Abstract
Infertility is a worldwide reproductive health problem and there are still many unknown etiologies of infertility. In recent years, increasing evidence emerged and confirmed that epigenetic regulation played a leading role in reproduction. However, the function of m6A modification in infertility remains unknown. Here we report that METTL3-dependent m6A methylation plays an essential role in female fertility via balancing the estrogen and progesterone signaling. Analysis of GEO datasets reveal a significant downregulation of METTL3 expression in the uterus of infertile women with endometriosis or recurrent implantation failure. Conditional deletion of Mettl3 in female reproductive tract by using a Pgr-Cre driver results in infertility due to compromised uterine endometrium receptivity and decidualization. m6A-seq analysis of the uterus identifies the 3’UTR of several estrogen-responsive genes with METTL3-dependent m6A modification, like Elf3 and Celsr2, whose mRNAs become more stable upon Mettl3 depletion. However, the decreased expression levels of PR and its target genes, including Myc, in the endometrium of Mettl3 cKO mice indicate a deficiency in progesterone responsiveness. In vitro, Myc overexpression could partially compensate for uterine decidualization failure caused by Mettl3 deficiency. Collectively, this study reveals the role of METTL3-dependent m6A modification in female fertility and provides insight into the pathology of infertility and pregnancy management.
Similar content being viewed by others
Introduction
Infertility is a universal health issue, with an estimated 15% of couples experiencing infertility worldwide [1]. Endometrial disorders, including endometriosis and chronic inflammation, are the major reasons for female infertility. Endometriosis afflicts more than 10% of reproductive women, and those with stage III/IV endometriosis experience significantly lower rates of implantation and pregnancy [2, 3]. In addition, women with severe endometriosis have lower pregnancy rates compared with women with mild endometriosis [4]. Despite multiple IVF (in vitro fertilization) treatments, 10–15% of women still fail to achieve pregnancy, which is defined as recurrent implantation failure (RIF) [5]. The underlying mechanisms behind endometrial disorders remain unclear.
A growing body of evidence shows that epigenetic aberrations, including DNA methylation [6], histone acetylation [7], and noncoding RNAs [8, 9], might contribute to endometrial disorders. m6A, the most prevalent RNA modification in eukaryotes, plays important roles in RNA splicing, translocation, stability, and translation [10]. The functional effects of m6A are mediated by “writer”, “eraser”, and “reader” proteins [11]. The writer complex, consisting of a core METTL3-METTL14 m6A methyltransferase along with regulatory subunits, such as KIAA1429, RBM15, RBM15B, WTAP, and ZC3H13, catalyzes the m6A methylation of mRNA [10]. The eraser enzymes, including FTO and ALKBH5, mediate the reversal of this methylation. m6A methylated transcripts are recognized by reader proteins including the YTH family (YTHDF1/2/3 and YTHDC1/2), HNRNPA2/B1, HNRNPC, HNRNPG (RBMX), the insulin-like growth factor 2 mRNA-binding protein family IGF2BP1/2/3 [10]. Several studies have revealed the association of m6A modification with gametogenesis and fertility for both sexes [12, 13]. m6A modification has also been implicated in the pathogenesis of endometrium-related diseases. Reduced levels of m6A appeared to play an oncogenic role in patients with endometrial cancer by activating the AKT pathway [14]. Through extensive mining of public databases, Zhai et al. revealed that m6A levels are reduced in the endometrium and myometrium of women suffering from adenomyosis compared with endometrium from healthy candidates [15]. The dysregulation of m6A regulators has also been observed in endometriosis [16]. METTL3-dependent m6A is engaged in the maturation of primary microRNA126 mediated by DGCR8, which further increased the migration and invasion of endometrial stromal cells in endometriosis [17]. However, the function of m6A modification in infertility and uterine biology remains unknown.
Through extensive public database mining, we observed in this study that the mRNA of METTL3 was decreased in endometrium from infertile women with endometriosis or RIF. Using uterine-specific Mettl3-deficient mice, we demonstrated that METTL3-mediated m6A modification is critical for implantation and decidualization. Mechanistically, loss of the m6A-modified sites might lead to estrogen dominance and progesterone resistance. We found that loss of Mettl3 stabilizes the mRNA of estrogen target genes, such as Elf3 and Celsr2, leading to hyperactivation of estrogen response. We also provide evidence in favor of METTL3 playing an important role in maintaining PR level and then driving c-Myc expression which is conducive to decidualization. Collectively, our findings contribute to the understanding of the etiology of female infertility, providing a molecular framework that might be useful for diagnostic and therapeutic strategies for infertility.
Results
METTL3 expression is reduced in endometrium of infertile women
To address whether m6A regulators play a role in endometriosis-related infertility, we acquired gene expression profiles of endometrial tissue from the Gene Expression Omnibus (GEO) database. From the analysis of the expression level of m6A regulators in dataset GSE120103, which includes fertile women with stage IV endometriosis and infertile women with stage IV endometriosis, we found that most m6A regulators, including METTL3, CBLL1, ALKBH5, FTO, YTHDC1, YTHDF2, IGF2BP2, HNRNPA2B1, HNRNPC, and LRPPRC, were significantly downregulated, while ZC3H13 and IGF2BP1 were significantly upregulated in infertile group versus fertile group (Fig. 1A, and Supplementary Fig. 1A). From the analysis of the expression level of m6A regulators in the endometrium of women with RIF using dataset GSE58144, we found that METTL3, YTHDC2, YTHDF3, HNRNPC, and FMR1 were significantly decreased, while ZC3H13 were significantly increased in RIF patients compared with healthy controls (Fig. 1B, and Supplementary Fig. 1B). These results suggest that m6A regulators play an important role in both endometriosis-related infertility and RIF. Further analysis of the correlation between decreased METTL3 expression and disease using receiver operating characteristic (ROC) curves demonstrated that the area under the curve (AUC) for reduced endometrial METTL3 expression was 0.9136 (95% confidence interval (CI), 77.5% to 100%) in infertile patients with stage IV endometriosis (Fig. 1C), indicating downregulation of endometrial METTL3 could well distinguish infertile from fertile endometriosis patients. Similarly, the AUC value for RIF was 0.6321 (95% CI, 53% to 73.5%) (Fig. 1D), indicating that reduced endometrial METTL3 could well distinguish RIF patients from healthy controls. Collectively, these results suggested that the reduction of METTL3 might be associated with infertility.
A Expression landscape of METTL3 in the endometrium of fertile (n = 9) and infertile (n = 9) women with stage IV endometriosis in dataset GSE120103. Data are presented as mean ± SD, ***P < 0.001, relative to “Fertile stage IV Endometriosis” group. B Expression landscape of METTL3 in the endometrium of women with RIF following in vitro fertilization (IVF) treatment (n = 43) and healthy control women (n = 72) 7 days after the putative luteinizing hormone surge (GSE58144). Data are presented as mean ± SD, *P < 0.05, relative to control. ROC curve evaluation of the relationship between decreased expression of METTL3 and infertile stage IV endometriosis (C), or RIF (D).
Ablation of Mettl3 results in infertility due to embryo implantation failure
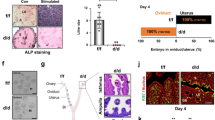
Mouse models allow us to study the sequence of events involved in the occurrence and progression of diseases. To study the role of METTL3 in the uterus during pregnancy, we generated a mouse model with conditional deletion of Mettl3 in Pgr-positive cells (Fig. 2A). To do so, we started with mice carrying homozygous alleles of Mettl3 with loxP sites placed in exon 2 and exon 3. Mettl3flox/flox mice were mated to mice carrying a Pgr-Cre allele, in which Cre is knocked into the Pgr locus allowing Cre protein expression to be driven by the native Pgr promoter [18]. We produced Mettl3flox/floxPgrcre/+ (Mettl3 cKO) mice, eliminating the expression of Mettl3 in tissues expressing the progesterone receptor (PR), i.e., epithelium, stroma, and myometrium of the uterus [18]. Ablation of Mettl3 in the uterus was confirmed by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) (Fig. 2B), and immunofluorescence (Fig. 2C). We observed vaginal plugs in Mettl3 cKO mice indicating that mating behavior was normal in these females. Female fertility was assessed by mating Mettl3 cKO and control females with wild-type (WT) males continuously for 6 months and tracking the number of litters and pups produced by each female. The control females were fertile (average number of pups/litter: 6.6 ± 0.4), whereas Mettl3 cKO females were sterile and did not produce any pups during the 6 months mating trial (Fig. 2D). Four stages of mouse estrous cycle were observed in Mettl3 cKO mice by vaginal smears (Fig. 2E). The examination of the reproductive duct in Mettl3 cKO mice did not exhibit any significant alterations compared with the controls (Fig. 2F). Also, no significant histology differences were found in the uterus (Fig. 2G), vagina (Fig. 2H), ampullar oviduct (Fig. 2I), or ovary (Fig. 2J) of Mettl3 cKO mice compared with the control mice. And serum levels of estradiol-17β (E2) and progesterone (P4) in Mettl3-deficient mice and control mice were comparable (Fig. 2K, L). Superovulation of 6-week-old mice showed no significant difference in the quantity of oocytes released between Mettl3 cKO and control mice (Fig. 2M). These results suggest that infertility in Mettl3 cKO mice is primarily due to a uterine functional defect.
