Abstract
The new coronavirus SARS-CoV-2 is the causative agent of the COVID-19 pandemic, which so far has caused over 6 million deaths in 2 years, despite new vaccines and antiviral medications. Drug repurposing, an approach for the potential application of existing pharmaceutical products to new therapeutic indications, could be an effective strategy to obtain quick answers to medical emergencies. Following a virtual screening campaign on the most relevant viral proteins, we identified the drug raloxifene, a known Selective Estrogen Receptor Modulator (SERM), as a new potential agent to treat mild-to-moderate COVID-19 patients. In this paper we report a comprehensive pharmacological characterization of raloxifene in relevant in vitro models of COVID-19, specifically in Vero E6 and Calu-3 cell lines infected with SARS-CoV-2. A large panel of the most common SARS-CoV-2 variants isolated in Europe, United Kingdom, Brazil, South Africa and India was tested to demonstrate the drug’s ability in contrasting the viral cytopathic effect (CPE). Literature data support a beneficial effect by raloxifene against the viral infection due to its ability to interact with viral proteins and activate protective estrogen receptor-mediated mechanisms in the host cells. Mechanistic studies here reported confirm the significant affinity of raloxifene for the Spike protein, as predicted by in silico studies, and show that the drug treatment does not directly affect Spike/ACE2 interaction or viral internalization in infected cell lines. Interestingly, raloxifene can counteract Spike-mediated ADAM17 activation in human pulmonary cells, thus providing new insights on its mechanism of action. A clinical study in mild to moderate COVID-19 patients (NCT05172050) has been recently completed. Our contribution to evaluate raloxifene results on SARS-CoV-2 variants, and the interpretation of the mechanisms of action will be key elements to better understand the trial results, and to design new clinical studies aiming to evaluate the potential development of raloxifene in this indication.
Similar content being viewed by others
Introduction
Coronaviruses are the causative agents of multiple respiratory and intestinal infection in humans and animals [1, 2], and SARS-CoV-2 is able to cause severe acute respiratory illness, multi-organ failure up to the death [3, 4]. In addition, prolonged prothrombin times, mild thrombocytopenia and elevated D-dimer values were observed together with lymphocytopenia, also suggested as a predictor of prognosis [5, 6].
As of March 24th, 2022, SARS-CoV-2 infection led to more than 6 million deaths worldwide (https://covid19.who.int/) and 7.5% of COVID-19 cases reported by EU/EEA countries to the European Surveillance System (TESSy) were hospitalized.
The current COVID-19 pandemic is believed to have initially developed in an animal host: SARS-CoV-2 shares high genome similarity with betacoronaviruses isolated from a bat (RaTG13) and from a Malayan pangolin. Therefore, SARS-CoV-2 has been hypothesized to have originated in bats and gone through multiple recombination events while migrating through other mammals [7]. It is increasingly evident that animals are an important epidemiological part of this pandemic in transmission and appearance of viral variants [8, 9] well as in the new mechanisms of infection that gradually emerge [10]. To date, notwithstanding the advent of antivirals, vaccine programs [11] and social distancing interventions, it is believed that the virus will most likely become endemic [12]. In addition, the emerging of SARS-CoV-2 variants raises great concern for vaccine efficacy, reinfection events, increased transmissibility and disease severity. As the virus started to spread, a mutated Spike SARS-CoV-2 variant (D614G) emerged which was associated with increased infectivity, becoming the predominant variant in Europe and worldwide without any increase in disease severity [13, 14]. During the last year, other variants were defined as “variants of concern” (VOC). Some of them are considered of high clinical relevance like B.1.1.7 (UK, alpha), B.1.351 (South African, beta), B.1.1.28 (Brazilian P.1, gamma), B.1.427 and B.1.429 (Californian, epsilon), characterized by increased transmissibility, immune evasion and higher virulence [15,16,17,18]. In May 2021, the B.1.617.2 (Indian, Delta) was added to the WHO list of VOCs and described as more transmissible (up to 50% than the alpha variant), able to escape adaptive immunity, and to cause sharp rises in infections in many countries, including those with relatively high vaccination coverage [19, 20]. As of November 26th 2021, the newly emerged B.1.1.529 variant, firstly identified in South-Africa, was classified as “omicron” VOC (https://www.who.int/news/item/28-11-2021-update-on-omicron), characterized by higher transmissibility than other VOCs but milder symptoms, mainly of the upper respiratory tract [21]. The growing relevance of new VOCs stimulated further investigations and new impetus to develop broad-spectrum drugs or vaccines for long-term prevention, treatment and control of COVID-19. The virus entry machinery has been considered a privileged target to identify potential therapeutic targets and in this context several preclinical studies and clinical trials are ongoing [22]. Recent evidence shows that Nuclear Receptors (NRs), and in particular sex hormone receptors like estrogen (ER) and androgen (AR) receptors, could be involved in determining the outcome of COVID-19 infection because able to regulate the viral entry protein expression and activity [23,24,25]. A protective effect of estrogens in the progression of COVID-19 infection has been associated with their role in the regulation of innate and adaptive immune responses, as well as in the control of the cytokine storm [26,27,28,29,30,31,32], whereas the activation of androgen receptors seems to correlate with the worse COVID-19 clinical outcome observed in men [25, 33,34,35]. Recently, in the context of the H2020 project EXSCALATE4CoV (E4C), an extensive virtual screening campaign on SARS-CoV-2 target proteins based on the EXSCALATE platform, a tool for drug repurposing [36,37,38], allowed to identify several molecules active against SARS-CoV-2. Raloxifene, a well-known SERM [39,40,41,42], was selected following this approach. Besides a direct inhibition of viral protein functions, several papers propose the use of raloxifene and other SERMs as potential therapeutic strategy to treat paucisymptomatic COVID-19 due to the multiple links between ER modulation and host response to viral infections [43] that suggest beneficial effects both in controlling viral replication in early post-infection phase, and in preventing/attenuating the cytokine storm and inflammation in more severe COVID-19 [44]. Further, a large body of literature points to a protective role of the ER-signaling as relevant in the observed gender sensitivity, as additional boost to develop SERMs, specifically raloxifene, in this indication [43]. Raloxifene is a drug registered for the treatment and prevention of osteoporosis and risk of invasive breast cancer in postmenopausal women [45], ER agonist in the bone, liver and cardiovascular system, and ER antagonist in reproductive tissues [46]. This tissue specificity allows for its use in postmenopausal osteoporosis and prevention of breast cancer without increasing the risk of endometrial cancer [47, 48]. Raloxifene was also studied in men for treatment of diseases as schizophrenia, prostate cancer and osteoporosis [49, 50]. Interestingly, raloxifene effect was characterized against Ebola [51, 52], Hepatitis C [53, 54], Hepatitis B [55], Zika [56], Influenza A viruses [57], and as adjuvant antiviral treatment of chronic Hepatitis C in women [58]. Raloxifene in vitro activity on SARS-CoV-2 was previously reported [59], and the drug was observed to reduce infectivity in a dose-dependent manner (IC50 value of 7.1 μM). Here we report a full characterization of the antiviral activity of raloxifene on SARS-CoV-2 in two cellular contexts, Vero E6 and Calu-3 cell lines, testing the efficacy of the treatment on all the most relevant VOCs, and clearly showing the independence of raloxifene activity from viral variant specificity. Furthermore, mechanistic studies shed light on its mechanism of action supporting the concept that a pleiotropic mechanism may account for its inhibition of viral replication. In particular, the ability of raloxifene to modulate the Spike-induced ADAM17 expression may account for a potential protection of pulmonary damage in consideration of the key role of ADAM-17 in virus entry and replication, and pathophysiological consequences in the lungs due to its excessive activation in SARS-CoV-2 infection [60].
