Abstract
The interaction between LncRNA and RNA-binding protein (RBPs) plays an essential role in the regulation over the malignant progression of tumors. Previous studies on the mechanism of SNHG1, an emerging lncRNA, have primarily focused on the competing endogenous RNA (ceRNA) mechanism. Nevertheless, the underlying mechanism between SNHG1 and RBPs in tumors remains to be explored, especially in prostate cancer (PCa). SNHG1 expression profiles in PCa were determined through the analysis of TCGA data and tissue microarray at the RNA level. Gain- and loss-of-function experiments were performed to investigate the biological role of SNHG1 in PCa initiation and progression. RNA-seq, immunoblotting, RNA pull-down and RNA immunoprecipitation analyses were utilized to clarify potential pathways with which SNHG1 might be involved. Finally, rescue experiments were carried out to further confirm this mechanism. We found that SNHG1 was dominantly expressed in the nuclei of PCa cells and significantly upregulated in PCa patients. The higher expression level of SNHG1 was dramatically correlated with tumor metastasis and patient survival. Functionally, overexpression of SNHG1 in PCa cells induced epithelial–mesenchymal transition (EMT), accompanied by down-regulation of the epithelial marker, E-cadherin, and up-regulation of the mesenchymal marker, vimentin. Increased proliferation and migration, as well as accelerated xenograft tumor growth, were observed in SNHG1-overexpressing PCa cells, while opposite effects were achieved in SNHG1-silenced cells. Mechanistically, SNHG1 competitively interacted with hnRNPL to impair the translation of protein E-cadherin, thus activating the effect of SNHG1 on the EMT pathway, eventually promoting the metastasis of PCa. Our findings demonstrate that SNHG1 is a positive regulator of EMT activation through the SNHG1-hnRNPL-CDH1 axis. SNHG1 may serve as a novel potential therapeutic target for PCa.
Similar content being viewed by others
Introduction
Prostate cancer (PCa) is the frequency life-threatening tumor in male genitourinary system1. The malignant transformation of the prostate follows a multi-step process, starting with prostatic intraepithelial neoplasia, Then there is localized prostate cancer, followed by locally invasive advanced prostatic adenocarcinoma, which eventually develops into metastatic PCa2. The patient eventually developed metastatic castration resistant prostate cancer due to castration resistance and metastasis, resulting in death. There’s growing evidence that epithelial–mesenchymal transition (EMT) emerges a great role in the metastasis of various cancers, including PCa3,4,5. The progression of PCa is closely related to the proliferation and invasion phenotype of cancer cell6. However, the cellular and molecular mechanisms responsible for the metastasis PCa are incompletely understood.
Long non-coding RNAs (LncRNAs) are RNAs with a length of more than 200 nucleotides without protein-coding function. In recent years, the expansion of knowledge has revealed that lncRNAs play an important role in multiple biological processes, such as alternative splicing, nuclear import, imprinting, cell differentiation and RNA decay7. The abnormal expression of lncRNAs have also been reported to contribute to tumorigenic processes of many human malignancies, including PCa8,9. In PCa, recent studies have revealed that LncRNA DLX6-AS1 enhances PCa Malignant Phenotype and Lymph Node Metastasis10, and LncRNA AC245100.4 promotes the proliferation of PCa through binding to HSP9011. Small nucleolar RNA host gene 1(SNHG1), a novel LncRNA, has shown to be aberrantly high expression and oncogenic characteristics in various cancers12. Previous studies have illustrated that SNHG1 was upregulated in PCa and was associated with PCa proliferation through the namely competing endogenous RNA (ceRNA) mechanism13,14. While, the specific function of SNHG1 have not been well investigated in the context of PCa metastasis and the underlying mechanism are also needed to be elucidated.
In this study, we sought to determine the expression and the biological function of SNHG1 in PCa, especially its role in metastasis. Expression levels of SNHG1 were determined in the PCa primary tumor tissues and its correlations with clinicopathological parameters were also analyzed. We further investigated the effects of SNHG1 on the aggressive phenotypes of PCa cell lines in vitro and in vivo. The regulatory role of SNHG1 on CDH1 were also explored to elucidate the potential mechanisms. Taken together, our results have demonstrated that SNHG1 played a critical role in the progression of PCa.