A Illustration of the Mettl3 conditional allele with loxP sites placed flanking exon 2 and exon 3. B Relative Mettl3 mRNA level in the uteri of Mettl3 cKO (n = 7) and control (n = 8) mice on GD4. Data are presented as mean ± SD, ***P < 0.001, relative to control. C Immunofluorescence of METTL3 and CK8 in the uteri of Mettl3 cKO and control mice on GD4. Nuclei were stained with DAPI. Scale bars: 75 μm. D Female fertility was assessed. Data are presented as mean ± SD. E Vaginal smear assays of Mettl3 cKO mice confirmed each stage of the normal estrous cycle. Scale bars: 100 μm. F Gross morphology of female reproductive tracts in Mettl3 cKO and control mice at 8 weeks of age. HE staining of the cross-sections of the uterus (G), vagina (H), oviduct (I), and ovary (J) in control and Mettl3 cKO mice at 8 weeks of age. Serum concentrations of E2 (K) or P4 (L) in Mettl3 cKO (n ≥ 9) and control mice (n ≥ 5) were analyzed and shown as mean ± SD. M Number of oocytes collected from superovulated Mettl3 cKO (n = 6) and control females (n = 8). N Representative photographs of control uterus (n = 6) with implantation sites and Mettl3 cKO uterus (n = 5) without blue bands on GD5. Scale bars, 1 cm. O The number of implantation sites in (N) were counted and are reported as mean ± SD, ***P < 0.001, relative to control.
Blastocyst implantation into the uterus is an essential step for the establishment of pregnancy. To identify the stage-specific failure of pregnancy in Mettl3 cKO females, we subsequently analyzed the implantation status in Mettl3 cKO females. Chicago Blue dye was injected to visualize the number and location of implanted embryos in the uterus on gestation day (GD) 5. The uterine horns of WT control mice had an average of 7.83 ± 0.99 implantation sites that appeared normally spaced per pregnant female, whereas Mettl3 cKO mice had no grossly visible implantation sites on GD5 (Fig. 2N, O). These results clearly indicate that uterine METTL3 is indispensable for normal embryo implantation.
Mettl3 deficiency impairs uterine receptivity and decidualization
To reveal the underlying causes accounting for the defective implantation failure in Mettl3 cKO mice, we investigated whether ablation of Mettl3 alters uterine receptivity and decidualization. We first examined the proliferation versus differentiation status of uterine cells using Ki67 immunostaining at pre-implantation. As shown in Fig. 3A, in control mice, cell proliferation was reduced in epithelial cells before embryo attachment and increased in stromal cells in preparation for implantation on GD4 (Fig. 3A, B). However, the proliferative responses in stromal compartments of the uterus from Mettl3 cKO mice were significantly reduced on GD4 compared with the control mice (Fig. 3A, B). E-cadherin, a cell polarity and cell junction marker, showed higher apical expression in Mettl3 cKO mice compared with the controls (Fig. 3C, D). These observations collectively indicated abnormal uterine receptivity in Mettl3 cKO mice in peri-implantation.
Representative immunofluorescence images (A) and the percentage (B) of Ki67+ epithelial cells and stromal cells in uterine of Mettl3 cKO and control mice on GD4. Data were calculated using 11 images from 3 control mice and 14 images from 3 Mettl3 cKO mice. Representative immunofluorescence images (C) and quantification of E-cadherin (D) in uterine epithelial cells of Mettl3 cKO and control mice on GD4. Fluorescence intensities of uterine epithelial E-cadherin were calculated using 6 images from 3 control mice and 12 images from 3 Mettl3 cKO mice. Representative immunofluorescence images (E) and the percentage (F) of Ki67+ epithelial cells and stromal cells in uterine of Mettl3 cKO and control mice following induction of artificial pregnancy (pollard experiment). Data were calculated using 12 images from 3 control mice and 7 images from 3 Mettl3 cKO mice. Representative immunofluorescence images (G) and quantification of E-cadherin (H) in uterine epithelial cells of Mettl3 cKO and control mice following induction of artificial pregnancy. Fluorescence intensities of uterine epithelial E-cadherin were calculated using 12 images from 3 control mice and 15 images from 3 Mettl3 cKO mice. A, C, E, G Nuclei were stained with DAPI. Scale bars: 75 μm. I Immunohistochemistry of METTL3 with paraffin sections from pregnant females on GD0 and GD5. Sections are counterstained with hematoxylin. Brown staining denotes METTL3+ cells. Scale bars: 100 μm. J Representative pictures showing the gross morphology of oil-treated uterine horns (left horn) and untreated uterine horns (right horn) from control (n = 6) and Mettl3 cKO mice (n = 6) collected 5 days after oil injection. Scale bars: 1 cm. K The weight per unit length of oil-injected and untreated uterine horns in (J). Data are presented as mean ± SD, **P < 0.01. L Histology of control and Mettl3 cKO mice uterus in (J), as measured by HE staining, Scale bars 200 μm. M Relative mRNA levels of decidualization marker genes (Wnt4, Bmp8a, Bmp2, Prl8a2) in the uterine horns of control and Mettl3 cKO mice in (J). B, D, F, H, M Data are presented as mean ± SD, *P < 0.05, **P < 0.01, ***P < 0.001, relative to control and unstimulated.
To further ascertain the role of METTL3 in uterine receptivity, E2 and P4 were administered to ovariectomized control and Mettl3 cKO mice to mimic early pregnancy (pollard experiment) (Supplementary Fig. 2A). As expected, sequential E2 and P4 treatment induced uterine stromal cell proliferation in control mice (Fig. 3E, F). However, neither uterine stromal cells nor epithelial cells displayed a proliferative response in Mettl3-deficient mice (Fig. 3E, F). Increased E-cadherin expression was also detected in the luminal epithelium of Mettl3 cKO mice compared with the controls in the pollard experiment (Fig. 3G, H). These data support the hypothesis that Mettl3 deficiency causes endometrial receptivity abnormalities.
In response to implantation, stromal cells surrounding the mucosal crypt where the embryo resides proliferate extensively and differentiate into polyploid decidual cells. As METTL3 protein level in the uterus was substantially increased from GD0 to GD5 (Fig. 3I), we next examined the impact of Mettl3 ablation on decidualization using an artificial decidualization model (Supplementary Fig. 2B). The control mice displayed a decidual uterine horn that responded well to the artificial induction. However, Mettl3 ablation entirely preclude decidualization (Fig. 3J, K), which was further confirmed by histological analysis (Fig. 3L). We also observed a significant decrease in the expression of decidualization markers, including Wnt4, Bmp8a, Bmp2, and Prl8a2 in the uterus of Mettl3 cKO mice (Fig. 3M). Taken together, these data suggest that Mettl3 cKO mice were sterile as a result of defective uterine receptivity and decidualization.
Ablation of Mettl3 results in an abnormal uterine transcriptome
To understand the molecular basis of the implantation failure phenotype in Mettl3 cKO mice, we performed an RNA-seq analysis of the uterine tissue of Mettl3 cKO and control mice on GD4. Global gene expression profiles of Mettl3 cKO versus control mice were analyzed. 922 genes were differentially expressed, including 572 up-regulated genes and 350 down-regulated genes in the uterus of Mettl3 cKO mice compared with control mice (Fig. 4A). GO enrichment analysis showed that the up-regulated genes were mainly enriched in the regulation of epithelium, such as “Regulation of morphogenesis of an epithelium”, “Morphogenesis of a branching epithelium”, “Mesonephric epithelium development”, and “Cell junction maintenance” (Fig. 4B), while down-regulated genes were mainly involved in “Wnt signaling pathway”, “Regulation of actin filament-based process”, “Regulation of actin cytoskeleton organization” (Fig. 4C). GSEA analysis of MSigDB gene sets was performed (Fig. 4D, E). Specifically, the term “REACTOME REPRODUCTION” was observed negatively enriched in Mettl3 cKO mice compared with the controls (Fig. 4D). And the term “REACTOME CELL CELL JUNCTION ORGANIZATION” was enriched in Mettl3 cKO mice compared with the controls (Fig. 4E). These results suggested that METTL3 regulates luminal epithelial remodeling during the window of implantation, which is consistent with enhanced E-cadherin expression in the uterus of Mettl3 cKO mice (Fig. 3C, G). Moreover, cell proliferation-related terms, including “WP CELL CYCLE”, “REACTOME DNA REPLICATION”, “HALLMARK E2F TARGETS” were suppressed in Mettl3 cKO mice (Fig. 4E), which is consistent with the decreased proliferation in uterine stromal cells (Fig. 3A, E). Interestingly, among the significantly dysregulated genes, 42 genes associated with uterine receptivity and implantation were shown in the heatmap (Fig. 4F), including E2-responsive genes (Igfbp5, Greb1, Ltf, Wnt4, Lcn2, Sprr2f, Celsr2, Muc1, Elf3, Fzd10), P4-responsive genes (Mmp9, Lamc3, Fst, Maob, Srd5a1, Myd88, Pfkfb3, Lrp2, Hdc, Gldc, Jam2, Runx1, Cebpb, Cebpd, Osmr, Mmp11, Sox17, Ihh, Cldn3, Msx1, Msx2, Cited4, Sox7, Galnt12), LE-specific genes (Hdc, Jam2, Cldn7), GE-specific genes (Foxa2), hedgehog signaling-associated genes (Ptch2, Ihh), Pan-uterine epithelial-associated genes (Lamc3, Maob, Srd5a1, Myd88, Pfkfb3, Lrp2, Gldc, Ltf, Sox17, Ihh, Cldn3, Cited4, Sprr2f, Muc1), other uterine receptivity- and implantation-related genes (Irf4, Cyp3a59, Foxo1, Gadd45a, Cyp3a57). We believe that deregulated E2/P4-responsive genes in Mettl3 cKO mice might be responsible for compromised uterine receptivity and decidualization.
