Abstract
Kidney disease progression can be affected by Na+ abundance. A key regulator of Na+ homeostasis is the ubiquitin ligase NEDD4-2 and its deficiency leads to increased Na+ transport activity and salt-sensitive progressive kidney damage. However, the mechanisms responsible for high Na+ induced damage remain poorly understood. Here we show that a high Na+ diet compromised kidney function in Nedd4-2-deficient mice, indicative of progression toward end-stage renal disease. Injury was characterized by enhanced tubule dilation and extracellular matrix accumulation, together with sustained activation of both Wnt/β-catenin and TGF-β signaling. Nedd4-2 knockout in cortical collecting duct cells also activated these pathways and led to epithelial–mesenchymal transition. Furthermore, low dietary Na+ rescued kidney disease in Nedd4-2-deficient mice and silenced Wnt/β-catenin and TGF-β signaling. Our study reveals the important role of NEDD4-2-dependent ubiquitination in Na+ homeostasis and protecting against aberrant Wnt/β-catenin/TGF-β signaling in progressive kidney disease.
Similar content being viewed by others
Introduction
Chronic kidney disease (CKD), defined as a decline in kidney function over time, affects nearly 10% of the global population1. Mounting evidence suggests that excessive Na+ consumption can hasten the progression of CKD and contribute to end-stage renal disease (ESRD) via several mechanisms, including increased arterial pressure caused by hypertension2. High levels of Na+ can also influence CKD by compromising endothelial cell health and affecting tissue remodeling and fibrosis3.
The reabsorption of Na+ from dietary salt is facilitated by multiple channels and transporters in the kidney. The final stage of this process occurs in the distal tubule and the cortical collecting duct (CCD), controlled by the epithelial sodium channel (ENaC)4. The amount of functional ENaC on the plasma membrane is regulated by multiple factors including ubiquitination by the ubiquitin ligase NEDD4-25. Genetic variation of Nedd4L (human Nedd4-2) is associated with developmental disorders, hypertension, and end-stage renal disease6. Furthermore, reduced expression of Nedd4L has been recently identified in diabetic nephropathy7, highlighting its relevance to kidney disease. Previously, kidney-specific deletion of Nedd4-2 in mice has been shown to result in CKD-like pathology due to elevated ENaC levels driving increased uptake of Na+8. Feeding these mice a high Na+ diet during early postnatal stages results in aberrant ENaC regulation and increased kidney damage9. However, the mechanisms that are responsible for this Na+-sensitive renal damage remain unknown.
A number of signaling pathways have been implicated in the progression of CKD10. Wnt/β-catenin and TGF-β signaling11 are key events activated in response to high Na+12,13. TGF-β signaling, through activation of SMAD proteins, is a strong stimulator of extracellular matrix (ECM) production and fibrosis12. Similarly, sustained activation of Wnt/β-catenin signaling can drive CKD progression from acute kidney injury (AKI) by activating interstitial fibroblasts and ECM production14. Of note, the extent of Wnt signaling is important in influencing kidney disease, as it has also been shown to play a protective role in healing and repair15. Both of these signaling pathways are potent inducers of epithelial–mesenchymal transition (EMT)16, however significant debate as to the contribution of EMT-induced fibrosis to kidney disease has arisen in recent years17.
Here, we demonstrate that a short-term high Na+ diet fed to adult kidney-specific Nedd4-2-deficient mice promotes progression toward ESRD in the absence of hypertension. Sustained activation of Wnt/β-catenin and TGF-β signaling and resultant fibrotic deposition are key drivers of renal disease in mice. Similarly, these pathways are activated in Nedd4-2-deficient CCD cells in vitro, leading to EMT. Importantly, we show that a low Na+ diet ameliorates kidney disease in Nedd4-2-deficient mice by inhibiting Wnt/β-catenin and TGF-β signaling. We conclude that in addition to proper regulation of ENaC, NEDD4-2 mediated ubiquitination-dependent control of Wnt/β-catenin and TGF-β signaling is critical in preventing Na+-induced CKD progression.
Results
High dietary Na+ drives progression towards ESRD in Nedd4-2 Ksp1.3 mice
Mice deficient for Nedd4-2 specifically in the renal tubules (Nedd4-2Ksp1.3) develop mild kidney disease due to upregulation of ENaC8,9. To evaluate the consequences of increased Na+ on kidney function, Nedd4-2Ksp1.3 adult mice were fed a standard Na+ (0.2%) or high Na+ (3.1%) diet for 17 days. Beyond this, significant weight loss required termination of the experiment. On the standard Na+ diet, there was no difference between the glomerular filtration rate (GFR) of control and Nedd4-2Ksp1.3 mice (Fig. 1A). However, after a high Na+ diet, Nedd4-2Ksp1.3 mice had a significant reduction in GFR. In addition, serum creatinine levels were significantly increased in Nedd4-2Ksp1.3 mice after high Na+ (Fig. 1B). Blood urea was increased on the standard Na+ diet, and further increased after high Na+ diet feeding (Fig. 1C). Hence, the underlying kidney injury present in Nedd4-2Ksp1.3 mice is exacerbated by high Na+ feeding, resulting in progression toward ESRD.
A GFR calculated from 24 h urine and blood collection after 17 days of standard (Std.) or high Na+ diet. B, C Serum creatinine and urea levels after varied diets. D BP measured by noninvasive tail-cuff averaged over 3 days for each time point. Pre: before the commencement of diet, Post: at the conclusion of varied Na+ diet. SBP systolic blood pressure, DBP diastolic blood pressure. A–C Significance determined using an unpaired two-tailed Student’s t test (means ± SEM). D Significance determined using one-way ANOVA of repeated measurements (means ± SEM, n = 9–11). *P < 0.05, **P < 0.01, ****P < 0.0001.
At 2 months of age, there was no significant difference in starting (Pre) blood pressure (BP) between control and Nedd4-2Ksp1.3 mice (Fig. 1D). On a standard Na+ diet, there was no change in the systolic BP or diastolic BP of mice of either genotype. Post high Na+ feeding, there was again no change in BP of control mice, however, Nedd4-2Ksp1.3 mice showed a significant decrease in BP (Fig. 1D). These results suggest that the reduction in kidney function caused by lack of Nedd4-2 is not due to hypertension.