Taken together, this evidence supports the concept that raloxifene may exert a protective effect on SARS-CoV-2 infected patients through a pleiotropic mechanism of action. A Phase 2/3 clinical trial (NCT05172050) in outpatients with mild to moderate COVID-19 has been recently completed (Nicastri E. and et al., 2022 eClinicalMedicine; accepted for publication), and the results will hopefully give additional insights on the therapeutic potential of raloxifene.
Materials and methods
All information is reported in Supplementary Information.
Results
In vitro effects of raloxifene on metabolism and SARS-CoV-2 infection in different cell lines
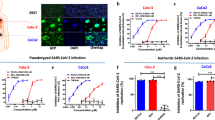
Vero E6 cells were cultured for 48 h in the absence or presence of different raloxifene concentrations. Raloxifene-treated cells showed a normal surface-adherent phenotype until the concentration of 15μM (Fig. 1A). A drug-dependent cytopathic effect (CPE) was evident at 20μM, involving the entire monolayer at 25μM and 30μM. At the same time, raloxifene showed a slight effect on the extent of cellular ATP accumulation at a concentration of 1.25 μM to 15 μM (87% and 70%, respectively). At higher doses, raloxifene showed a dose-dependent effect on ATP accumulation, reaching 56%, 35% and 0.6% at 20 µM, 25 µM and 30 µM, respectively (Fig. 1B). The half-maximal cytotoxic concentration (CC50) in Vero E6 cells was determined to be 18.4 µM.
Vero E6 cells were cultured for 48 h in the absence or in the presence of raloxifene at different concentrations. A 10× bright‐field images of Vero E6 cells after incubation for 48 h at 37 °C with the indicated raloxifene concentrations. B CellTiter-Glo was used to measure the antimetabolic effect of raloxifene. C-F Vero E6 cells were infected with SARS-CoV-2 and cultured in the absence or in the presence of different doses of raloxifene. C Viral yield in cell supernatants was quantitated by qRT-PCR. D Viral titer in cell supernatants was evaluated by plaque assay and plotted as percentage of plaque reduction compared to SARS-CoV-2. E Quantitation of SARS-CoV-2 genomes at the intracellular level by qRT-PCR. F NP expression in infected cells was analyzed by western blot (left panel). Densitometric analysis of western blot is shown in the right panel. Graph represents the percentage of NP expression. All the experiments were performed at least in three independent replicates and pictures shown are representative. Data are presented as the mean ± standard error of the mean *P < 0.05; **P < 0.01; ****P < 0.0001.
Further, an apoptosis assay by flow cytometry was conducted at different doses of raloxifene (1.25, 2.5, 5, 10, 15, 20, 25 and 30 µM) in VERO E6 cells. As shown in Fig. S3 (Supplementary Information), raloxifene did not induce apoptosis at concentrations ranging from 1.25 to 20 μM, while it triggered a significant induction of apoptosis at 25 and 30 μM (36% and 45%, respectively).
Preliminary experiments were conducted to evaluate the effect of raloxifene in the viral pre-entry, entry or post-entry phases. Raloxifene displayed antiviral activity in the post-entry phase only (Fig. S1). Next, Vero E6 cells were infected with SARS-CoV-2 (B.1 lineage) at a multiplicity of infection (MOI) of 0.05, and 1 h p.i. (postinfection) cultured in the absence or presence of different drug concentrations. Raloxifene efficiently inhibited viral replication. Viral genome copy numbers evaluated by qRT-PCR at 48 h p.i., showed a significant reduction of the virus yield already at a concentration of 2.5μM (2.9-fold reduction), with a maximal reduction at 15 µM (1400-fold reduction) (Fig. 1C). Raloxifene also displayed a dose-dependent inhibition of viral replication, as determined by infectious viral titers, exhibiting a 70% reduction of viral titer at a concentration of 5 µM, with 94% to 100% inhibition at 10 µM and 15 µM, respectively (Fig. 1D). Its efficacy was then confirmed at intracellular level; quantification of viral RNA in SARS-CoV-2-infected cells showed a significant reduction of intracellular viral genome copy number at 10 µM, and a 99-fold reduction at 15 µM (Fig. 1E). Accordingly, western blot (WB) analysis showed a viral dose-dependent inhibition upon raloxifene treatment with 43% reduction of NP viral protein expression at a concentration of 5 µM, with 65% and 97% reduction at 10 µM and 15 µM, respectively (Fig. 1F and S2). The half-maximal inhibitory concentration (IC50) was calculated to be 3.3 µM, while the selectivity index (SI) was calculated to be 5.6.