A Volcano plot depicting genes upregulated (red) or downregulated (blue) 1.5-fold or more in the uterus of Mettl3 cKO mice vs control on GD4. A heatmap of differentially expressed genes is shown on the right (n = 3 mice per group). GO terms of the upregulated genes (B) and downregulated genes (C) in the uterus of Mettl3 cKO mice compared with that of control mice. D GSEA of “REACTOME_REPRODUCTION” gene set in the uterus of Mettl3 cKO mice relative to control mice. E GSEA analysis of the indicated gene sets in the uterus of Mettl3 cKO mice relative to control mice. F Heatmap of differentially expressed uterine receptivity-related genes between Mettl3 cKO and control mice on GD4 generated from RNA-seq data.
Loss of Mettl3 leads to the activation of estrogen-driven transcriptional response by stabilizing mRNAs of the estrogen-regulated genes
The implantation window in mouse uterus occurs between GD4 and GD5 and is characterized by a transition from an E2-dominant proliferative state to a P4-responsive state [19,20,21]. GSEA analysis revealed that “HALLMARK ESTROGEN RESPONSE EARLY”, “HALLMARK ESTROGEN RESPONSE LATE” were positively enriched in the uterus of Mettl3 cKO mice (Fig. 4E). Concomitantly, the majority of E2 responsive genes were found to be upregulated in the uterus of Mettl3 cKO mice compared with the controls (Fig. 5A). And RT-qPCR results further indicated that both E2-target epithelial genes (Muc1, Ltf, Elf3, Celsr2) and stromal genes (Wnt4, Fzd10) were significantly upregulated in the Mettl3-deficient uterus (Fig. 5B). We then measured uterine ER protein level in mice on GD4 using immunohistochemistry (Fig. 5C) and western blot (Fig. 5D) and in the pollard experiment using immunofluorescence staining (Fig. 5E), and found that uterine ER level in Mettl3 cKO mice was not significantly altered compared with control mice (Fig. 5C–E).
A Heatmap of uterine E2-responsive genes in Mettl3 cKO and control mice on GD4 generated from RNA-seq data. B Relative mRNA levels of E2-regulated genes in control (n = 8) and Mettl3 cKO (n = 7) females on GD4. C Immunohistochemistry of hormone receptor ER with uterine sections from Mettl3 cKO and control females on GD4. Sections are counterstained with hematoxylin. Brown staining denotes ER+ cells. Scale bars: 100 μm. D Immunoblotting analysis was conducted to compare uterine ER protein levels in control mice (n = 3) and Mettl3 cKO mice (n = 3) on GD4. The experiments were repeated three times. α/β-tubulin was used as the loading control. Values are expressed as the mean ± SD. E Representative immunofluorescence images and quantification of ER in the uterus of Mettl3 cKO and control mice following induction of artificial pregnancy. Nuclei were stained with DAPI. Scale bars: 75 μm. Fluorescence intensities of uterine ER were calculated using 10 images from 3 control mice and 14 images from 3 Mettl3 cKO mice. Results are representative of 3 independent experiments. Integrative Genomics Viewer (IGV) tracks displaying transcripts and m6A peaks distribution of Elf3 (F) and Celsr2 (H) mRNAs in m6A-seq. The high-confidence m6A site is marked as an arrow. m6A enrichment in Elf3 (G) and Celsr2 (I) mRNA in the uterus of Mettl3 cKO (n = 3) and control mice (n = 3), as determined by m6A-RIP-qPCR. RNA stability assay. The relative mRNA levels of Elf3 (J) and Celsr2 (L) were detected by RT-qPCR. The remaining mRNAs were normalized to t = 0. (n = 3 per group, biological repeated 3 times). RT-qPCR analysis of Elf3 (K) and Celsr2 (M) mRNA abundance. Relative expression was normalized to t = 0 of control uterine cells (n = 3 per group, biological repeated 3 times). Data are presented as mean ± SD, *P < 0.05, **P < 0.01, ***P < 0.001.
To define the possible targets regulated by m6A modification involved in the functional maintenance of endometrial receptivity and female fertility, m6A-seq of the uteri of Mettl3 cKO mice and the control mice were conducted. The abundance of m6A modifications in CDS, 3′UTR, 5′UTR, start codon, and stop codon were profiled (Supplementary Fig. 3A, B). After identifying the representative consensus of the m6A motif in the uterus of Mettl3 cKO and control mice (Supplementary Fig. 3C), global hypomethylation of m6A at the transcription level (Supplementary Fig. 3D), we found that approximately 80% of methylated mRNAs contained 1 peak (Supplementary Fig. 3E). And we defined 319 hypo-methylated m6A genes whose mRNA transcripts were identified as down-regulated (p < 0.05, Hypo-down) and 1020 hypo-methylated m6A genes along with up-regulated mRNA transcript (p < 0.05, Hypo-up) in Mettl3 cKO mice compared with WT controls (Supplementary Fig. 3F). Notably, the m6A level at 3’UTR of estrogen-responsive genes Elf3 and Celsr2 mRNA had substantially enriched m6A peaks in control uterus, but not in Mettl3 cKO mice (Fig. 5F–I). To ascertain the role of METTL3-mediated m6A modification in regulating the mRNA levels of Elf3 and Celsr2, mRNA decay assays were performed, and the results demonstrated that loss of m6A modification did appreciably inhibit the decay of Elf3 and Celsr2 mRNAs (Fig. 5J–M). Collectively, these results indicate that deficiency of METTL3-dependent m6A modification might increase the mRNA stability of E2-responsive genes, such as Elf3 and Celsr2, leading to the overactivation of estrogen signaling in the uterus.
Mettl3-deficient uteri show progesterone resistance due to reduced expression of PR and its downstream genes
The P4 and E2-responsive signaling pathways are tightly regulated in the endometrium. E2 drives uterine epithelial proliferation. During pre-implantation, E2 level is low, P4 initiates stromal cell proliferation, and primes the uterus to be receptive. P4 resistance and E2 dominance are most likely to happen when the balance between P4 and E2 signaling is lost [22]. Apart from overactivated estrogen signaling, Mettl3 deficiency also hampers uterine P4 response. Most of the P4-responsive genes were found to be significantly downregulated in the uterus of Mettl3 cKO mice as illustrated in Fig. 6A. Some of these genes were confirmed by RT-qPCR. The expression of P4-target molecules such as Lrp2 in the epithelium and Hoxa10, Fst, and Il13ra2 in the stroma was markedly downregulated in Mettl3 cKO mice compared with the control (Fig. 6B). Furthermore, GSEA analysis revealed a negative enrichment of “WILCOX RESPONSE TO PROGESTERONE UP” pathway in the uterus of Mettl3 cKO mice compared with their counterpart controls (Fig. 4E). Interestingly, although serum P4 levels were comparable in Mettl3-deficient mice and control mice (Fig. 2L), P4 and E2 co-treatment did induce uterine stromal cell proliferation in OVX WT control mice but not in Mettl3 cKO mice (Fig. 3E), indicating that the loss of progesterone response in Mettl3 cKO mice was not caused by decreased progesterone levels. P4 exerts its regulatory role by binding to its cognate receptor PR. To elucidate the possible reasons for progesterone resistance in Mettl3 cKO mice, we examined the expression of uterine PR in GD4 pregnant mice (Fig. 6C) and in the pollard model (Fig. 6D), and found that the expression of PR was markedly decreased in the uteri of Mettl3 cKO mice compared with the controls (Fig. 6C, D). Taken together, these observations suggest that METTL3 affects the endometrial response to progesterone during pregnancy by maintaining the expression level of PR.