Weight loss, polyuria and electrolyte imbalances are associated with renal disease
Nedd4-2Ksp1.3 mice lost significant weight after high Na+ feeding (Fig. 2A). This loss primarily occurred after the first day of high Na+ (Supplementary Fig. 1). Food intake and feces output were similar, suggesting that the weight loss was not caused by a compromised appetite (Fig. 2B). Nedd4-2Ksp1.3 mice had a significant increase in water intake (polydipsia) and urine (polyuria), which was further increased after a high Na+ diet (Fig. 2C). In the collecting duct, Aquaporin-2 (AQP2) is the primary channel responsible for water reabsorption that allows concentration of urine, with loss of this channel causing polyuria18. Consistent with this, there was a trend toward a reduction in AQP2 (unglycosylated and glycosylated forms) in Nedd4-2Ksp1.3 kidneys on a standard Na+ diet, which was significantly reduced after high Na+ feeding (Fig. 2D, E). Similarly, AQP3 levels were reduced in Nedd4-2Ksp1.3 kidneys, and further reduced after high Na+ (Fig. 2D, E). Immunostaining of kidney sections revealed AQP2 localization in the apical membrane of primarily cortical collect duct cells, marked by Dolichos Biflorus Agglutinin (DBA) (Fig. 2F), as expected19. A dramatic reduction in AQP2 expression in Nedd4-2Ksp1.3 kidneys was observed in both diet conditions (Fig. 2F). AQP3 was greatly reduced in Nedd4-2Ksp1.3 kidneys on standard Na+ and further reduced by high Na+. These results suggest that water imbalance and osmotic stress are key features of salt-sensitive kidney disease in Nedd4-2-deficient mice.
A Weight change at the conclusion of 17 days of the diet. B Food intake and feces produced or C water intake and urine output, over 24 h in metabolic cages. D Immunoblot analysis of AQP2 and AQP3 with GAPDH as a loading control. Arrow indicates unglycosylated form, arrowhead indicates highest glycosylated form. The area within the bracket represents quantitated region. E Quantitation of AQP immunoblots normalized to GAPDH loading control. F Immunostaining of AQP2 or AQP3 (green) with DBA marker for collecting ducts (red). DNA is stained by Hoechst (blue). Scale bar: 50 μm. Data presented as means ± SEM and significance determined using one-way ANOVA of repeated measurements (A), unpaired two-tailed Student’s t test (B, C), or presented as means ± SD with significance determined using unpaired two-tailed Student’s t test (E). *P < 0.05, **P < 0.01, ***P < 0.005, ****P < 0.0001.
In support of this, compared to control mice, urine of Nedd4-2Ksp1.3 mice had significantly decreased osmolarity and creatinine and increased urea levels after the high Na+ diet (Table 1). Despite the maintenance of urinary Na+, both K+ and Ca2+ levels were elevated. Blood serum levels of K+ and Cl− were decreased, and Ca2+ increased in Nedd4-2Ksp1.3 mice after high Na+ compared to control mice (Table 1). Furthermore, additional indicators of kidney function including sodium bicarbonate (HCO3−), globulin, and total protein were increased in Nedd4-2Ksp1.3 mice. As previously reported in these mice8, significantly lower aldosterone levels were observed under standard Na+, further exacerbated by high dietary Na+.
Renal injury is characterized by ECM accumulation and fibrosis
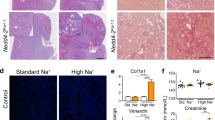
Nedd4-2Ksp1.3 kidneys from mice fed a standard or high Na+ diet had an uneven appearance, as observed by gross morphology (Supplementary Fig. 2A). Haematoxylin and Eosin (H&E) staining of kidney sections revealed that the extent of injury in Nedd4-2Ksp1.3 kidneys consisting of dilated tubules, mesenchymal infiltration, and cellular debris was severely exacerbated after high Na+ (Fig. 3A, Supplementary Fig. 2B). The expression of Kim-1, a proximal tubule marker for kidney injury, was significantly increased in standard Na+ fed Nedd4-2Ksp1.3 mice and further increased by high Na+ (Fig. 3B). Additional markers of kidney injury, Vimentin, and Collagen (Col1a1) were increased in Nedd4-2Ksp1.3 kidneys, with similar levels after both diets (Fig. 3B, Supplementary Fig. 2C). An increase of ECM deposition and fibrosis after high Na+ was observed by vimentin immunostaining (Fig. 3C), and picrosirius red staining of connective tissue (Fig. 3D).
A Haematoxylin and Eosin staining, scale bar: 100 μm, 1 mm for inset. B qRT-PCR analysis of Kim-1, Vimentin, and Collagen 1 (Col1a1) relative to TATA-box binding protein (TBP). C Immunostaining of vimentin (green) with DBA marker for collecting ducts (red). DNA is stained by Hoechst (blue). Scale bar: 50 μm. D Picrosirius red staining of collagen, scale bar: 100 μm. E Immunostaining of αSMA (green) and E-cadherin (red). DNA is stained by Hoechst (blue). Arrows indicate regions of αSMA positive interstitium. Scale bar: 50 μm. F Immunoblot analysis of E-cadherin and αSMA with GAPDH as a loading control. Arrow indicates full-length E-cadherin, the arrowhead is 30 kDa cleaved form. Data in B presented as means ± SEM and significance determined using unpaired two-tailed Student’s t test, n = 3–4. *P < 0.05, **P < 0.01, ***P < 0.005.
The myofibroblast marker, α-smooth muscle actin (αSMA), is normally present only surrounding the blood vessels in the kidney (Fig. 3E) but is upregulated during kidney injury20. Some αSMA positive cells were present surrounding the glomeruli and in the interstitium in Nedd4-2Ksp1.3 kidneys, which was apparent to a much greater extent following the high Na+ diet (Fig. 3E). This staining was distinct from E-cadherin positive epithelial tubular cells, suggesting a mesenchymal nature of these cells (Fig. 3E and lower magnification in Supplementary Fig. 2D). Loss of E-cadherin, a classical Ca2+-dependent cell adhesion molecule, is known to been associated with EMT and fibrosis. In contrast, here Nedd4-2Ksp1.3 mice had increased tubular basolateral E-cadherin staining which was more widespread following high Na+ with some staining on the apical membrane, suggesting a loss of polarity (Fig. 3E). Similarly, the total protein levels of αSMA and E-cadherin were elevated in Nedd4-2Ksp1.3 kidneys, and to a greater extent by the high Na+ diet (Fig. 3F, quantitated in Supplementary Fig. 2E). Interestingly, an increase in a 30 kDa E-cadherin fragment was observed in Nedd4-2Ksp1.3 kidneys (Fig. 3F), similar to that previously reported in the ligated kidney21.
Damage to parenchyma involves sustained activation of Wnt/β-catenin and TGF-β signaling
As Wnt and TGF-β signaling are well known to play a role in CKD progression22, we examined if kidney injury in Nedd4-2-deficient mice is caused by defective Wnt and TGF-β signaling. The expression of Wnt1, which is activated early after kidney injury14 was upregulated in Nedd4-2Ksp1.3 kidneys on the standard Na+ diet, however, was comparable to controls following the high Na+ diet (Fig. 4A). A similar pattern was observed for Wnt3 (Supplementary Fig. 3A). Wnt4, which has been demonstrated to have sustained upregulation after severe kidney injury, was upregulated in Nedd4-2Ksp1.3 kidneys on the standard Na+ diet and maintained after high Na+ (Fig. 4A). NEDD4-2 has been implicated in Wnt signaling by promoting Disheveled 2 (DVL2) ubiquitination and degradation leading to inhibition of β-catenin degradation22,23. Levels of DVL2 were increased in Nedd4-2Ksp1.3 kidneys on standard Na+ and further after high Na+ diet (Fig. 4B, quantitated in Supplementary Fig. 3B). Consistently, Nedd4-2Ksp1.3 kidneys had increased total β-catenin on standard and high Na+ diets (Fig. 4B). Immunostaining revealed an accumulation of cytoplasmic β-catenin in Nedd4-2Ksp1.3 kidneys after both diets, with evidence of translocation into the nucleus (Fig. 4C). This was not restricted to DBA-positive collecting ducts which lack NEDD4-2 expression, suggesting a nonautonomous role of Nedd4-2-deficiency in activating this signaling pathway.
