transfection approach, as described before16. Briefly, siRNA at a final concentration of 30 nM was diluted in 50 µl of Opti-MEM (11058-021; Gibco, Carlsbad, CA, USA), and 1 µl of Lipofectamine 2000 (catalogue no. 52758; Invitrogen) was diluted in 50 µl OPTI-MEM. The diluted siRNA and Lipofectamine 2000 were mixed and incubated at ambient temperature for 20 min. Twenty microliters of transfection mixture was added to the tissue culture plate, and subsequently 10,000 cells in 60 μl transfection medium (complete DMEM without antibiotics) were added to each well. Twenty-four hours later, the transfection cocktail was replaced with complete DMEM. Small-molecule inhibitor targeting CXCR4 (WZ811) was purchased from Selleckchem Inc. (Houston, TX) and was used at 1.0 µM.
Survival analysis and bioinformatics
Kaplan–Meier plots for overall survival in different cancer RNASEQ datasets were conducted as described before17. Network analysis was conducted as described before18. Gene expression data for MCF7, FaDU, MDA-MB-231, PC-3, HT-29, and MDA-MB-468 from the Cell Line Encyclopedia (CCLE) project were retrieved from the Gene Expression Omnibus (GEO) depository under accession number GSE36133. Data were subsequently analyzed in R as described before19.
Statistical analyses
Statistical analysis and graphing were performed using Microsoft EXCEL 2016 and GraphPad Prism 6.0 software (GraphPad software, San Diego, CA, USA). P values were calculated using the two-tailed t-test and p value < 0.05 was considered significant.
References
Junttila, M. R. & de Sauvage, F. J. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature 501, 346–354 (2013).
Mishra, P. J. et al. Carcinoma-associated fibroblast-like differentiation of human mesenchymal stem cells. Cancer Res. 68, 4331–4339 (2008).
Klopp, A. H., Gupta, A., Spaeth, E., Andreeff, M. & Marini, F. 3rd Concise review: dissecting a discrepancy in the literature: do mesenchymal stem cells support or suppress tumor growth? Stem Cells 29, 11–19 (2011).
He, N. et al. MSCs inhibit tumor progression and enhance radiosensitivity of breast cancer cells by down-regulating Stat3 signaling pathway. Cell Death Dis. 9, 1026 (2018).
Al-toub, M. et al. Pleiotropic effects of cancer cells’ secreted factors on human stromal (mesenchymal) stem cells. Stem Cell Res. Ther. 4, 114 (2013).
Al-toub, M. et al. CDH1 and IL1-beta expression dictates FAK and MAPKK-dependent cross-talk between cancer cells and human mesenchymal stem cells. Stem Cell Res. Ther. 6, 135 (2015).
Karnoub, A. E. et al. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature 449, 557–563 (2007).
Zhang, X. H. et al. Selection of bone metastasis seeds by mesenchymal signals in the primary tumor stroma. Cell 154, 1060–1073 (2013).
Vandercappellen, J., Van Damme, J. & Struyf, S. The role of CXC chemokines and their receptors in cancer. Cancer Lett. 267, 226–244 (2008).
Molyneaux, K. A. et al. The chemokine SDF1/CXCL12 and its receptor CXCR4 regulate mouse germ cell migration and survival. Development 130, 4279–4286 (2003).
Burns, J. M. et al. A novel chemokine receptor for SDF-1 and I-TAC involved in cell survival, cell adhesion, and tumor development. J. Exp. Med. 203, 2201–2213 (2006).
Khakoo, A. Y. et al. Human mesenchymal stem cells exert potent antitumorigenic effects in a model of Kaposi’s sarcoma. J. Exp. Med. 203, 1235–1247 (2006).
Al-Nbaheen, M. et al. Human stromal (mesenchymal) stem cells from bone marrow, adipose tissue and skin exhibit differences in molecular phenotype and differentiation potential. Stem. Cell Rev. 9, 32–43 (2013).
Zhao, Q. et al. Role of CXCR7 as a common predictor for prognosis in solid tumors: a meta-analysis. J. Cancer 9, 3138–3148 (2018).
Simonsen, J. L. et al. Telomerase expression extends the proliferative life-span and maintains the osteogenic potential of human bone marrow stromal cells. Nat. Biotechnol. 20, 592–596 (2002).
Vishnubalaji, R. et al. Genome-wide mRNA and miRNA expression profiling reveal multiple regulatory networks in colorectal cancer. Cell Death Dis. 6, e1614 (2015).
Gyorffy, B. et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res. Treat. 123, 725–731 (2010).
Alajez, N. M. Significance of BMI1 and FSCN1 expression in colorectal cancer. Saudi J. Gastroenterol. 22, 288–293 (2016).
Hamam, D. et al. microRNA-320/RUNX2 axis regulates adipocytic differentiation of human mesenchymal (skeletal) stem cells. Cell Death Dis. 5, e1499 (2014).
Acknowledgments
This project was funded by the National Plan for Science, Technology and Innovation (MAARIFAH), King Abdulaziz City for Science and Technology, Kingdom of Saudi Arabia, award number (11-MED-1942-02). The Qatar National Library funded the publication of this article.
Authors’ contributions
M.A.-t, M.A., and R.V., performed the experiments; M.A.-t, A.A., and M.K. participated in concept and design and in reviewing the manuscript; N.M.A. participated in concept and decision, obtained funding, and contributed to manuscript writing and editing.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by M. V. Nikilson Chirou
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Al-toub, M., Almohawes, M., Vishnubalaji, R. et al. CXCR7 signaling promotes breast cancer survival in response to mesenchymal stromal stem cell-derived factors. Cell Death Discov. 5, 87 (2019). https://doi.org/10.1038/s41420-019-0169-3
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41420-019-0169-3
This article is cited by
-
Overexpression of CXCR7 accelerates tumor growth and metastasis of lung cancer cells
Respiratory Research (2020)
-
Concurrent targeting of BMI1 and CDK4/6 abrogates tumor growth in vitro and in vivo
Scientific Reports (2019)