We then tested raloxifene on Calu-3 cells. The CC50 value was found to be 24.4 µM (Fig. 2A). Next, Calu-3 cells were infected as described above, and supernatants collected at 48 h p.i. and tested by qRT-PCR. The treatment significantly reduced the virus yield (Fig. 2B). Raloxifene displayed a dose-dependent inhibition of viral replication, as determined by infectious viral titers, exhibiting a 67% reduction of viral titer at a concentration as low as 10 µM, with 96% and 98% inhibition at drug concentrations of 15 µM and 25 µM, respectively. The efficacy of the treatment was confirmed at intracellular level by qRT-PCR and WB on NP (Fig. 2C-E and S3). The IC50 was calculated and found to be 9 µM, while SI was calculated to be 2.7.
Calu-3 cells were cultured for 48 h in absence or in the presence of raloxifene at different concentrations. A CellTiter-Glo was used to measure the antimetabolic effect of raloxifene. B–E Cells were infected with SARS-CoV-2 and cultured in the absence or in the presence of different doses of raloxifene. B Viral yield in cell supernatants was quantitated by qRT-PCR. C Viral titer in cell supernatants was evaluated by plaque assay and plotted as percentage of plaque reduction compared to SARS-CoV-2. D Quantitation of SARS-CoV-2 genomes at the intracellular level by qRT-PCR. E Nucleocapsid (NP) protein expression in infected cells was analyzed by western blot (left panel). Densitometric analyses of western blot results are shown. Graph represents the percentage of NP protein expression. All the experiments were performed at least in three independent replicates and pictures shown are representative. Data are presented as the mean + standard error of the mean *P < 0.05; **P < 0.01; ****P < 0.0001.
Raloxifene exerts antiviral activity on SARS-CoV-2 variants
We then performed a systematic study of the antiviral efficacy of raloxifene on the most common variants in Vero E6 cells (Fig. 3). Different viral strains were used: the wild type (WT) isolated in January 2020 from Chinese patient, two different isolates for the D614G spike variants (named GV and D614G), and the main VOCs isolated in UK, Brazil, South Africa and India (VOC B1.1.7/alpha, VOC P.1/gamma, VOC B1.351/beta, and VOC B1.617.2/delta, respectively). We first determined the time window for each variant in which CPE appeared. It was evident at 48 h for all the tested strains but VOC B1.1.7 and VOC P.1, for which evident CPE appeared later (56 h and 72 h, respectively). In parallel, uninfected cells were cultured in presence of different doses of raloxifene to evaluate possible treatment-related cytotoxicity. In cells treated with 15μM drug, we observed a reduced percentage of viable cells (82.9 + /−8.69% and 76.3 + /−5.84%, at 48 h and 72 h respectively, compared to 100% in untreated cells). To determine antiviral efficacy of raloxifene, CPE was measured in infected cells treated with serial dilutions of raloxifene (15 to 0.23 μM) using the time windows identified for each strain. The drug was able to recover cell viability in Vero E6 cells infected with all the tested viral strains. The IC50 calculated on recovering of cell viability ranged from 4.50 to 7.99 μM depending on the strain (Fig. 3) with strong antiviral activity against all the tested variants.
The graph shows the inhibition of CPE observed at different concentration of raloxifene on different VOCs. The IC50 values calculated by nonlinear regression are shown in the table. Percentage of viable cells calculated on not treated not infected = 100%; not treated SARS-CoV-2 infected cells= 0%. Bars indicate SD.
System Biology screening to investigate polypharmacological effects of raloxifene against SARS-CoV-2 infection
Aiming to gain insights on relevant pathways potentially modulated by raloxifene in the context of viral infection we built a molecular network connecting the human-virus interactome and proteins involved in COVID-19 pathogenesis (SI section). The resulting network was used to generate a list of proteins used as a probe for screening all papers on raloxifene in which proteins relevant for SARS-CoV-2 infection are cited. Three functional groups of human genes involved in the biology of viral infection that could be modulated by raloxifene were identified (Figure S2):
-
1.
Genes connected to inflammation, including those modulated by the raloxifene molecular target ER2;
-
2.
Genes expressed in the lungs which are modulated by ER receptors whose downregulation results associated with severe asthma, and whose unfavorable variants cause worse respiratory consequences, according to GWAS studies;
-
3.
Genes directly modulated by the virus, both during the cell entry and the replication phase, including proteins upstream or downstream some of raloxifene-controlled pathways.
Within the first group of genes the cytokine Interleukin-6 (IL-6) [61] was identified, one of the clinically validated targets of the anti COVID-19 therapy. A raloxifene-mediated downregulation of IL-6 inflammatory signal and expression was found in a clinical setting [62]. Other serum cytokines (i.e. TNF-α and TGF-β1) involved in SARS-CoV-2 cytokine storm are also regulated by raloxifene.
The second group includes genes that regulate the production of nitric oxide involved in the vascular and respiratory response to the viral infection. Raloxifene can upregulate the expression of eNOS (NOS3) in rat thoracic aorta, and it is expected to exert a potentially important vasculo-protective effect, and to contribute to clinical improvements in ARDS and pulmonary hypertension [63], concept already tested in clinical trials also with other compounds [64].
As for the third group, a direct antiviral action of raloxifene in inhibiting viral replication and/or infection was found in different contexts like Ebola Virus [51, 52, 65], HBV [56], HCV [53], Influenza Virus A [57], and also in a clinical trial against HCV [58]. We found a group of genes directly modulated by the virus, both during the cell entry phase and the replication phase, which also include raloxifene-controlled pathways, ACE2, TMPRSS2 and ADAM17 [66]. ER mRNA levels are associated with these gene transcripts in the atrium, suggesting a role for ER in modulating the expression of proteins relevant for viral entry and expression.