A qRT-PCR analysis of Wnt1 and Wnt4 genes relative to the TBP housekeeping gene. B Immunoblot analysis of β-catenin and DVL2 with GAPDH as a loading control. C Immunostaining of β-catenin (green) with DBA marker for collecting ducts (red). DNA is stained by Hoechst (blue). Arrows indicate nuclear staining. Scale bar: 50 μm. D Immunoblot analysis of pSMAD2/3 and total SMAD2/3 with GAPDH as a loading control. E qRT-PCR analysis of genes relative to TBP. Data presented as means ± SEM and significance determined using unpaired two-tailed Student’s t test, n = 3–4. *P < 0.05, **P < 0.01.
In addition to Wnt, TGF-β signaling has well-established links to CKD, with TGF-βR1 and SMAD2/3 components of this pathway regulated by NEDD4-2-dependent ubiquitination24,25. Whilst no increase in TGF-β1 ligand, or its receptor, TGF-βR1, was observed in Nedd4-2Ksp1.3 kidneys (Supplementary Fig. 3A, C), levels of phosphorylated (p) SMAD2 and SMAD3, known NEDD4-2 substrates24, were increased on the standard Na+ diet, and pSMAD2 was further increased after high Na+ (Fig. 4D, quantitated in Supplementary Fig. 3B). Increased expression of Fibronectin and Serpine, two Wnt/TGF-β target genes involved in the fibrotic response15, were observed in Nedd4-2Ksp1.3 kidneys, reaching significance on either standard Na+ diet (Serpine), or after high Na+ (Fibronectin) (Fig. 4E). However, targets genes of the cell cycle and proliferation, cMyc and Cyclin D1 were not changed (Supplementary Fig. 3D). Interestingly, genes promoting EMT in kidney disease, Snai1, and Twist were significantly increased in Nedd4-2Ksp1.3 kidneys only after high Na+, suggesting that EMT may play a role in the severity of this disease (Fig. 4E).
Nedd4-2 KO in CCD cells confirms the direct role of NEDD4-2 in Wnt/β-catenin/TGF-β signaling
To further investigate the roles of Wnt/β-catenin and TGF-β signaling in kidney disease resulting from Nedd4-2 deficiency, we generated Nedd4-2 knockout (KO) in a CCD cell line. Sequencing confirmed a frameshift mutation (Supplementary Fig. 4A), that resulted in a complete loss of NEDD4-2 protein expression (Fig. 5A, Supplementary Fig. 4B) and increased vimentin (Fig. 5A, B). Nedd4-2 KO resulted in increased sensitivity to Wnt and TGF-β signaling as treatment with TGF-β ligand or the Wnt activator LiCl26 further increased Vimentin expression (Fig. 5B). Interestingly, TGF-β exposure resulted in reduced endogenous NEDD4-2 levels as well as increased vimentin protein (Fig. 5C). Similar to Nedd4-2Ksp1.3 mice, β-catenin levels were elevated in Nedd4-2 KO cells (Fig. 5C). β-catenin was increased in the nuclear fraction of Nedd4-2 KO cells and was localized to the nucleus as well as the cell membrane, particularly after TGF-β stimulation (Fig. 5D, E and Supplementary Fig. 4C). DVL2 levels were unchanged (Supplementary Fig. 4D) indicating that the increase in β-catenin is independent of DVL2-dependent Wnt signaling. In addition, pSMAD2 and 3 levels were increased in KO cells and this was exacerbated by TGF-β treatment (Fig. 5F). Inhibition of TGF-βR1 using LY-364947 significantly reduced Vimentin levels in Nedd4-2 KO cells (Fig. 5G), even after stimulation with TGF-β1 (Supplementary Fig. 4E), confirming the contribution of TGF-β signaling to the Nedd4-2 KO phenotype.
CRISPR–Cas9 was used to generate CCD-N21 clones lacking NEDD4-2 or wild-type control clones. A Immunoblot analysis of NEDD4-2 and Vimentin with GAPDH as a loading control. B qRT-PCR analysis of Vimentin relative to TBP in untreated cells, 2.5 ng/ml TGF-β1 or 10 mM LiCl treated for 72 h, n = 4 for 4 independent clones each. C Immunoblot analysis of NEDD4-2, vimentin, and β-catenin with GAPDH as a loading control, untreated or after 72 h TGF-β1 stimulation. Numbers indicate quantitation of β-catenin bands relative to untreated wild type lane, normalized to GAPDH. D Immunoblot analysis of β-catenin after separation of cytoplasmic (marked by α-tubulin) and nuclear (marked by Histone 3, H3) fractions. Numbers indicate quantitation of β-catenin bands relative to untreated wild-type lane, normalized to α-tubulin for a cytoplasmic fraction or H3 for nuclear fraction. E Immunostaining of β-catenin (green) with and without TGF-β1 treatment. DNA is stained by Hoechst (blue). Scale bar: 50 μm. F Immunoblot analysis of pSMAD2/3 and total SMAD2/3 with GAPDH as a loading control. Numbers indicate quantitation of pSMAD bands relative to untreated wild-type lane, normalized to β-actin. G qRT-PCR analysis of Vimentin relative to TBP in untreated or 72 h 500 nM LY-364947 treated Nedd4-2 KO cells, n = 4. Data presented as means ± SEM (B, G) and significance determined using unpaired two-tailed Student’s t test. *P < 0.05, **P < 0.01. Immunoblots are representative of three experiments with similar results.
We next analyzed Nedd4-2 KO cells for evidence of EMT as Wnt/β-catenin and TGF-β signaling are implicated in this process. Wild-type cells displayed a classic epithelial cobblestone morphology, whereas Nedd4-2 KO cells were elongated with cellular protrusions, often positive for vimentin expression (Fig. 6A, B). Vimentin has been shown to promote cellular migration and a migratory phenotype is a characteristic of mesenchymal cells27. Consistent with this, Nedd4-2 KO CCD cells demonstrated significantly increased migration compared to wild-type cells (Fig. 6C, D). In addition, Nedd4-2 KO cells displayed an increase in N-cadherin, further exacerbated by TGF-β1 stimulation (Fig. 6E). Total E-cadherin levels remained stable after 3 days, although cytoplasmic accumulation was observed in response to the dissolution of adherens junctions (Fig. 6F, G). A further 3 days of TGF-β1 treatment showed a marked reduction in E-cadherin protein abundance and membrane localization. Together, these data demonstrate that loss of NEDD4-2 in CCD cells activates Wnt/β-catenin and TGF-β signaling, which is sufficient to drive partial EMT.
