Abstract
Lung adenocarcinoma (LUAD) has high incidence and mortality rates worldwide; however, its detailed molecular pathology remains unclear. Although circRNAs have gradually been identified as molecules that are differentially expressed in tumors and play key roles in tumor progression, their role in LUAD is poorly understood. Through microarray analysis, we obtained the circRNA expression profile of LUAD and found that circ-HMGA2 (hsa_circ_0027446), a novel RNA, is highly expressed in LUAD. The high expression of circ-HMGA2 was further verified in 36 paired LUAD and adjacent normal tissues. Functionally, circ-HMGA2 promoted LUAD cell metastasis in vitro and in vivo. The luciferase reporter assay and FISH results showed that circ-HMGA2 interacts with miR-1236-3p and that miR-1236-3p interacts with ZEB1. In addition, miR-1236-3p was expressed at low levels in LUAD, inhibited LUAD cell metastasis, and suppressed the function of circ-HMGA2. ZEB1 is an EMT-promoting transcription factor. The PCR and WB analysis results showed that circ-HMGA2 promotes both ZEB1 expression and EMT. MiR-1236-3p had the opposite effect, reversing the promotive effect of circ-HMGA2 on EMT. In summary, circ-HMGA2 promotes LUAD cell metastasis through the miR-1236-3p/EMT axis, indicating that it could be a therapeutic target in LUAD.
Similar content being viewed by others
Introduction
Lung cancer is the leading cause of cancer death, and its prognosis remains poor worldwide1. Lung adenocarcinoma (LUAD) is the most common type of lung tumor, accounting for 40% of lung tumors2. The average survival rate of patients with LUAD is very low, ranging from 4 to 17%3. Although surgery and other treatments for LUAD have greatly improved in recent years, the 5-year relative survival rate of LUAD is still very low4. Tumor metastasis is the leading cause of death in most cancer patients, including those with LUAD5,6. Therefore, discovering the mechanism of LUAD metastasis to improve early treatment and prolong the survival time of patients is an urgent need.
The mechanism of tumor metastasis is complex and poorly understood7. Epithelial-mesenchymal transition (EMT), during which epithelial cells gradually lose cell adhesion, undergo alterations in the cytoskeletal composition, and acquire mesenchymal traits, is an important component of the metastatic process8,9. Recently, accumulating evidence has shown that EMT is closely related to LUAD metastasis10. Zinc finger E-box binding homeobox 1 (ZEB1) has been reported to be a transcription factor (TF) that induces EMT and the metastasis of cells in diverse types of cancers11. ZEB1 orchestrates EMT by suppressing the expression of epithelial genes and upregulating mesenchymal markers12. In LUAD, expression of ZEB1 is an early critical event in tumor metastasis, indicating a reduced therapeutic response and poor prognosis13,14. To enhance the understanding of the roles of ZEB1 and EMT in LUAD, more in-depth study is needed.
Circular RNA (circRNA), as a novel form of RNA, has recently received increasing attention in tumor research. CircRNAs are highly conserved and stable because of their closed-loop structure and lack of 5′–3′ polarity15. CircRNAs are abundantly and differentially expressed in various diseases or stages of cancer; thus, some have been considered potential diagnostic biomarkers16. In tumors, some circRNAs act as oncogenes, affecting the occurrence and development of tumors and thus providing new therapeutic targets for the future17. Since circRNAs were reported to act as miRNA sponges18,19, an increasing number of studies have shown that they affect tumor occurrence and progression by affecting the function of one or more miRNAs20. For example, circ_0026344 suppresses the metastasis of human colorectal cancer cells by sponging miR-18321, and the circRNA hsa_circ_0001178 facilitates the invasion and metastasis of colorectal cancer cells by sponging multiple miRNAs to upregulate ZEB1 expression22. In addition, circRNAs have been found to act by interacting with proteins, and some circRNAs can even be translated into proteins23,24. Recently, circ-SOX4 was reported to promote LUAD progression through the miR-1270/PLAGL2/WNT axis25. Although the functions of some circRNAs in LUAD have been elucidated, the functions of numerous circRNAs in LUAD have not been identified, and this topic deserves further exploration.
In this study, circ-HMGA2 (hsa_circ_0027446), a newly identified circRNA, was found to be significantly overexpressed in LUAD. Both in functional studies in cells and in animal experiments, circ-HMGA2 was found to promote the metastasis of LUAD cells. Furthermore, research showed that circ-HMGA2 promotes the metastasis of LUAD cells through the miR-1236-3p/EMT axis.
Materials and methods
Specimens
A total of 36 pairs of LUAD and adjacent normal tissue samples were provided by the Cancer Hospital Affiliated with Guangzhou Medical University. All tissues were analyzed by quantitative real-time PCR (qRT-PCR). The study protocol was approved by the Ethics Committee of the Cancer Hospital Affiliated with Guangzhou Medical University, and all patients provided informed consent before enrollment. Tissues were stored at −80 °C until use.