A Heatmap of uterine P4-responsive genes in Mettl3 cKO and control mice on GD4 generated from RNA-seq data. B Relative expression of mRNA for P4-regulated genes in control (n = 8) and Mettl3 cKO (n = 7) females on GD4. C Immunohistochemistry of hormone receptor PR with uterine sections from Mettl3 cKO and control females on GD4. Sections are counterstained with hematoxylin. Brown staining denotes PR+ cells. Scale bars: 100 μm. D Representative immunofluorescence images and quantification of PR in the uterus of Mettl3 cKO and control mice following induction of artificial pregnancy. Nuclei were stained with DAPI. Scale bars: 75 μm. Fluorescence intensities of uterine PR were calculated using 14 images from 3 control mice and 15 images from 3 Mettl3 cKO mice. E RT-qPCR of Myc mRNA expression in the uterus of control (n = 8) and Mettl3 cKO mice (n = 7). F Representative immunofluorescence images and quantification of c-Myc in uterine epithelial and stromal cells of Mettl3 cKO and control mice on GD4. Nuclei were stained with DAPI. Scale bars: 75 μm. Fluorescence intensities of uterine c-Myc were calculated using 11 images from 3 control mice and 16 images from 3 Mettl3 cKO mice. Results are representative of 3 independent experiments. G GSEA analysis of “MYC-responsive genes” gene set in the uterus of Mettl3 cKO mice relative to that in the uterus of control mice. H The isolated uterine stromal cells were stained with vimentin, and the purity was analyzed by flow cytometry. I RT-qPCR analysis was performed to examine the expression of Myc, Prl, Prl8a2, Rrm2, and Ldha in uterine stromal cells of Mettl3 cKO mice after transfection of Myc overexpression plasmid or control plasmid for in vitro decidualization (biological repeated three times). Data are presented as mean ± SD, *P < 0.05, **P < 0.01, ***P < 0.001, relative to control.
To further confirm the repressed PR signaling in Mettl3 cKO mice, we detected the expression level of PR target gene Myc [23]. c-Myc is a transcription factor involved in cell proliferation and is required for uterine stromal cell proliferation during peri-implantation [24,25,26]. We found that Myc mRNA and protein levels were significantly reduced in GD4 Mettl3 cKO uterus compared with WT controls (Fig. 6E, F). We also noticed a decrease in the c-Myc protein level in the uterus of Mettl3 cKO mice compared with control mice in the pollard model (Supplementary Fig. 4A). Using several MYC-target gene sets published previously [27, 28], GSEA revealed a significant decrease in MYC-target gene signatures in the uterus of Mettl3-deficient mice (Fig. 6G and Supplementary Fig. 4B, C), indicating Mettl3 deficiency resulted in a compromised c-Myc signaling pathway. Furthermore, under in vitro decidualization, overexpression of Myc in Mettl3-deficient stromal cells could significantly upregulate the expression of reliable marker genes for decidualization, including Prl, Prl8a2, Rrm2 [26] and Ldha [29] (Fig. 6H, I). These results indicated that Mettl3-deficient uteri show progesterone resistance due to reduced expression of PR and its downstream genes.
The levels of P4 and E2-dependent genes PR, MYC, ELF3 are correlated with METTL3 in human endometrium
To investigate whether METTL3 functions through regulating the balance between E2 and P4 signaling in human endometrium, we analyzed the correlation between METTL3 and PGR, MYC, or ELF3 in human endometrium from two independent cohorts, GSE58144 and GSE4888. PGR and METTL3 mRNA levels were positively correlated in human endometrium (Fig. 7A), as well as the levels of MYC and METTL3 (Fig. 7B). However, ELF3 and METTL3 mRNA levels were negatively correlated in human endometrium as indicated (Fig. 7C). The results above indicated that the expression of PR, MYC, ELF3, and METTL3 is conserved between mouse and human. With this, a reasonable assumption might be that P4 resistance and E2 dominance could be related to the decrease of METTL3 in human endometrium in some disease conditions.
A Pearson’s correlation of mRNA levels between PGR and METTL3 in human endometrium (GSE58144 and GSE4888). B Pearson’s correlation of mRNA levels between MYC and METTL3 in human endometrium (GSE58144 and GSE4888). C Pearson’s correlation of levels between ELF3 and METTL3 in human endometrium (GSE58144 and GSE4888).
Discussion
m6A methylation, regulating RNA stability, degradation, translation, alternative splicing, and gene expression, plays critical roles in various biological processes. m6A levels are elevated in the mouse uterus throughout pregnancy [30], suggesting m6A modifications are regulated by hormones. m6A demethylase FTO expression can be induced by estrogen in endometrial cancer via activation of the PI3K-Akt and MAPK pathways [31]. Our study reveals that METTL3 expression is significantly decreased in endometrium from infertile women with endometriosis or recurrent implantation failure, suggesting that m6A modifications might be involved in the pathogenesis of infertility. However, the molecular mechanisms of METTL3 attenuation in the etiology and pathophysiology of infertility remain undetermined. Using Mettl3 cKO mouse model, we provide evidence in favor that Mettl3 loss causes infertility due to implantation failure.
The implantation window in mice occurs between GD4 and GD5, when the uterus becomes receptive [32,33,34], i.e., a reduction in the proliferation and polarity of uterine epithelial cells, as well as the induction of proliferation and decidualization of uterine stromal cells. E-cadherin, typically downregulated in the receptive epithelium, had stronger apical expression in the Mettl3 cKO mice compared with the controls, indicating the abnormality of luminal epithelium differentiation during implantation. Meanwhile, persistent epithelial proliferation during the preimplantation stage results in implantation failure in several knockout mouse studies [35, 36]. In our study, a few of Ki67-positive cells were observed in the uterine epithelium of Mettl3 cKO mice on GD4. Intriguingly, a significant reduction of Ki67-positive cells in the uterine stroma of the Mettl3 cKO mice on GD4 was observed, indicating a defect in stromal cell proliferation in Mettl3 cKO mice. Decidualization involves the differentiation of uterine stromal fibroblasts to decidual cells after embryo implantation, playing an important role in maintaining pregnancy. Decidualization defect of endometrial stromal cells has been regarded as a primary cause of endometriosis-related infertility [37, 38]. And our findings from decidualization experiments in Mettl3 cKO mice support this notion.
A receptive uterus is characterized by a uterine transition from an E2-dominant proliferative state to a P4-responsive state [19,20,21]. To ascertain the role of METTL3 in E2 and P4 responses, we performed GSEA analysis based on MSigDB gene sets, and found that “HALLMARK ESTROGEN RESPONSE EARLY”, “HALLMARK ESTROGEN RESPONSE LATE” were positively enriched, whereas “WILCOX RESPONSE TO PROGESTERONE UP” was negatively enriched in Mettl3 cKO mice compared with the controls. We also profiled the level of E2 and P4 responsive genes in the uterus on GD4, and found that most of the E2 responsive genes were sharply upregulated, while most P4 responsive genes were significantly decreased. These results established the critical role of METTL3 as a mediator of both estrogen and progesterone signaling in the uterus.
For the E2 responsive genes, we found that Elf3 mRNA level is significantly increased in Mettl3 cKO mice, and its m6A enrichment at 3′UTR is sharply decreased. Further mRNA decay assays demonstrated an increase in Elf3 mRNA stability in the absence of METTL3-dependent m6A modification. ELF3 is an m6A reader, which can directly bind to the m6A site at the 5′UTR of mRNAs participating in the translation initiation of specific genes. In addition, ELF3 binds to ERα in the absence of E2, but dissociates with ERα upon E2 treatment in a dose- and time-dependent manner [39], suggesting a possible feedback loop between ERα signaling and ELF3. Another E2-responsive gene CELSR2 is negatively associated with the overall survival time of endometrial cancer patients [40]. CELSR2 deficiency impaired cell proliferation of hepatocytes [41], liver cancer cells [42], and Schwann cells [43]. Interestingly, uterine Celsr2 in Mettl3 cKO mice reflects a similar mRNA expression and m6A modification pattern to Elf3. Based on our mRNA decay assays, deficiency of METTL3-dependent m6A might increase the mRNA stability of Celsr2.
E2 and P4 play a regulatory role by binding to their cognate receptors, ER and PR, respectively. Moreover, the level of hormone receptors can be regulated by m6A modification. ERα mRNA m6A methylation is significantly upregulated by R-2HG due to FTO degradation, which may then result in suppressing ERα protein expression via translational regulation, hence reducing cholangiocarcinoma [44]. The elevated Estrogen Related Receptor γ in chemoresistant cancer cells can be attributed to m6A-dependent splicing of precursor ESRRG mRNA [45]. However, we did not observe METTL3-dependent m6A methylation in Esr1, Esr2, Esrra, and Esrrb mRNAs in mouse uterus (data not shown). In note, uterine ER level was not altered in Mettl3 cKO mice compared with the controls. Endometrial stromal PR is mediated by E2/ER signaling [46]. And prior to conceptus implantation, PR expression is downregulated in the uterine epithelial compartment and is upregulated in uterine stromal cells [47]. Insufficient PR signaling hampers intimate stromal-epithelial crosstalk and thus uterine receptivity [48, 49]. We also found a sharp reduction of uterine stromal PR protein level in Mettl3 cKO mice on GD4, and in the pollard experiment, when compared with the control mice. The results above indicated that METTL3 may regulate the P4 response of uterine stroma during pregnancy by upregulating PR.
