Abstract
SEMA6A is a multifunctional transmembrane semaphorin protein that participates in various cellular processes, including axon guidance, cell migration, and cancer progression. However, the role of SEMA6A in clear cell renal cell carcinoma (ccRCC) is unclear. Based on high-throughput sequencing data, here we report that SEMA6A is a novel target gene of the VHL-HIF-2α axis and overexpressed in ccRCC. Chromatin immunoprecipitation and reporter assays revealed that HIF-2α directly activated SEMA6A transcription in hypoxic ccRCC cells. Wnt/β-catenin pathway activation is correlated with the expression of SEMA6A in ccRCC; the latter physically interacted with SEC62 and promoted ccRCC progression through SEC62-dependent β-catenin stabilization and activation. Depletion of SEMA6A impaired HIF-2α-induced Wnt/β-catenin pathway activation and led to defective ccRCC cell proliferation both in vitro and in vivo. SEMA6A overexpression promoted the malignant phenotypes of ccRCC, which was reversed by SEC62 depletion. Collectively, this study revealed a potential role for VHL-HIF-2α-SEMA6A-SEC62 axis in the activation of Wnt/β-catenin pathway. Thus, SEMA6A may act as a potential therapeutic target, especially in VHL-deficient ccRCC.
Similar content being viewed by others
Introduction
Renal cell carcinoma (RCC) is the second leading cause of death in patients with urinary cancer worldwide, with a frequency of 1/30,000, accounting for 2–3% of all adult cancers [1], and one of the tenth most common cancers globally [2]. The most common subtype of RCC is clear cell renal cell carcinoma (ccRCC) [3], which accounts for about 75% of all RCC cases [4]. It is characterized by malignant epithelial cells with clear cytoplasm, dense nest-like growth, and complicated dendritic blood vessels [5]. Currently, surgery is the main strategy for the treatment of ccRCC [6, 7]. However, the cancer is not sensitive to postoperative radiotherapy and chemotherapy, accompanied by high metastasis and recurrence rates [8]. In addition, clinically effective methods for early prevention, early diagnosis, post-diagnosis treatment, and prognostic evaluation of ccRCC are yet lacking [9]. Therefore, clarifying the molecular mechanisms of ccRCC will provide clues for the early diagnosis and targeted therapy of ccRCC.
Interestingly, ccRCC is triggered by the biallelic inactivation of the tumor suppressor gene Von Hippel-Lindau (VHL) in renal epithelial cells, which deregulates the hypoxia-inducible factors (HIFs), including HIF-1a and HIF-2α [10, 11]. VHL can form a classical E3 ubiquitin ligase complex with elonginB, elonginC, Cullin2, and Rbx1 [12]. This complex can target HIF-2α for degradation through the ubiquitin-proteasome pathway [13]. Under normoxia, the proline residues of HIF-2α are hydroxylated by prolyl hydroxylase, and the hydroxylated HIF-2α is then recognized by this E3 ubiquitin ligase complex, followed by sequential degradation [14]. Conversely, VHL failed to recognize HIF-2α under hypoxia, resulting in its accumulation [15]. The accumulated HIF-2α is associated with the constitutively expressed HIF-1β to form a heterodimer, which is then transferred to the nucleus. Subsequently, the expression of the downstream genes, vascular endothelial growth factor A (VEGFA) and cyclin D1 (CCND1) [16], is activated, and several physiological processes, such as angiogenesis, cell metabolism, cell proliferation, and cell apoptosis are regulated [17,18,19]. VHL mutations are observed in ~70–80% of ccRCC [20], and the Cullin2-VHL E3 ubiquitin ligase complex cannot be formed, resulting in the stabilization of the HIF-2α protein irrespective of the cellular oxygen supply [21]. The stabilized HIF-2α, together with HIF-1β, causes the abnormal activation of the downstream oncogenes and promotes the occurrence and development of ccRCC [22].
Herein, the transcriptional profiling from HIF-2α silenced, and VHL overexpressed ccRCC cells was integrated, which facilitated, which in turn led to the identification of SEMA6A as a direct downstream target gene of the VHL-HIF-2α axis. Next, we revealed that SEMA6A is required for the integrity of the HIF-2α-β-catenin signaling pathway in ccRCC, and the genetic deletion of SEMA6A resulted in impaired proliferation ability in ccRCC cells by repressing SEC62-dependent β-catenin stabilization and activation.
Materials and methods
Data collection
GSE149005, GSE32297, GSE68417, and GSE79683 were downloaded from the gene expression omnibus (GEO) database on the NCBI website. (The R software limma package was used to perform differential expression analysis, and p < 0.05 and |logFC| > 1.2 were set as the cutoff criteria. Venn diagrams were drawn to obtain intersecting genes, and Gene Ontology (GO) and Kyoto Encyclopedia of Genomes (KEGG) pathway analyses were performed. The mutation frequencies and copy number alterations in the SEMA6A gene were observed by genomic analysis of renal clear cell carcinoma (The Cancer Genome Atlas Firehose Legacy, 538 samples) through the cBioPortal online tool (http://www.cbioportal.org/). The transcript levels of SEMA6A were obtained from the GEPIA database (http://gepia.cancer-pku.cn) between normal kidney tissue and KIRC. The expression of SEMA6A in different subtypes of kidney renal clear cell carcinoma (KIRC) was analyzed via the UALCAN online tool (http://ualcan.path.uab.edu/).
Cell culture and tissue samples
All cell lines used in this study were purchased from American Type Culture Collection (ATCC) and cultured in an appropriate medium containing 10% fetal bovine serum (FBS, Gibco, Grand Island, NY, USA) and supplemented with penicillin (100 U/ml) and streptomycin (100 µg/ml), in a 37 °C incubator with 5% CO2. All cell lines were negative for mycoplasma during the experiment. 6 paired fresh human ccRCC samples and adjacent normal kidney tissues were obtained from the Second People’s Hospital of Lianyungang. The study was approved by the Medical Ethical Committee of the Second People’s Hospital of Lianyungang, and informed consent was obtained from all subjects or their relatives.