CircRNA microarray hybridization and data analysis
Total RNA was digested with RNase R (Epicentre, USA) to remove linear RNAs and enrich circRNAs. Then, the enriched circRNAs were amplified and transcribed to fluorescent cRNA utilizing a random priming method (Arraystar Super RNA Labeling Kit; Arraystar; USA). The labeled cRNAs were hybridized to an Arraystar Human circRNA Array (8x15K, Arraystar). After washing the slides, the arrays were scanned with an Agilent G2505C scanner. Agilent Feature Extraction software (version 11.0.1.1) was used to analyze the acquired array images. Quantile normalization and subsequent data processing were performed using the limma package in R software. CircRNAs with statistically significant differential expression between the two groups were identified through volcano plot filtering. Differentially expressed circRNAs between two samples were identified through fold change filtering. Hierarchical clustering was performed to visualize the distinguishable circRNA expression patterns among samples.
Total RNA extraction and quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from frozen tissues and cells using RNAiso Plus (Takara, Japan) according to the manufacturer’s instructions. RNA was quantified using a NanoDrop 2000 (Thermo Fisher Scientific, USA). cDNA was synthesized using PrimeScript RT Master Mix (Takara, China). qPCR was performed with TB Green Premix Ex Taq II (Takara, China) with the following thermal cycling program: 95 °C for 30 s and 40 cycles at 95 °C for 5 s, 60 °C for 30 s, and a dissociation step. The 2−ΔΔCt method was used to calculate the relative expression level of each gene. The primer sequence is synthesized by Sangon Biotech (Supporting Table S1).
Cell culture
The human lung adenocarcinoma (LUAD) cell lines used in this study (H1299 and A549) were purchased from ATCC (USA). All cells were cultured in RPMI 1640 medium (Gibco, USA) supplemented with 10% fetal bovine serum (FBS; Gibco, USA). All cell lines were incubated at 37 °C in a humidified atmosphere containing 5% CO2.
Evaluation of the circular character of circ-HMGA2
The circular character of circ-HMGA2 was demonstrated by Sanger sequencing. The circ-HMGA2 sequencing results were compared with the sequences provided by circBASE26. To prove the stability of circular RNA, circ-HMGA2 and HMGA2 were treated with RNase R (Epicentre, USA), and the content of both was assessed by PCR and agarose gel electrophoresis. In addition, LUAD cells were treated with actinomycin D, and the stability of circ-HMGA2 and HMGA2 was verified by RT-PCR.
Cell transfection
Three siRNAs targeting the back-splice site of circ-HMGA2 and the negative control siRNA were designed and synthesized by RiboBio (Guangzhou, China). The target sequences of the si-circ-HMGA2 constructs were as follows: si-has-1, 5′-CCTAGGAAATGGGAACCAA-3′; si-circ-2, 5′-AAATGGGAACCAACCGGTG-3′; si-circ-3, 5′-ATGGGAACCAACCGGTGAG-3′. The circ-HMGA2 overexpression plasmid was synthesized by GeneChem (Shanghai, China). MiR-1236-3p mimics/inhibitors were designed by and purchased from RiboBio (Guangzhou, China). Before transfection, cells were cultured to 60-70% confluence in 6-well plates. Lipofectamine 3000 was used for transfection according to the manufacturer’s protocols.
Migration and invasion assays
For the migration assay, RPMI 1640 medium containing 20% FBS was added to the lower chambers of a 24-well transwell plate, and transfected cells were resuspended in serum-free medium and seeded in the upper chambers (approximately 4 × 104 cells in a 200 µl volume per chamber). After 24 h of co-culture, a portion of the cells had migrated through the membrane insert (8 µm pore size) from the upper chambers into the lower chambers. Cells in the lower chambers were fixed with 4% paraformaldehyde for 25 min, stained with 0.1% crystal violet for 25 min, and finally counted under a microscope (LEICA, Germany) at ×50 magnification. In contrast to the migration experiment, the invasion assay required the addition of Matrigel (Corning, USA) to the upper chambers and an incubation time of 48 h. All other steps were consistent with those in the migration assay.
Wound healing assay
LUAD cells were seeded in 6-well plates. When the cells were confluent in the plate, a linear wound was created in the center of the plates with a 200 µl pipette tip. In addition, the serum concentration was changed from 10 to 1%. Wound healing was evaluated with a microscope (LEICA, USA) at 0, 24, and 48 h, and was quantified as follows: wound healing rate = width of wound closure/width of original wound × 100%.
Animal study
The animal study was authorized by the Animal Ethics Committee of Zhujiang Hospital of Southern Medical University. Female BALB/c nude mice were purchased from Guangdong Experimental Animal Center (Guangzhou, China). A lung metastasis model was randomly established by injecting 2 × 106 H1299 cells into 4-week-old nude mice via the tail vein (n = 5 mice per group). Five weeks after injection, the mice were sacrificed. Their lungs were removed and subjected to hematoxylin and eosin (HE) staining. The lung metastatic nodules were carefully counted.
Luciferase reporter assay
Before co-transfection, 293T cells were seeded in 96-well plates at a density of 1 × 104 cells per well for 24 h. Lipofectamine 2000 (Invitrogen, USA) was used to transfect cells with the indicated transcript. Twenty-four hours after transfection, cells were lysed, and luciferase activity was analyzed by a dual-luciferase reporter assay system (Promega, USA).