ChIP-seq analysis identified PR-binding sites within Myc in the uterus of mice treated with P4 [23], implying that c-Myc is a target of PR. The Myc family proteins contain three well-defined members: c-Myc, N-Myc, and L-Myc. NDRG2 [50] and NDRG4 [51], downstream-regulated genes of N-Myc, are upregulated at implantation sites during early pregnancy in mice, and their downregulation inhibit the decidualization process of mouse endometrial stromal cells. c-Myc, a well-known oncoprotein, is a transcription factor involved in ribosome biogenesis, protein translation, cell-cycle progression, and metabolism, orchestrating a broad range of biological functions, such as cell proliferation, and differentiation [52]. On day 4 of pregnancy, an increase in c-Myc expression in uterine stromal cells was identified, accompanied by a robust proliferation of stromal cells [24]. E2 administration raised epithelial c-Myc levels and DNA synthesis rapidly [24, 25], and P4 administration elevated stromal c-Myc levels in vivo and in vitro [24, 26]. We detected a decrease in the protein levels of c-Myc in the uterus of Mettl3 cKO mice, and Myc overexpression was able to partially restore the deficit of decidualization in vitro, further confirming that METTL3 may regulate the hormone response of uterine epithelial and stromal cells by influencing the expression of PR and its target genes.
In summary, this study provides evidence in favor of the critical role of METTL3-dependent m6A methylation in maintaining a balanced estrogen and progesterone signaling pathway, which is conducive to endometrial receptivity and female fertility, thereby providing insightful information for the pathology of infertility and pregnancy management.
Materials and methods
GEO data processing
RIF gene expression profiles (GSE58144) and endometriosis gene expression profiles (GSE120103, GSE4888) with clinical information were downloaded from GEO database (https://www.ncbi.nlm.nih.gov/geo/) using “GEOquery” package (v2.64.2) in R software (v4.2.1). For data processing, the “limma” package (v3.52.2) was applied for background correction and quantile normalization of all the raw data files, and the expression values were then obtained. The probe set with max-average value was chosen as the expression value for the same gene with multiple probe sets. Barplots, ROC curves, and correlation analysis were drawn using “ggplot2” package (v3.4.0). The code used for GEO data processing is available at https://github.com/wansh007/data-process.
Mice
C57BL/6N-Mettl3em1cyagen (Mettl3flox/flox) mice were generated by Cyagen Biosciences Inc (Guangzhou, China) using the CRISPR–Cas9-based genome-editing system. Pgr-Cre mice were obtained from Cyagen Biosciences Inc (Guangzhou, China). To generate Mettl3 cKO mice, Mettl3flox/flox mice were bred to mice carrying the Pgr-Cre knock-in allele to obtain Mettl3flox/flox PgrCre/+ mice (Mettl3 cKO mice). The females were chosen for the indicated experiments. To reduce the effect of genetic background variability, littermate floxed and gene-deleted mice were used in the same experiments. All mice used in this study were housed in specific pathogen-free animal facilities in the Animal Resource Center at Jinan University, following the ethical guidelines of the Animal Ethics Committee of Jinan University (IACUC-20220418-03).
HE, immunofluorescence and immunohistochemistry staining
Uterine tissues were fixed in Paraformaldehyde Fix Solution (G1101-500ML, Servicebio) for 24 h and embedded in paraffin, and sectioned at 5 μm. Sections were dried at 60 °C for 30 min and stored at room temperature. Sections were deparaffinized and rehydrated in xylene, 100% ethanol, 90% ethanol, 85% ethanol, 75% ethanol and double distilled water, before HE, immunofluorescence and immunohistochemistry staining.
For HE staining, sections were stained with eosin (G1100, Solarbio) and hematoxylin (G1140, Solarbio). Images were captured using an Olympus BX53 microscope.
For immunofluorescence staining, antigen retrieval was carried out by heating the sections in sodium citrate buffer (10 mM sodium citrate, 0.05% Tween 20, pH 6.0) at 95 °C for 15 min. The sections were immersed in 3% H2O2 for 45 min at room temperature to quench endogenous peroxidase, permeabilized in 0.2% Triton-100 in PBS for 45 min, and then blocked with 1% (w/v) BSA Fraction V (ST023, Beyotime) and 10% goat serum (v/v) (B900780, Proteintech) in PBS before the primary antibodies were added. Then the sections were incubated with the following primary antibodies against METTL3 (ab195352, Abcam), CK8 (DSHB, TROMA-I), Ki67 (ab15580, Abcam), E-Cadherin (3195S, Cell Signaling Technology), PR (8757 S, Cell Signaling Technology), ER-alpha (ab32063, Abcam), c-MYC (1:200, 10828-1-AP, Proteintech) overnight at 4 °C. Followed by incubation with the secondary antibodies: Alexa Fluor 488-conjugated affiniPure Goat anti-Rabbit IgG (H + L) (1:400, 115-585-146, Jackson ImmunoResearch), Alexa Fluor 594-conjugated affiniPure Donkey Anti-Rat IgG (H + L) (1:400, 712-585-153, Jackson ImmunoResearch), Alexa Fluor 594-conjugated affiniPure Goat anti-Mouse IgG (H + L) (1:400, 111-545-144, Jackson ImmunoResearch) for 1 h, and nuclei-staining with DAPI (1:1000, D9542, Sigma) for 10 min at room temperature. Images were taken using a Leica TCS SP8 confocal microscope.
For immunohistochemistry, antigen retrieval was performed and endogenous peroxide was blocked as aforementioned. After being blocked with 1% (w/v) BSA Fraction V (ST023, Beyotime) and 10% goat serum (v/v) (B900780, Proteintech) in PBS for 1 h, the primary antibodies were added. The sections were incubated with the following primary antibodies against METTL3 (1:800, ab195352, Abcam), PR (1:400, 9856S, Cell Signaling Technology), ER-alpha (1:400, ab32063, Abcam), Ki67 (1:2000, ab15580, Abcam) overnight at 4 °C. On the following day, the sections were incubated with biotinylated goat anti-Rabbit IgG (1:400, BA-1000-1.5, Vector Laboratories) for 1 h at room temperature. After several rinses in PBS, the sections were incubated with the Vectastain Elite ABC reagent (PK-6100, Vector Laboratories) for 30 min, and immunoreactive signals were developed using ImmPACT DAB EqV Peroxidase (HRP) Substrate (SK-4103, Vector Laboratories), and counterstained with hematoxylin. Images were captured using an Olympus BX53 microscope.
Immunofluorescence images were analyzed using ImageJ (National Institutes of Health, v1.8.0_345). The level of E-Cadherin in uterine epithelial cells was determined by calculating the mean intensity of E-Cadherin signals. Similarly, the levels of ER, PR, and c-Myc in the uteri were determined by calculating the mean intensities of ER, PR, and c-Myc signals. The number of Ki67-positive (Ki67+) cells and the total number of epithelial or stromal cells were manually counted in several uterine sections from three mice in each group. Percentage of Ki67+ epithelial cells and percentage of Ki67+ epithelial cells were then calculated.
Protein extraction and western blot analysis
Protein extraction and western blot analysis were performed as previously described [53]. Western blotting experiments were analyzed using the following antibodies against ER-alpha (1:1000, ab32063, Abcam), α/β-Tubulin (1:4000, 2148S, Cell Signaling Techology), goat anti-rabbit IgG-HRP (1:2500, AS006, Asbio Technology). The experiments were performed with 3 replicates.
Fertility analyses
Female mice at least 8 weeks old were mated with fertile wild-type males to induce pregnancy (vaginal plug = day 1 of pregnancy). Uteri on day 4 post-mating were collected fixed in Paraformaldehyde Fix Solution (G1101-500ML, Servicebio) for histology or flash-frozen for RT-qPCR analysis. Successful pregnancy was confirmed by flushing embryos from the uteri on GD4. Tail intravenous injection with 0.1 mL of 1% Chicago blue dye (C8679, Sigma-Aldrich) was applied to identify implantation sites on GD5. Female fertility was assessed by mating cohorts of Mettl3 cKO (n = 12) and control (n = 10) mice individually with WT males proven breeders continuously for 6 months. The numbers of pups per litter per dam were recorded as mean ± SD.
Crystal violet staining of vaginal smear for mouse estrous cycle staging identification
The estrous cycle phases of Mettl3 cKO and control mice (8–10 week) were determined by crystal violet staining of vaginal smears, as previously described [54]. Briefly, the vaginal cells were washed with 100 μL PBS and transferred to a dry glass slide using a pipette. The slide was air-dried and stained for 1 min with 1% crystal violet (V5265-500ML, Sigma), followed by three 1-min rinses in water. The slides were coated with neutral balsam (G8590-100ml, Solarbio) and viewed with an Olympus BX53 microscope.