A Light microscopy of wild type and Nedd4-2 KO cells, scale bar: 200 μm (with higher magnification inset) or immunostaining of vimentin (green) and Phalloidin (red) with Hoechst staining of DNA (blue). Scale bar: 50 μm. B Number of vimentin-positive cells per field of view, n = 4 clones, each with three fields of view. C Light microscopy of cells showing migration distance after scratch, scale bar: 200 μm. D Migration distance quantitated in four clones, n = 3 replicates each. E Immunoblot analysis of N-cadherin with GAPDH as a loading control, untreated or after 72 h 2.5 ng/ml TGF-β1 stimulation. Numbers indicate quantitation of N-cadherin bands relative to untreated wild-type lane, normalized to GAPDH. F Immunoblot analysis of E-cadherin with β-actin as a loading control, untreated or after 3 or 6 days of TGF-β1 stimulation. Numbers indicate quantitation of E-cadherin bands relative to untreated wild type lane, normalized to β-actin. G Immunostaining of E-cadherin (green) after 0, 3, and 6 days of TGF-β1 treatment. DNA is stained by Hoechst (blue). Scale bar: 50 μm. Data presented as means ± SEM (B, D) and significance determined using unpaired two-tailed Student’s t test. ****P < 0.0001.
Low Na+ diet inhibits Wnt/β-catenin/TGF-β signaling and kidney disease in Nedd4-2 Ksp1.3 mice
To investigate whether low dietary Na+ could rescue Wnt/β-catenin/TGF-β signaling and kidney damage in adult mice, a low Na+ (0.05%) diet was fed to adult mice after the renal disease had been established. Levels of DVL2, β-catenin, pSMAD2, and pSMAD3 were similar in control and Nedd4-2Ksp1.3 kidneys (Fig. 7A). Similarly, immunostaining showed comparable levels of E-cadherin, αSMA, and β-catenin in Nedd4-2Ksp1.3 and control kidneys after the low Na+ diet (Fig. 7B). Correlating with reduced collagen and ECM deposition, vimentin was no longer increased (Fig. 7B). Whilst Kim-1 expression was still increased in Nedd4-2Ksp1.3 kidneys, this was to a much lower extent when compared with mice fed a low Na+ diet (Figs. 7C and 3B). Vimentin and Collagen levels were no longer significantly different from controls. Importantly, polydipsia and polyuria were no longer present (Fig. 7D), and histological analysis revealed correction of renal pathology (Fig. 7E). GFR was not significantly different between control and Nedd4-2Ksp1.3 mice. Blood creatinine and urea levels were similar in both genotypes (Fig. 7F), with no other serum differences in electrolytes or kidney function (Table 2). Other than decreased creatinine and increased Ca2+, urine electrolyes were stablized. Collectively, these data demonstrate that kidney disease in Nedd4-2Ksp1.3 mice is rescued by low dietary Na+, due at least in part to the inhibition of Wnt/β-catenin and TGF-β signaling.
Mice were fed a low Na+ (0.05%) diet for 17 days. A Immunoblot analysis of DVL2, β-catenin, pSMAD2/3, and total SMAD2/3 with GAPDH as loading controls. B Immunostaining of αSMA (green) with E-cadherin (red), β-catenin (green) with DBA (red), and vimentin (green) with DBA (red). DNA is stained by Hoechst (blue). Scale bar: 50 μm. C qRT-PCR analysis of genes relative to TBP. D Water intake and urine output over 24 h in metabolic cages. E Haematoxylin and Eosin staining of kidneys, scale bar: 100 μm, 1 mm for inset. F GFR calculated from 24 h urine and blood collection, and serum creatinine and urine levels. Data presented as means ± SEM and significance determined using unpaired two-tailed Student’s t test, n = 3-4. *P < 0.05.
Discussion
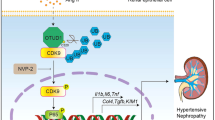
Variants of human NEDD4L are associated with renal disease in humans28 and loss of Nedd4-2 in mice results in CKD-like pathology8. In this study, we demonstrate that high dietary Na+ fed to adult Nedd4-2Ksp1.3 mice drives disease toward ESRD, associated with increased Wnt/β-catenin/TGF-β signaling and resulting in renal damage and fibrosis (Fig. 8). Nedd4-2 KO in CCD cells confirms the direct involvement of NEDD4-2 in regulating Wnt/β-catenin/TGF-β signaling as well as the role of NEDD4-2 in EMT, potentially contributing to the phenotype in mice.
NEDD4-2 regulates several molecular processes that contribute to the progression of kidney disease, including Na+ transport and fibrotic signaling pathways. This occurs through ubiquitination of substrates such as ENaC (and other channels and transporters) as well as downstream components of Wnt/β-catenin/TGF-β1 signaling pathways. In the absence of NEDD4-2, increased Na+ contributes to sustained tubular damage with a mild reduction in kidney function. Sustained signaling through Wnt and TGF-β1 pathways, particularly as a result of high dietary Na+, drives increased kidney damage and severely decreased renal function suggesting progression from CKD to ESRD. Figure created with BioRender.com.
GFR and other parameters of renal function were significantly altered by high dietary Na+ in Nedd4-2Ksp1.3 mice. Together with the weight loss and lethargy of these animals, this is indicative of progression towards ESRD. Increased Na+ has been shown to produce a significant rise in BP which can subsequently induce renal damage29. However renal damage can be exacerbated by high Na+ in the absence of hypertension30,31. Given that hypertension is common in several models of Nedd4-2 deficiency6 and NEDD4L polymorphisms in humans6, it was hypothesized that high dietary Na+ may exacerbate kidney damage via increased BP in Nedd4-2Ksp1.3 mice. In this study, although a trend towards increased BP in Nedd4-2Ksp1.3 mice on standard Na+ was evident, high Na+ diet feeding resulted in decreased BP. This eliminates the possibility that exacerbated kidney disease is caused by Na+-induced hypertension, but rather may reflect volume depletion caused by osmotic diuresis. Both polydipsia and polyuria in Nedd4-2Ksp1.3 mice were exacerbated by high Na+, reminiscent of nephrogenic diabetes insipidus, often associated with kidney damage and mutations in aquaporin water channels32,33,34. Indeed, Nedd4-2Ksp1.3 kidneys displayed a reduction in the water channels AQP2 and AQP3. Hence, osmotic diuresis and subsequent volume and hemodynamic changes are likely to contribute to kidney damage in Nedd4-2Ksp1.3 mice.