Fluorescence in situ hybridization (FISH)
The FISH probe was designed by RiboBio (Guangzhou, China). For the fluorescent probes, Cy3 was used as the marker for circ-HMGA2, and FAM was used as the marker for miR-1236-3p. 4′,6-Diamidino-2-phenylindole (DAPI) was used to stain nuclei. FISH was performed according to the instructions of the FISH kit (GenePharma, China). The stained cells were then visualized using a confocal microscope (Leica, Germany).
Western blot (WB) analysis
RIPA buffer was added to cells and incubated with shaking on ice for more than half an hour. The lysate supernatant was collected, and a protease inhibitor (1%; ComWin Biotech, Beijing, China) was added. The supernatant was centrifuged, and the protein concentration was measured using a BCA Protein Assay Kit (Beyotime, Shanghai, China). Proteins were separated by 10% SDS-PAGE and were then transferred to PVDF membranes (Millipore, USA). After blocking for 1 h with 5% skim milk powder, membranes were incubated with the primary antibody at 4 °C overnight. The following primary antibodies were used: anti-ZEB1 (1:1000 dilution,; Proteintech, USA), anti-E-cadherin (1:1000 dilution, Cell Signaling Technology (CST), USA), anti-N-cadherin (1:1000 dilution; CST, USA), anti-vimentin (1:1000 dilution; Bioss, China), and anti-GAPDH (1:5000 dilution; Bioss, China). Membranes were washed three times for 10 min each in TBST containing 0.1% Tween and were then incubated with goat anti-rabbit or goat anti-mouse secondary antibodies (1:8000 dilution, Bioss, China) for 1 h at room temperature. Membranes were washed three times with TBST containing 0.1% Tween. Protein bands were detected with SuperSignal West Femto Agent (Millipore) and visualized with the Chemical Mp Imaging System (Bio-Rad).
Immunohistochemistry (IHC)
IHC was performed as described previously27. Serial sections of tissue were incubated with antibodies specific for EMT-related proteins at 4 °C overnight and then with secondary antibodies for 1 h at room temperature.
Statistical analyses
The student’s t-test was performed with SPSS 19.0 software to determine whether the differences between the two experimental groups were significant. P < 0.05 was used as the criterion for significant differences.
Results
Differentially expressed circRNAs in LUAD and adjacent normal tissues
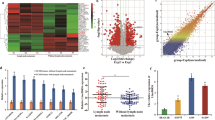
To filter the differentially expressed circRNAs in LUAD tissues and adjacent normal tissues, we performed a high-throughput human circular RNA microarray analysis of three pairs of LUAD and adjacent normal tissues. The analysis identified 12,444 distinct circRNAs, among which 85.7% were derived from exons, and the rest were derived from sense overlapping, intergenic, intronic, and antisense transcripts (Supporting Fig. S1A). A fold change of >2.0 and a P value of <0.05 were used as effective criteria to select significantly differentially expressed circRNAs. A volcano plot was constructed to visualize the differential expression patterns between tumor and nontumor tissues (Fig. 1A). It shows that 184 circRNAs were significantly aberrantly expressed in the circRNA microarray. Among these circRNAs, 103 were significantly upregulated and 81 were significantly downregulated in LUAD tissues compared with the paired adjacent normal tissues. In addition, the analysis identified 150 overlapping circRNAs and 30 novel circRNAs with respect to circBase26.
A According to the criteria of fold change >2.0 and P < 0.05, the significantly aberrantly expressed circRNAs were selected from the volcano plot. B The heat map shows the top 10 circRNAs with the greatest increases and decreases in expression in 3 LUAD tissues compared to the paired adjacent normal tissues.
The hierarchical clustering results showed a distinguishable circRNA expression profile among the samples (Supporting Fig. S1B). To further narrow the field of circRNAs, we selected 10 of the most strongly upregulated and downregulated circRNAs for the preliminary experiment (Fig. 1B). Eventually, we found that hsa_circ_0027446 was related to LUAD metastasis.
Characterization of hsa_circ_0027446 (circ-HMGA2) in LUAD
Circ-HMGA2, located on chromosome 12q14.3, is derived from exons 2–3 of the HMGA2 gene through back-splicing (Fig. 2A). To prove its circular structure, the PCR products of circ-HMGA2 were analyzed by Sanger sequencing. The sequencing results were consistent with the sequence of the back-spliced region in circBase (Fig. 2B). Due to its circular structure, circRNA is more stable and resistant to RNase R digestion than linear RNA. The RNase R digestion experiment showed that the linear transcripts of HMGA2 were degraded, while the circular transcripts of HMGA2 were retained (Fig. 2C). In addition, treatment with actinomycin D demonstrated that circ-HMGA2 was more stable than the linear HMGA2 transcript (Fig. 2D). Moreover, the expression of circ-HMGA2 was significantly higher in the 36 LUAD tissues than in the paired adjacent normal tissues (Fig. 2E). Compared with BEAS-2B cells, circ-HMGA2 was also significantly highly expressed in five LUAD cells (Fig. 2F). The FISH results revealed that circ-HMGA2 was localized mainly in the cytoplasm (Fig. 2G). Taken together, these data showed the basic characteristics of circ-HMGA2 in LUAD cells.