Results
LncRNA SNHG1 is upregulated in PCa tissues and sub-located in the nucleus
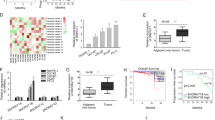
To identify the expression and localization of SNHG1 in PCa, we utilized bioinformatics to analyze the expression and prognosis of SNHG1 in PRAD in TCGA database. As shown in (Fig. 1A–C), we identified SNHG1 transcript level is overexpression in prostate cancer. Meanwhile, we evaluated the correlation between SNHG1 level and clinical outcomes from TCGA database using Kaplan–Meier analysis and log-rank tests. In the PRAD group, the prognosis of 247 PCa patients with high transcriptional level of SNHG1 was significantly worse than that of 248 patients with low expression of SNHG1.
A SNHG1 is overexpressed in TCGA PCa data. B, C Survival analysis of SNHG1 (TCGA PRAD) showed that both overall and disease-free survival were worse in the highly expressed group. D FISH analysis of PCa tissues confirmed that SNHG1 was highly expressed in PCa tissues and located in the nucleus (G means Gleason grade group). E SNHG1 expression was analyzed by qRT-PCR in five PCa cancer cell lines (DU145, PC-3, 22RV1, LNCaP, and C4-2), compared with the human Prostatic immortalized epithelial cells (RWPE-1). F Distribution of SNHG1 in PCa cells detected by fractionation of DU145 and C4-2 cells followed by qRT-PCR. G FISH analysis of DU145 cell. The nuclei were stained with DAPI and 18 S was used as a positive control for cytoplasmic staining. GS means Gleason Score. Scale bar, 20 μm. All data are shown as the mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001 by two-tailed Student’s t test.
Next, the expression and localization of SNHG1 was validated in 67 PCa tissues and 14 adjacent normal tissues via RNA FISH. we divided the sample into high (++, +++) and low (−, +) SNHG1 expression group according to the positive cell nuclear stain percentage of SNHG1 and then explored the correlation between the clinicopathological characters of prostate cancer patients and SNHG1 expressions. SNHG1 was significantly overexpressed in 67.16% (45 of 67 PCa tissues) and low expression in all (14 of 14 normal adjacent tissues) normal samples and 32.84% (22 of 67) PCa tissues (Table 1 and Fig. 1D). Besides, as shown in Table 1, SNHG1 transcript level was positively associated with Gleason Score but it may not be related to the patient’s age and clinical TNM stage in our study by Chi-square tests.
Furthermore, we validated the higher expression level of SNHG1 in DU145, PC3, LNCaP, c4-2, and other PCa cell lines compared to that in RWPE-1 cell line (Fig. 1E). Then, we cultured PCa cell DU145 and C4-2 as cell models of high and low SNHG1 expression in PCa. Through cellular fractionation assays (Fig. 1F) and RNA fluorescence in situ hybridization (FISH) (Fig. 1G), we demonstrated that SNHG1 was mainly distributed in the nucleus of DU145 cells and cytoplasm of C4-2 cells. Taken together, our data confirmed that SNHG1 is highly expressed in PCa tissues and cell lines, which might potentially serve as a novel independent predictor of overall survival in PCa.
LncRNA SNHG1 is essential for promoting PCa cells proliferation, migration in Vitro
To figure out the potential role of SNHG1 in promoting prostate cancer progression, siRNA or lentivirus shRNA were used to knock down the endogenous expression of SNHG1 in DU145 and C4-2 cells and used an overexpression plasmid system to upregulate SNHG1 in DU145 and C4-2 cells (Fig. S1A). CCK-8 assays, colonies formation and EDU assays demonstrated that upregulated SNHG1 remarkably enhanced PCa cells proliferation, while SNHG1 knockdown significantly blocked the proliferative abilities of PCa cells (Fig. 2A–C). Notably, SNHG1 knockdown impaired the migration ability of PCa cells, while upregulated SNHG1 transcription level promoted cell migration (Fig. 2D–E). These results suggested that SNHG1 promotes cell proliferation, migration in PCa cells. Besides, SNHG1 overexpression did not affect the migration abilities of the immortalized prostatic epithelial cell line RWPE-1 cells (Fig. S1B–E).