High dietary Na+ led to increased areas of vimentin-positive interstitial ECM deposition throughout the renal parenchyma. Vimentin has also been observed in injured tubular cells during tubular degeneration, and during regeneration in acute injury35,36; however, was not observed in tubules of Nedd4-2Ksp1.3 kidneys. Interestingly, E-cadherin staining was elevated in Nedd4-2Ksp1.3 kidneys, with a pronounced basolateral and partial apical localization, in contrast to many models of kidney disease where a reduction of E-cadherin is associated with EMT and fibrosis. This is similar to increased E-cadherin in the ligated kidney following ureteric obstruction, particularly in dilated distal tubules surrounded by inflamed interstitium37. Hypotonic stress can act as an activator of mechanical stretch-induced E-cadherin in cells38, suggesting that increased tubular pressure may contribute to elevated E-cadherin. Furthermore, an increase in a 30 kD E-cadherin fragment was also observed, which may reflect apoptotic cells as caspase-3 cleaves the C terminus to leave a fragment of this size21.
Transient activation of Wnt/β-catenin signaling has been associated with kidney injury and recovery, whereas sustained activation of this signaling contributes to renal fibrosis and disease progression, via activation of key fibrosis-related genes14,15,39. In the Nedd4-2Ksp1.3 model of kidney disease, mRNA expression of Wnt ligands were generally increased, with activation of distinct subsets of ligands sustained by high Na+, culminating in β-catenin upregulation. This is similar to the sustained activation of Wnt ligands observed in the progression of ischemia-/reperfusion-induced AKI to CKD14. In vitro, NEDD4-2 has been shown to regulate Wnt signaling by targeting DVL2 for degradation23. Although not observed in CCD cells in vitro, levels of DVL2 were also upregulated in the kidneys of Nedd4-2Ksp1.3 mice, further exaggerated by high Na+. Hence, whilst DVL2 may also be regulated by other components of the pathway, our work provides evidence of DVL2 regulation by NEDD4-2 in vivo. Inhibition of WNT signaling by several factors has been shown to protect against kidney fibrosis39. The identification of a role for NEDD4-2 in regulating in this pathway in the kidney adds to this understanding, contributing to the identification of potential future therapeutic targets to halt CKD progression.
TGF-β1-induced fibrosis is a key feature of progressive renal disease40, and high dietary Na+ can increase levels of TGF-β1 in the glomerulus and tubules12. NEDD4-2 directly binds to TGF-β activated pSMAD2/3 to ubiquitinate and target these proteins for degradation24. Conditional KO of Nedd4-2 in the lung results in elevated TGF-β signaling via increased pSMAD2/3, which culminates in progressive pulmonary fibrosis41. Upregulation of pSMAD2 and pSMAD3 in this study implicates this pathway in kidney disease in Nedd4-2Ksp1.3 mice, particularly after high Na+.
In addition to ECM deposition, both TGF-β and Wnt/β-catenin signaling have been associated with EMT in kidney disease. High dietary Na+ has also been implicated in EMT in rats42,43. However, the contribution of EMT to renal fibrosis has been highly debated35, and direct evidence of EMT was not observed in Nedd4-2Ksp1.3 kidneys. Nevertheless, KO of Nedd4-2 in the CCD cell line resulted in an elongated morphology, increased vimentin and migration rate, and loss of E-cadherin after TGF-β1 stimulation. This implies a partial EMT phenotype after the loss of NEDD4-2 and a sensitivity of these cells to EMT induction by TGF-β1. Importantly, low dietary Na+ ameliorated kidney disease in Nedd4-2Ksp1.3 mice, due at least in part to repressed Wnt/β-catenin/TGF-β signaling. Thus, TGF-β and Wnt/β-catenin signaling is important in the Na+-induced progression of renal disease, and in parenchymal damage caused by fibrosis, inflammation and tubular cell death. In conclusion, we propose that NEDD4-2 prevents CKD and its progression to ESRD via targeting key proteins involved in Na+ homeostasis and fibrotic signaling (Fig. 8). Given the growing association of NEDD4L variants and expression levels with human kidney diseases, this study sheds understanding on how known signaling pathways may be aberrantly regulated in disease and provides a novel potential diagnostic and therapeutic target.
Materials and methods
Ethics approval
All animal studies were approved by the institutional ethics and biosafety committees of the University of South Australia and were carried out according to the National Health and Medical Research Council of Australia guidelines.
Mouse lines and sample collection
Kidney-specific Nedd4-2-deficient mice (Nedd4-2Ksp1.3) were generated in our laboratory previously8,44 and bred at the University of South Australia core animal facility (Adelaide, Australia) under specific pathogen-free conditions. Male mice at 6–8 weeks of age were fed standard sodium chow (0.2% Na+), high sodium chow (3.1% Na+) or low sodium chow (0.05% Na+) (Specialty Feeds, WA, Australia) for 17 days. At the time of collection, after the final metabolic cage, mice were anesthetized, blood collected by cardiac puncture, and organs dissected after cervical dislocation. Capsules were removed from the kidneys and placed into a Histochoice reagent (ProSciTech, Kirwan, QLD, Australia) for histological analysis of paraffin-embedded or frozen samples. For paraffin samples, kidneys were transferred to 70% ethanol and then embedded in paraffin. Kidneys for frozen sectioning were soaked in 30% sucrose overnight before being embedded in OCT (ProSciTech, Kirwan, QLD, Australia). The remaining kidney was snap-frozen in liquid nitrogen for immunoblot or mRNA analysis. Nine mice of each genotype, for each diet condition, were analyzed.
Histological analysis
Sections (5 μm) were cut using a paraffin microtome, de-paraffinized with xylene, and dehydrated through a graded series of ethanol. Slides were stained with Hematoxylin and Eosin using standard protocols. To evaluate collagen deposition using picrosirius red, slides were stained for 1 h in saturated picric acid with 0.1% Direct Red 80 (Sigma-Aldrich), then washed in 0.01 N hydrochloric acids for 2 min. Digital images were acquired by using a NanoZoomer (Hamamatsu).
BP measurements
BP was measured by a tail-cuff system (CODA Monitor; Kent Scientific Corporation, Torrington, CT, USA) essentially as described previously8. Briefly, at the same time each morning, BP was recorded for 5 acclimation cycles followed by 30–35 measurement cycles. The three highest and lowest readings were removed and an average of the remaining recordings used to calculate the daily BP. An average of 3 days was used to calculate the BP at day 0 before the diet was changed, and an average of BP at post-diet days 13, 14, and 16 used to calculate the final BP. Heating pads were used to keep the mice warm throughout the experiment to ensure sufficient blood flow to the tail.
Blood and urine analyses
Electrolytes and other parameters of kidney function in blood were carried out by SA Pathology (Adelaide, Australia). Urine osmolality was measured using an Advanced 3320 osmometer (Advanced Instruments) and other parameters using an Advia 2400 chemistry system (Siemens). Serum aldosterone levels were measured using an Aldosterone Elisa Kit (Abcam, ab136933) and analyzed on a Spark 10 M microplate reader (Tecan, Switzerland).