A The schematic illustration showing that circ-HMGA2 is cyclized from exons 2-3 of HMGA2. B Sanger sequencing showed the back-splice junction sites in circ-HMGA2. C Gel electrophoresis was performed to analyze RNase R digestion. Circ‐HMGA2 was resistant to RNase R digestion. D Upon exposure to actinomycin D, circ-HMGA2 was more stable than its linear form. E The relative expression levels of circ-HMGA2 in LUAD tissues and adjacent normal tissues were determined by qRT-PCR. F Circ-HMGA2 was significantly highly expressed in LUAD cells. G Circ-HMGA2 was localized mainly in the cytoplasm of H1299 cells, as verified by FISH. Scale bar: 25 μm. *P < 0.05.
Circ-HMGA2 promotes LUAD cell metastasis by inducing EMT
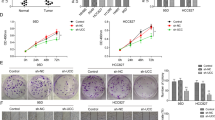
To investigate the biological function of circ-HMGA2 in LUAD, we knocked down the expression of circ-HMGA2 in H1299 and A549 cells. The qRT-PCR results showed that the transfection was successful (Fig. 3A). However, knockdown of circ-HMGA2 expression did not affect the expression of the linear HMGA2 transcript (Supporting Fig. S2). The results of the transwell assay showed that the migration and invasion abilities of H1299 and A549 cells were significantly reduced when the expression of circ-HMGA2 was inhibited (Fig. 3B, C). The wound healing experiments led to the same conclusion (Fig. 4D). But the proliferation of the cells has not been significantly affected (Supporting Fig. S3A). Similarly, we constructed a circ-HMGA2 overexpression vector and verified a significant increase in circ-HMGA2 expression by qRT-PCR (Fig. 3E). The migration and invasion abilities of H1299 and A549 cells with circ-HMGA2 overexpression were also enhanced, as shown by the results of the corresponding transwell assays (Fig. 3F, G).
A The expression of circ-HMGA2 in H1299 and A549 cell lines was significantly reduced after transfection with siRNA. B, C The results of transwell assays showed that the metastatic ability of H1299 and A549 cells was decreased after the circ‐HMGA2 knockdown. Scale bar: 200 μm. D The results of wound healing experiments showed that the migration of H1299 and A549 cells was decreased after circ‐HMGA2. Scale bar: 200 μm. E Circ-HMGA2 was significantly overexpressed in H1299 and A549 cells. F, G The metastatic ability was increased when the expression of circ-HMGA2 was enhanced. Scale bar: 200 μm. H Circ-HMGA2 had a promotive effect on EMT. *P < 0.05, **P < 0.01.
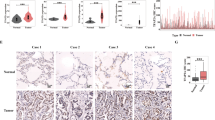
A In the lung metastasis model, knocking down the expression of circ-HMGA2 resulted in a significant reduction in the number of metastases on the lung surface. B, C HE staining showed that mice in the circ-HMGA2 knockdown group showed significantly fewer metastases. Scale bar: 500 μm. D, E The results of IHC showed that knocking down the expression of circ-HMGA2 also inhibited the progress of EMT in vivo. Scale bar: 30 μm. **P < 0.01.
EMT is an important phenotype in tumor metastasis. Therefore, we investigated whether circ-HMGA2 affects EMT in LUAD cells. WB analysis showed that overexpression of circ-HMGA2 in LUAD cells decreased the expression of the epithelial marker E-cadherin and increased the expression of the mesenchymal markers Vimentin and N-cadherin (Fig. 3H). It has the opposite result when knocking down the expression of circ-HMGA2 in the cells (Supporting Fig. S3B). These findings indicated that circ-HMGA2 promotes LUAD cell metastasis by inducing EMT.
Circ-HMGA2 facilitates the metastasis of LUAD cells in vivo
To validate whether circ-HMGA2 positively affects metastasis in vivo, we established lung metastasis models by intravenously injecting H1299 cells into nude mice. After six weeks, lung colonization was evaluated. The lung metastasis model showed that the number of metastatic nodules on the lung surface of the knockdown circ-HMGA2 group was significantly less than that of the control group. (Fig. 4A). The HE staining results further verified knocking down the expression of circ-HMGA2 inhibits the metastasis ability of H1299 cells in vivo (Fig. 4B, C). In addition, the IHC results showed that inhibiting the expression of circ-HMGA2 can inhibit the process of EMT in tumor tissues (Fig. 4D, E). These results were consistent with our findings in vitro.
Circ-HMGA2 acts as a sponge for miR-1236-3P
We found that circ-HMGA2 was composed of exons and was localized mainly in the cytoplasm. These results suggested that circ-HMGA2 may function by acting as a miRNA sponge28. To investigate miRNAs potentially involved in the function of circ-HMGA2, we used the miRanda database to identify the five miRNAs most likely to bind to circ-HMGA2 (Fig. 5A). The qRT-PCR data showed that miR-1236-3p expression was significantly lower in the 36 LUAD specimens than in the paired adjacent normal tissues (Fig. 5B). A prior study showed that miR-1236-3p can influence cancer metastasis29. We experimentally verified that miR-1236-3p inhibits the metastasis of LUAD cells (Fig. 5C). It also found that miR-1236-3p inhibits the progression of EMT (Fig. 5D). These results suggested that circ-HMGA2 may function by sponging miR-1236-3p. For the luciferase reporter assay, two luciferase reporter vectors containing circ-HMGA2 with wild-type and mutated miR-1236-3p binding sites were constructed (Fig. 5E). The luciferase reporter assay results showed that luciferase activity was negatively correlated with the expression of miR-1236-3p in cells transfected with the luciferase reporter construct containing wild-type circ-HMGA2. However, luciferase activity did not change significantly in cells transfected with the luciferase reporter construct containing mutant circ-HMGA2 was transfected into cells (Fig. 5F). Furthermore, the FISH results showed that miR-1236-3p was also localized mainly in the cytoplasm (Fig. 5G). Overall, these results indicated that circ-HMGA2 acts as a sponge for miR-1236-3p.