Measurement of serum estradiol and progesterone levels
Mouse blood samples were collected on GD4 in the morning and serum progesterone (P4), as well as estradiol-17β (E2) levels, were measured by Estradiol 2 Assay Kit (H102-1, NanJingJianCheng Bioengineering Institute, Nanjing, China) and Progesterone Assay Kit (H089, NanJingJianCheng Bioengineering Institute, Nanjing, China) according to the manufacturer’s recommendations.
Hormone treatments
Mice were ovariectomized at 6 weeks of age and rested for 2 weeks to remove endogenous ovarian hormone in most experiments. For evaluating the effects of E2, a subcutaneous injection of 100 ng E2 (E8875, Sigma) was given to OVX mice and sacrificed 24 h after the last injection. The hormonal profile of pregnancy at the time of implantation was simulated using a “pollard” experiment scheme [55]. Briefly, mice were treated with daily subcutaneous injections of E2 (100 ng) for 2 days. Following this treatment and after 2 days of rest, the mice received a daily injection of P4 (1 mg) for 3 days. On the fourth day, mice were treated with 100 ng of E2 and 1 mg of P4. The mice were sacrificed 16 h after the E2 + P4 injection. The uteri were harvested for RT-qPCR and histology analysis.
Superovulation
Superovulation studies were conducted to assess ovarian function. To induce superovulation, 6-week-old mice were administrated 7.5 IU of pregnant mare serum gonadotropin (PMSG, hor-272, ProSpec). Human chorionic gonadotropin (hCG, 230734, sigma) (7.5 IU) was injected subcutaneously 48 h after PMSG injection. At 14 h post hCG injection, the ovaries and oviducts were surgically removed, and the cumulus-oocyte complexes mass was recovered from the oviduct and collected into M2 medium (Sigma) containing 1 mg/mL of hyaluronidase (H3506, Sigma) to dissociate the cumulus cells from oocytes. The numbers of oocytes were counted and recorded.
In vivo decidualization assay
In vivo artificial decidual response was conducted similarly to that described previously [56]. Briefly, the ovariectomized mice were given daily subcutaneous injections of 100 ng of E2 (E8875, Sigma) prepared in sesame oil for 3 days (day 1–3). On day 6–8, mice were given a subcutaneous injection of 1 mg of P4 and 6.7 ng of E2 dissolved in sesame oil. Artificial decidualization was processed by intraluminal injection of 50 μL of sesame oil into the right uterine horn 6 h after the last injection of E2 and P4, with the left uterine horn acting as a negative control. Mice received daily E2 and P4 administration for 5 more days and sacrificed on day 14.
Isolation and culture of uterine cells
Uterine cells were isolated from day 4 pseudopregnant mice as previously described [57]. The uterine horns of control and Mettl3 cKO mice were cut longitudinally, washed with HBSS, and digested with 1% (w/v) pancreatin (P7545, Sigma) and 6 mg/ml dispase II (Sigma) in HBSS for 1 h at 4 °C followed by 1 h at room temperature and 10 min at 37 °C. The tissues were rinsed with HBSS for three times, and the supernatant was filtered through a 100-μm nylon cell strainer and collected as epithelial cells. And the remaining tissues were further digested within 0.15 mg/ml collagenase I (17100017, Invitrogen) in HBSS at 37 °C for 30 min. The tissues were washed 3 times with HBSS. And the supernatant above was filtered through a 100-μm nylon cell strainer for the collection of uterine stromal cells. The uterine cells were cultured in DMEM/F12 (319-080-CL, Wisentbio) containing 2% charcoal-stripped FBS (04-201-1A, Biological Industries) and 15 mM HEPES (15630080, Thermo Fisher).
In vitro decidualization assay
To induce stromal cells to undergo decidualization, the harvested stromal cells were cultured for 30 min, the medium was changed to remove unattached cells. And the cell culture was continued by adding fresh medium supplemented with P4 (1 μM) and E2 (10 nM) dissolved in ethanol for different time points. Transfection of Myc overexpression plasmid in Mettl3 cKO uterine stromal cells was performed according to Lipo3000 protocol (L3000015, Life Technologies). For the six-well culture plate, 2 μg Myc overexpression plasmid (EX-Mm30812-M02, GeneCopoeia) or 2 μg control plasmid (EX-NEG-M02-B, GeneCopoeia) was used for the transfection. The cells were cultured with P4 (1 μM) and E2 (10 nM) for 2 days, and the mRNA levels of Myc and of decidualization-related genes were analyzed by RT-qPCR.
RNA extraction and RT-qPCR analysis
Total RNA was extracted from uterine tissues using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. The complementary DNAs (cDNAs) were synthesized with PrimeScript RT Master Mix (RR036A, Takara) by using 500 ng total RNAs according to the manufacturer’s instructions. Quantitative real-time PCR was performed to assess the expression of genes of interest with SYBR Green (RR820A, Takara) on a CFX Connect Real-Time PCR Detection System (Biorad). Experimental gene expression data were normalized to Actb. The RT-qPCR primers are listed in Table 1.
RNA-seq and data analysis
The uteri of Mettl3 cKO and control mice on GD4 were obtained for RNA-seq analysis. Total RNA was extracted from the uteri using TRIzol reagent (Invitrogen) following the manufacturer’s protocol. RNA purity and quantification were evaluated using the NanoDrop 2000 spectrophotometer (Thermo Scientific, USA). RNA integrity was assessed using the Agilent 2100 Bioanalyzer (Agilent Technologies, USA). The libraries were constructed using TruSeq Stranded mRNA LT Sample Prep Kit (Illumina, USA) and sequenced on an Illumina Novaseq6000 platform (OE Biotech Co., Ltd., Shanghai, China). The clean reads were mapped to the mouse reference genome (GRCm38.p6) using HISAT2. FPKM of each gene was calculated using Cufflinks, and the read counts of each gene were obtained by HTSeq-count. Differential expression analysis was performed using the DESeq (v3.8) R package. p value < 0.05 and foldchange > 1.5 or foldchange <2/3 was set as the threshold for significantly differential expression. GO, KEGG, and GSEA analyses were conducted by clusterprofiler R package (v4.4.4). All the heatmaps were drawn by pheatmap R package (version 1.0.12). Z-score were calculated and scaled by row or column.
m6A sequencing and data processing
For m6A sequencing, the poly(A) RNA was fragmented into small pieces using Magnesium RNA Fragmentation Module (NEB, cat.e6150, USA) under 86 °C 7 min. The cleaved RNA fragments were incubated for 2 h at 4 °C with anti-m6A antibody (202003, Synaptic Systems) in IP buffer (50 mM Tris-HCl, 750 mM NaCl and 0.5% IGEPAL CA-630). The IP RNA was reverse-transcribed to create the cDNA by SuperScript™ II Reverse Transcriptase (1896649, Invitrogen), which was next used to synthesize U-labeled second-stranded DNAs with E. coli DNA polymerase I (m0209, NEB), RNase H (m0297, NEB) and dUTP Solution (R0133, Thermo Fisher). An A-base is then added to the blunt ends of each strand, preparing them for ligation to the indexed adapters. Each adapter contains a T-base overhang for ligating the adapter to the A-tailed fragmented DNA. Single- or dual-index adapters are ligated to the fragments, and size selection was performed with AMPureXP beads. After the heat-labile UDG enzyme (m0280, NEB) treatment of the U-labeled second-stranded DNAs, the ligated products are amplified with PCR by the following conditions: initial denaturation at 95 °C for 3 min; 8 cycles of denaturation at 98 °C for 15 s, annealing at 60 °C for 15 s, and extension at 72 °C for 30 s; and then final extension at 72 °C for 5 min. The average insert size for the final cDNA library was 300 ± 50 bp. At last, we performed the 2 × 150 bp paired-end sequencing (PE150) on an Illumina Novaseq6000 (LC-Bio Technology Co., Ltd., Hangzhou, China).
For the bioinformatic analysis of m6A-seq, fastp software (v0.19.4) was used to remove the reads that contained adapter contamination, low-quality bases and undetermined bases with default parameters. The sequence quality of IP and input samples were also verified using fastp. We used HISAT2 (v2.0.4) to map reads to the reference genome of mouse (GRCm38.p6). Mapped reads of IP and input libraries were provided for R package exomePeak (v2.13), which identifies m6A peaks. The bigwig format was converted from the bam format by bamcoverage tool in deeptools (version 2.5.4), and was adapted for visualization on the IGV software (v2.14.0). RPGC (reads per genomic content) method was used for normalization. MEME (v4.12.0) and HOMER (v4.10) were used for de novo and known motif finding followed by localization of the motif with respect to peak summit. Called peaks were annotated by intersection with gene architecture using R package ChIPseeker (v3.8). Then StringTie (v2.1.2) was used to perform expression level for all mRNAs from input libraries by calculating FPKM (total exon fragments/mapped reads (millions) × exon length (kB)). The differentially expressed mRNAs were selected with log2(fold change) >1 or log2(fold change) <−1 and P < 0.05 by R package edgeR (v3.38.4).
m6A-RIP-qPCR
Purified mRNAs of the uteri of Mettl3 cKO mice and control mice were prepared and fragmented into ~100 nt by RNA fragmentation reagents (e6150, NEB). Immunoprecipitation was performed using anti-m6A antibody (202003, Synaptic Systems) as described previously. The enrichment of m6A was measured with quantitative RT-PCR. Primers for m6A-RIP-qPCR are listed in Table 2.
mRNA stability assay
To assess mRNA stability, primary uterine cells were treated with actinomycin D (Sigma) at a final concentration of 5 μg/mL for 0 h, 2 h, and 4 h. Total RNA samples were extracted and subjected to RT-qPCR analysis. Results were normalized to the expression of Actb. Fold differences in expression levels were calculated according to the 2−ΔΔCT method.