A, B CCK-8 assay and colony-formation assay were used to determine the proliferation ability of PCa cells by knocking down or overexpressing LncRNA SNHG1. C EdU assays were used to determine the proliferation ability of PCa cells. D Transwell assays showed that SNHG1 knockdown or overexpression could inhibit or promote PCa cell migration. E SNHG1 knockdown attenuated cell migration in DU145 and C4-2 cells, while up-regulation of SNHG1 showed the opposite results. *P < 0.01, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Knockdown LncRNA SNHG1 inhibited PCa cells tumorigenesis in vivo
For the purpose of validating the potential impact of SNHG1 depletion on the tumorigenesis of PCa in vivo, DU145 cells transfected with sh-SNHG1 or Negative Control lentivirus vector were injected subcutaneously in male nude mice. Tumors in mice implanted in DU145/sh-SNHG1 cells were remarkably smaller than those in the control cells (Fig. 3A, B). Likewise, SNHG1 knockdown cells grew slower in compared with the control cohort, the average tumor volumes and weight in the ultimate experiment were significantly lower in the DU145/sh-SNHG1 cohort versus the negative control lentivirus vector cohort (Fig. 3C, D). IHC staining of ki-67 antigen confirmed that subcutaneous tumor proliferation was significantly reduced in the sh-SNHG1 group (Fig. 3E).
A, B Negative control or sh-SNHG1was stably transfected into DU145 cells, which were injected into the nude mice. C Tumor volumes were calculated after injection every 4 days. D Tumor weights were represented as means of tumor weights ± SD. E IHC analyses of ki-67 confirmed that the proliferation of subcutaneous tumor was inhibited in sh-SNHG1 group. ***P < 0.001, ****P < 0.0001.
Related genes of lncRNA SNHG1 in PCa cells
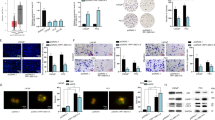
To validate the potential related genes that may be regulated by SNHG1 in PCa cells, RNA transcriptome sequencing was carried out in DU145/NC or DU145/sh-SNHG1 cells. As a result of silencing SNHG1, a total of 837 genes greater than 1.5-fold change increased, whereas 208 genes exhibited a decrease in abundance (<−1.5 fold change) (Fig. 4A). Meanwhile, A thorough study of the oncology analysis highlighted the most obvious biological phenomena of overexpression associated with the pathways in cell adhesion molecules (CAMs) (Fig. 4B–D), and the RNA sequencing data were verified by qRT-PCR (Fig. 4E). However, the mechanism by which SNHG1 regulates CAMs is still unclear.
A Hierarchical clustering of 1045 transcripts altered (≥1.5-fold change) in NC-treated cells and shRNA SNHG1-treated cells with three repeats. B, C KEGG analysis and differential genes GSEA enrichment analysis demonstrated that cell adhesion molecules are the potential targets of SNHG1 pathway. D Volcanic map analysis of differential genes. E The altered mRNA levels of genes were selectively confirmed by qRT-PCR in knockdown SNHG1. F SDS-PAGE silver staining of RNA pull-down protein samples showed a significant difference in protein band from 55 to 70 kDa. G Differential protein band mass spectrometry showed that the protein was hnRNPL. H The interaction possibilities of hnRNPL and SNHG1 were detected in RPIseq, and the results showed that hnRNPL could well bind with SNHG1 well (RPISeq). I Pull-down assays showed that biotinylated SNHG1 could retrieve hnRNPL in DU145 cells by western blot(flow-through was a input control). J RNA immunoprecipitation revealed that hnRNPL could also specifically bind to SNHG1. K, L The western blotting results after SNHG1 knockdown in DU145 and C4-2 cells showed that the expression level of hnRNPL did not change, while E-cad and Claudin-1 were significantly up-regulated, Vimentin, Slug and ZEB1 were significantly decreased. overexpression of SNHG1 showed an opposite effect of these protein in in DU145 and C4-2 cells. Error bars indicate means ± SD. ***P < 0.001, ****P < 0.0001.
SNHG1 activates the EMT pathway in PCa cells through interaction with hnRNPL
In order to further clarify the pathway of SNHG1 regulating network, we used RNA-protein pull-down to analyze the protein molecules interacting with SNHG1. By silver staining of pull-down protein SDS-PAGE gel, we found that SNHG1 was specifically bound to 60-70kda protein bands (Fig. 4F), the main protein of SNHG1 interaction was identified as hnRNPL by mass spectrometry and western blot (Fig. 4G–I). Other potential interactors of SNHG1 were shown in Table S1. Besides, the interaction between hnRNPL and SNHG1 was further predicted by RPISeq (Fig. 4H), and the binding relationship was double checked by hnRNPL RIP-PCR (Fig. 4J). Furthermore, western blot showed that SNHG1 knockdown and overexpression did not affect the hnRNPL protein level, and it can regulate EMT pathway (Fig. 4K–L). Finally, our previous work confirmed that SNHG1 triggers EMT pathway through directly binding to hnRNPL in the development of prostate cancer.