Metabolic cage studies
Mice were housed in metabolic cages for a 6 h training session, followed by a 24 h session the following day. Mice were provided ad libitum access to deionized drinking water and pelleted chow. At the end of the experiment, urine was collected, and blood and tissue samples were taken as described above.
Generation of Nedd4-2 KO CCD cell lines
Single guide RNA targeting a region in exon 10 of mouse Nedd4-2 was cloned into the Px459 vector (Addgene), following the Zhang Genome engineering CRISPR–Cas9 protocol45. The following primers were used: F: 5′ CACCGACCGACGCTTCCGCTCTCGG 3′, R: 5′ AAACCCGAGTGCGGAAGCGTCGGTC 3′. The plasmid was transfected using Lipofectamine 3000 reagent (Invitrogen), with an empty Px459 vector serving as a control to generate wild-type clones. After 24 h, media was replaced and supplemented with 2.5 μg/mL puromycin (Sigma-Aldrich) for a further 24 h to select for transfected cells. Surviving cells were passaged at low seeding density and single colonies selected and propagated. Deletion of Nedd4-2 was confirmed by sequencing of the region in exon 10 and immunoblotting for NEDD4-2 protein. Four clonal cell lines were selected for analysis for both NEDD4-2 KO and wild-type (empty Px459 vector) controls.
Cell studies
CCD-N21 cells were grown in DMEM/F12 media (Gibco) supplemented with 2% fetal calf serum, 1% ITS, 1 nM 3,3′,5-triiodo-L-thyronin, 10 ng/mL EGF and 50 nM dexamethasone (all from Sigma-Aldrich). Cells were grown at 37 °C in a humidified atmosphere of 5% CO2 and passaged twice a week. For experiments in which cells were treated, 1 × 105 cells per well were seeded in 6-well plates for 72 h, or 6 days, before collection and grown in the presence or absence of 2.5 ng/ml recombinant mouse TGF-β1 (R&D Systems, 7666-MB), 10 mM LiCl (Sigma-Aldrich, 746460) or 500 nM LY-364947 (Sigma-Aldrich, L6293). For subcellular fractionation, cells were lysed in ice-cold cytoplasmic extraction buffer (10 mM HEPES, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, 0.05% NP40, and HALT protease and phosphatase inhibitor cocktail [Thermo Fisher Scientific] pH 7.9) for 10 min on ice, centrifuged at 3000 rpm for 10 min and supernatant reserved. The nuclear protein-containing pellet was then resuspended in ice-cold nuclear extraction buffer (5 mM HEPES, 1.5 mM MgCl2, 300 mM NaCl, 0.2 mM EDTA, 0.5 mM DTT, and 26% glycerol, pH 7.9), sonicated, and left on ice for 30 min before centrifuging at 16,000 rpm for 20 min with supernatant reserved. For wound closure (scratch) assay, cells were grown to full confluency before a vertical scratch in the cell layer was performed using a 20 μl pipette tip. Cells were washed with fresh media added and images were taken at 0, 8, and 24 h post scratch. The average distance migrated from each leading edge was measured for each time point.
Immunoblotting
For mouse samples, half of each kidney was lysed in ice-cold extraction buffer at pH 7.5 (50 mM Tris-HCl pH7.5, 1 mM EDTA, 1 mM EGTA, 0.27 M sucrose, 0.1% β-mercaptoethanol and HALT protease and phosphatase inhibitor cocktail [Thermo Fisher Scientific]). Tissue was homogenized, frozen in liquid nitrogen, immediately thawed, and incubated at 4 °C on a nutator for 30 min and centrifuged at 13,000 rpm for 5 min. For cell studies, cells were lysed in Verhagen lysis buffer (150 mM NaCl, 20 mM Tris-HCl pH 7.5, 2 mM EDTA, 10% glycerol, and 1% Triton-X) and lysed as above. For pSMAD2/3 immunoblots, cells were lysed in sodium dodecyl sulfate (SDS) lysis buffer (150 mM NaCl, 10 mM Tris-HCl pH 8.0, 2% SDS), boiled for 5 min then centrifuged at 13,000 rpm for 5 min. Supernatant protein (25 μg) was combined with protein load buffer (100 mM Tris-HCl pH 6.8, 200 mM DTT, 4% SDS, 0.2% bromophenol blue, 20% glycerol), heated at 37 °C for 30 min, loaded onto 4–20% precast sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels (Bio-Rad) and transferred to PVDF membrane using the Trans-blot Turbo instrument (Bio-Rad). Membranes were blocked with 5% skim milk in TBS-T (Tris-buffered saline, 0.05% Tween 20) and primary antibodies added; anti-AQP2 (Abcam, ab65837, 1:1000), anti-AQP3 (Abcam, ab125219, 1:1000), anti-vimentin (Abcam, #ab92547, 1:1000), anti-αSMA (Abcam, ab7817, 1:2000), anti-E-cadherin (Cell Signaling Technology, #24E10, 1:1000), anti-GAPDH (Cell Signaling Technology, 14C10, 1:10000), anti-β-catenin (Cell Signaling Technology, 6B3, 1:1000), anti-pSMAD2 (Thermo Fisher, 44244 G, 1:1000), anti-pSMAD3 (Abcam, ab52903, 1:1000), anti-SMAD2/3 (Cell Signaling Technology, 8685P, 1:1000), anti-TGF-βR1 (Invitrogen, PA5-32631, 1:1000), anti-DVL2 (Cell Signaling Technology, 3216, 1:1000), anti-NEDD4-2 (in house46, 1:1000), anti-α-Tubulin (Abcam, ab4074, 1:1000), anti-H3 (Abcam, ab176842, 1:1000), anti-β-actin (Sigma, AM4302, 1:2000), anti-N-cadherin (Cell Signaling Technology, 14215, 1:1000). Horseradish peroxidase secondary antibodies (Millipore) was added at 1:2000 and developed with ECL Prime (GE Healthcare) or West Femto (Thermo Scientific). GAPDH was developed using Cy5 secondary antibody (GE Healthcare). Images were acquired on a ChemiDoc Touch Imager (BioRad). Quantitation was conducted using Image Lab Software (BioRad), with each band normalized to GAPDH or β-actin.
Immunostaining
Paraffin sections (5 μm) were deparaffinized and hydrated in a graded ethanol series. Heat-mediated antigen retrieval was carried out by boiling for 10 min in 10 mM citric acid solution (pH 6). Tissue sections were blocked with 10% goat serum. Primary antibodies used were as above (all at 1:200): AQP2, AQP3, Vimentin, and E-cadherin. Additional antibodies were β-catenin (BD Transduction Labs, 610154); rhodamine-labeled DBA (Vector Laboratories, RL-1032); FITC-conjugated mouse anti-αSMA (Sigma-Aldrich, F3777). For immunostaining of cells, cells were grown on coverslips and fixed for 15 min in 4% PFA, then blocked for 1 h in 1% BSA with 0.5% Triton-X. Sections were incubated with primary antibody (as above) and Alexa Fluoro 594 Phalloidin (Thermo Fisher, A12381) overnight at 4 °C and with secondary for 1 h at room temperature.
