Abstract
UPF1 is proved to dysregulate in multiple tumors and influence carcinogenesis. However, the role of UPF1 in oxaliplatin resistance in colorectal cancer (CRC) remains unknown. In our study, UPF1 is upregulated in CRC in mRNA and protein levels and overexpression of UPF1 predicts a poor overall survival (OS) and recurrence-free survival (RFS) in CRC patients and is an independent risk factor for recurrence. UPF1 promotes chemoresistance to oxaliplatin in vitro and in vivo. UPF1-induced oxaliplatin resistance can be associated with interaction between zinc finger of UPF1 and Toprim of TOP2A and increasing phosphorylated TOP2A in a SMG1-dependent manner. Moreover, UPF1 maintains stemness in a TOP2A-dependent manner in CRC. Taken together, UPF1 was overexpressed and predicted a poor prognosis in CRC. UPF1 enhanced chemoresistance to oxaliplatin in CRC, which may result from regulation of TOP2A activity and maintenance of stemness. Our findings could provide a new therapy strategy for chemoresistance to oxaliplatin in CRC patients.
Similar content being viewed by others
Introduction
Colorectal cancer (CRC) is the third most frequent for incidence and the second most frequent for mortality worldwide1. However, in China, CRC is the fourth most common and the fifth leading cause of cancer death2. Surgical resection is the potential radical treatment for CRC. Other than surgery, radiochemotherapy, targeted therapy, and immunotherapy can be applied to CRC patients with lymph node metastases or distant metastases3,4,5. However, the acquisition of drug resistance is a major hurdle for good clinical prognosis6.
Oxaliplatin, a third-generation platinum coordination complex, can be used for treatment in multiple cancers7,8,9. Oxaliplatin-based combined chemotherapy is routinely applied in advanced and metastatic CRC and significantly increases the overall survival (OS) rate and metastases resection rate10. Unfortunately, acquired or intrinsic resistance brings failure to treatment. The underlying mechanisms of oxaliplatin resistance include DNA damage response and repair, inhibition of cell death, cellular transport, detoxification, and epigenetic alteration11.
UPF1, an mRNA surveillance factor, is a RNA-dependent ATPase and helicase for nonsense-mediated decay of mRNAs containing premature stop codons12. In addition, UPF1 is proved to dysregulate and influence carcinogenesis in multiple tumors. UPF1 is commonly mutated in pancreatic adenosquamous carcinoma and there is little or no UPF1 expression in many adenosquamous carcinoma tumors compared to adjacent normal tissue13. It is revealed that UPF1 is downregulated in hepatocellular carcinoma and inhibits the tumor progression14,15. However, the role of UPF1 in oxaliplatin resistance in CRC remains unclear.
Topoisomerases are enzymes responsible for overcoming topological problems in the process of DNA replication, transcription and repair, and proved therapeutic targets of anticancer16. Human topoisomerase II-α (TOP2A) plays an important role in organizing genome structure and promoting chromosome segregation17. Growing lines of evidence indicate that TOP2A is overexpressed in a variety of tumors, such as lung cancer, hepatocellular cancer, and colorectal cancer. Elevated expression of TOP2A is associated with advanced stage of disease, aggressive phenotype of tumor, and poor prognosis18. Targeting TOP2A is a promising approach in cancer therapy and some agents have been approved in clinical use, such as etoposide and teniposide18. As an enzyme, the activity of TOP2A can be regulated by protein–protein interactions and post-translational modifications (phosphorylation, ubiquitination, and SUMOylation)19,20,21.
In our study, we demonstrated that UPF1 was overexpressed in CRC tissues and high expression of UPF1 predicted a poor prognosis of CRC patients. UPF1 promoted oxaliplatin resistance both in vitro and in vivo in CRC. UPF1-induced oxaliplatin resistance can be associated with elevated phosphorylation of TOP2A and maintenance of stemness. These findings contribute to improving our understanding of oxaliplatin resistance, and may provide a new therapy strategy for chemoresistance to oxaliplatin in CRC patients.
Results
UPF1 is upregulated and predicts a poor prognosis in CRC
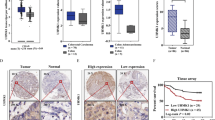
To investigate the expression of UPF1 in CRC tissues, we performed bioinformatics analysis of UPF1 in mRNA level using the public RNA sequencing datasets from The Cancer Genome Atlas (TCGA)22 and found that UPF1 was overexpressed (P = 0.043, Fig. 1a). We also detected it by IHC staining of tissue microarrays using a retrospective cohort containing 76 patients with paired CRC tumor and normal tissues. The representative IHC images of UPF1 staining were shown in Fig. 1b. Similarly, the results verified that UPF1 was dramatically upregulated in CRC tissues (P < 0.001, Fig. 1c). In these patients, UPF1-positive was detected in 52 (68.4 %) of the tumor tissues, whereas only 4 (5.3 %) of the normal tissues were UPF1-positive (Table 1, P < 0.001).
a UPF1 was overexpressed in CRC tissues in mRNA level from TCGA datasets. b Images of UPF1 in IHC staining in CRC tissue microarrays, scale bar, 200 μm and 100 μm. c UPF1 was overexpressed in CRC tissues in protein level by IHC staining in tissue microarrays. d–e UPF1 positive predicted worse OS and RFS in CRC patients. f–g Univariate and multivariate COX regression analysis revealed that UPF1 was an independent risk factor for recurrence in CRC patients.