SNHG1/hnRNPL complex regulate E-cadherin expression
Next, to investigate the exact binding site between SNHG1 and hnRNPL, CatRAPID fragment prediction was used to reveal that the sequence of SNHG1 at 400–600 bp have a significant binding possibility with hnRNPL (Fig. 5A). Then, we constructed SNHG1-mut truncated mutant overexpression vector with 400–607 bp deletion of SNHG1 sequence compared with the SNHG1-wt (Fig. 5B). In DU145/sh-SNHG1 cells, Vector, SNHG1-mut and SNHG1-wt plasmids were transfected, and the over-expression efficiency of plasmids was verified by qRT-PCR (Fig. 5C). Meanwhile, the results of hnRNPL RIP-PCR in aforesaid cells confirmed that 400–607 bp truncated SNHG1-mut could not bind to hnNRPL protein. In addition, hnRNPL also lost the ability to capture CDH1 to a great extent when it could not specifically bind to SNHG1 compared with SNHG1-wt overexpressed cell lines (Fig. 5D). Meanwhile, Transwell and wound healing assays demonstrated that overexpressing mutant SNGH1 could not promote cell migration ability in PCa cells, when compared with the wild type SNHG1 (Fig. 5E–F). As hnRNPL is essential for mRNA stability, we wondered if the SNHG1/hnRNPL stabilizes certain unknown downstream effector proteins, such as E-cadherin. Therefore, using siRNA targeting on hnRNPL, we effectively inhibited its expression in DU145 cells. Surprisingly, we found that E-cadherin was remarkably downregulated both in mRNA and protein levels, while SNHG1 did not show obvious difference (Fig. 5G–H). We further found that knockdown of hnRNPL significantly reduced the mRNA stability of CDH1(E-cadherin) (Fig. 5I). Therefore, we demonstrated that SNHG1/hnRNPL complex could break the balance between hnRNPL and E-cadherin mRNA, causing to E-cadherin protein downregulation.
A CatRAPID fragment prediction revealed that the sequence of SNHG1 at 400–600 bp had a significant binding possibility with hnRNPL (http://service.tartaglialab.com/page/catra-pid_group). B Schematic diagram of 400–607 bp truncated mutation in SNHG1. C Relative quantitative detection of truncated mutation and wild-type SNHG1 overexpression by qRT-PCR. D HnRNPL RIP confirmed that knockdown SNHG1 reduced the binding ability of hnRNPL to CDH1, and the overexpression of wild-type SNHG1 could restore the binding ability, while the SNHG1-mut with poor binding ability of hnRNPL could not rescue hnRNPL to capture CDH1. E, F Effects of overexpression of wild type SNHG1 or mutant SNHG1 on cell migration by wound healing and migration assays. G, H Knockdown of hnRNPL suppressed the mRNA and protein expression of CDH1(E-cadherin). I HnRNPL knockdown in DU145 cells downregulated CDH1(E-cadherin) mRNA abundance, while ActinomycinD (2.0 μg/ml) were used to inhibit RNA synthesis. All data are shown as the mean ± SD. **P < 0.01, ***P < 0.001, ****P < 0.0001 by two-tailed Student’s t test.
HnRNPL knockdown reverses the carcinogenic effects of SNHG1 promotion on cell metastasis
To explore whether SNHG1 exerts its biological function by binding to hnRNPL, a rescue assay between SNHG1 and hnRNPL was performed. Transwell and wound healing assays revealed that hnRNPL knockdown significantly reversed the SNHG1 overexpression-induced cell migration ability in PCa cells (Fig. 6A, B). Meanwhile, western blot showed that forced expression of hnRNPL reversed the SNHG1 upregulating-induced inhibition of E-cadherin protein (Fig. 6C). Moreover, Survival analysis revealed that PCa patients with both low SNHG1 and HnRNPL expression levels had better overall survival and disease-free survival with GEPIA (Fig. 6D). In general, we illustrated that LncRNA SNHG1 contributed to PCa growth and metastasis by directly binding to hnRNPL to coregulate E-cadherin, which activates PCa EMT (Fig. 6E).