A Analysis of the miRanda database indicated that circ-HMGA2 contains a binding site for miR-1236-3p. B The relative expression levels of miR-1236-3p in LUAD tissues and adjacent normal tissues were determined by qRT-PCR. C Transwell assays were performed in H1299 and A549 cells after transfection of the miR-1236-3p inhibitor. Scale bar: 200 μm. D miR-1236-3p inhibited EMT progression in LUAD cells. E Schematic illustration of circ-HMGA2 luciferase reporter vectors containing the wild-type (WT) or mutant (Mut) putative miR-1236-3p binding sites. F Luciferase activity was analyzed in 293T cells after co-transfection with miR-1236-3p mimics, miR-NC or miR-1236-3p inhibitors, and the WT or Mut luciferase reporter vectors. The relative luciferase activity was presented by the relative hRluc/hluc ratio. G The FISH results showed that circ-HMGA2 and miR-1236-3p are co-localized in the cytoplasm. Scale bar: 8 μm. *P < 0.05, **P < 0.01.
Circ-HMGA2 inhibits LUAD metastasis through the circ-HMGA2/miR-1236-3p/EMT axis
We found that circ-HMGA2 and miR-1236-3p are involved in the metastasis of LUAD and that circ-HMGA2 sponges miR-1236-3p. However, whether circ-HMGA2 acts through miR-1236-3p in LUAD required further investigation. Our experimental results demonstrated that the miR-1236-3p mimic inhibited the effect of circ-HMGA2 on LUAD metastasis (Fig. 6A, B). Via miRanda, we predicted that ZEB1 is downstream of miR-1236-3p. ZEB1 has been reported to be a transcription factor that induces EMT; thus, circ-HMGA2 may eventually lead to the metastasis of LUAD cells through promoting EMT30. To investigate the relationship between miR-1236-3p and ZEB1, two luciferase reporter vectors containing ZEB1 with wild-type and mutated miR-1236-3p binding sites were constructed(Fig. 6C). The luciferase reporter assay results showed that co-transfection of miR-1236-3p mimics and wild-type ZEB1 but not co-transfection of miR-1236-3p mimics and mutant ZEB1 significantly affected luciferase activity (Fig. 6D). Moreover, overexpression of circ-HMGA2 increased luciferase activity, but co-transfection of circ-HMGA2 and miR-1236-3p did not induce a change in luciferase activity (Fig. 6E). Next, overexpression of circ-HMGA2 was found to increase the expression of ZEB1 mRNA and protein and to promote EMT (Fig. 6F, Fig. 3H). However, transfection of miR-1236-3p mimics inhibited the ZEB1 expression and suppressed EMT (Fig. 6G, Fig. 4C). In addition, circ-HMGA2 blocked the effects of miR-1236-3p mimics on ZEB1 and EMT (Fig. 6H, I). It was also blocked the effects of si-ZEB1 on EMT (Supporting Fig. S4). In summary, these results indicated that circ-HMGA2 promotes the metastasis of LUAD cells by inhibiting the suppressive effect of miR-1236-3p on EMT.
A, B The results of transwell assays showed that miR-1236-3p inhibited the promotive effect of circ-HMGA2 on LUAD metastasis. Scale bar: 200 μm. C Schematic of luciferase reporter vectors containing wild-type (WT) and mutant (Mut) ZEB1. D Luciferase activity was analyzed in 293T cells after co-transfection with miR-1236-3p mimics, miR-NC or miR-1236-3p inhibitors, and the WT or Mut luciferase reporter vectors. E Luciferase activity was analyzed in 293T cells after co-transfection with miR-1236-3p mimics, circ-HMGA2-NC or circ-HMGA2-over, and the WT or Mut luciferase reporter vectors. The relative luciferase activity was presented by the relative hRluc/hluc ratio. F The qRT-PCR results showed that circ-HMGA2 significantly increased the expression of ZEB1. G The qRT-PCR results showed that miR-1236-3p significantly inhibited the expression of ZEB1. H, I The mRNA and protein levels of ZEB1 and EMT was not changed after co-transfection with miR-1236-3p mimics and circ-HMGA2. **P < 0.01.
Discussion
Recently, accumulating evidence has shown that circRNAs are related to tumor occurrence and development and could have a future use as tumor biomarkers20. Some studies have shown that circRNAs play an important role in lung cancer. For example, upregulation of circ_0016760 promotes NSCLC cell progression through the miR-1287/GAGE1 axis31. Silencing of circ_0067934 suppresses the progression of NSCLC cells and correlates with unfavorable prognosis in NSCLC32. However, few studies on circRNAs in LUAD have been conducted. Currently, circ-HMGA2 was a novel circRNA never been reported before. Our study investigated circ-HMGA2 for the first time and its function in LUAD. We found that circ-HMGA2 was significantly highly expressed in LUAD through microarray, and its high expression was again verified by RT-PCR in LUAD cells and 36 pairs of clinical samples. In addition, its downstream miR-1236-3P has been reported to be related to the metastasis of a variety of cancers, and we verified its significantly low expression in LUAD tissues by RT-PCR. These suggest that the expression of circ-HMGA2 and miR-1236-3P may have important significance in the diagnosis of LUAD and may become biomarkers of LUAD in the future. This also implies that circ-HMGA2 has a potential role in LUAD.