Virtual screening on SARS-CoV-2 proteins and SPR analysis: raloxifene among the best-scored binders
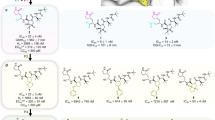
High-Performance Computing (HPC) simulation was conducted to generate a profile of raloxifene against SARS-CoV-2 proteins. The simulation was performed with LiGen™ (Ligand Generator) software. Figure 4A reports the docking scores obtained on the selected targets. To further investigate the virus entry machinery, we decided to experimentally validate by Surface Plasmon Resonance (SPR) the binding between raloxifene and the Spike protein (S). We describe for the first time that raloxifene has direct binding affinity for S, its S1 domain and the Receptor Binding Domain (RBD). The affinity was assessed by flowing the compound on the three proteins immobilized on parallel surfaces of the same sensor chip. S, S1 and RBD were immobilized by direct coupling amine chemistry or captured via a Fc-tag. Raloxifene binds the target proteins in a specific and concentration-dependent manner (Fig. 4B). The sensorgrams (black lines) fit well (red lines) with the Langmuir equation, thus allowing to calculate the binding parameters; raloxifene binds S with a dissociation constant (KD) ranging from 45.7 to 32.0 µM, depending on immobilization, while KD is 41.7 and 48.5 µM for S1 and RBD, respectively. SPR experiments confirmed a specific and dose-dependent binding of raloxifene to S.
A LiGen™ docking score values that predict the binding affinity of the molecules in the protein binding site are reported in shades of green: dark green corresponds to higher values. Scores are also reported (the higher, the better). B SPR experiments showed that S (upper panels), S1 (lower left panel) and RBD (lower right panel) bind raloxifene. The sensorgrams (black) were obtained after subtraction of the signal observed on the reference (empty) surface, to show the specific binding signal. For each target protein, the entire sensorgrams (i.e. association and dissociation phases) obtained with three raloxifene concentrations, were globally fitted using the 1:1 Langmuir model. Red lines show the resulting fitting while the corresponding binding parameters are shown in the insets: kon and koff are the association and dissociation rate constants, respectively, while KD is the equilibrium dissociation constants.
Raloxifene does not interfere with SARS-CoV-2 entry
We used different approaches to identify a possible role of raloxifene in controlling ACE2-S interaction [67, 68]. In the SPR experiments, ACE2 and S were preincubated in the absence or presence of the drug, and then flowed over the corresponding immobilized target proteins (S and ACE2, respectively); no influence of raloxifene in ACE2-S binding was observed (Fig. 5A). Raloxifene was not also able to interfere with ACE2-S1 binding observed in the FRET-based assay (Fig. 5B). Although we cannot completely exclude that raloxifene interferes with the entry mechanisms in other conditions, these data may suggest that other S-mediated processes could be involved in raloxifene-S binding.
A SPR experiments showed that raloxifene does not interfere with either S binding to immobilized ACE2 (left panel) or with ACE2 binding to immobilized S (right panel). Solutions were injected in triplicate; black lines show the sensorgrams with the proteins alone, while red lines show the sensorgrams with the proteins preincubated with raloxifene. B TR-FRET ACE2-SpikeS1 interaction assay showed that raloxifene is not able to interfere with the protein binding.
In vitro effect of raloxifene on S-induced ADAM17 mRNA expression
To test the hypothesis that raloxifene can interfere with S-induced signalling involved in viral replication without affecting S/ACE2 binding, the direct effect of S and raloxifene on ADAM17 in pulmonary cells, such as Calu-3 an A549 cells, was checked. Treatment with recombinant S and raloxifene on ADAM17 modulation was monitored at transcriptional level for 48 h to verify a possible effect of raloxifene alone in unstimulated cells (Fig. 6A, B). Interestingly, in Calu-3 cells (Fig. 6A) in the presence of S we observed an upward trend in ADAM17 mRNA expression levels, significantly inhibited by treatment with raloxifene (− 40%), as compared to S. The modulation of ADAM17 gene expression in A549 cells by treatments confirmed the observed trend. In fact, the treatment with S protein (Fig. 6B) induced an increase of ADAM17 levels which was significantly counteracted by raloxifene.
Effect of 48 hours treatment on modulation of ADAM17 mRNA in Calu-3 (A) and A549 (B) cell line treated with S protein (S 10 ng/ml), raloxifene (Raloxifene 20μΜ) and combined treatments (S 10 ng/ml + Raloxifene 20μΜ). Results are expressed as mean ± SD of 3 independent experiments. *p < 0.05, **p < 0.01, ***P < 0.001.