Sections were then incubated with the corresponding fluorescently tagged secondary antibody (AlexaFluor-488 or AlexaFluor-568, Thermo Fisher Scientific), counterstained with Hoechst 33342 (Thermo Fisher) and mounted in Prolong Gold Antifade reagent (Invitrogen). Stained samples were imaged using an LSM 800 confocal microscope using Zen 2011 (Black Edition) version 8.1.5.484 (Carl Zeiss Microscopy, Jena, Germany). Image analysis was conducted using Adobe image suite software.
Quantitative real-time PCR
Total RNA was isolated from half of each kidney using TRIzol Reagent (Life Technologies) and RNA was reverse-transcribed with a High Capacity cDNA reverse transcription kit (Applied Biosciences). qRT-PCR was performed and analyzed as described47, where all data are normalized to TBP (TATA box binding protein) levels. Primer sequences are Collagen-1 (Col1a1) F: CGGAGAAGAAGGAAAACGAGGAG, R: CACCATCAGCACCAGGGAAAC. Vimentin, F: CGGCTGCGAGAGAAATTGC, R: CCACTTTCCGTTCAAGGTCAAG. Kidney injury molecule 1 (Kim-1), F: TGGTTGCCTTCCGTGTCTCT, R: TCAGCTCGGGAATGCACAA, Wnt1, F: TCAGAACCGCAGCACAGAAC, R: TTCACGATGCCCCACCATC. Wnt3, F: GGGGCGTATTCAAGTAGCTG, R: GTAGGGACCTCCCATTGGAT. Wnt4, F: CGAGCAATTGGCTGTACCTGG, R: CAGGCCTTTGAGTTTCTCGC. Fibronectin, F: GAAGACAGATGAGCTTCCCCA, R: GGTTGGTGATGAAGGGGGTC. Serpine, F: ACAACCCGACAGAGACAATCC, R: TCGTCCCAAATGAAGGCGTC. Twist, F: GAGGTCTTGCCAATCAGCCA, R: CCAGTTTGATCCCAGCGTTT. cMyc, F: TGTTCTCTGCCTCTGCCCG, R: GCATCGTCGTGGCTGTCTG. Cyclin D1, F: CCTCTCCTGCTACCGCACAA, R: TTGTTTAGCCAGAGGCCGGT. TGF-β1, F: GATACGCCTGAGTGGCTGTC, R: AAGCCCTGTATTCCGTCTCC. TATA-box binding protein (TBP), F: CAAACCCAGAATTGTTCTCCTT, R: ATGTGGTCTTCCTGAATCCCT.
Statistical analysis of data
Statistical analysis was performed using GraphPad Prism software (v6.0). A Mann–Whitney test for non-parametric data was used to assess changes in blood/urine parameters in Tables 1 and 2, and one-way ANOVA used to assess repeated measurements for BP and weight changes. The remaining data were analyzed using an unpaired two-tailed Student’s t test. A P value of ≤0.05 was considered significant. All values are presented as mean ± SEM (or ±SD for immunoblot quantitation), as indicated in the figure legends.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Cockwell, P. & Fisher, L. A. The global burden of chronic kidney disease. Lancet 395, 662–664 (2020).
McMahon, E. J., Campbell, K. L., Mudge, D. W. & Bauer, J. D. Achieving salt restriction in chronic kidney disease. Int. J. Nephrol. 2012, 720429 (2012).
Oppelaar J. J. & Vogt L. Body fluid-independent effects of dietary salt consumption in chronic kidney disease. Nutrients 11, 2779 (2019).
Butterworth, M. B. Regulation of the epithelial sodium channel (ENaC) by membrane trafficking. Biochim. Biophys. Acta 1802, 1166–1177 (2010).
Goel, P., Manning, J. A. & Kumar, S. NEDD4-2 (NEDD4L): the ubiquitin ligase for multiple membrane proteins. Gene 557, 1–10 (2015).
Manning, J. A. & Kumar, S. Physiological functions of Nedd4-2: lessons from knockout mouse models. Trends Biochem. Sci. 43, 635–647 (2018).
Wilson, P. C. et al. The single-cell transcriptomic landscape of early human diabetic nephropathy. Proc. Natl Acad. Sci. USA 116, 19619–19625 (2019).
Henshall, T. L. et al. Deletion of Nedd4-2 results in progressive kidney disease in mice. Cell Death Differ. 24, 2150–2160 (2017).
Manning J. A. et al. Dietary sodium modulates nephropathy in Nedd4-2-deficient mice. Cell Death Differ. 27, 1832–1843 (2020).
Zaidan M. et al. Signaling pathways predisposing to chronic kidney disease progression. JCI Insight 5, e126183 (2020).
Zhou, B. et al. Interactions between beta-catenin and transforming growth factor-beta signaling pathways mediate epithelial-mesenchymal transition and are dependent on the transcriptional co-activator cAMP-response element-binding protein (CREB)-binding protein (CBP). J. Biol. Chem. 287, 7026–7038 (2012).
Hovater, M. B. & Sanders, P. W. Effect of dietary salt on regulation of TGF-beta in the kidney. Semin. Nephrol. 32, 269–276 (2012).
Wei, S. Y. et al. Multiple mechanisms are involved in salt-sensitive hypertension-induced renal injury and interstitial fibrosis. Sci. Rep. 7, 45952 (2017).
Xiao, L. et al. Sustained activation of Wnt/beta-catenin signaling drives AKI to CKD progression. J. Am. Soc. Nephrol. 27, 1727–1740 (2016).
Tan, R. J., Zhou, D., Zhou, L. & Liu, Y. Wnt/beta-catenin signaling and kidney fibrosis. Kidney Int. Suppl. (2011) 4, 84–90 (2014).
Zhang, J., Tian, X. J. & Xing, J. Signal transduction pathways of EMT induced by TGF-beta, SHH, and WNT and their crosstalks. J. Clin. Med. 5, 41 (2016).
Sheng, L. & Zhuang, S. New insights into the role and mechanism of partial epithelial-mesenchymal transition in kidney fibrosis. Front. Physiol. 11, 569322 (2020).
Krishnan, R., Eley, L. & Sayer, J. A. Urinary concentration defects and mechanisms underlying nephronophthisis. Kidney Blood Press. Res. 31, 152–162 (2008).
Takata, K., Matsuzaki, T., Tajika, Y., Ablimit, A. & Hasegawa, T. Localization and trafficking of aquaporin 2 in the kidney. Histochem. Cell Biol. 130, 197–209 (2008).
Saratlija Novakovic, Z. et al. The interstitial expression of alpha-smooth muscle actin in glomerulonephritis is associated with renal function. Med. Sci. Monit. 18, CR235–CR240 (2012).