Cell proliferation, colony formation, and migration assays
For cell counting kit-8 (CCK-8) analysis, 5 × 104 cells were seeded in a 96-well plate and CCK-8 reagent was added to the wells; the absorbance was measured at 450 or 490 nm. For bromodeoxyuridine (BrdU) assay, the cells were seeded in a 96-well plate and incubated for 24 h. Then, the cell culture medium was discarded, the cell fixative solution was added to each well, and the cells were fixed, denatured, and incubated with anti-BrdU and TMB substrate. The absorbance was measured at 450 nm on a microplate reader. For colony formation assay, about 1000 cells were seeded in each well of a 12-well plate, and the seeded 12-well plate was cultured for about 2 weeks in a 5% CO2 and 37 °C incubator. Cells were washed with phosphate-buffered saline (PBS), fixed with paraformaldehyde for 15–30 min, and stained with 0.05% crystal violet or Giemsa to count the number of colonies. To assess the cell migration ability, 5 × 105 cells were seeded in 12-well plates for 24 h. When the cell density was about 80%, a 10-μl pipette tip was used to draw a line perpendicular to the bottom of each well, washed three times with PBS to remove the floating cells. Images were taken under the microscope at the same position at 0, 24, and 48 h after scratching, and the migration area was calculated using ImageJ software.
Structure modeling and protein docking
Full-length sequences of SEMA6A and SEC62 were uploaded onto the I-TASSER webserver, and the modeling was run with default settings. Five structure clusters were obtained as results for each protein, the final models were selected according to the overall conformations, and the C-scores were calculated by the server. The protein-protein docking was performed using the Cluspro online server. Ligplot software was used to analyze the binding force between the two proteins, and Pymol software was used for three-dimensional (3D) protein-binding conformation imaging.
Immunoprecipitation (IP) and glutathione-S-transferase (GST)-pull-down
The IP protocol has been described previously [23]. Briefly, cells were lysed with IP buffer (100 mM NaCl, 20 mM Tris-HCl pH 8.0, 0.5 mM EDTA, 0.5% (v/v) Nonidet P-40) with protease inhibitor cocktail and phosphatase inhibitor for 30 min on ice. The cells were sonicated, and the lysate was collected by centrifugation at 14,000 rpm for 10 min. For endogenous IP, the filtered supernatant was incubated with either anti-SEMA6A or anti-SEC62 or IgG and protein A/G beads at 4 °C overnight on a rotating wheel. The immunoprecipitates were washed three times with IP buffer and denatured at 95 °C for 5 min. The final sample was separated by 12% SDS-PAGE and detected by immunoblotting. The interaction between SEMA6A and SEC62 was assessed by GST-pull-down assay. Briefly, GST or GST-SEC62 was expressed in BL21 cells and purified using glutathione-Sepharose beads. Equivalent amounts of GST or GST fusion proteins were resuspended in reaction buffer (20 mM HEPES, pH 7.5; 5 mM MgCl2, 0.5 mM EDTA, 0.05% NP40, 1 mM DTT and 130 mM KCl,) containing 0.2 mg/ml bovine serum albumin (BSA). Then, 1 mg FLAG-SEMA6A-transfected cell lysate was added to each mixture, followed by rotation at room temperature for 1 h. Subsequently, the beads were boiled in a sample buffer to elute the bound proteins. The final sample was separated by SDS-PAGE and detected by immunoblotting.
Western blot
Cells were harvested and lysed with SDS-containing lysis buffer (100 mM Tris-HCl, pH 6.8, 100 mM DTT, 1% SDS, 10% glycerol). An equivalent amount of protein from each sample was separated by SDS-PAGE and transferred to polyvinylidene fluoride (PVDF) membranes. After blocking with 5% nonfat milk for 1 h, the membranes were incubated with the indicated primary antibodies at 4 °C overnight. After washing with Tris buffered saline Tween (TBST) buffer (50 mM Tris-HCl, 0.15 M NaCl and 0.05% Tween 20), the membrane was incubated with the secondary antibody at 25 °C for 1 h. Then, the immunoreactive bands were developed using enhanced chemiluminescence reagent with β-Actin as a loading control. The primary antibodies were as follows: anti-HIF-2α (sc-8712, Santa Cruz), anti-SEMA6A (sc-398302, Santa Cruz), anti-SEC62(ab244335, abcam), and anti-β-Actin(sc-8432, Santa Cruz).
RNA inhibition and quantitative real-time polymerase chain reaction (qRT-PCR)
Lentiviral human shRNA plasmids targeting SEMA6A or HIF-2α were purchased from Sigma. The lentiviral particles were used to infect ccRCC cells and screened by puromycin for ~2 weeks. RNA was extracted from the samples using the RNeasy extraction kit and reverse transcribed into cDNA using the reverse transcription kit according to the manufacturer’s instructions. RT-PCR was performed using Light Cycler 480, and β-actin was used as an internal reference gene. The relative expression of the target gene was analyzed using the 2−ΔΔCt method.
Luciferase activity and chromatin immunoprecipitation (ChIP) assays
The TOP/FOP flash reporters were purchased from Beyotime. The promoter region of human SEMA6A gene was amplified from the genomic DNA of A498 cells and cloned into the pGL4.15 vector (Promega, Madison, WI, USA). After transfection, the cells were harvested and lysed, and the luciferase activity was assayed using the Dual Luciferase Assay System (Promega). The luciferase activity was normalized to Renilla luciferase activity. For ChIP assay, cells were fixed with formaldehyde, and DNA was sheared to fragments at 100–500 bp by sonication. Then, the supernatant was incubated with antibodies against HIF-2α or normal serum IgG at 4 °C overnight.
Xenograft assays
For subcutaneous xenografts, 3 × 106 cells in 300 μl PBS were injected subcutaneously into the flanks of 6-week-old male nude mice. Tumors were measured every 7 days after injection, the tumor volume was calculated according to the formula (length × width2)/2, and the mice were sacrificed 4 weeks after inoculation. All animal experiments were approved by the Institutional Animal Ethics Committee of Jiangsu Ocean University, and animal care was in accordance with the institutional guidelines.
Statistical analyses
All experiments were repeated at least three times. Data were expressed as mean ± standard deviation (SD). The statistical analysis was performed using GraphPad Prism 9.0 software (USA, San Diego, CA). The differences between the groups were calculated using either the Student’s t test or one-way analysis of variance using Tukey’s test. Statistical significance was expressed as *p < 0.05, **p < 0.01, and ***p < 0.001, respectively.