Discussion
Raloxifene was proposed as potential candidate for the treatment of COVID-19 due to the in silico predicted possibility to interfere with viral replication and disease progression via multiple mechanisms of action, both ER-dependent and independent [43]. Based on these observations, a clinical study was conducted (NCT05172050) in patients with mild to moderate COVID-19 whose results are being published (Nicastri E. et al., 2022 eClinicalMedicine, accepted for publication). This study compared the approved pharmacological dosage of raloxifene 60 mg/day, used to treat and prevent osteoporosis in postmenopausal women, with the dosage of 120 mg/day, also extensively evaluated in clinical trials, to confirm the safety and tolerability of the drug in the indication, and to compare the efficacy of the two doses with the aim of gaining further information on possible ER-dependent or independent mechanisms of action. Previous data from screening campaigns reported the antiviral activity of raloxifene [59]; in this paper we report an in-depth in vitro characterization of the antiviral activity of the drug against the WT and the most common variants of SARS-CoV-2. First of all, we confirmed the anti-CPE of the drug in two relevant experimental systems, Vero E6 monkey kidney cells, commonly used to study coronavirus infection [69,70,71] and human pulmonary Calu-3 cells, a predictive model of airway epithelial origin [72]. Interestingly, raloxifene was tested against all the most common circulating SARS-CoV-2 VOCs, confirming that the drug retains a high and consistent activity, thus reinforcing the interest on its potential use as antiviral agent in COVID-19. In the two cellular systems here described, raloxifene CC50 was set at high micromolar range, far from the low micromolar range of the observed antiviral activity, and the SI was superimposable, ranging from 2 to 7, in line with the expected characteristics for translation into human trials [73, 74]. Among SERMs, raloxifene has a unique benefit/risk profile obtained from its extensive safety database due to its use in cancer patients, in postmenopausal women, and in men [75, 76]. The occurrence of thromboembolic events, even though rare, must be regarded with caution, due to the risk of thromboembolic manifestations in COVID-19 patients, thus a short duration of treatment is recommended and avoidance of treating patients with concomitant risks of thromboembolic events. The potential of SERMs, in particular raloxifene, for the treatment of COVID-19 found a promising confirmation in a recent retrospective study in a population of cancer patients in which protection from SARS-CoV-2 infection and significant reduction in severity and duration of the infection were observed in the subpopulation of patients treated with raloxifene [77]. To strengthen the hypothesis that raloxifene could exert a polypharmacological action in COVID-19 patients, the results of a system biology study strongly suggest the positive influence of the drug during the viral infection due to the modulation of several human genes playing key roles in the biology of the viral infection. The study matched the available information on genes and pathways regulated by raloxifene against a Cytoskape-generated human SARS-CoV-2 interactome network. Data analysis confirmed a putative antiviral activity of raloxifene, highlighting a potential anti-inflammatory action, a vasculo-protective activity through upregulation of eNOS expression (NOS3) [63, 78], and an antiviral activity by modulating key pathways of viral entry and replication, such as ACE2, TMPRSS2 and ADAM17 [66]. Modulation of ER-activated pathways is likely to be only partially responsible for its antiviral activity, since it has been found conserved within the SERM class, but with non-superimposable structure-activity relationship. Several mechanisms have been proposed to support the antiviral action of raloxifene [79, 80]. In agreement with EXSCALATE predictions [43], raloxifene was reported to inhibit SARS-CoV-2 3C-like protease (3 CLpro) viral protein with an IC50 value of 5.61 μM [81] that could partially explain the observed in vitro activity. We also checked in this paper its affinity for S and found a low affinity although a specific binding, that could account at least in part for the observed antiviral mechanism. In fact, even if the direct affinity of raloxifene towards S alone is relatively low, we cannot exclude that this binding becomes relevant in case of protein-protein interaction. Surprisingly, and in apparent contradiction with previous studies [81] (https://opendata.ncats.nih.gov/covid19/databrowser), we did not observe a significant inhibition of ACE2-S interaction. Even if we can’t completely rule out the possibility that in other experimental conditions raloxifene can block the viral entry process, our results are perfectly aligned with the observation that the anti-viral activity on Vero E6 and Calu-3 cells occurs only in the post-entry conditions. To evaluate possible alternative effects of raloxifene on S-induced biological activities, we investigated in Calu-3 and A549 cells the direct effect of S on the expression of ADAM17, known for playing a key role in ACE2 shedding, and whose inhibition previously resulted to have a protective effect on SARS-CoV infection [82]. The observed increase of ADAM17 mRNA expression induced by S in both Calu-3 and A549 cells, and its corresponding reduction in presence of raloxifene, represent a first evidence of the drug ability to interfere with S-induced biological activity, downstream the interaction with the membrane ACE2 receptor. ADAM17 modulation may explain the observed in vitro effects but can also suggest a positive in vivo regulation of the anti/pro-inflammatory process balance, tissue regeneration and remodeling [60, 83]. These results confirm that raloxifene may exert a pharmacological action useful to prevent disease progression and exacerbation by direct interaction with viral proteins and/or modulation of ER signaling. One of the main goals of this study was the characterization of raloxifene on the most frequent circulating viral variants because VOCs pose a threat to the ongoing efforts to control the COVID-19 pandemic due to the hypothesis that the infection, originated in bats underwent multiple recombination events during migration through other mammals [7]. Evidence that the new VOCs have acquired the ability to expand species tropism to murines [9] suggests the possibility that these species act as a zoonotic viral reservoir. VOCs diffusion and evolution confirm the need to address multiple mechanisms to inhibit viral replication of the new emerging SARS-CoV-2 variants, and our results, remarkably, highlight the independence of raloxifene activity from individual viral variants. Based on our knowledge, raloxifene is one of the first examples of treatment efficacious against the main circulating SARS-CoV-2 variants, and our results suggest that it could represent a useful additional option to the therapeutic armamentarium for the management of COVID-19 patients in the next years.
Data availability
All data generated or analysed during this study are included in this published article and in article supplementary material.
References
Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17:181–92.
Hu B, Guo H, Zhou P, Shi ZL. Characteristics of SARS-CoV-2 and COVID-19. Nat Rev Microbiol. 2021;19:141–54.
Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 2020;395:565–74.
Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020;395:497–506.
Tan L, Wang Q, Zhang D, Ding J, Huang Q, Tang YQ, et al. Lymphopenia predicts disease severity of COVID-19: a descriptive and predictive study. Signal Transduct Target Ther. 2020;5:33.
Della-Morte D, Pacifici F, Ricordi C, Massoud R, Rovella V, Proietti S, et al. Low level of plasminogen increases risk for mortality in COVID-19 patients. Cell Death Dis. 2021;12:773.
Salian VS, Wright JA, Vedell PT, Nair S, Li C, Kandimalla M, et al. COVID-19 Transmission, Current Treatment, and Future Therapeutic Strategies. Mol Pharm. 2021;18:754–71.
Michelitsch A, Wernike K, Ulrich L, Mettenleiter TC, Beer M. SARS-CoV-2 in animals: From potential hosts to animal models. Adv Virus Res. 2021;110:59–102.
Wei C, Shan KJ, Wang W, Zhang S, Huan Q, Qian W. Evidence for a mouse origin of the SARS-CoV-2 Omicron variant. J Genet Genomics. 2021;48:1111–21.
Pan T, Chen R, He X, Yuan Y, Deng X, Li R, et al. Infection of wild-type mice by SARS-CoV-2 B.1.351 variant indicates a possible novel cross-species transmission route. Signal Transduct Target Ther. 2021;6:420.
Creech CB, Walker SC, Samuels RJ. SARS-CoV-2 Vaccines. Jama 2021;325:1318–20.
Torjesen I. Covid-19 will become endemic but with decreased potency over time, scientists believe. Bmj 2021;372:n494.
Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, et al. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell 2020;182:812–27. e19
Mascola JR, Graham BS, Fauci AS. SARS-CoV-2 Viral Variants-Tackling a Moving Target. Jama 2021;325:1261–2.
Funk T, Pharris A, Spiteri G, Bundle N, Melidou A, Carr M, et al. Characteristics of SARS-CoV-2 variants of concern B.1.1.7, B.1.351 or P.1: data from seven EU/EEA countries, weeks 38/2020 to 10/2021. Euro surveillance: bulletin Europeen sur les maladies transmissibles = European communicable disease bulletin. 2021;26.
Davies NG, Abbott S, Barnard RC, Jarvis CI, Kucharski AJ, Munday JD, et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. 2021;372:eabg3055.
Tegally H, Wilkinson E, Giovanetti M, Iranzadeh A, Fonseca V, Giandhari J, et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature 2021;592:438–43.
Faria NR, Mellan TA, Whittaker C, Claro IM, Candido DDS, Mishra S, et al. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science 2021;372:815–21.
Twohig KA, Nyberg T, Zaidi A, Thelwall S, Sinnathamby MA, Aliabadi S, et al. Hospital admission and emergency care attendance risk for SARS-CoV-2 delta (B.1.617.2) compared with alpha (B.1.1.7) variants of concern: a cohort study. Lancet Infect Dis. 2021;22:35–42.
Aleem A, Akbar Samad AB, Slenker AK. Emerging Variants of SARS-CoV-2 And Novel Therapeutics Against Coronavirus (COVID-19). StatPearls. Treasure Island (FL) 2021.
Ao D, Lan T, He X, Liu J, Chen L, Baptista-Hon DT, et al. SARS-CoV-2 Omicron variant: Immune escape and vaccine development. MedComm. 2022;3:e126. (2020)
Zhang Q, Xiang R, Huo S, Zhou Y, Jiang S, Wang Q, et al. Molecular mechanism of interaction between SARS-CoV-2 and host cells and interventional therapy. Signal Transduct Target Ther. 2021;6:233.
Qiao Y, Wang XM, Mannan R, Pitchiaya S, Zhang Y, Wotring JW, et al. Targeting transcriptional regulation of SARS-CoV-2 entry factors ACE2 and TMPRSS2. Proc Natl Acad Sci U S A. 2020;118:e2021450118.
Bravaccini S, Fonzi E, Tebaldi M, Angeli D, Martinelli G, Nicolini F, et al. Estrogen and Androgen Receptor Inhibitors: Unexpected Allies in the Fight Against COVID-19. Cell Transplant. 2021;30:963689721991477.
Strope JD, Chau CH, Figg WD. Are sex discordant outcomes in COVID-19 related to sex hormones? Semin Oncol. 2020;47:335–40.
Khan D, Ansar Ahmed S. The Immune System Is a Natural Target for Estrogen Action: Opposing Effects of Estrogen in Two Prototypical Autoimmune Diseases. Front Immunol. 2015;6:635.
Straub RH. The complex role of estrogens in inflammation. Endocr Rev. 2007;28:521–74.
Kovats S. Estrogen receptors regulate innate immune cells and signaling pathways. Cell Immunol. 2015;294:63–9.
Stilhano RS, Costa AJ, Nishino MS, Shams S, Bartolomeo CS, Breithaupt-Faloppa AC, et al. SARS-CoV-2 and the possible connection to ERs, ACE2, and RAGE: Focus on susceptibility factors. FASEB J: Off Publ Federation Am Societies Exp Biol. 2020;34:14103–19.
Mauvais-Jarvis F, Klein SL, Levin ER. Estradiol, Progesterone, Immunomodulation, and COVID-19 Outcomes. Endocrinology. 2020;161:bqaa127.
Mishra R, Behera LM, Rana S. Binding of raloxifene to human complement fragment 5a ((h)C5a): a perspective on cytokine storm and COVID19. J Biomol Struct Dyn 2022;40:982–94.
Perricone C, Triggianese P, Bartoloni E, Cafaro G, Bonifacio AF, Bursi R, et al. The anti-viral facet of anti-rheumatic drugs: Lessons from COVID-19. J Autoimmun. 2020;111:102468.
Haitao T, Vermunt JV, Abeykoon J, Ghamrawi R, Gunaratne M, Jayachandran M, et al. COVID-19 and Sex Differences: Mechanisms and Biomarkers. Mayo Clin Proc. 2020;95:2189–203.
Abate BB, Kassie AM, Kassaw MW, Aragie TG, Masresha SA. Sex difference in coronavirus disease (COVID-19): a systematic review and meta-analysis. BMJ open. 2020;10:e040129.
Lipsa A, Prabhu JS. Gender disparity in COVID-19: Role of sex steroid hormones. Asian Pac J tropical Med. 2021;14:5–9.
Gervasoni S, Vistoli G, Talarico C, Manelfi C, Beccari AR, Studer G, et al. A Comprehensive Mapping of the Druggable Cavities within the SARS-CoV-2 Therapeutically Relevant Proteins by Combining Pocket and Docking Searches as Implemented in Pockets 2.0. Int J Mol Sci. 2020;21:5152.
Grottesi A, Besker N, Emerson A, Manelfi C, Beccari AR, Frigerio F, et al. Computational Studies of SARS-CoV-2 3CLpro: Insights from MD Simulations. International journal of molecular sciences. 2020;21:5346.
Manelfi C, Gossen J, Gervasoni S, Talarico C, Albani S, Philipp BJ, et al. Combining Different Docking Engines and Consensus Strategies to Design and Validate Optimized Virtual Screening Protocols for the SARS-CoV-2 3CL Protease. Molecules. 2021;26:797.
Muchmore DB. Raloxifene: A selective estrogen receptor modulator (SERM) with multiple target system effects. oncologist. 2000;5:388–92.
Goldstein SR. Selective estrogen receptor modulators and bone health. Climacteric: the journal of the International Menopause Society. 2021:1–4.