A, B HnRNPL knockdown reversed the SNHG1 overexpression-induced cell migration ability in PCa cells. C western blot showed that forced expression of hnRNPL reversed the SNHG1 upregulating-induced inhibition of E-cadherin protein. D Survival analysis revealed that PCa patients with both low SNHG1 and hnRNPL expression levels had better overall survival and disease-free survival (GEPIA, http://gepia.cancer-pku.cn/). E Schematic diagram displaying the mechanism underlying SNHG1 could competitively bind to hnRNPL leading to downregulating of E-cadherin. **P < 0.01, ***P < 0.001.
Discussion
In recent years, the incidence of prostate cancer has been on the rise in China. Despite that the diagnosis rate of prostate cancer has been improved by diagnostic methods such as screening PSA and imaging, the mortality caused by PCa remains unchanged, and metastasis is a critical factor leading to the death of prostate cancer15,16. Therefore, to reveal the pathogenesis of PCa may help us find specific biomarkers, which bears great significance for the diagnosis, treatment and prognosis of patients with PCa.
LncRNAs participates in the development of cancer through a variety of mechanisms that regulate transcription, translation and protein function17. LncRNA prostate cancer antigen 3 (PCA3) has been approved by the U.S. FDA as a marker for the diagnosis of PCa, and has also been included in “Guidelines for Diagnosis and Treatment of Urological Diseases in China”18, announcing tremendous potential of lncRNA as a biomarker for the diagnosis and treatment of PCa, which is worthy of in-depth study. Although several studies have proved that SNHG1 promoted PCa proliferation via serving as “miRNA sponeges”11,12, the functions and potential mechanisms in PCa remains unclear. We identified lncRNA SNHG1 was overexpressed in prostate cancer tissues and cells, and associated with patient’s clinical characteristics. Results of gain and loss of function shown that lncRNA SNHG1 promoted PCa cell proliferation and migration in vitro, and loss-of SNHG1 inhibited vivo tumor growth, suggesting that SNHG1 is essential in PCa progression.
It is well known that LncRNAs mainly act as ceRNA to backout the inhibiting effects of miRNAs on their downstream targets19,20. SNHG1 was also reported to regulate miR-199a-3p and miR-337-3p/AKT2 axis, leading to promotion in PCa cells11,12. However, recently studies revealed that RNA binding protein plays a crucial role in LncRNA regulating network. Peng et al. revealed that Lnc-FAM84B-4 interacts with protein hnRNPK to restrain MAPK pathway, leading to CRC progression21. Klingenberg et al. indicated that CASC9/hnRNPL serves as a lncRNA/protein complex associated with clinically relevant viability and affects AKT signaling in HCC22. Therefore, RNA-Pulldown and RIP was constructed to establish the SNHG1 and hnRNPL interaction in PCa cells. hnRNPL belongs to the hnRNP protein family, which participates in the malignant progression of tumors though mediating alternative splicing of precursor mRNA, strengthening mRNA stability, regulating mRNA translation etc23,24,25,26. Linked to our results, we demonstrated that the binding of SNHG1 to hnRNPL is responsible for a part of its cancer-promoting features in PCa.
The EMT is a dynamic and critical process that enable the transition from early cancer to aggressive cancer, characterized via a decrease of epithelial markers and a upregulation in mesenchymal markers and EMT-related TFs27, but the upper mechanisms regulating EMT require fully understanding. The major epigenetic events, such as DNA methylation, histone modification, nucleosome-reshaping, and noncoding RNAs, play vital roles in activation or suppression of the initiation and sustainability of EMT28,29,30. CDH1, also known as E-cadherin, Notably, loss of E-cadherin was reckoned as the crucial step to initiate EMT that sustained PCa metastasis. Our works exhibited that the interaction of SNHG1 with hnRNPL is responsible for a part of EMT through next generation sequencing and RIP assays in PCa cells. Furthermore, the SNHG1/hnRNPL complex lead to a downstream of E-cadherin via binding to its mRNA, which promotes EMT in prostate cancer. These results also provide a new sight in SNHG1 regulating network.