To explore the clinical significance of UPF1 in CRC, we analyzed the correlation between the expression of UPF1 and clinicopathological characteristics of CRC patients (Table 1). A significant association was observed between UPF1 positive and TOP2A positive (P = 0.012, 58.3% in UPF1-negative group vs 84.6% in UPF1-positive group). High UPF1 expression (P = 0.035) and high TNM stage (P = 0.040) more likely result in recurrence in CRC patients. However, high expression of UPF1 and recurrence in CRC patients had no correlation with gender, age, tumor location, tumor size, lymphovascular invasion, perineural invasion, and carcinoembryonic antigen level. Additionally, Kaplan–Meier curves showed a strong correlation between UPF1 higher expression and a poorer OS (P = 0.045, Fig. 1d). UPF1-positive patients also had a shorter recurrence-free survival (RFS) (P = 0.029, Fig. 1e). In addition, univariate and multivariate COX regression analysis suggested that high expression of UPF1 (HR = 3.719; 95% CI = 1.313 to 10.529; P = 0.013) and poor differentiation (HR = 2.927; 95% CI = 1.186 to 7.222; P = 0.020) were independent prognostic risk factors for recurrence in CRC (Fig. 1f, g). In summary, UPF1 is aberrantly upregulated in CRC and high expression of UPF1 predicts a poor prognosis in CRC patients.
UPF1 promotes oxaliplatin resistance in CRC in vitro
The baseline expression of UPF1 in CRC cell lines and normal colonic epithelial cell line was detected by immunoblot. Notably, UPF1 expression in the normal cell line, NCM460, was remarkably less compared with CRC cell lines (Fig. 2a). We chose DLD1 to overexpress UPF1 due to its relative low baseline expression. And based on the same rule, UPF1 was knocked down in HCT116 using three small hairpin RNAs (shRNAs). Overexpression and knockdown were confirmed by Western blotting and SH2 was the most efficient in UPF1 knockdown (Fig. 2b, c). HA and FLAG tags were used for co-immunoprecipitation (co-IP) assay. Drug cytotoxicity assay showed that overexpression of UPF1 decreased oxaliplatin sensitivity in DLD1 while knockdown of UPF1 increased oxaliplatin sensitivity in HCT116 (Fig. 2d, e). In apoptosis assay, overexpression and knockdown of UPF1 had no influence on apoptosis rate in DLD1 and HCT116, respectively. Whereas after treatment for 48 h, oxaliplatin-induced apoptosis was significantly reduced in DLD1-UPF1 and raised in HCT116-shUPF1 (Fig. 2f–i). Clone formation assay also affirmed that UPF1 promoted oxaliplatin resistance in CRC in vitro. The number of colonies were more in DLD1-UPF1 group and less in HCT116- shUPF1 group compared with control group with treatment of oxaliplatin for 48 h (Fig. 2j, k). However, the size of colonies remained no obvious change between different groups, which indicated that UPF1 may have no effect on cell proliferation. CCK-8 assay also showed that UPF1 did not influence proliferation in DLD1 and HCT116 cell lines (Fig. S1a, b).
a The baseline expression of UPF1 in CRC cell lines and normal colonic epithelial cell line detected by Western blot. b–c Overexpression and knockdown of UPF1 in DLD1 and HCT116, respectively. d–e Cell viability of stable CRC cell lines treated with a gradient concentration of oxaliplatin assessed by CCK-8 assay. f–i Apoptosis rate of stable CRC cell lines treated with or without oxaliplatin assessed by flow cytometry. j–k Clone formation of stable CRC cell lines treated with oxaliplatin.
UPF1 promotes oxaliplatin resistance in CRC in vivo
Nude mice were divided into 2 groups and injected with HCT116-shNC and HCT116-shUPF1 cell lines, respectively. One half of each group was treated with 5% glucose solution and the other treated with oxaliplatin (5 mg/kg). The tumor xenografts were shown in Fig. 3a. In the groups treated with glucose solution, there is no significant difference between HCT116-shNC and HCT116-shUPF1 groups. Whereas, xenografts in HCT116-shNC group grew faster and were much larger and heavier after treatment with oxaliplatin (Fig. 3b, c). In line, these findings were further confirmed by TUNEL assay (Fig. 3d). Apoptosis rate in tissues remained no change in HCT116-shUPF1 but after being treated with oxaliplatin, apoptosis rate remarkably raised compared with control. Xenografts assay in vivo revealed that UPF1 promotes oxaliplatin resistance in CRC.
a Xenografts of HCT116-shNC and HCT116-shUPF1 treated with or without oxaliplatin. b Tumor volume in nude mice bearing HCT116 cells in different groups. Tumor volume equaled length × width2 × 0.5 and was measured twice a week. c Tumor weight in nude mice bearing HCT116 cells in different groups. d TUNEL assay was performed to detect tumor cell apoptosis in xenograft tumor tissues, scale bar, 40 μm.