Hernandez E, Valera R, Alonzo E, Bajares-Lilue M, Carlini R, Capriles F, et al. Effects of raloxifene on bone metabolism and serum lipids in postmenopausal women on chronic hemodialysis. Kidney Int. 2003;63:2269–74.
Lewis JS, Jordan VC. Selective estrogen receptor modulators (SERMs): mechanisms of anticarcinogenesis and drug resistance. Mutat Res. 2005;591:247–63.
Allegretti M, Cesta MC, Zippoli M, Beccari A, Talarico C, Mantelli F, et al. Repurposing the estrogen receptor modulator raloxifene to treat SARS-CoV-2 infection. Cell Death Differ. 2021;29:156–66.
Calderone A, Menichetti F, Santini F, Colangelo L, Lucenteforte E, Calderone V. Selective Estrogen Receptor Modulators in COVID-19: A Possible Therapeutic Option? Front Pharmacol. 2020;11:1085.
Messalli EM, Scaffa C. Long-term safety and efficacy of raloxifene in the prevention and treatment of postmenopausal osteoporosis: an update. Int J women’s health. 2010;1:11–20.
Clemett D, Spencer CM. Raloxifene: a review of its use in postmenopausal osteoporosis. Drugs 2000;60:379–411.
Vogel VG, Costantino JP, Wickerham DL, Cronin WM, Cecchini RS, Atkins JN, et al. Effects of tamoxifen vs raloxifene on the risk of developing invasive breast cancer and other disease outcomes: the NSABP Study of Tamoxifen and Raloxifene (STAR) P-2 trial. Jama 2006;295:2727–41.
DeMichele A, Troxel AB, Berlin JA, Weber AL, Bunin GR, Turzo E, et al. Impact of raloxifene or tamoxifen use on endometrial cancer risk: a population-based case-control study. J Clin Oncol. 2008;26:4151–9.
Weickert TW, Weinberg D, Lenroot R, Catts SV, Wells R, Vercammen A, et al. Adjunctive raloxifene treatment improves attention and memory in men and women with schizophrenia. Mol psychiatry. 2015;20:685–94.
Smith MR, Fallon MA, Lee H, Finkelstein JS. Raloxifene to prevent gonadotropin-releasing hormone agonist-induced bone loss in men with prostate cancer: a randomized controlled trial. J Clin Endocrinol Metab. 2004;89:3841–6.
Montoya MC, Krysan DJ. Repurposing Estrogen Receptor Antagonists for the Treatment of Infectious Disease. mBio. 2018;9:e02272–18.
Yoon YS, Jang Y, Hoenen T, Shin H, Lee Y, Kim M. Antiviral activity of sertindole, raloxifene and ibutamoren against transcription and replication-competent Ebola virus-like particles. BMB Rep. 2020;53:166–71.
Takeda M, Ikeda M, Mori K, Yano M, Ariumi Y, Dansako H, et al. Raloxifene inhibits hepatitis C virus infection and replication. FEBS open bio. 2012;2:279–83.
Murakami Y, Fukasawa M, Kaneko Y, Suzuki T, Wakita T, Fukazawa H. Selective estrogen receptor modulators inhibit hepatitis C virus infection at multiple steps of the virus life cycle. Microbes Infect. 2013;15:45–55.
Lamontagne J, Mills C, Mao R, Goddard C, Cai D, Guo H, et al. Screening and identification of compounds with antiviral activity against hepatitis B virus using a safe compound library and novel real-time immune-absorbance PCR-based high throughput system. Antivir Res. 2013;98:19–26.
Eyre NS, Kirby EN, Anfiteatro DR, Bracho G, Russo AG, White PA, et al. Identification of Estrogen Receptor Modulators as Inhibitors of Flavivirus Infection. Antimicrobial agents and chemotherapy. 2020;64:e00289–20.
Peretz J, Pekosz A, Lane AP, Klein SL. Estrogenic compounds reduce influenza A virus replication in primary human nasal epithelial cells derived from female, but not male, donors. Am J Physiol Lung Cell Mol Physiol. 2016;310:L415–25.
Furusyo N, Ogawa E, Sudoh M, Murata M, Ihara T, Hayashi T, et al. Raloxifene hydrochloride is an adjuvant antiviral treatment of postmenopausal women with chronic hepatitis C: a randomized trial. J Hepatol. 2012;57:1186–92.
Imamura K, Sakurai Y, Enami T, Shibukawa R, Nishi Y, Ohta A, et al. iPSC screening for drug repurposing identifies anti-RNA virus agents modulating host cell susceptibility. FEBS Open Bio. 2021;11:1452–64.
Zipeto D, Palmeira JDF, Arganaraz GA, Arganaraz ER. ACE2/ADAM17/TMPRSS2 Interplay May Be the Main Risk Factor for COVID-19. Front Immunol. 2020;11:576745.
Aziz M, Fatima R, Assaly R. Elevated interleukin-6 and severe COVID-19: A meta-analysis. J Med Virol. 2020;92:2283–5.
Ozmen B, Kirmaz C, Aydin K, Kafesciler SO, Guclu F, Hekimsoy Z. Influence of the selective oestrogen receptor modulator (raloxifene hydrochloride) on IL-6, TNF-alpha, TGF-beta1 and bone turnover markers in the treatment of postmenopausal osteoporosis. Eur Cytokine Netw. 2007;18:148–53.
Rahimian R, Dube GP, Toma W, Dos Santos N, McManus BM, van Breemen C. Raloxifene enhances nitric oxide release in rat aorta via increasing endothelial nitric oxide mRNA expression. Eur J Pharmacol. 2002;434:141–9.
Morrell ED, Tsai BM, Crisostomo PR, Hammoud ZT, Meldrum DR. Experimental therapies for hypoxia-induced pulmonary hypertension during acute lung injury. Shock. 2006;25:214–26.
Kouznetsova J, Sun W, Martinez-Romero C, Tawa G, Shinn P, Chen CZ, et al. Identification of 53 compounds that block Ebola virus-like particle entry via a repurposing screen of approved drugs. Emerg Microbes Infect. 2014;3:e84.