Results
Identification of the potential target genes of the VHL-HIF-2α axis in ccRCC cells
In order to explore new substrate genes of HIF-2α in ccRCC, we first analyzed the transcriptional profiling of HIF-2α-regulated genes in ccRCC cells using a previously published RNA sequencing (RNA-seq) dataset (GSE149005). GSE149005 contains a transcriptional gene expression profile of 786-O cells treated with sgRNAs to HIF-2α or scrambled control. p < 0.05 and |logFC| > 1.2 were set as the cutoff criteria. A total of 118 genes were upregulated, and 144 genes were downregulated after HIF-2α was depleted (Supplementary Tables 1 and 2). The differentially expressed genes (DEGs) were exhibited by the heatmap and volcano map, respectively (Fig. 1A–C). The KEGG analysis showed that these downregulated genes were significantly enriched in the PI3K-AKT signaling pathway, RCC, HIF-1 signaling pathway, and p53 signaling pathway (Fig. 1D). The gene-concept network generated by the R package clusterProfiler showed that VEGFA, CCND1, and CDKN1A were the key genes linked to these enriched pathways (Fig. 1E). Thus, these results highlighted the critical roles of HIF-2α and its target genes in the regulation of these pathways. As the tumor suppressor VHL protein promotes the ubiquitination and degradation of HIF-2α, we speculated that forced expression of VHL in ccRCC cells might indirectly inhibit the expression of HIF-2α target genes. Interestingly, the overexpression of VHL inhibited the expression of CXCR4, a downstream gene of HIF-2α. In order to further determine the genes regulated by VHL, we used a previously published GEO dataset to analyze the transcription profile of VHL-regulated genes in ccRCC cells. Using the same cutoff criteria, we found that 81 genes were upregulated and 49 genes were downregulated in the presence of exogenous VHL protein (Fig. 1F, G and Supplementary Tables 3 and 4). Next, we identified that the genes were downregulated after knocking out HIF-2α and VHL overexpression. As depicted in Fig. 1H, 15 genes were identified using these screening criteria. Together, our screening strategy showed that these genes might contain new target genes of the VHL-HIF-2α axis in ccRCC cells.
A Heatmap of the DEGs expression regulated by HIF-2α in ccRCC cells in GSE149005. p < 0.05, | log2FC| > 1.2. B Volcano map of DEGs expression regulated by HIF-2α in GSE149005. C Number of up- and downregulated DEGs in GSE149005. D KEGG pathway analysis of the downregulated genes in GSE149005 through the clusterProfiler R package. E Gene network diagram generated by the clusterProfiler R package showed the key genes associated with the enrichment pathway. F Volcano map of the DEGs in GSE32297. p < 0.05, |log2FC| > 1.2. G Number of up- and downregulated DEGs in GSE32297. H Venn diagram shows the intersection between the downregulated genes in GSE149005 and GSE32297 databases.
SEMA6A is a direct target gene of HIF-2α
The correlation between the mRNA expression of HIF-2α and these 15 potential target genes in ccRCC was identified from the TCGA database (Fig. 2A), including EGLN3, VEGFA, SEMA6A, and ADM. Among these, VEGFA is a well-known HIF-2α target gene and EGLN3 positively regulates the mRNA expression of HIF-2α [24]. Therefore, we focused on SEMA6A, a protein known as the regulator of axonal guidance [25], because it might be another target gene of HIF-2α in ccRCC. We first compared the correlation of SEMA6A expression with two previously reported sets of hypoxia metagenes [26, 27]. We found that the expression of SEMA6A showed a positive correlation with these two groups of hypoxic metagenes in the TCGA-KIRC database, suggesting that SEMA6A may also have some expression characteristics of hypoxia metagenes (Fig. 2B, C and Supplementary Tables 5 and 6). Interestingly, the mRNA expression level of SEMA6A was also significantly correlated with both CCND1 and NDRG1 (Fig. 2D, E), two known HIF-2α target genes, suggesting that SEMA6A is associated with the HIF-2α signaling pathway in ccRCC. Moreover, both hypoxia and hypoxia mimics significantly induce the expression of SEMA6A in both A498 and 786-O cells (Fig. 2F–I and Supplementary Fig. 1A–D). Then, we utilized a HIF-2α antagonist PT2385 to treat A498 cells and found that PT2385 treatment reduced both the mRNA and protein levels of SEMA6A (Fig. 2J, K). Silencing the expression of HIF-2α by two different shRNAs consistently decreased the levels of NDRG1 and SEMA6A proteins (Fig. 2L). These data suggested that SEMA6A might be a putative HIF-2α downstream target gene in ccRCC. Notably, a classic hypoxia response element (HRE) was detected in the promoter region of the human SEMA6A gene (Fig. 2M), prompting us to test whether this site is essential for HIF-2α recognition and SEMA6A expression. In a luciferase assay, we found that HIF-2α significantly promotes the luciferase activity of the SEMA6A wild-type (WT) promoter (Fig. 2N). Moreover, ChIP analysis showed that HIF-2α binds to this putative HRE site in the SEMA6A promoter (Fig. 2O, P). These data indicated that SEMA6A is a direct target gene of HIF-2α in ccRCC.
A mRNA correlation between the mRNA level of HIF-2α and its 15 putative target genes in TCGA KIRC dataset was analyzed using the GEPIA platform (http://gepia2.cancer-pku.cn/). B The mRNA correlation between SEMA6A and 95 hypoxia metagenes in the TCGA KIRC database. C The mRNA correlation between SEMA6A and 25 hypoxia metagenes in the TCGA KIRC database. D mRNA correlation between CCND1 and SEMA6A in the TCGA KIRC database. E mRNA correlation between NDRG1 and SEMA6A in the TCGA KIRC database. F, G Relative mRNA and protein levels of SEMA6A in A498 cells under either normoxia or hypoxia conditions. ***p < 0.001. H, I Relative mRNA and protein levels of SEMA6A in A498 cells in response to CoCl2 treatment. **p < 0.01. J, K Relative mRNA and protein levels of SEMA6A in A498 cells in response to PT2385 treatment. **p < 0.01. L Protein levels of HIF-2α, SEMA6A, and NDRG1 in A498 cells transfected with control shRNA or shRNAs targeting HIF-2α. M Schematic representation of the human SEMA6A gene promoter and the putative HIF-2α binding site. TSS transcription start site. N Human SENA6A promoter contains a potential HIF-2α binding site, highlighted in red. Mutant sites are marked with a cross. FLAG-HIF-2α was co-transfected with the indicated plasmids into 293T cells for 36 h. Luciferase activity was then measured. ***p < 0.001. O ChIP-qPCR assay showed the enrichment of HIF-2α in the putative HIF-2α binding site of the SEMA6A promoter region. ***p < 0.001. P ChIP-PCR assay showed the occupation of HIF-2α in the putative HIF-2α binding site of the SEMA6A promoter region.