UPF1-induced oxaliplatin resistance is involved with interaction with TOP2A and increase of phosphorylated TOP2A in a SMG1-dependent manner
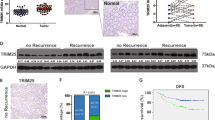
167 kinds of proteins were identified by silver staining (Fig. 4a) and mass spectrometry (MS) which were listed in additional file S1. MS score represents the reliability of detected proteins to a certain extent. UPF1 was identified with the highest MS score (3992.20). Known interacting partners of UPF1, such as UPF2 (MS score = 36.92) and SMG1 (MS score = 0.00)23 were also identified. TOP2A (MS score = 132.22), TOP1 (MS score = 61.59), CCAR2 (MS score = 53.75) and XRCC6 (MS score = 95.64) were selected as candidates for UPF1 interacted proteins according to pathway analysis and biological functions. TOP2A was proved to be upregulated in mRNA and protein levels in CRC tissues (Fig. S1c–e). Enrichment analysis exhibited related pathways and TOP2A was unearthed to be involved in platinum-resistant pathway (Fig. S1f). The co-IP assay showed the interaction between UPF1 and TOP2A, but not TOP1, CCAR2 and XRCC6 (Fig. 4b). Similarly, UPF1 was also precipitated using an anti-FLAG resin in HEK-293T cell line transfected into TOP2A-FLAG (Fig. 4c). We further found that expression of total TOP2A remained no significant change but phosphorylated TOP2A at the site of Ser1106 was increased after upregulation of UPF1. Correspondingly, silencing UPF1 had no influence on total TOP2A expression but attenuated phosphorylated TOP2A at the same site (Fig. 4d). UPF1 shows protein phosphokinase activity neither in published papers nor in its biological functions. SMG1, a member of the PI3K (phosphoinositide 3-kinase related kinases) family, is also a key factor in NMD. SMG1 could interact with UPF1 and directly phosphorylates UPF124,25,26. In our study, SMG1 was also identified in mass spectrometry (additional file S1). Co-IP assay and ICC demonstrated that SMG1 could interact with UPF1 (Fig. S2). Phosphorylated TOP2A was elevated in DLD-UPF1 compared with control. By knockdown of SMG1 in DLD1-UPF1, elevation of phosphorylated TOP2A was brought to a halt (Fig. 4e, f). SMG1 may play a role in UPF1-induced phosphorylation of TOP2A. In DLD1 cell line transfected into UPF1-FLAG, the results of immunocytochemistry indicated the colocalization of UPF1 and TOP2A (Fig. 4g).
a Signature bands of DLD1-UPF1 cell line emerged after silver staining. The location of TOP2A, TOP1, CCAR2, and XRCC6 were marked. b–c Interaction of UPF1 and TOP2A proved in co-IP assay. (d) UPF1 had no influence on the total expression of TOP2A but raised phosphorylated TOP2A at the site of Ser1106. e SMG1 was silenced using small hairpin RNAs. f UPF1 raised phosphorylated TOP2A and the elevation was brought to a halt by knockdown of SMG1. g Images of immunocytochemistry assay showed interaction between UPF1 and TOP2A, scale bar, 25 μm.
To identify the binding region between UPF1 and TOP2A, we generated the truncated mutants of UPF1 and TOP2A to perform co-IP assays. The full length of TOP2A contains 1531 amino acids and six truncated mutants were generated (Fig. 5a, b). Co-IP assay revealed that Toprim domain may be the binding region with UPF1 (Fig. 5c). The full length of UPF1 contains 1129 amino acids and five truncated mutants were generated (Fig. 5d, e). The results elucidated that the zinc finger domain of UPF1 may interact with TOP2A (Fig. 5f).
a Schematic diagram of key domains in TOP2A. b Six truncated mutants of TOP2A fused with FLAG tag were generated according to its key domains. c Co-IP assays were performed of the lysates from HEK-293T cells transfected with HA-tagged UPF1 and FLAG-tagged truncated mutants of TOP2A. d Schematic diagram of key domains in UPF1. e Five truncated mutants of UPF1 fused with HA tag were generated according to its key domains. f Co-IP assays were performed of the lysates from HEK-293T cells transfected with FLAG-tagged TOP2A and HA-tagged truncated mutants of UPF1.
To verify the function of TOP2A in UPF1-induced oxaliplatin resistance, TOP2A was silenced using three small hairpin RNAs. Sh1 was selected for following experiments due to its biggest efficiency (Fig. 6a, b). Of note, drug cytotoxicity assay demonstrated that UPF1-induced resistant cells regained the sensitivity to oxaliplatin after knockdown of TOP2A (Fig. 6c). As shown in clone formation assay, DLD1-Ctrl, DLD1-UPF1, and DLD1-UPF1-shTOP2A were treated by oxaliplatin for 48 h. We can see that UPF1 overexpression significantly increased the number of colonies while the effect was abolished by silencing the TOP2A gene in the cells. The size of colonies had no difference among different groups (Fig. 6d, e). The results in apoptosis assay came to the same conclusion (Fig. 6f, g). In vivo, xenografts in DLD1-UPF1 group were much larger and heavier under treatment with oxaliplatin. The trend of chemoresistance was brought to a halt with TOP2A knockdown (Fig. 6h). In same, TUNEL assay testified that inhibition of apoptosis in tissue induced by UPF1 was deactivated by silencing TOP2A (Fig. 6i). Altogether, these data strongly suggested that TOP2A played an essential role in the UPF1-induced chemoresistance to oxaliplatin in CRC.
a, b TOP2A was silenced using small hairpin RNAs. c Drug cytotoxicity assay, d, e clone formation assay, and f, g flow cytometry showed abrogation of chemoresistance to oxaliplatin induced by UPF1 by knockdown of TOP2A. h Xenografts of DLD1-UPF1, DLD1-Ctrl, and DLD1-UPF1-shTOP2A treated with oxaliplatin and tumor volume and weight of xenografts in different groups. i TUNEL assay was performed to detect tumor cell apoptosis in xenograft tumor tissues, scale bar, 40 μm.
UPF1 maintains stemness in CRC in a TOP2A-dependent manner
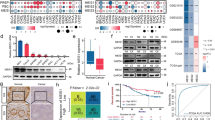
The ability of mammosphere formation was enhanced after upregulation of UPF1 in DLD1 and was attenuated after knockdown of UPF1 in HCT116 (Fig. 7a, b). EpCAM has been proposed as one of the cancer stem cell (CSC) markers27,28,29. In our study, the percentage of EpCAM-positive cells was raised in DLD1-UPF1 and diminished in HCT116-shUPF1 (Fig. 7c, d). Notably, UPF1-induced enhancement of stemness was abated by silencing the expression of TOP2A in mammosphere formation assay (Fig. 7e, f). Meanwhile, UPF1-induced change of EpCAM+ cell rates was abolished by knockdown of TOP2A (Fig. 7g, h).
a–b The ability of mammosphere formation was enhanced after upregulation of UPF1 in DLD1 and was attenuated after knockdown of UPF1 in HCT116, scale bar, 100 μm. c, d The percentage of EpCAM-positive cells was raised in DLD1-UPF1 and diminished in HCT116-shUPF1 compared with control. e–f Silencing TOP2A abrogated enhancement of the ability of mammosphere formation induced by UPF1, scale bar, 100 μm. g, h Silencing TOP2A down regulated the rise of the percentage of EpCAM-positive cells induced by UPF1.