Current research shows that circRNA is not simply a non-functional product of pre-mRNA splicing, as historically believed, and numerous circRNAs have been reported to play important roles in various diseases33. Especially in the occurrence and development of tumors, differentially expressed circRNAs can often act as oncogenes34. Our research proved that circ-HMGA2 can promote the metastasis of LUAD and the progression of EMT through in vivo and in vitro functional experiments. This is the first report of the function of circ-HMGA2 in LUAD. These results suggest that circ-HMGA2 may play an important role in assessing the progression of LUAD and provide a basis for becoming a potential therapeutic target in the future.
Previous reports indicate that circRNAs can act as miRNA sponges, thereby inhibiting the effects of miRNAs on their targeted mRNAs35. Through in-depth research, we found that circ-HMGA2 acts as a miRNA sponge to regulate LUAD metastasis. Bioinformatic analysis predicted that circ-HMGA2 binds to miR-1236-3p, and our study confirmed that circ-HMGA2 binds to miR-1236-3p and showed that both are localized in the cytoplasm. Substantial evidence indicates that miR-1236-3p can inhibit tumor metastasis in colorectal cancer, glioma, and gastric cancer29,36,37. Our research confirmed that miR-1236-3p can suppress metastasis and EMT and can inhibit the effect of circ-HMGA2 in LUAD. This study shows for the first time that circ-HMGA2 and miR-1236-3P are combined, and circ-HMGA2 promotes LUAD metastasis through miR-1236-3P. These results also suggest that miR-1236-3P plays an important role in the metastasis of LUAD and may become a potential therapeutic target in the future.
In addition, we found that miR-1236-3p binds ZEB1. ZEB1 is a well-known transcription factor that plays an important role in the metastasis of LUAD cells38. The relationship between ZEB1 and epigenetic regulation of EMT genes was discovered only recently. ZEB1 promotes epigenetic silencing of E-cadherin by recruiting multiple chromatin remodeling enzymes from the E-cadherin promoter39. Therefore, ZEB1 promotes the progress of EMT. Our study found that miR-1236-3p inhibits the expression of ZEB1, circ-HMGA2 promotes the expression of ZEB1, and miR-1236-3P would inhibit the promotive effect of circ-HMGA2 on ZEB1 expression. Further research showed that upon mutation of the miR-1236-3p binding site in circ-HMGA2, circ-HMGA2 no longer affected the expression of ZEB1. These results indicate that circ-HMGA2 acts as a sponge for miR-1236-3p to regulate the ZEB1/EMT pathway and promote LUAD cell metastasis. These also emphasize the important functions of circ-HMGA2 and miR-1236-3P in LUAD metastasis.
Conclusions
Our study provides the first evidence that circ-HMGA2 is highly expressed in patients with LUAD and promotes LUAD cell metastasis and EMT in vivo and in vitro. Mechanistically, the circ-HMGA2/miR-1236-3p/ZEB1 axis promotes EMT and metastasis of LUAD cells (Fig. 7). These results indicate that circ-HMGA2 could be a biomarker and an important therapeutic target in LUAD.
References
Wang, C., Tan, S., Liu, W., Lei, Q., Qiao, W. & Wu, Y. et al. RNA-Seq profiling of circular RNA in human lung adenocarcinoma and squamous cell carcinoma. Mol. Cancer 18, 134 (2019).
Du, J., Zhang, G., Qiu, H., Yu, H. & Yuan, W. The novel circular RNA circ-CAMK2A enhances lung adenocarcinoma metastasis by regulating the miR-615-5p/fibronectin 1 pathway. Cell Mol. Biol. Lett. 24, 72 (2019).
Hirsch, F. R., Scagliotti, G. V., Mulshine, J. L., Kwon, R., Curran, W. J. & Wu, Y. L. et al. Lung cancer: current therapies and new targeted treatments. Lancet 389, 299–311 (2017).
Walter, J. E., Heuvelmans, M. A., de Jong, P. A., Vliegenthart, R., van Ooijen, P. & Peters, R. B. et al. Occurrence and lung cancer probability of new solid nodules at incidence screening with low-dose CT: analysis of data from the randomised, controlled NELSON trial. Lancet Oncol. 17, 907–916 (2016).
Altorki, N. K., Markowitz, G. J., Gao, D., Port, J. L., Saxena, A. & Stiles, B. et al. The lung microenvironment: an important regulator of tumour growth and metastasis. Nat. Rev. Cancer 19, 9–31 (2019).
Xie, L., Chen, Y., Chen, J., Zhang, H., Liao, Y. & Zhou, Y. et al. Anti-tumor effects and mechanism of GA-13315, a novel gibberellin derivative, in human lung adenocarcinoma: an in vitro and in vivo study. Cell Mol. Biol. Lett. 24, 6 (2019).