SEMA6A is overexpressed in ccRCC patients
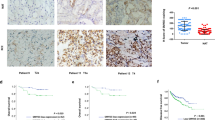
Next, we used the online cBioPortal database to investigate the expression and mutation of SEMA6A in ccRCC and found that the gene was mutated in 58/446 (13%) samples. The main genetic changes included gene amplification (50 cases, 11.2%) and mutations (8 cases, 1.79%) (Fig. 3A). The copy number and mutation types are shown in Fig. 3B. We also explored the mRNA expression of SEMA6A in ccRCC from TCGA using the online GEPIA database. Compared to 100 normal renal tissues, the mRNA expression of SEMA6A in 523 ccRCC tissues was significantly increased (Fig. 3C). Also, the mRNA expression of SEMA6A in different subtypes of ccRCC was significantly higher than that in the normal renal tissues (Fig. 3D). Moreover, searching the online UALCAN database revealed that the mRNA expression level of SEMA6A in different pathological stages and tumor grades of ccRCC was higher than that in normal renal tissues (Fig. 3E, F). Consistent with these observations, higher mRNA expression of SEMA6A in ccRCC than in normal and benign tissues was confirmed in an independent cohort of ccRCC patients (GSE68417) (Fig. 3G). We also found that SEMA6A is highly expressed in kidney cancer cell lines compared to most other cancer types exploring the Cancer Cell Line Encyclopedia (CCLE) database (Fig. 3H, I). We then collected 6 paired fresh human ccRCC samples and adjacent normal kidney tissues, and then performed qRT-PCR and western blot experiments. Our results showed that the mRNA and protein levels of SEMA6A were higher in most ccRCC samples when compared with adjacent normal tissues (Fig. 3J, K). Taken together, these data show that SEMA6A is overexpressed in ccRCC patients.
A Mutation of SEMA6A in ccRCC was obtained through the online cBioPortal database (http://www.cbioportal.org/). B Copy number and mutation types of the expression of SEMA6A in ccRCC. (TCGA, Firehose Legacy; 538 total samples; Genome map selection: Mutations, Putative copy-number alterations from GISTIC, mRNA expression z-scores relative to diploid samples (RNA-Seq V2 RSEM). C mRNA expression of SEMA6A in the TCGA database obtained through the GEPIA website (http://gepia.cancer-pku.cn). Red represents ccRCC tissue and black represents normal tissue. *p < 0.05. D Correlation between SEMA6A expression and ccRCC subtypes from TCGA database obtained through the GEPIA website (http://gepia.cancer-pku.cn). Red represents ccRCC tissue and black represents normal tissue. *p < 0.05. E Correlation between SEMA6A expression and the tumor stage of ccRCC patients was obtained through the UALCAN database (http://ualcan.path.uab.edu). F Correlation between SEMA6A expression and the tumor grade of ccRCC patients was obtained through the UALCAN database (http://ualcan.path.uab.edu). G Box plot of the difference in the expression level of SEMA6A in the GSE68417. Red represents normal tissues, green represents benign tissues, blue represents low-grade ccRCC tissues, and purple represents high-grade ccRCC tissues. **p < 0.01, ***p < 0.001. H mRNA expression of SEMA6A in cell lines from different cancer types was obtained through the CCLE website (https://sites.broadinstitute.org/ccle). I Bar graph of SEMA6A mRNA expression in several kidney cancer cell lines. J, K The mRNA and protein levels of SEMA6A in six paired ccRCC tissues and adjacent normal kidney tissues.
Targeting SEMA6A inhibits ccRCC proliferation
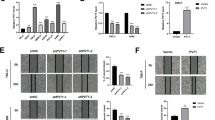
To evaluate the biological function of SEMA6A in ccRCC, we suppressed SEMA6A expression in ccRCC cells. A498 cells were treated by lentiviral infections with five independent shRNAs against SEMA6A. Although all shRNAs could inhibit the expression of SEMA6A, #1 and #3 showed the best inhibitory effect (Fig. 4A). Silencing SEMA6A by #1 and #3 shRNAs also significantly decreased the level of SEMA6A protein (Fig. 4B). The CCK-8 assay showed that silencing SEMA6A can significantly inhibit the proliferation of A498 cell line (Fig. 4C). Consistently, SEMA6A-depleted cells show decreased numbers of BrdU-incorporating cells (Fig. 4D). Moreover, SEMA6A knockdown cells showed enhanced Caspase3/7 activity, suggesting increased apoptosis in these cells (Fig. 4E). Downregulation of SEMA6A also impaired the ability of A498 cells to form colonies (Fig. 4F). In addition, wound healing assays showed that SEMA6A knockdown significantly reduces the migratory and invasion abilities of A498 cells (Fig. 4G, H). A similar phenotype of decreased malignancy was observed in 786-O and Caki-1 cells with SEMA6A knockdown (Supplementary Figs. 2 and 3). To analyze the consequences of SEMA6A silencing in vivo, we subcutaneously engrafted A498 cells expressing control and SEMA6A shRNAs into nude mice and monitored tumor growth. Consistent with the in vitro results, SEMA6A depletion decreased the subcutaneous tumor volume, size, and weight (Fig. 4I–L). Taken together, these findings demonstrated that SEMA6A depletion inhibits ccRCC cell growth both in vitro and in vivo.
A Relative expression levels of SEMA6A mRNA in A498 cells transfected with indicated shRNAs. *p < 0.05, **p < 0.01, ***p < 0.001. B Western blot analysis of SEMA6A protein in A498 cells stably expressing indicated shRNAs. C Proliferation ability of A498 cells from B was determined by CCK-8 assay. **p < 0.01. D Relative number of BrdU-incorporated cells in A498 cells from B. **p < 0.01. E Relative Caspase3/7 activity in A498 cells from B **p < 0.01. F Colonies of A498 cells from B. *p < 0.05, **p < 0.01. G Migration ability of A498 cells from B. **p < 0.01, ***p < 0.001. H Invasion ability of A498 cells from B. **p < 0.01, ***p < 0.001. I–L A498 cells stably expressing indicated shRNAs were injected subcutaneously into 6-week-old male nude mice (n = 5), and tumor growth curves were drawn weekly (I). After 4 weeks, mice were sacrificed by cervical dislocation. Then, the tumors were removed, photographed (J), weighed (K), and subjected to Western blot analysis to detect SEMA6A expression (L). **p < 0.01, ***p < 0.001.