Discussion
CRC maintains a high rate of incidence and mortality worldwide, as well as that in China1,2. Stage II CRC exhibits a recurrence rate of about 20% after radical resection30. For patients with stage III disease, recurrence rate can be as high as more than 50%31. Chemotherapy resistance is a major cause of recurrence and poor prognosis in CRC patients32.
Oxaliplatin, as a DNA interacting agent, results in DNA damage and induction of DNA strand breaks thus activates apoptosis signal pathway in cancer cells33. Oxaliplatin is the first platinum drug with proven activity against colorectal tumors. Unfortunately, drug resistance brings failure to treatment, which becomes a pressing problem in clinical treatment in CRC patients.
It is reported that UPF1 can play a role in regulated mRNA and protein decay34. In addition, UPF1 is proved to dysregulate in multiple tumors and influence carcinogenesis13,14,15. However, there is lack of detailed study of UPF1 in CRC. It is reported that UPF1 might play a part in the selection of target gene mutations with a functional role in MSI-H carcinogenesis35. In line, Ada Collura et al. ascertained that inhibition of nonsense mediated decay in vivo using amlexanox reduced MSI tumor growth36. Currently, the function of UPF1 and potential mechanisms remain unclear in CRC.
Firstly, we demonstrated that UPF1 was aberrantly overexpressed in CRC tissues. And high UPF1 expression more likely resulted in recurrence and predicted a worse OS in CRC patients. In addition, UPF1-positive was an independent prognostic risk factor for recurrence in CRC.
Herein, how UPF1 acts as a oncogene in CRC needed deep study. CCK−8, clone formation assays and flow cytometry indicated that UPF1 may have no effect on cell proliferation and apoptosis. While assays in vitro showed that overexpression of UPF1 weakened sensitivity to oxaliplatin in DLD1 while knockdown of UPF1 reinforced sensitivity to oxaliplatin in HCT116. In vivo, we reached the same conclusion that UPF1 promotes oxaliplatin resistance in CRC.
Afterwards, the underlying mechanism of UPF1-induced oxaliplatin resistance remained to be revealed. Mass spectrometry analysis identified 166 kinds of proteins that may interact with UPF1. Enrichment analysis exhibited related pathways and TOP2A was unearthed to be involved in platinum-resistant pathway. Kyoto Encyclopedia of Genes and Genomes pathway map showed the detailed signal network in platinum drug resistance (map01524)37. Oxaliplatin-induced DNA-adducts activated nucleotide excision repair through Topoisomerase II, which could attenuate DNA damage and result in oxaliplatin resistance38. TOP2A, DNA topoisomerase, is an enzyme that controls and alters the topologic states of DNA during transcription39. TOP2A is upregulated and could induce tumor development and progression in multiple tumors40,41,42,43 and is proved important therapeutic target of anticancer agents44. Etoposide, as an inhibitor of TOP2A, is a kind of cell cycle specific antitumor drug and applied in multiple tumors. The catalytic activity of TOP2A can be modulated by interactions with various proteins44. It is reported that phosphorylation of serine1106 in the catalytic domain of TOP2A regulates enzymatic activity and drug sensitivity45,46. In CRC tissues, TOP2A was proved to be upregulated in mRNA and protein levels. In the IHC staining of tissue microarrays, a significant association was observed between UPF1 positive and TOP2A positive. TOP1 could modulate colorectal cancer response to irinotecan47. CCAR2 overexpression decreased the chemosensitivity to oxaliplatin in CRC48. XRCC6, as a DNA repair gene, may participate in platinum resistance by modulating the DNA repair capacity49. TOP2A, TOP1, CCAR2, and XRCC6 were selected as candidates for UPF1 interacted proteins according to pathway analysis and biological functions. Forward and reverse co-IP assays proved the interaction of UPF1 with TOP2A, instead of TOP1, CCAR2, and XRCC6. In DLD1 transfected into UPF1-FLAG, the results of immunocytochemistry assay also indicated the colocalization of UPF1 and TOP2A. To identify the binding region between UPF1 and TOP2A, we generated the truncated mutants of UPF1 and TOP2A to perform co-IP assays. Toprim in TOP2A is a catalytic domain involved in DNA strand breakage and rejoining and may be the binding region with UPF1. The zinc fingers vary widely in structure, as well as in function. And zinc finger domain of UPF1 may interact with Toprim in TOP2A. YacG is a bacterial type chromosome-encoded and zinc-finger containing protein. A crystal structure of the YacG-DNA gyrase complex revealed that the interaction between zinc-finger of YacG with the Toprim domain regulated TOP2A activity50,51. In our study, interaction with UPF1 may modulate the enzyme activity of TOP2A in a similar manner. We further found that expression of total TOP2A remained no significant change after upregulation of UPF1. While the interaction with UPF1 increased phosphorylated TOP2A at the site of Ser1106. However, UPF1 shows protein phosphokinase activity neither in published papers nor in its biological functions. The mechanism of phosphorylation of TOP2A remains unknown. SMG1, a member of the PI3K (phosphoinositide 3-kinase related kinases) family, is also a key factor in NMD. SMG1 could interact with UPF1 and directly phosphorylate UPF1. The formation of SMG1-UPF1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers UPF1 phosphorylation and NMD24,25,26. In our study, SMG1 was also identified in mass spectrometry. Co-IP assay and ICC demonstrated that SMG1 could interact with UPF1. Phosphorylated TOP2A was elevated in DLD-UPF1 compared with control. By knockdown of SMG1 in DLD1-UPF1, elevation of phosphorylated TOP2A was brought to a halt. SMG1 may play a role in UPF1-induced phosphorylation of TOP2A. Of note, UPF1-mediated oxaliplatin resistance was abolished by silencing the TOP2A gene in vitro and in vivo.