Statistics
Statistical analyses were performed using 2-tailed Student’s t test. Data are presented as mean ± SD. P values less than 0.05 were considered statistically significant. *P < 0.05, **P < 0.01 and ***P < 0.001.
Data availability
The RNA-seq data were deposited in the GEO repository at NCBI under the accession number GSE208096. The m6A-seq data were available from the corresponding author upon request. The code used for GEO data process is available at https://github.com/wansh007/data-process.
References
Agarwal A, Mulgund A, Hamada A, Chyatte MR. A unique view on male infertility around the globe. Reprod Biol Endocrinol. 2015;13:37. https://doi.org/10.1186/s12958-015-0032-1
Zondervan KT, Becker CM, Missmer SA. Endometriosis. N Engl J Med. 2020;382:1244–56. https://doi.org/10.1056/NEJMra1810764
Kuivasaari P, Hippeläinen M, Anttila M, Heinonen S. Effect of endometriosis on IVF/ICSI outcome: stage III/IV endometriosis worsens cumulative pregnancy and live-born rates. Hum Reprod. 2005;20:3130–5. https://doi.org/10.1093/humrep/dei176
Barnhart K, Dunsmoor-Su R, Coutifaris C. Effect of endometriosis on in vitro fertilization. Fertil Steril. 2002;77:1148–55. https://doi.org/10.1016/s0015-0282(02)03112-6
Pantos K, Grigoriadis S, Maziotis E, Pistola K, Xystra P, Pantou A, et al. The role of interleukins in recurrent implantation failure: a comprehensive review of the literature. Int J Mol Sci. 2022,23. https://doi.org/10.3390/ijms23042198
Izawa M, Taniguchi F, Terakawa N, Harada T. Epigenetic aberration of gene expression in endometriosis. Front Biosci (Elite Ed). 2013;5:900–10. https://doi.org/10.2741/e669
Kim TH, Yoo JY, Choi KC, Shin JH, Leach RE, Fazleabas AT, et al. Loss of HDAC3 results in nonreceptive endometrium and female infertility. Sci Transl Med. 2019, 11. https://doi.org/10.1126/scitranslmed.aaf7533
Xu H, Zhou M, Cao Y, Zhang D, Han M, Gao X, et al. Genome-wide analysis of long noncoding RNAs, microRNAs, and mRNAs forming a competing endogenous RNA network in repeated implantation failure. Gene. 2019;720:144056. https://doi.org/10.1016/j.gene.2019.144056
Ghafouri-Fard S, Shoorei H, Taheri M. Role of non-coding RNAs in the pathogenesis of endometriosis. Front Oncol. 2020;10:1370. https://doi.org/10.3389/fonc.2020.01370
Murakami S, Jaffrey SR. Hidden codes in mRNA: control of gene expression by m(6)A. Mol Cell. 2022;82:2236–51. https://doi.org/10.1016/j.molcel.2022.05.029
Zaccara S, Ries RJ, Jaffrey SR. Reading, writing and erasing mRNA methylation. Nat Rev Mol cell Biol. 2019;20:608–24. https://doi.org/10.1038/s41580-019-0168-5
Xu K, Yang Y, Feng GH, Sun BF, Chen JQ, Li YF, et al. Mettl3-mediated m(6)A regulates spermatogonial differentiation and meiosis initiation. Cell Res. 2017;27:1100–14. https://doi.org/10.1038/cr.2017.100
Mu H, Li H, Liu Y, Wang X, Mei Q, Xiang W. N6-Methyladenosine modifications in the female reproductive system: roles in gonad development and diseases. Int J Biol Sci. 2022;18:771–82. https://doi.org/10.7150/ijbs.66218
Liu J, Eckert MA, Harada BT, Liu SM, Lu Z, Yu K, et al. m(6)A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nat Cell Biol. 2018;20:1074–83. https://doi.org/10.1038/s41556-018-0174-4
Zhai J, Li S, Sen S, Opoku-Anane J, Du Y, Chen ZJ, et al. m(6)A RNA methylation regulators contribute to eutopic endometrium and myometrium dysfunction in adenomyosis. Front Genet. 2020;11:716. https://doi.org/10.3389/fgene.2020.00716
Jiang L, Zhang M, Wu J, Wang S, Yang X, Yi M, et al. Exploring diagnostic m6A regulators in endometriosis. Aging. 2020;12:25916–38. https://doi.org/10.18632/aging.202163
Li X, Xiong W, Long X, Dai X, Peng Y, Xu Y, et al. Inhibition of METTL3/m6A/miR126 promotes the migration and invasion of endometrial stromal cells in endometriosis. Biol Reprod. 2021;105:1221–33. https://doi.org/10.1093/biolre/ioab152
Soyal SM, Mukherjee A, Lee KY, Li J, Li H, DeMayo FJ, et al. Cre-mediated recombination in cell lineages that express the progesterone receptor. Genes. 2005;41:58–66. https://doi.org/10.1002/gene.20098
Wang H, Dey SK. Roadmap to embryo implantation: clues from mouse models. Nat Rev Genet. 2006;7:185–99. https://doi.org/10.1038/nrg1808
Wetendorf M, DeMayo FJ. The progesterone receptor regulates implantation, decidualization, and glandular development via a complex paracrine signaling network. Mol Cell Endocrinol. 2012;357:108–18. https://doi.org/10.1016/j.mce.2011.10.028
Wang X, Wu SP, DeMayo FJ. Hormone dependent uterine epithelial-stromal communication for pregnancy support. Placenta. 2017;60:S20–s26. https://doi.org/10.1016/j.placenta.2017.07.003.
Marquardt RM, Kim TH, Shin JH, Jeong JW. Progesterone and estrogen signaling in the endometrium: what goes wrong in endometriosis? Int J Mol Sci. 2019,20. https://doi.org/10.3390/ijms20153822
Rubel CA, Lanz RB, Kommagani R, Franco HL, Lydon JP, DeMayo FJ. Research resource: genome-wide profiling of progesterone receptor binding in the mouse uterus. Mol Endocrinol. 2012;26:1428–42. https://doi.org/10.1210/me.2011-1355
Huet-Hudson YM, Andrews GK, Dey SK. Cell type-specific localization of c-myc protein in the mouse uterus: modulation by steroid hormones and analysis of the periimplantation period. Endocrinology. 1989;125:1683–90. https://doi.org/10.1210/endo-125-3-1683
Li SY, Yan JQ, Song Z, Liu YF, Song MJ, Qin JW, et al. Molecular characterization of lysyl oxidase-mediated extracellular matrix remodeling during mouse decidualization. FEBS Lett. 2017;591:1394–407. https://doi.org/10.1002/1873-3468.12645
Lei W, Feng XH, Deng WB, Ni H, Zhang ZR, Jia B, et al. Progesterone and DNA damage encourage uterine cell proliferation and decidualization through up-regulating ribonucleotide reductase 2 expression during early pregnancy in mice. J Biol Chem. 2012;287:15174–92. https://doi.org/10.1074/jbc.M111.308023
Zeller KI, Jegga AG, Aronow BJ, O’Donnell KA, Dang CV. An integrated database of genes responsive to the Myc oncogenic transcription factor: identification of direct genomic targets. Genome Biol. 2003;4:R69. https://doi.org/10.1186/gb-2003-4-10-r69
Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformation. 2011;27:1739–40. https://doi.org/10.1093/bioinformatics/btr260
Zuo RJ, Gu XW, Qi QR, Wang TS, Zhao XY, Liu JL, et al. Warburg-like glycolysis and lactate shuttle in mouse decidua during early pregnancy. J Biol Chem. 2015;290:21280–91. https://doi.org/10.1074/jbc.M115.656629
Zhao S, Lu J, Chen Y, Wang Z, Cao J, Dong Y. Exploration of the potential roles of m6A regulators in the uterus in pregnancy and infertility. J Reprod Immunol. 2021;146:103341. https://doi.org/10.1016/j.jri.2021.103341
Zhang Z, Zhou D, Lai Y, Liu Y, Tao X, Wang Q, et al. Estrogen induces endometrial cancer cell proliferation and invasion by regulating the fat mass and obesity-associated gene via PI3K/AKT and MAPK signaling pathways. Cancer Lett. 2012;319:89–97. https://doi.org/10.1016/j.canlet.2011.12.033
Vasquez YM, DeMayo FJ. Role of nuclear receptors in blastocyst implantation. Semin Cell Dev Biol. 2013;24:724–35. https://doi.org/10.1016/j.semcdb.2013.08.004
Large MJ, DeMayo FJ. The regulation of embryo implantation and endometrial decidualization by progesterone receptor signaling. Mol Cell Endocrinol. 2012;358:155–65. https://doi.org/10.1016/j.mce.2011.07.027