SEMA6A activated the Wnt/β-catenin pathway via β-catenin stabilization
To further determine the mechanism of action of SEMA6A in ccRCC, we analyzed the transcriptional profiles of SEMA6A-regulated genes in ccRCC cells using the GSE79683 dataset. The dataset consists of the transcriptional gene expression profiles from SEMA6A WT and SEMA6A knockout (KO) mice. p < 0.05 and |logFC| > 1.2 were set as the cutoff criteria, and the DEGs were exhibited in both heatmap (Fig. 5A) and volcano map (Fig. 5B). Compared to WT control, 124 genes were upregulated, and 50 were downregulated in SEMA6A KO mice tissue (Fig. 5C and Supplementary Tables 7 and 8). The pathway enrichment analysis of these downregulated genes revealed that the Wnt signaling pathway is significantly suppressed in SEMA6A KO mice tissue (Fig. 5D). Then, we divided the sample data of KIRC in the TCGA database into high- and low-expression groups according to SEMA6A mRNA expression levels, and conducted GSEA. Compared to the low SEMA6A expression group, the Wnt/β-catenin signaling pathway was significantly enriched in the high SEMA6A expression (Fig. 5E). In agreement, the KIRC dataset from TCGA revealed that the mRNA level of SEMA6A was positively correlated with both FZD1 and Myc, two well-known downstream target genes of the Wnt/β-catenin signaling pathway (Fig. 5F, G). These data prompted us to explore whether SEMA6A could regulate the Wnt/β-catenin signaling pathway. Thus, we conducted a TOPFlash/FOPFlash assay to assess whether SEMA6A affects the transcriptional activity of β-catenin/TCF complex. We also found that the ectopic expression of SEMA6A significantly increases TOPflash activity, but not FOPflash activity, compared to control vector expression (Fig. 5H). Conversely, silencing SEMA6A decreased the TOPflash activity without affecting the FOPflash activity (Fig. 5I). The decreased TOPflash activity could be attributed to the impaired transcriptional activity of the β-catenin/TCF complex, as observed by the declined mRNA expression of two β-catenin/TCF target genes: CCND1 and C-Myc (Fig. 5J). Surprisingly, the mRNA level of β-catenin was unchanged in SEMA6A-depleted cells (Fig. 5J), while the immunoblotting results showed that inhibition of SEMA6A expression decreased the protein levels of both β-catenin and CCND1 (Fig. 5K), suggesting that SEMA6A regulates the stability of β-catenin. In agreement, both the proteasome inhibitor MG132 and the pan-cullin inhibitor MLN4924 prevented the decline of β-catenin protein levels in SEMA6A-depleted cells (Fig. 5L), indicating that this decline was caused by enhanced proteolysis. Furthermore, depletion of SEMA6A increased the ubiquitination of β-catenin protein (Fig. 5M). Together, these results suggested that SEMA6A activates the Wnt/β-catenin signaling pathway by promoting the stability of β-catenin protein. Typically, the administration of ICG-001, a small molecule compound that selectively antagonizes β-catenin/TCF-mediated transcriptional activity, significantly reversed SEMA6A-induced TOPflash activation (Fig. 5N). In addition, ICG-001 administration also significantly reversed SEMA6A-induced A498 cell proliferation (Supplementary Fig. 6A), Caspase3/7 activation (Supplementary Fig. 6B) and colony formation in vitro (Supplementary Fig. 6C), suggesting SEMA6A-regualted β-catenin activation is related to ccRCC progression. Consistent with previous findings, HIF-2α depletion significantly reduced the transcriptional activity of β-catenin/TCF complex, which could be partially restored by SEMA6A overexpression (Fig. 5O), indicating that SEMA6A mediates HIF-2α-induced activation of the Wnt/β-catenin signaling pathway. In conclusion, the current results indicated that SEMA6A activates the Wnt/β-catenin signaling pathway in ccRCC.
A Heatmap showing DEGs expression regulated by SEMA6A in ccRCC cells. B Volcano plot showing differential genes regulated by SEMA6A in GSE149005. C Number of genes regulated by SEMA6A includes 124 up- and 50 downregulated genes. D Correlative signaling pathways enriched in genes downregulated by SEMA6A. E GSEA plot of SEMA6A enrichment in HALLMARK_WNT_BETA_CATENIN_SIGNALING signaling pathway. F mRNA correlation of C-Myc and SEMA6A in the TCGA KIRC database. G mRNA correlation of FZD1 and SEMA6A in the TCGA KIRC database. H TOPFlash or FOPFlash were co-transfected with empty vector plasmid or SEMA6A into 293T cells, and the luciferase activity was measured after 36 h. **p < 0.01. I TOPFlash or FOPFlash were co-transfected with indicated shRNAs into 293T cells, and the luciferase activity was measured after 36 h. **p < 0.01, ***p < 0.001. J Relative mRNA levels of Cyclin D1, C-Myc, and CTNNB1 in cells stably expressing the indicated shRNAs. ***p < 0.001. K Western blot analysis of lysates from cells in J. L Western blot analysis of A498 cells with or without SEMA6A depletion, followed by treatment with DMSO, MG132, and MLN4924 for 4 h, respectively. M A498 cells with or without SEMA6A depletion were treated with MG132 for 4 h. Ubiquitinated proteins were enriched using tandem ubiquitin-binding entity 1 (TUBE1) resin and subjected to immunoblot analysis using the indicated antibodies. N TOPFlash or FOPFlash was co-transfected with or without SEMA6A into A498 cells and then treated with or without ICG-001. The luciferase activity was measured after 36 h. ***p < 0.001. O TOPFlash or FOPFlash was co-transfected with indicated shRNAs into A498 cells, and then luciferase activity was measured after 36 h. *p < 0.05, **p < 0.01.