Cancer stem cells (CSCs) are defined by their functional properties and could be able to self-renew and propagate the tumor52. CSC-like properties or stemness have an essential role in the drug resistance in CRC53,54. In our study, the stemness was progressed in UPF1-upregulation cell line and was impeded in UPF1-knockdown cell line. CSCs are characterized by specific markers, such as EpCAM. Flow cytometry indicated that the percentage of EpCAM-positive cells was diminished after silencing UPF1 and increased after upregulating UPF1. TOP2A is abundant in pluripotent embryonic stem cells and inhibition of TOP2A noticeably decreases pluripotency and differentiation potential. TOP2A-inactivated embryonic stem cells failed to generate embryonic bodies and thus do not differentiate55. Thus, we concluded that TOP2A could regulate stemness in a certain manner. Notably, UPF1-induced CSC-like properties was abated by silencing the expression of TOP2A. Altogether, these data strongly suggested that TOP2A played an essential role in the UPF1-induced chemoresistance of CRC cells in response to oxaliplatin in vitro and in vivo and TOP2A could be a therapeutic target in UPF1-overexpressed CRC patients.
There are some limitations in this study that could be addressed in future research. We have not figured out how interaction between UPF1 and TOP2A changes the enzyme activity. And we wonder whether combination with the drugs targeted for TOP2A could make synergistic effect in patients with highly expressed UPF1.
Conclusions
UPF1 was overexpressed and predicted a poor prognosis in CRC. UPF1 enhanced chemoresistance to oxaliplatin in CRC, which may result from regulation of TOP2A activity and maintenance of stemness. Our findings could provide a new therapy strategy for chemoresistance to oxaliplatin in CRC patients (Fig. 8).
Materials and methods
Patients and specimens
76 patients were pathologically diagnosed with CRC and received surgical therapy at Fudan University Shanghai Cancer Center. Patients who received neoadjuvant radiochemotherapy were excluded from our study. Specimens from the 76 patients were made into tissue microarrays. Our study was carried out in accordance with the requirements of the Biomedical Ethics Committee of Fudan University Shanghai Cancer Center.
Cell lines and culture
CRC cell lines (DLD1, HT29, LOVO, HCT116, RKO, SW480, SW620) and HEK−293T cell line were purchased from Type Culture Collection Cell Bank, Chinese Academy of Sciences. Normal colonic epithelial cell line (NCM460) was purchased from American Type Culture Collection. All cell lines were cultured in Dulbecco’s Modified Eagles Medium with 10% fetal bovine serum, penicillin (107 U/L) and streptomycin (10 mg/L) and incubated at 37 °C in a humidified atmosphere containing 5% CO2 in Fudan University Shanghai Cancer Center.
Lentivirus production and transfection
Core plasmids of UPF1 were purchased from OBiO Technology (Shanghai, China) and core plasmids of TOP2A were purchased from Genechem (Shanghai, China). The detailed information of core plasmids was shown in additional file S2. Core plasmids were co-transfected with psPAX2 and pMD2.G into HEK−293T cells using Hieff TransTM Liposomal Transfection Reagent (Yeasen, Shanghai, China). 48 h later, virus supernatant was collected. 3 × 105 cells were cultured in a 6-well plate. After incubation in virus supernatant for 48 h, stable cell lines were selected with puromycin or flow cytometry and transfected efficiency was evaluated by immunoblot.
Transient transfection
To construct truncated mutants of UPF1 and TOP2A, specific primers (additional file S3) were designed to subclone corresponding sequences into pLVX-IRES-Puro vector. Small interfering RNAs (siRNAs) of SMG1 were purchased from Biomics Biotech (Jiangsu, China). The sequences of siRNA were shown in additional file S2. Plasmids overexpressing UPF1-HA and FLAG-tagged TOP2A truncated mutants or overexpressing TOP2A-FLAG and HA-tagged UPF1 truncated mutants were co-transfected into HEK-293T cells using Hieff TransTM Liposomal Transfection Reagent (Yeasen, Shanghai, China). 48 h later, cells were lysed for co-immunoprecipitation (Co-IP) assay. siRNAs were transfected into cells using Lipofectamine 3000 Transfection Reagent (Invitrogen, Carlsbad, USA). 48 h later, cells were lysed for Western blotting.
Flow cytometry
Apoptosis assay was conducted using Annexin V-PE/7-AAD apoptosis detection kit (Yeasen, Shanghai, China). EpCAM-positive cells were detected using FITC-conjugated EpCAM antibody (Sino Biological, Beijing, China). Briefly, 3 × 105 cells were cultured in a 6-well plate with or without being treated with oxaliplatin for 48 h. Adherent cells and cells in supernatant were collected and washed twice. Cells suspended with 1× binding buffer with the addition of 5 μL Annexin V and 10 μL 7-AAD or 10 μL EpCAM antibody. After incubation protecting from light for 15 min, apoptosis rate or EpCAM-positive cells were detected by Cytomics FC 500 MPL Flow cytometer (Beckman, USA).