Tasdogan, A., Faubert, B., Ramesh, V., Ubellacker, J. M., Shen, B. & Solmonson, A. et al. Metabolic heterogeneity confers differences in melanoma metastatic potential. Nature 577, 115–120 (2020).
Singh, M., Yelle, N., Venugopal, C. & Singh, S. K. EMT: mechanisms and therapeutic implications. Pharm. Ther. 182, 80–94 (2018).
Brabletz, T., Kalluri, R., Nieto, M. A. & Weinberg, R. A. EMT in cancer. Nat. Rev. Cancer 18, 128–134 (2018).
Diepenbruck, M. et al. miR-1199-5p and Zeb1 function in a double-negative feedback loop potentially coordinating EMT and tumour metastasis. Nat. Commun. 8, 1168 (2017).
Title, A. C., Hong, S. J., Pires, N. D., Hasenohrl, L., Godbersen, S. & Stokar-Regenscheit, N. et al. Genetic dissection of the miR-200-Zeb1 axis reveals its importance in tumor differentiation and invasion. Nat. Commun. 9, 4671 (2018).
Tan, X., Banerjee, P., Liu, X., Yu, J., Gibbons, D. L. & Wu, P. et al. The epithelial-to-mesenchymal transition activator ZEB1 initiates a prometastatic competing endogenous RNA network. J. Clin. Invest. 128, 3198 (2018).
Chen, L., Gibbons, D. L., Goswami, S., Cortez, M. A., Ahn, Y. H. & Byers, L. A. et al. Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nat. Commun. 5, 5241 (2014).
Larsen, J. E., Nathan, V., Osborne, J. K., Farrow, R. K., Deb, D. & Sullivan, J. P. et al. ZEB1 drives epithelial-to-mesenchymal transition in lung cancer. J. Clin. Invest. 126, 3219–35 (2016).
Su, M., Xiao, Y., Ma, J., Tang, Y., Tian, B. & Zhang, Y. et al. Circular RNAs in cancer: emerging functions in hallmarks, stemness, resistance and roles as potential biomarkers. Mol. Cancer 18, 90 (2019).
Patop, I. L., Wust, S. & Kadener, S. Past, present, and future of circRNAs. EMBO J. 38, e100836 (2019).
Naeli, P., Pourhanifeh, M. H., Karimzadeh, M. R., Shabaninejad, Z., Movahedpour, A. & Tarrahimofrad, H. et al. Circular RNAs and gastrointestinal cancers: epigenetic regulators with a prognostic and therapeutic role. Crit. Rev. Oncol. Hematol. 145, 102854 (2020).
Hansen, T. B., Jensen, T. I., Clausen, B. H., Bramsen, J. B., Finsen, B. & Damgaard, C. K. et al. Natural RNA circles function as efficient microRNA sponges. Nature 495, 384–8 (2013).
Memczak, S., Jens, M., Elefsinioti, A., Torti, F., Krueger, J. & Rybak, A. et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495, 333–8 (2013).
Verduci, L., Strano, S., Yarden, Y. & Blandino, G. The circRNA-microRNA code: emerging implications for cancer diagnosis and treatment. Mol. Oncol. 13, 669–680 (2019).
Shen, T., Cheng, X., Liu, X., Xia, C., Zhang, H. & Pan, D. et al. Circ_0026344 restrains metastasis of human colorectal cancer cells via miR-183. Artif. Cells Nanomed. Biotechnol. 47, 4038–4045 (2019).
Ren, C., Zhang, Z., Wang, S., Zhu, W., Zheng, P. & Wang, W. Circular RNA hsa_circ_0001178 facilitates the invasion and metastasis of colorectal cancer through upregulating ZEB1 via sponging multiple miRNAs. Biol. Chem. 401, 487–496 (2020).
Zang, J., Lu, D. & Xu, A. The interaction of circRNAs and RNA binding proteins: an important part of circRNA maintenance and function. J. Neurosci. Res 98, 87–97 (2020).
Li, R., Jiang, J., Shi, H., Qian, H., Zhang, X. & Xu, W. CircRNA: a rising star in gastric cancer. Cell Mol. Life Sci. 77, 1661–1680 (2020).
Gao, N. & Ye, B. Circ-SOX4 drives the tumorigenesis and development of lung adenocarcinoma via sponging miR-1270 and modulating PLAGL2 to activate WNT signaling pathway. Cancer Cell Int. 20, 2 (2020).
Glazar, P., Papavasileiou, P. & Rajewsky, N. circBase: a database for circular RNAs. RNA 20, 1666–70 (2014).
Zeng, K., Wang, Z., Ohshima, K., Liu, Y., Zhang, W. & Wang, L. et al. BRAF V600E mutation correlates with suppressive tumor immune microenvironment and reduced disease-free survival in Langerhans cell histiocytosis. Oncoimmunology 5, e1185582 (2016).
Zhao, W., Cui, Y., Liu, L., Qi, X., Liu, J. & Ma, S. et al. Splicing factor derived circular RNA circUHRF1 accelerates oral squamous cell carcinoma tumorigenesis via feedback loop. Cell Death Differ. 27, 919–933 (2020).