SEMA6A enhances the binding between SEC62 and β-catenin
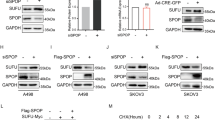
To further investigate how SEMA6A regulates the stability of β-catenin protein, we searched for SEMA6A-interacting protein. A total of 82 putative SEMA6A-interacting proteins were identified using the BioGRID dataset (Supplementary Fig. 4), among which SEC62 formed a protein complex with β-catenin and inhibited the degradation of β-catenin protein [28]. To evaluate whether SEC62 is a binding partner of SEMA6A, we carried out structure-based computational analyses to show the interaction between SEMA6A and SEC62. Since the crystal structures for these two proteins are currently not available, we used the I-TASSER webserver for structural modeling. The protein-protein docking was performed using the Cluspro online server, a rigid protein docking algorithm based on a fast Fourier transform. Ligplot software was used to analyze the binding force between the two proteins, and Pymol software was used for 3D protein-binding conformation imaging. The modeling results revealed that many hydrogen bonding forces were formed between both proteins (Fig. 6A and Supplementary Fig. 5), suggesting that SEMA6A might form a tight protein complex with SEC62. Subsequently, we purified endogenous SEMA6A protein from A498 cell lysate and detected the presence of SEC62 protein (Fig. 6B). Moreover, the reciprocal interaction was validated by IP-Western blot analysis using an anti-SEC62 antibody (Fig. 6C). To determine whether this interaction could be reconstituted in vitro, we performed a glutathione S-transferase (GST) pull-down assay using GST-SEC62 and GST as baits and found that GST-tagged SEC62, but not GST alone, efficiently pulled down SEMA6A protein, suggesting a direct interaction between SEC62 and SEMA6A (Fig. 6D). Reportedly, SEC62 has competitively disrupted the interaction between β-catenin and APC to inhibit the β-catenin destruction complex assembly, driving us to assess whether SEMA6A affected the interaction between SEC62 and β-catenin. To test this hypothesis, we purified the SEC62 protein complex from A498 cells with or without SEMA6A depletion and then detected the presence of β-catenin by immunoblotting. Consistent with previous findings, SEC62 was bound to β-catenin in A498 cells. However, in the absence of SEMA6A, the interaction between SEC62 and β-catenin was impaired (Fig. 6E). On the contrary, overexpression of exogenous SEMA6A significantly enhanced this interaction (Fig. 6F). Together, these results indicated that SEMA6A promoted the interaction between SEC62 and β-catenin to prevent the degradation of β-catenin protein, thereby indicating that SEC62 might be critical for SEMA6A-induced β-catenin activation and biological functions. Luciferase assay showed that SEC62 knockdown decreased the transcriptional ability of β-catenin induced by SEMA6A (Fig. 6G). Furthermore, SEC62 depletion significantly reversed SEMA6A-induced A498 cell proliferation (Fig. 6H), BrdU incorporation (Fig. 6I), Caspase3/7 activation (Fig. 6J), colony formation (Fig. 6K) and invasion in vitro (Fig. 6L). Importantly, SEC62 depletion also significantly reversed SEMA6A-induced A498 cell proliferation in nude mice in vivo (Fig. 6M–O). Together, our results demonstrated that HIF-2α-induced SEMA6A expression promoted ccRCC progression through SEC62-dependent β-catenin stabilization and activation (Fig. 7).
A Structural model of the protein-protein interaction between SEMA6A (Cyan) and SEC62 (Green). The yellow dotted lines represent hydrogen bonds. B Cell lysates from A498 cells were immunoprecipitated with IgG or anti-SEMA6A antibody and subjected to immunoblot analysis with indicated antibodies. C Cell lysates from A498 cells were immunoprecipitated with IgG or anti-SEC62 antibody and subjected to immunoblot with indicated antibodies. D GST Sepharose beads coated with E. coli produced GST or GST-SEC62 were incubated with purified FLAG-SEMA6A. The beads were washed, and the bound proteins were analyzed by Western blotting with indicated antibodies. E Cell lysates from A498 cells stable expressing con-shRNA or SEMA6A-shRNA were immunoprecipitated with SEC62 antibody and then subjected to immunoblotting with indicated antibodies. F Cell lysates from A498 cells stable expressing vector control or FLAG-SEMA6A were immunoprecipitated with SEC62 antibody and subjected to immunoblotting with indicated antibodies. G A498 cells were co-transfected with TOPflash plasmid and the indicated shRNAs for 36 h. Then, the luciferase activity was measured. *p < 0.05. H Proliferation ability of A498 cells stable expressing the indicated shRNAs was determined by CCK-8 assay. **p < 0.01. I BrdU incorporation in A498 cells from H *p < 0.05. J Relative Caspase3/7 activity in A498 cells from H **p < 0.01. K Images of colony formation of A498 cells from H. L Migration ability of A498 cells from H. *p < 0.05. M–O A498 cells stably expressing indicated shRNAs were injected subcutaneously into 6-week-old male nude mice (n = 5), and tumor growth curves were drawn weekly (M). After 4 weeks, mice were sacrificed by cervical dislocation. The tumors were removed, photographed (N), and weighed (O). ***p < 0.001.
In this working model, we showed that hypoxia-dependent HIF-2α protein stabilization and activation-induced SEMA6A expression in ccRCC, which in turn bound to SEC62 protein to inhibit the ubiquitination and degradation of β-catenin, thereby promoting the expression of multiple tumor-promoting genes and tumor progression.
Discussion
In the present study, we screened the substrates of the VHL-HIF-2α axis based on the available transcriptome data. Next, we hypothesized that the mRNA levels of potential substrates of the VHL-HIF-2α axis are decreased in VHL-overexpressing cells and increased in HIF-2α-deficient cells. Moreover, the mRNA levels of these potential substrates are positively correlated with the activity of the HIF-2α signaling pathway. According to these screening criteria, 4 genes were identified, including SEMA6A and 3 known canonical target genes of HIF-2α. SEMA6A is a multifunctional protein that participates in various cellular processes, including vascular development and angiogenesis [29]. SEMA6A can form a complex with plexins to regulate the actin cytoskeleton, motility, and cell proliferation and is involved in cancer development [30]. Next, we showed that SEMA6A was elevated under physical hypoxia or pharmacologically-simulated hypoxia conditions, whereas genetic ablation or pharmacological inhibition of HIF-2α significantly decreased SEMA6A expression in ccRCC cells, suggesting that hypoxia-induced SEMA6A expression requires the transcriptional activity of HIF-2α. Typically, ChIP and luciferase assays revealed that HIF-2α directly activates SEMA6A transcription in hypoxic ccRCC cells. The pooled TCGA and GEO data showed that SEMA6A mRNA was significantly increased in ccRCC compared to normal tissues and benign kidney tissues, suggesting that SEMA6A is involved in ccRCC progression. Consistent with this finding, depletion of SEMA6A significantly restrained the proliferation and migration ability of ccRCC in vitro and limited the in vivo growth of cancer in nude mice. Together, these results highlighted an oncogenic role of SEMA6A in ccRCC.