Western blotting
Total protein was extracted with NuPAGE® LDS sample buffer (Thermo Fisher Scientific, Waltham, USA). A certain amount of protein was separated by SDS-PAGE gel and transferred to PVDF membranes. After blockage with non-fat milk powder and incubation with primary antibodies and horseradish peroxidase-conjugated secondary antibodies, images were captured with ImageQuant™ biomolecular imager (General Electric, USA). All the antibodies used in our study were listed in additional file S4.
Drug cytotoxicity assay
Drug cytotoxicity was evaluated by Cell Counting Kit-8 (CCK-8) assay. Cell suspension was seeded in a 96-well plate and cells were treated with a gradient concentration of oxaliplatin for 48 h after adhesion. Medium containing CCK-8 reagent (Yeasen, Shanghai, China) was added to each well of the plate. After two-hours’ incubation, absorbance at 450 nm was detected by microplate reader (BioTek, USA).
Clone formation assay
1000 cells were cultured in a 6-well plate and treated with oxaliplatin for 48 h after adhesion. The medium was replaced by the fresh every three days. 2 weeks later, cells were fixed with 4% paraformaldehyde for 30 min and stained with crystal violet for 30 min. Images were photographed and visible colonies were counted.
Co-immunoprecipitation (Co-IP) assays, silver staining, and mass spectrometry
Cells in culture dish were lysed with IP buffer containing protease inhibitors and phosphatase inhibitor cocktails (Topscience, Shanghai, China). IP buffer was a mixture of 1 mM EDTA, 20 mM HEPES, 150 mM NaCl, 0.05% sodium deoxycholate, and 0.05% NP-40. Lysate was spun at the speed of 12,000 rpm at 4 °C for 12 min. 60 μL was kept for input and the rest of the extract was immunoprecipitated with anti-HA beads or anti-FLAG resins at 4 °C for more than 3 h. After washing 5 times with IP buffer, beads or resins of protein bound were lysed with 1.25× SDS loading buffer and boiled at 100 °C for 10 min. Silver staining was conducted according to the manufacture of Fast Silver Stain Kit (Beyotime, Shanghai, China). Bands appeared after electrophoresis and silver staining in SDS-PAGE gel received mass spectrometry analysis (Wayen Biotechnologies, Shanghai, China) for protein identification. Interacted protein was further confirmed by immunoblot.
Mammosphere formation assay
Mammosphere formation assay was used to define stemness in vitro in our study and conducted as previously described56. Cells were digested by trypsin and washed twice. 300 cells suspended in 200 μL mammosphere medium were seeded in a 96-well plate with ultra-low attachment surface. The formulation of mammosphere medium was shown in additional file S5. 10 days later, images were photographed with an Olympus microscope system and mammospheres were counted.
Immunohistochemistry (IHC)
IHC was performed as previously described57. The results of IHC staining were determined by immunoreactive score, ranging from 0 to 12. Immunoreactive score equals the number of positive cells multiplied by the staining intensity. The number of cells stained were divided into 5 groups: 0, 1–10%, 11–50%, 51–80%, and 81–100%, corresponding to 0–4 score, respectively. And staining intensity has 4 levels, negative, weak, moderate, and strong, corresponding to 0–3 score, respectively.
Immunocytochemistry
Sterilized round coverslips were laid in a 24-well plate and cells were seeded on them. After 24 h incubation, cells were fixed with 4% paraformaldehyde for 30 min and permeabilized on ice with 0.25% Triton X−100. Then cells were blocked with bovine serum albumin and incubated with primary antibodies for 2 h and fluorescent secondary antibodies protecting from light for 1 h at room temperature. After mounting with DAPI Fluoromount mounting medium for 15 min, images were captured using Leica confocal system.
Xenograft experiments
5 × 106 cells were subcutaneously injected into four-week-old male BALB/c nude mice. 5 days later, mice were randomly divided into groups and treated with oxaliplatin (5 mg/kg) or 5% glucose solution by intraperitoneal injection, twice per week for three weeks, respectively. All mice were sacrificed and tumors were collected and weighed. Tumor volume equaled length × width2 × 0.5 and was measured twice a week. Xenografts were saved in 4% paraformaldehyde for following experiments. Apoptosis rates in the xenograft were determined by terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL) assay using TMR (red) TUNEL Cell Apoptosis Detection Kit (Servicebio, Wuhan, China). Xenograft experiments were approved by the Committee on Animals Handling of Fudan University Shanghai Cancer Center.
Statistical analysis
Data are shown in mean ± standard deviation. All analyses were performed by IBM SPSS 22.0 software. Quantitative variables were analyzed using Student’s t test. The Log-rank test in the Kaplan–Meier method and the COX regression model were used to assess patients’ survival outcome and prognostic factors. All the experiments were performed in triplicate. A two-tailed value of P < 0.05 was considered statistically significant (*P < 0.05; **P < 0.01; ***P < 0.001).
Ethics approval and consent to participate
Our study was carried out in accordance with the requirements of the Biomedical Ethics Committee of Fudan University Shanghai Cancer Center. Informed consent was obtained from all patients included in the study. Xenograft experiments were approved by the Committee on Animals Handling of Fudan University Shanghai Cancer Center.
Data availability
The data used to support the findings of this study are included within the article and the supplementary materials.
References
Bray, F. et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424 (2018).
Chen, W. et al. Cancer incidence and mortality in China, 2014. Chin. J. Cancer Res. 30, 1–12 (2018).
Ganesh, K. et al. Immunotherapy in colorectal cancer: rationale, challenges and potential. Nat. Rev. Gastroenterol. Hepatol. 16, 361–375 (2019).