Wang H, Sun X, J LV, Kon ND, Ferrario CM, Groban L. Estrogen receptors are linked to angiotensin-converting enzyme 2 (ACE2), ADAM metallopeptidase domain 17 (ADAM-17), and transmembrane protease serine 2 (TMPRSS2) expression in the human atrium: insights into COVID-19. Hypertens Res. 2021;44:882–4.
Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–8.
Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S, et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271–80. e8
Matsuyama S, Nao N, Shirato K, Kawase M, Saito S, Takayama I, et al. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc Natl Acad Sci USA. 2020;117:7001–3.
Tseng CT, Tseng J, Perrone L, Worthy M, Popov V, Peters CJ. Apical entry and release of severe acute respiratory syndrome-associated coronavirus in polarized Calu-3 lung epithelial cells. J Virol. 2005;79:9470–9.
Kaye M. SARS-associated coronavirus replication in cell lines. Emerg Infect Dis. 2006;12:128–33.
Sims AC, Burkett SE, Yount B, Pickles RJ. SARS-CoV replication and pathogenesis in an in vitro model of the human conducting airway epithelium. Virus Res. 2008;133:33–44.
Verma S, Twilley D, Esmear T, Oosthuizen CB, Reid AM, Nel M, et al. Anti-SARS-CoV Natural Products With the Potential to Inhibit SARS-CoV-2 (COVID-19). Front Pharmacol. 2020;11:561334.
Wei Y, Wang H, Xi C, Li N, Li D, Yao C, et al. Antiviral Effects of Novel 2-Benzoxyl-Phenylpyridine Derivatives. Molecules. 2020;25:1409.
Barrett-Connor E, Mosca L, Collins P, Geiger MJ, Grady D, Kornitzer M, et al. Effects of raloxifene on cardiovascular events and breast cancer in postmenopausal women. N. Engl J Med. 2006;355:125–37.
Vogel VG. The NSABP Study of Tamoxifen and Raloxifene (STAR) trial. Expert Rev anticancer Ther. 2009;9:51–60.
Montopoli M, Zorzi M, Cocetta V, Prayer-Galetti T, Guzzinati S, Bovo E, et al. Clinical outcome of SARS-CoV-2 infection in breast and ovarian cancer patients who underwent antiestrogenic therapy. Annals of oncology: official journal of the European Society for. Med Oncol. 2021;32:676–7.
Bouman A, Heineman MJ, Faas MM. Sex hormones and the immune response in humans. Hum Reprod update. 2005;11:411–23.
Smetana K Jr, Rosel D, BrAbek J. Raloxifene and Bazedoxifene Could Be Promising Candidates for Preventing the COVID-19 Related Cytokine Storm, ARDS and Mortality. Vivo. 2020;34:3027–8.
Hong S, Chang J, Jeong K, Lee W. Raloxifene as a treatment option for viral infections. J Microbiol. 2021;59:124–31.
Chiou WC, Hsu MS, Chen YT, Yang JM, Tsay YG, Huang HC, et al. Repurposing existing drugs: identification of SARS-CoV-2 3C-like protease inhibitors. J Enzym Inhib Med Chem. 2021;36:147–53.
Palau V, Riera M, Soler MJ. ADAM17 inhibition may exert a protective effect on COVID-19. Nephrol Dial Transpl. 2020;35:1071–2.
Scheller J, Chalaris A, Garbers C, Rose-John S. ADAM17: a molecular switch to control inflammation and tissue regeneration. Trends Immunol. 2011;32:380–7.
Acknowledgements
We thank AXXAM spa for providing the in vitro FRET-based assay to test S-ACE2 binding. We thank Dr. Rubina Novelli from Dompé farmaceutici s.p.a. for reviewing the paper. We thank Prof. Marcin Nowotny, Head of the Laboratory of Protein Structure of the IIMCB (Warsaw, Poland) for providing the Spike protein (S) for the experiments. This work was supported in part by EXSCALATE (EXaSCale smArt pLatform Against paThogEns) supercomputing platform funded by Horizon 2020 European Project “N.101003551-EXSCALATE4CoV”, and by “Progetto COVID 2020 12371675 Ricerca finalizzata” and “Linea 1 Ricerca Corrente COVID”, both funded by Italian Ministry of Health.
Author information
Authors and Affiliations
Contributions
All authors contributed equally in editing and proofreading of the manuscript. DI, ARB, MA and MCC jointly supervised this work; DI, MCC, MZ, AB, CT, LS, FC, MM and EMB wrote the manuscript and prepared the figures; CT and CM generated the virtual screening data; FC, AZ, AB, FF performed the antiviral characterization of raloxifene; LB and GM conducted the studies on raloxifene activity on SARS-CoV-2 variants, contributed in the writing of the manuscript and prepared the figures; LS contributed in the studies on raloxifene activity on SARS-CoV-2 variants; MG, MB and AP generated the SPR data; MM, VC and SB provided expression data; EMB performed the system biology screening; MA, MCC, DI and EMB edited the paper; MA, AC, DI, ARB, EMB and EN provided strategic input.
Corresponding author
Ethics declarations
Competing interests
The authors MA, MCC, MZ, CT, DI, CM, ARB are permanent employees of Dompé farmaceutici s.p.a. coordinator of the Horizon 2020 European project “N.101003551-EXSCALATE4CoV”, and sponsor of the running clinical trial conducted with raloxifene in COVID-19 patients. The authors EN, LB, GM and LS are employees of L. Spallanzani National Institute for Infectious Disease, IRCCS, Rome. The authors EMB, FC, AB, AZ, FF, MG, MB, AP, MM, VC, SB, and AC declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by Gerry Melino
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Iaconis, D., Bordi, L., Matusali, G. et al. Characterization of raloxifene as a potential pharmacological agent against SARS-CoV-2 and its variants. Cell Death Dis 13, 498 (2022). https://doi.org/10.1038/s41419-022-04961-z
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41419-022-04961-z
This article is cited by
-
Relevance of Spike/Estrogen Receptor-α interaction for endothelial-based coagulopathy induced by SARS-CoV-2
Signal Transduction and Targeted Therapy (2023)