Feng, W., Gong, H., Wang, Y., Zhu, G., Xue, T. & Wang, Y. et al. circIFT80 functions as a ceRNA of miR-1236-3p to promote colorectal cancer progression. Mol. Ther. Nucleic Acids 18, 375–387 (2019).
Peng, D. H., Kundu, S. T., Fradette, J. J., Diao, L., Tong, P. & Byers, L. A. ZEB1 suppression sensitizes KRAS mutant cancers to MEK inhibition by an IL17RD-dependent mechanism. Sci. Transl. Med. 11, eaaq1238 (2019).
Li, Y., Hu, J., Li, L., Cai, S., Zhang, H. & Zhu, X. et al. Upregulated circular RNA circ_0016760 indicates unfavorable prognosis in NSCLC and promotes cell progression through miR-1287/GAGE1 axis. Biochem. Biophys. Res. Commun. 503, 2089–2094 (2018).
Wang, J. & Li, H. CircRNA circ_0067934 silencing inhibits the proliferation, migration and invasion of NSCLC cells and correlates with unfavorable prognosis in NSCLC. Eur. Rev. Med. Pharm. Sci. 22, 3053–3060 (2018).
Yin, Y., Long, J., He, Q., Li, Y., Liao, Y. & He, P. et al. Emerging roles of circRNA in formation and progression of cancer. J. Cancer 10, 5015–5021 (2019).
Zhu, X. P., Wang, X. L., Ma, J., Fang, Y. F., Zhang, H. J. & Zhang, C. et al. Down-regulation of miR-1236-3p is correlated with clinical progression and unfavorable prognosis in gastric cancer. Eur. Rev. Med. Pharm. Sci. 22, 5914–5919 (2018).
Duan, X., Liu, D., Wang, Y. & Chen, Z. Circular RNA hsa_circ_0074362 promotes glioma cell proliferation, migration, and invasion by attenuating the inhibition of miR-1236-3p on HOXB7 expression. DNA Cell Biol. 37, 917–924 (2018).
Chen, L. L. & Yang, L. Regulation of circRNA biogenesis. RNA Biol. 12, 381–8 (2015).
Meng, S., Zhou, H., Feng, Z., Xu, Z., Tang, Y. & Li, P. et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol. Cancer 16, 94 (2017).
Ying, X., Zhu, J. & Zhang, Y. Circular RNA circ-TSPAN4 promotes lung adenocarcinoma metastasis by upregulating ZEB1 via sponging miR-665. Mol. Genet. Genom. Med. 7, e991 (2019).
Zhang, Y., Xu, L., Li, A. & Han, X. The roles of ZEB1 in tumorigenic progression and epigenetic modifications. Biomed. Pharmacother. 110, 400–408 (2019).
Acknowledgements
The invaluable support of all participants in this research is appreciated. This work is supported by grant from National Natural Science Foundation of China (No. 81974434), grants from the Science and Technology Foundation of Guangdong Province (No. 2018A0303130233, 2020A0505100038), grant from the Science and Technology Program of Guangzhou City (No. 201907010037), grant from the Affiliated Cancer Hospital and Institute of Guangzhou Medical University (No. 2020-YZ-01), and grant from Clinical Key Specialty Construction Project of Guangzhou Medical University (No. YYPT202017).
Author information
Authors and Affiliations
Contributions
Z.Y. and X.Z. performed the experiments and analyzed the data. Z.Y. wrote the manuscript. Y.W. provided some reagents for the experiments. Y.L. and Z.L. helped with in vivo and in vitro assays. Z.Y. makes the processing and stitching of pictures. Y.Z. was the principal investigator and provided the expenses for the experiments. M.L. helped with the final approval of the version to be published. M.L. made contributions to conception and design. L.Q. helped with in vivo assays. X.W. helped with in vitro assays. K.D. revising it critically for important intellectual content. L.L. helped with the analysis and interpretation of data.
Corresponding authors
Ethics declarations
Ethics statement
This study was approved by the Ethics Committee of Guangzhou Medical University and was conducted in accordance with the government policies and the Declaration of Helsinki.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by B. Rotblat
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yu, Z., Zhu, X., Li, Y. et al. Circ-HMGA2 (hsa_circ_0027446) promotes the metastasis and epithelial-mesenchymal transition of lung adenocarcinoma cells through the miR-1236-3p/ZEB1 axis. Cell Death Dis 12, 313 (2021). https://doi.org/10.1038/s41419-021-03601-2
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41419-021-03601-2
This article is cited by
-
Paclitaxel-induced inhibition of NSCLC invasion and migration via RBFOX3-mediated circIGF1R biogenesis
Scientific Reports (2024)
-
Circ_0027446 promotes malignant development of glioblastoma by interacting with miR-346 to up-regulate PGK1
Metabolic Brain Disease (2023)
-
Circ_0001715 Functions as a miR-1249-3p Sponge to Accelerate the Progression of Non-small Cell Lung Cancer via Upregulating the Level of FGF5
Biochemical Genetics (2023)
-
Regulation of Cervical Cancer Development by a Novel Circ_0000212/miR-1236-3p/GREM1 ceRNA Crosstalk
Molecular Biotechnology (2023)
-
Circ_0000808 promotes the development of non-small cell lung cancer by regulating glutamine metabolism via the miR-1827/SLC1A5 axis
World Journal of Surgical Oncology (2022)