Rosello, S., Papaccio, F., Roda, D., Tarazona, N. & Cervantes, A. The role of chemotherapy in localized and locally advanced rectal cancer: a systematic revision. Cancer Treat. Rev. 63, 156–171 (2018).
Zhai, Z. et al. Colorectal cancer heterogeneity and targeted therapy: clinical implications, challenges and solutions for treatment resistance. Semin. Cell Dev. Biol. 64, 107–115 (2017).
Hammond, W. A., Swaika, A. & Mody, K. Pharmacologic resistance in colorectal cancer: a review. Ther. Adv. Med. Oncol. 8, 57–84 (2016).
Edeline, J. et al. Gemcitabine and Oxaliplatin Chemotherapy or Surveillance in Resected Biliary Tract Cancer (PRODIGE 12-ACCORD 18-UNICANCER GI): a randomized phase iii study. J. Clin. Oncol. 37, 658–667 (2019).
Sessa, C., ten Bokkel Huinink, W. W. & du Bois, A. Oxaliplatin in ovarian cancer. Ann. Oncol. 10(Suppl 1), 55–57 (1999).
Suker, M. et al. FOLFIRINOX for locally advanced pancreatic cancer: a systematic review and patient-level meta-analysis. Lancet Oncol. 17, 801–810 (2016).
Hind, D. et al. The use of irinotecan, oxaliplatin and raltitrexed for the treatment of advanced colorectal cancer: systematic review and economic evaluation. Health Technol. Assess. 12, iii–ix, xi-162 (2008).
Martinez-Balibrea, E. et al. Tumor-related molecular mechanisms of oxaliplatin resistance. Mol. Cancer Ther. 14, 1767–1776 (2015).
Lykke-Andersen, J., Shu, M. D. & Steitz, J. A. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell 103, 1121–1131 (2000).
Liu, C. et al. The UPF1 RNA surveillance gene is commonly mutated in pancreatic adenosquamous carcinoma. Nat. Med. 20, 596–598 (2014).
Chang, L. et al. The human RNA surveillance factor UPF1 regulates tumorigenesis by targeting Smad7 in hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 35, 8 (2016).
Zhou, Y. et al. UPF1 inhibits the hepatocellular carcinoma progression by targeting long non-coding RNA UCA1. Sci. Rep. 9, 6652 (2019).
Pilati, P., Nitti, D. & Mocellin, S. Cancer resistance to type II topoisomerase inhibitors. Curr. Med Chem. 19, 3900–3906 (2012).
Lee, J. H. & Berger, J. M. Cell cycle-dependent control and roles of DNA topoisomerase II. Genes 10, 859 (2019).
Chen, T., Sun, Y., Ji, P., Kopetz, S. & Zhang, W. Topoisomerase IIalpha in chromosome instability and personalized cancer therapy. Oncogene 34, 4019–4031 (2015).
Watt, P. M. & Hickson, I. D. Structure and function of type II DNA topoisomerases. Biochem. J. 303, 681–695 (1994).
Shinagawa, H., Miki, Y. & Yoshida, K. BRCA1-mediated ubiquitination inhibits topoisomerase II alpha activity in response to oxidative stress. Antioxid. Redox Signal. 10, 939–949 (2008).
Azuma, Y., Arnaoutov, A. & Dasso, M. SUMO-2/3 regulates topoisomerase II in mitosis. J. Cell Biol. 163, 477–487 (2003).
From: https://www.cbioportal.org/study/summary?id=coadread_tcga_pan_can_atlas_2018.
Ivanov, P. V., Gehring, N. H., Kunz, J. B., Hentze, M. W. & Kulozik, A. E. Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO J. 27, 736–747 (2008).
Kashima, I. et al. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 20, 355–367 (2006).
Melero, R. et al. Structures of SMG1-UPFs complexes: SMG1 contributes to regulate UPF2-dependent activation of UPF1 in NMD. Structure 22, 1105–1119 (2014).
Yamashita, A., Kashima, I. & Ohno, S. The role of SMG-1 in nonsense-mediated mRNA decay. Biochim. Biophys. Acta 1754, 305–315 (2005).
Chaudhary, K., Poirion, O. B., Lu, L. & Garmire, L. X. Deep learning-based multi-omics integration robustly predicts survival in liver cancer. Clin. Cancer Res. 24, 1248–1259 (2018).
Ho, D. W. et al. Single-cell transcriptomics reveals the landscape of intra-tumoral heterogeneity and stemness-related subpopulations in liver cancer. Cancer Lett. 459, 176–185 (2019).
Zhu, Y. et al. miR-145 antagonizes SNAI1-mediated stemness and radiation resistance in colorectal cancer. Mol. Ther. 26, 744–754 (2018).
Marshall, J. L. Risk assessment in stage II colorectal cancer. Oncology 24, 9–13 (2010).
Meyerhardt, J. A. & Mayer, R. J. Systemic therapy for colorectal cancer. N. Engl. J. Med. 352, 476–487 (2005).
Yu, T. et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell 170, 548–563 e516 (2017).
Faivre, S., Chan, D., Salinas, R., Woynarowska, B. & Woynarowski, J. M. DNA strand breaks and apoptosis induced by oxaliplatin in cancer cells. Biochem. Pharmacol. 66, 225–237 (2003).
Feng, Q., Jagannathan, S. & Bradley, R. K. The RNA surveillance factor UPF1 represses myogenesis via its E3 ubiquitin ligase activity. Mol. Cell 67, 239–251 e236 (2017).
El-Bchiri, J. et al. Differential nonsense mediated decay of mutated mRNAs in mismatch repair deficient colorectal cancers. Hum. Mol. Genet. 14, 2435–2442 (2005).
Bokhari, A. et al. Targeting nonsense-mediated mRNA decay in colorectal cancers with microsatellite instability. Oncogenesis 7, 70 (2018).