Rubel CA, Jeong JW, Tsai SY, Lydon JP, Demayo FJ. Epithelial-stromal interaction and progesterone receptors in the mouse uterus. Semin Reprod Med. 2010;28:27–35. https://doi.org/10.1055/s-0029-1242990
Kim TH, Yoo JY, Wang Z, Lydon JP, Khatri S, Hawkins SM, et al. ARID1A is essential for endometrial function during early pregnancy. PLoS Genet. 2015;11:e1005537. https://doi.org/10.1371/journal.pgen.1005537
Kurihara I, Lee DK, Petit FG, Jeong J, Lee K, Lydon JP, et al. COUP-TFII mediates progesterone regulation of uterine implantation by controlling ER activity. PLoS Genet. 2007;3:e102. https://doi.org/10.1371/journal.pgen.0030102
Lessey BA, Kim JJ. Endometrial receptivity in the eutopic endometrium of women with endometriosis: it is affected, and let me show you why. Fertil Steril. 2017;108:19–27. https://doi.org/10.1016/j.fertnstert.2017.05.031
Yin X, Pavone ME, Lu Z, Wei J, Kim JJ. Increased activation of the PI3K/AKT pathway compromises decidualization of stromal cells from endometriosis. J Clin Endocrinol Metab. 2012;97:E35–43. https://doi.org/10.1210/jc.2011-1527
Gajulapalli VN, Samanthapudi VS, Pulaganti M, Khumukcham SS, Malisetty VL, Guruprasad L, et al. A transcriptional repressive role for epithelial-specific ETS factor ELF3 on oestrogen receptor alpha in breast cancer cells. Biochem J. 2016;473:1047–61. https://doi.org/10.1042/bcj20160019
Qiao Z, Jiang Y, Wang L, Wang L, Jiang J, Zhang J. Mutations in KIAA1109, CACNA1C, BSN, AKAP13, CELSR2, and HELZ2 are associated with the prognosis in endometrial cancer. Front Genet. 2019;10:909. https://doi.org/10.3389/fgene.2019.00909
Tan J, Che Y, Liu Y, Hu J, Wang W, Hu L, et al. CELSR2 deficiency suppresses lipid accumulation in hepatocyte by impairing the UPR and elevating ROS level. FASEB J. 2021;35:e21908. https://doi.org/10.1096/fj.202100786RR
Xu M, Zhu S, Xu R, Lin N. Identification of CELSR2 as a novel prognostic biomarker for hepatocellular carcinoma. BMC Cancer. 2020;20:313. https://doi.org/10.1186/s12885-020-06813-5
Zhou X, Zhan Z, Tang C, Li J, Zheng X, Zhu S, et al. Silencing Celsr2 inhibits the proliferation and migration of Schwann cells through suppressing the Wnt/β-catenin signaling pathway. Biochem Biophys Res Commun. 2020;533:623–30. https://doi.org/10.1016/j.bbrc.2020.09.015
Gao Y, Ouyang X, Zuo L, Xiao Y, Sun Y, Chang C, et al. R-2HG downregulates ERα to inhibit cholangiocarcinoma via the FTO/m6A-methylated ERα/miR16-5p/YAP1 signal pathway. Mol Ther Oncol. 2021;23:65–81. https://doi.org/10.1016/j.omto.2021.06.017
Chen Z, Wu L, Zhou J, Lin X, Peng Y, Ge L, et al. N6-methyladenosine-induced ERRγ triggers chemoresistance of cancer cells through upregulation of ABCB1 and metabolic reprogramming. Theranostics. 2020;10:3382–96. https://doi.org/10.7150/thno.40144
Kurita T, Lee K-j, Saunders PTK, Cooke PS, Taylor JA, Lubahn DB, et al. Regulation of progesterone receptors and decidualization in uterine stroma of the estrogen receptor-α knockout mouse1. Biol Reprod. 2001;64:272–83. https://doi.org/10.1095/biolreprod64.1.272
Tan J, Paria BC, Dey SK, Das SK. Differential uterine expression of estrogen and progesterone receptors correlates with uterine preparation for implantation and decidualization in the mouse. Endocrinology. 1999;140:5310–21. https://doi.org/10.1210/endo.140.11.7148
Lee K, Jeong J, Kwak I, Yu CT, Lanske B, Soegiarto DW, et al. Indian hedgehog is a major mediator of progesterone signaling in the mouse uterus. Nat Genet. 2006;38:1204–9. https://doi.org/10.1038/ng1874
Tranguch S, Cheung-Flynn J, Daikoku T, Prapapanich V, Cox MB, Xie H, et al. Cochaperone immunophilin FKBP52 is critical to uterine receptivity for embryo implantation. Proc Natl Acad Sci USA. 2005;102:14326–31. https://doi.org/10.1073/pnas.0505775102
Gu Y, Zhang X, Yang Q, Wang JM, He YP, Sun ZG, et al. Uterine NDRG2 expression is increased at implantation sites during early pregnancy in mice, and its down-regulation inhibits decidualization of mouse endometrial stromal cells. Reprod Biol Endocrinol. 2015;13:49. https://doi.org/10.1186/s12958-015-0047-7
Yang Q, Gu Y, Zhang X, Wang JM, He YP, Shi Y, et al. Uterine expression of NDRG4 is induced by estrogen and up-regulated during embryo implantation process in mice. PloS One. 2016;11:e0155491. https://doi.org/10.1371/journal.pone.0155491
Chen H, Liu H, Qing G. Targeting oncogenic Myc as a strategy for cancer treatment. Signal Transduct Target Ther. 2018;3:5. https://doi.org/10.1038/s41392-018-0008-7
Wan S, Sun Y, Fu J, Song H, Xiao Z, Yang Q, et al. mTORC1 signaling pathway integrates estrogen and growth factor to coordinate vaginal epithelial cells proliferation and differentiation. Cell Death Dis. 2022;13:862. https://doi.org/10.1038/s41419-022-05293-8
McLean AC, Valenzuela N, Fai S, Bennett SA. Performing vaginal lavage, crystal violet staining, and vaginal cytological evaluation for mouse estrous cycle staging identification. J Vis Exp. 2012:e4389. https://doi.org/10.3791/4389
Fullerton PT Jr, Monsivais D, Kommagani R, Matzuk MM. Follistatin is critical for mouse uterine receptivity and decidualization. Proc Natl Acad Sci USA. 2017;114:E4772–e4781. https://doi.org/10.1073/pnas.1620903114
Lydon JP, DeMayo FJ, Funk CR, Mani SK, Hughes AR, Montgomery CA Jr., et al. Mice lacking progesterone receptor exhibit pleiotropic reproductive abnormalities. Genes Dev. 1995;9:2266–78. https://doi.org/10.1101/gad.9.18.2266
Liang X-H, Deng W-B, Li M, Zhao Z-A, Wang T-S, Feng X-H, et al. Egr1 protein acts downstream of estrogen-leukemia inhibitory factor (LIF)-STAT3 pathway and plays a role during implantation through targeting Wnt4*. J Biol Chem. 2014;289:23534–45. https://doi.org/10.1074/jbc.M114.588897
Funding
Research reported in this publication was supported by the Natural Science Foundation of China (to MY, 82271754, and 82071737; to SW, 82101831, to JY, 32270981), 111 Project (B16201), and Kunming Medical Union Project (to JY, 202301AY070001-003).
Author information
Authors and Affiliations
Contributions
MY, ZY and SW designed the study. SW, YS, JZ, WM, JY, KC, and ZX conducted the experiments. SW, DG, and QZ provided technical support and discussions. MY, ZY and SW reviewed and edited the manuscript. All the authors commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics
All mice used in this study were housed in the specific pathogen-free animal facilities in the Animal Resource Center at Jinan University, following the ethical guidelines of the Animal Ethics Committee of Jinan University (IACUC-20220418-03).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by Mauro Piacentini
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wan, S., Sun, Y., Zong, J. et al. METTL3-dependent m6A methylation facilitates uterine receptivity and female fertility via balancing estrogen and progesterone signaling. Cell Death Dis 14, 349 (2023). https://doi.org/10.1038/s41419-023-05866-1
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41419-023-05866-1
This article is cited by
-
Bioinformatic Analysis of m6A Regulator-Mediated RNA Methylation Modification Patterns and Immune Microenvironment Characterization in Endometriosis
Biochemical Genetics (2024)
-
METTL3-regulated m6A modification impairs the decidualization of endometrial stromal cells by regulating YTHDF2-mediated degradation of FOXO1 mRNA in endometriosis-related infertility
Reproductive Biology and Endocrinology (2023)