Keller, S. H. & Nigam, S. K. Biochemical processing of E-cadherin under cellular stress. Biochem. Biophys. Res. Commun. 307, 215–223 (2003).
Wang, Y., Zhou, C. J. & Liu, Y. Wnt signaling in kidney development and disease. Prog. Mol. Biol. Transl. Sci. 153, 181–207 (2018).
Ding, Y., Zhang, Y., Xu, C., Tao, Q. H. & Chen, Y. G. HECT domain-containing E3 ubiquitin ligase NEDD4L negatively regulates Wnt signaling by targeting dishevelled for proteasomal degradation. J. Biol. Chem. 288, 8289–8298 (2013).
Gao, S. et al. Ubiquitin ligase Nedd4L targets activated Smad2/3 to limit TGF-beta signaling. Mol. Cell 36, 457–468 (2009).
Kuratomi, G. et al. NEDD4-2 (neural precursor cell expressed, developmentally down-regulated 4-2) negatively regulates TGF-beta (transforming growth factor-beta) signalling by inducing ubiquitin-mediated degradation of Smad2 and TGF-beta type I receptor. Biochem. J. 386, 461–470 (2005).
Clement-Lacroix, P. et al. Lrp5-independent activation of Wnt signaling by lithium chloride increases bone formation and bone mass in mice. Proc. Natl Acad. Sci. USA 102, 17406–17411 (2005).
Battaglia R. A., Delic S., Herrmann H., Snider N. T. Vimentin on the move: new developments in cell migration. F1000Res. 7 (2018).
Fouladkou, F. et al. A naturally occurring human Nedd4-2 variant displays impaired ENaC regulation in Xenopus laevis oocytes. Am. J. Physiol. Ren. Physiol. 287, F550–F561 (2004).
Grillo A., Salvi L., Coruzzi P., Salvi P. & Parati G. Sodium intake and hypertension. Nutrients 11, 1970 (2019).
Heerspink, H. J., Navis, G. & Ritz, E. Salt intake in kidney disease—a missed therapeutic opportunity? Nephrol. Dial. Transpl. 27, 3435–3442 (2012).
Washino, S., Hosohata, K., Jin, D., Takai, S. & Miyagawa, T. Early urinary biomarkers of renal tubular damage by a high-salt intake independent of blood pressure in normotensive rats. Clin. Exp. Pharm. Physiol. 45, 261–268 (2018).
Deen, P. M. Mouse models for congenital nephrogenic diabetes insipidus: what can we learn from them? Nephrol. Dial. Transpl. 22, 1023–1026 (2007).
Grunfeld, J. P. & Rossier, B. C. Lithium nephrotoxicity revisited. Nat. Rev. Nephrol. 5, 270–276 (2009).
Markowitz, G. S. et al. Lithium nephrotoxicity: a progressive combined glomerular and tubulointerstitial nephropathy. J. Am. Soc. Nephrol. 11, 1439–1448 (2000).
Kriz, W., Kaissling, B. & Le Hir, M. Epithelial-mesenchymal transition (EMT) in kidney fibrosis: fact or fantasy? J. Clin. Investig. 121, 468–474 (2011).
Grone, H. J., Weber, K., Grone, E., Helmchen, U. & Osborn, M. Coexpression of keratin and vimentin in damaged and regenerating tubular epithelia of the kidney. Am. J. Pathol. 129, 1–8 (1987).
Docherty, N. G. et al. Increased E-cadherin expression in the ligated kidney following unilateral ureteric obstruction. Kidney Int. 75, 205–213 (2009).
Kippenberger, S. et al. Hypotonic stress induces E-cadherin expression in cultured human keratinocytes. FEBS Lett. 579, 207–214 (2005).
Schunk, S. J., Floege, J., Fliser, D. & Speer, T. WNT-beta-catenin signalling—a versatile player in kidney injury and repair. Nat. Rev. Nephrol. 17, 172–184 (2021).
Meng, X. M., Tang, P. M., Li, J. & Lan, H. Y. TGF-beta/Smad signaling in renal fibrosis. Front. Physiol. 6, 82 (2015).
Duerr, J. et al. Conditional deletion of Nedd4-2 in lung epithelial cells causes progressive pulmonary fibrosis in adult mice. Nat. Commun. 11, 2012 (2020).
Pletinck, A. et al. Salt intake induces epithelial-to-mesenchymal transition of the peritoneal membrane in rats. Nephrol. Dial. Transpl. 25, 1688–1696 (2010).
Wang, Y. et al. Salt-induced epithelial-to-mesenchymal transition in Dahl salt-sensitive rats is dependent on elevated blood pressure. Braz. J. Med. Biol. Res. 47, 223–230 (2014).
Boase, N. A. et al. Respiratory distress and perinatal lethality in Nedd4-2-deficient mice. Nat. Commun. 2, 287 (2011).
Ran, F. A. et al. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013).
Konstas, A. A. et al. Regulation of the epithelial sodium channel by N4WBP5A, a novel Nedd4/Nedd4-2-interacting protein. J. Biol. Chem. 277, 29406–29416 (2002).
Foot, N. J., Gembus, K. M., Mackenzie, K. & Kumar, S. Ndfip2 is a potential regulator of the iron transporter DMT1 in the liver. Sci. Rep. 6, 24045 (2016).
Acknowledgements
We are grateful to Johannes Loffing for ENaC antibodies and the UniSA Core Animal Facility staff for animal advice and husbandry.
Funding
This work was supported by a National Health and Medical Research Council (NHMRC) of Australia Senior Principal Research Fellowship (GNT1103006), an NHMRC project grant (GNT1099307), and a research support package from the University of South Australia to S.K., as well as a Biomed City Project Grant from The Hospital Research Foundation (THRF) to S.K. and J.A.M.
Author information
Authors and Affiliations
Contributions
J.A.M., S.S.S., and S.K. conceived the project. J.A.M. designed and directed mouse studies. J.A.M. and S.S.S. executed experiments and analyzed the data. A.N. generated Nedd4-2 KO CCD cell lines. A.N. and T.L.H. characterized KO cell lines, and A.N., T.L.H., J.A.M., Y.K.G., and S.K. planned in vitro experiments and analyzed data. J.A.M. drafted the paper text and figures. All authors discussed and reviewed the paper.
Corresponding authors
Ethics declarations
Ethics statement
All animal studies were approved by the institutional ethics and biosafety committees of the University of South Australia and were carried out according to the National Health and Medical Research Council of Australia guidelines.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by M. Agostini
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Manning, J.A., Shah, S.S., Nikolic, A. et al. The ubiquitin ligase NEDD4-2/NEDD4L regulates both sodium homeostasis and fibrotic signaling to prevent end-stage renal disease. Cell Death Dis 12, 398 (2021). https://doi.org/10.1038/s41419-021-03688-7
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41419-021-03688-7