From: https://www.genome.jp/dbget-bin/www_bget?pathway:map01524.
Tsai-Pflugfelder, M. et al. Cloning and sequencing of cDNA encoding human DNA topoisomerase II and localization of the gene to chromosome region 17q21-22. Proc. Natl Acad. Sci. USA 85, 7177–7181 (1988).
Riccio, A. A., Schellenberg, M. J. & Williams, R. S. Molecular mechanisms of topoisomerase 2 DNA-protein crosslink resolution. Cell Mol. Life Sci. 77, 81–91 (2020).
Labbe, D. P. et al. TOP2A and EZH2 provide early detection of an aggressive prostate cancer subgroup. Clin. Cancer Res. 23, 7072–7083 (2017).
Pei, Y. F., Yin, X. M. & Liu, X. Q. TOP2A induces malignant character of pancreatic cancer through activating beta-catenin signaling pathway. Biochim. Biophys. Acta 1864, 197–207 (2018).
Zeng, S. et al. Prognostic value of TOP2A in bladder urothelial carcinoma and potential molecular mechanisms. BMC Cancer 19, 604 (2019).
Zhang, R., Xu, J., Zhao, J. & Bai, J. H. Proliferation and invasion of colon cancer cells are suppressed by knockdown of TOP2A. J. Cell Biochem. 119, 7256–7263 (2018).
Delgado, J. L., Hsieh, C. M., Chan, N. L. & Hiasa, H. Topoisomerases as anticancer targets. Biochem. J. 475, 373–398 (2018).
Chikamori, K. et al. Phosphorylation of serine 1106 in the catalytic domain of topoisomerase II alpha regulates enzymatic activity and drug sensitivity. J. Biol. Chem. 278, 12696–12702 (2003).
Grozav, A. G. et al. Casein kinase I delta/epsilon phosphorylates topoisomerase IIalpha at serine-1106 and modulates DNA cleavage activity. Nucleic Acids Res. 37, 382–392 (2009).
Meisenberg, C. et al. Clinical and cellular roles for TDP1 and TOP1 in modulating colorectal cancer response to irinotecan. Mol. Cancer Ther. 14, 575–585 (2015).
Kim, H. J., Moon, S. J., Kim, S. H., Heo, K. & Kim, J. H. DBC1 regulates Wnt/beta-catenin-mediated expression of MACC1, a key regulator of cancer progression, in colon cancer. Cell Death Dis. 9, 831 (2018).
Dimberg, J., Skarstedt, M., Slind Olsen, R., Andersson, R. E. & Matussek, A. Gene polymorphism in DNA repair genes XRCC1 and XRCC6 and association with colorectal cancer in Swedish patients. APMIS 124, 736–740 (2016).
Sengupta, S. & Nagaraja, V. YacG from Escherichia coli is a specific endogenous inhibitor of DNA gyrase. Nucleic Acids Res. 36, 4310–4316 (2008).
Vos, S. M. et al. Direct control of type IIA topoisomerase activity by a chromosomally encoded regulatory protein. Genes Dev. 28, 1485–1497 (2014).
Nassar, D. & Blanpain, C. Cancer stem cells: basic concepts and therapeutic implications. Annu. Rev. Pathol. 11, 47–76 (2016).
Kreso, A. et al. Variable clonal repopulation dynamics influence chemotherapy response in colorectal cancer. Science 339, 543–548 (2013).
Meacham, C. E. & Morrison, S. J. Tumour heterogeneity and cancer cell plasticity. Nature 501, 328–337 (2013).
Thakurela, S. et al. Gene regulation and priming by topoisomerase IIalpha in embryonic stem cells. Nat. Commun. 4, 2478 (2013).
Mani, S. A. et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 133, 704–715 (2008).
Zhu, C. C. et al. CCR6 promotes tumor angiogenesis via the AKT/NF-kappaB/VEGF pathway in colorectal cancer. Biochim. Biophys. Acta 1864, 387–397 (2018).
Acknowledgements
I would like to extend my deep gratitude to Pro. Ming Yao and Pro. Xianghuo He for their guidance and support for our study. The financial support of our study was from the Science and Technology Commission of Shanghai Municipality (19511121202, 20DZ1100101).
Author information
Authors and Affiliations
Contributions
Y.X. and W.S. conceived the experiments and helped write the manuscript. C.Z. and L.Z. conducted the experiments. C.Z., L.Z., S.Z. and W.D. analyzed and interpreted the data. C.Z., Y.X. and Y.Z. prepared the figures. C.Z. wrote the first draft and H.Z. revised the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by B. Zhivotovsky
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhu, C., Zhang, L., Zhao, S. et al. UPF1 promotes chemoresistance to oxaliplatin through regulation of TOP2A activity and maintenance of stemness in colorectal cancer. Cell Death Dis 12, 519 (2021). https://doi.org/10.1038/s41419-021-03798-2
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41419-021-03798-2
This article is cited by
-
Deubiquitinase USP7 stabilizes KDM5B and promotes tumor progression and cisplatin resistance in nasopharyngeal carcinoma through the ZBTB16/TOP2A axis
Cell Death & Differentiation (2024)
-
Mechanisms of Actinidia chinensis Planch in treating colon cancer based on the integration of network pharmacology, molecular docking, and experimental verification
Hereditas (2023)
-
PVT1/miR-136/Sox2/UPF1 axis regulates the malignant phenotypes of endometrial cancer stem cells
Cell Death & Disease (2023)
-
Nonsense-mediated RNA decay: an emerging modulator of malignancy
Nature Reviews Cancer (2022)
-
UPF1 contributes to the maintenance of endometrial cancer stem cell phenotype by stabilizing LINC00963
Cell Death & Disease (2022)