The transcriptome data analysis of SEMA6A KO mice tissue showed that the Wnt/β-catenin pathway is significantly downregulated in case of SEMA6A depletion. TCGA data analysis demonstrated that the mRNA expression of SEMA6A is closely correlated to the activation of Wnt/β-catenin pathway in ccRCC. The Wnt/β-catenin pathway is implicated in numerous signaling pathways, including embryogenesis, cell proliferation, migration and invasion, apoptosis, and organogenesis [31]. The abnormal activation of the signaling pathway induces tumorigenesis [32]. The current results showed that depletion of SEMA6A decreases the Wnt/β-catenin activity, as shown by impaired TOPflash luciferase activity and reduced Wnt/β-catenin downstream target gene expression. Conversely, the overexpression of SEMA6A activated the Wnt/β-catenin pathway, which could be reversed by pharmacological β-catenin inhibition. Also, β-catenin is a multifunctional protein encoded by the CTNNB1 gene that mediates signal transduction and exerts a core role in promoting tumor proliferation and metastasis in various malignancies [33]. Although SEMA6A knockdown inhibited the activity of the Wnt/β-catenin pathway, the mRNA level of β-catenin was not affected. Interestingly, the protein level of β-catenin is downregulated in SEMA6A-depleted cells, suggesting that SEMA6A regulates β-catenin at the posttranscriptional level. Consistent with this phenomenon, we showed that decreased β-catenin protein level in SEMA6A-depleted cells was caused by enhanced ubiquitination and degradation. The turnover of β-catenin is critical for the inactivation of the Wnt/β-catenin signaling pathway [34]. Reportedly, SEC62, a component of the protein translocation tool on the endoplasmic reticulum membrane, competitively disrupted the interaction between β-catenin and APC to prevent β-catenin destruction [28]. Interestingly, SEC62 is found to be a putative interaction protein of SEMA6A. Several technical methods at different levels confirmed the interaction between SEMA6A and SEC62 and found a critical role of SEMA6A in SEC62-mediated β-catenin stabilization and activation. Thus, our finding elucidated the molecular mechanism through which SEMA6A stabilizes β-catenin. We also found that depletion of SEC62 significantly reverses SEMA6A-induced malignant phenotypes in ccRCC both in vitro and in vivo, indicating a critical role of the SEMA6A-SEC62 axis in maintaining the malignant phenotypes of ccRCC.
Collectively, SEMA6A is identified as a novel target gene of VHL-HIF-2α with a cancer-promoting role in ccRCC. SEMA6A activates the Wnt/β-catenin signaling pathway by inhibiting the β-catenin ubiquitination and degradation, suggesting that SEMA6A acts as a potential therapeutic target in ccRCCs.
Data availability
All data generated or analyzed in this study are included in this paper and can be obtained from the corresponding author according to formal requirement.
References
Li RG, Deng H, Liu XH, Chen ZY, Wan SS, Wang L. Histone methyltransferase G9a promotes the development of renal cancer through epigenetic silencing of tumor suppressor gene SPINK5. Oxid Med Cell Longev. 2021;2021:6650781.
Solarek W, Czarnecka AM, Escudier B, Bielecka ZF, Lian F, Szczylik C. Insulin and IGFs in renal cancer risk and progression. Endocr Relat Cancer. 2015;22:R253–64.
Barth DA, Slaby O, Klec C, Juracek J, Drula R, Calin GA, et al. Current concepts of non-coding RNAs in the pathogenesis of non-clear cell renal cell carcinoma. Cancers. 2019;11:1580.
Liu J, Zhang W, Lin K, Ma W, Li W, Yao X. Immunological perspective on the malignant progression of renal clear cell carcinoma. Ann Transl Med. 2021;9:1544.
Low G, Huang G, Fu W, Moloo Z, Girgis S. Review of renal cell carcinoma and its common subtypes in radiology. World J Radio. 2016;8:484–500.
Lin E, Liu X, Liu Y, Zhang Z, Xie L, Tian K, et al. Roles of the dynamic tumor immune microenvironment in the individualized treatment of advanced clear cell renal cell carcinoma. Front Immunol. 2021;12:653358.
Ingels A, Campi R, Capitanio U, Amparore D, Bertolo R, Carbonara U, et al. Complementary roles of surgery and systemic treatment in clear cell renal cell carcinoma. Nat Rev Urol. 2022;19:391–418.
Pan X, Huang B, Ma Q, Ren J, Liu Y, Wang C, et al. Circular RNA circ-TNPO3 inhibits clear cell renal cell carcinoma metastasis by binding to IGF2BP2 and destabilizing SERPINH1 mRNA. Clin Transl Med. 2022;12:e994.
Majer W, Kluzek K, Bluyssen H, Wesoły J. Potential approaches and recent advances in biomarker discovery in clear-cell renal cell carcinoma. J Cancer. 2015;6:1105–13.
Guo K, Wei Y, Wang Z, Zhang X, Zhang X, Liu X, et al. Deubiquitylase OTUD6B stabilizes the mutated pVHL and suppresses cell migration in clear cell renal cell carcinoma. Cell Death Dis. 2022;13:97.
Xie H, Zhou J, Liu X, Xu Y, Hepperla AJ, Simon JM, et al. USP13 promotes deubiquitination of ZHX2 and tumorigenesis in kidney cancer. Proc Natl Acad Sci USA. 2022;119:e2119854119.
Cardote TAF, Gadd MS, Ciulli A. Crystal structure of the Cul2-Rbx1-EloBC-VHL ubiquitin ligase complex. Structure. 2017;25:901–11.e3.
Briston T, Stephen JM, Thomas LW, Esposito C, Chung YL, Syafruddin SE, et al. VHL-mediated regulation of CHCHD4 and mitochondrial function. Front Oncol. 2018;8:388.
Dong X, Su X, Yu J, Liu J, Shi X, Pan Q, et al. Homology modeling and molecular dynamics simulation of the HIF2α degradation-related HIF2α-VHL complex. J Mol Graph Model. 2017;71:116–23.
Yu T, Tang B, Sun X. Development of inhibitors targeting hypoxia-inducible factor 1 and 2 for cancer therapy. Yonsei Med J. 2017;58:489–96.
Martínez-Sáez O, Gajate Borau P, Alonso-Gordoa T, Molina-Cerrillo J, Grande E. Targeting HIF-2 α in clear cell renal cell carcinoma: a promising therapeutic strategy. Crit Rev Oncol Hematol. 2017;111:117–23.
Dell’Atti L, Bianchi N, Aguiari G. New therapeutic interventions for kidney carcinoma: looking to the future. Cancers. 2022;14:3616.
Wang R, Ma Y, Zhan S, Zhang G, Cao L, Zhang X, et al. B7-H3 promotes colorectal cancer angiogenesis through activating the NF-κB pathway to induce VEGFA expression. Cell Death Dis. 2020;11:55.
Fearnley GW, Latham AM, Hollstein M, Odell AF, Ponnambalam S. ATF-2 and Tpl2 regulation of endothelial cell cycle progression and apoptosis. Cell Signal. 2020;66:109481.
Sun J, Tang Q, Gao Y, Zhang W, Zhao Z, Yang F, et al. VHL mutation-mediated SALL4 overexpression promotes tumorigenesis and vascularization of clear cell renal cell carcinoma via Akt/GSK-3β signaling. J Exp Clin Cancer Res. 2020;39:104.
Cho H, Kaelin WG. Targeting HIF2 in clear cell renal cell carcinoma. Cold Spring Harb Symp Quant Biol. 2016;81:113–21.
Choi WSW, Boland J, Lin J. Hypoxia-inducible factor-2α as a novel target in renal cell carcinoma. J Kidney Cancer VHL. 2021;8:1–7.
Ji J, Shen J, Xu Y, Xie M, Qian Q, Qiu T, et al. FBXO2 targets glycosylated SUN2 for ubiquitination and degradation to promote ovarian cancer development. Cell Death Dis. 2022;13:442.
Miikkulainen P, Högel H, Seyednasrollah F, Rantanen K, Elo LL, Jaakkola PM. Hypoxia-inducible factor (HIF)-prolyl hydroxylase 3 (PHD3) maintains high HIF2A mRNA levels in clear cell renal cell carcinoma. J Biol Chem. 2019;294:3760–71.
Chen LH, Liao CY, Lai LC, Tsai MH, Chuang EY. Semaphorin 6A attenuates the migration capability of lung cancer cells via the NRF2/HMOX1 Axis. Sci Rep. 2019;9:13302.
Eustace A, Mani N, Span PN, Irlam JJ, Taylor J, Betts GN, et al. A 26-gene hypoxia signature predicts benefit from hypoxia-modifying therapy in laryngeal cancer but not bladder cancer. Clin Cancer Res. 2013;19:4879–88.
Winter SC, Buffa FM, Silva P, Miller C, Valentine HR, Turley H, et al. Relation of a hypoxia metagene derived from head and neck cancer to prognosis of multiple cancers. Cancer Res. 2007;67:3441–9.
Liu X, Su K, Sun X, Jiang Y, Wang L, Hu C, et al. Sec62 promotes stemness and chemoresistance of human colorectal cancer through activating Wnt/β-catenin pathway. J Exp Clin Cancer Res. 2021;40:132.
Zhao J, Tang H, Zhao H, Che W, Zhang L, Liang P. SEMA6A is a prognostic biomarker in glioblastoma. Tumour Biol. 2015;36:8333–40.
Rozbesky D, Verhagen MG, Karia D, Nagy GN, Alvarez L, Robinson RA, et al. Structural basis of semaphorin-plexin cis interaction. EMBO J. 2020;39:e102926.
Vallée A. Arterial stiffness and the canonical WNT/β-catenin pathway. Curr Hypertens Rep. 2022;24:499–507.
Wang Z, Zhao T, Zhang S, Wang J, Chen Y, Zhao H, et al. The Wnt signaling pathway in tumorigenesis, pharmacological targets, and drug development for cancer therapy. Biomark Res. 2021;9:68.
Zhou Y, Xu J, Luo H, Meng X, Chen M, Zhu D. Wnt signaling pathway in cancer immunotherapy. Cancer Lett. 2022;525:84–96.
Shang S, Hua F, Hu ZW. The regulation of β-catenin activity and function in cancer: therapeutic opportunities. Oncotarget. 2017;8:33972–89.
Funding
This study was supported by the National Natural Science Foundation of China (82273167, 82104174 and 81602626), Jiangsu Province Basic Research Program Natural Science Foundation (Outstanding Youth Fund Project, BK20220063), the Key Program of Basic Science (Natural Science) of Jiangsu Province (22KJA350001), “Huaguo Mountain Talent Plan” of Lianyungang City (Innovative Talents Liu Bin), Qing Lan Project of Jiangsu Universities (Outstanding young backbone teachers, Ji Jing), the Natural Science Foundation of Jiangsu Higher Education Institutions of China (No. 20KJB350008), Scientific Research Foundation for Returned Scholars of Tongji Hospital (project 2022hgry021), Jiangsu Key Laboratory of Marine Pharmaceutical Compound Screening (HY202103), Priority Academic Program Development of Jiangsu Higher Education Institutions.
Author information
Authors and Affiliations
Contributions
BL, WL and SL designed the study, supervised the process, and wrote the paper. JJ, YX, MX and XH were responsible for cellular and animal experiments. DR, TQ, WL, ZC, WS, ZZ, XW, WW, JM, QQ, AJ and ZS were responsible for bioinformation analysis. XM, JQ, YD, TG and JY analyzed the data. All authors collaborated to interpret results and develop the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by Professor Stephen Tait
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ji, J., Xu, Y., Xie, M. et al. VHL-HIF-2α axis-induced SEMA6A upregulation stabilized β-catenin to drive clear cell renal cell carcinoma progression. Cell Death Dis 14, 83 (2023). https://doi.org/10.1038/s41419-023-05588-4
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41419-023-05588-4