Based on previous observations, we provide a strong evidence that SNHG1 interacts with RNA-binding protein hnRNPL to activate EMT pathway, promoting the malignant progression of PCa, which is expected to provide a comprehensive understanding of the way SNHG1 acts on cancers. This thorough comprehension of the interaction relationship of SNHG1 and hnRNPL may lead to more valid strategies to diagnose and treat PCa.
Conclusion
In conclusion, we found that SNHG1 promotes the proliferation and progression of PCa, and high expression of both SNHG1 and hnRNPL can shorten the overall and disease-free survival of PCa patients. Further experiments demonstrated that SNHG1 can interact with hnRNPL and ultimately leads to down-regulation of E-cadherin expression and promote EMT progression, leading to tumor metastasis in PCa.
Materials and methods
Patients and clinical samples
Formalin fixed paraffin—embedded prostate cancer specimens were obtained from patients that undergo radical prostatectomy (RP) at zhujiang hospital from 2015 to 2018. The pathological types of the control group (total 14 specimens) were benign prostatic hyperplasia (BPH) or adjacent prostatic tissues. Patients’ clinical information was obtained by reviewing the follow-up records of their electronic medical records. The median age of the enrolled patients was 67.5 years and average age was 63.3 (range: 22–82 years). Clinical TNM staging and Gleason scores of patient specimens were based on the American Joint Committee on Cancer Eighth Edition (2017) and the 2016 World Health Organization classification of genitourinary tumors. All patients agreed to participate in this study by signing an informed consent in accordance with the ethical scheme formulated by the ethics committee of Zhujiang Hospital, Southern Medical University. We were blinding to the groups when assessing the outcome.
Survival analysis
Association of LncRNA SNHG1 and hnRNPL level on clinical events was evaluated using The Cancer Genome Atlas (TCGA) prostate adenocarcinoma cohort. Survival and expression data were acquired from TCGA data portal. Gene Expression Profiling Interactive Analysis (GEPIA) was utilized to analyze Overall survival and disease-free survival. Statistical significance was evaluated by Kaplan–Meier analysis and log-rank test.
Fluorescence in situ hybridization (FISH)
Fluorescence in situ hybridization was constructed to identify the intracellular expression and sublocalization of SNHG1 according to the protocol described before31, with minor modifications. PCa cells were grown on 15 mm confocal dishes (JET BIOFIL, Guangzhou, China). Probe sequences targeting homo-SNHG1 were designed and generated by Ribo Bio (Guangzhou, China). Positive control probe sequences targeting homo-18S (cy3, Ribo Bio, Guangzhou, China), All operations were carried out in accordance with the Ribo™ Fluorescent In Situ Hybridization kit instructions, and minor modifications were made during the prostate tissue FISH assay. The samples were captured in Olympus laser scanning microscope FV3000 (Olympus Corporation, Japan). Images were processed in ImageJ through 3 major steps: background subtraction, set a threshold to label all the cells and label the SNHG1 positive cells.
Cytoplasmic and nuclear RNA isolation
Cytoplasmic and nuclear RNA was extracted using a Nuclear/Cytoplasmic Isolation reagents (No. 78833, Thermo Fisher Scientific, United States) according to the manufacturer’s instructions. And qRT-PCR was applied for further detection.
Cell culture and cell lines
We used five human prostate cancer cell lines and one immortalized prostatic epithelial cell line, all the cells including RWPE-1, LNCaP, 22Rv1, PC-3, DU145 were obtained from Stem Cell Bank, Chinese Academy of Sciences. RPMI-1640 medium containing 10% fetal bovine serum (FBS, Hyclone) was used to culture all prostate cancer cells, and the Keratinocyte Serum Free Medium (KSFM) (Gibco, No. 10744-019) added in 5 ng/mL epidermal growth factor (EGF) (Gibco, No. 10450-013) was suitable for culturing RWPE-1 cells. A Wild environment (37°C in 5% CO2) was set for maintaining all cells.
Construction of cells with stable knockdown of SNHG1
We designed and synthesized three small—interfering RNAs targeting SNHG1 (NR_003098.2) and negative control (NC) siRNA negative control sequence without a specific target, all the siRNAs were synthesized by GeneChem (Shanghai, China). The sequence of siRNAs targeting SNHG1 is as follows, si-h-SNHG1_001: CAGCA GTTGA GGGTT TGCTG TGTAT; si-h-SNHG1_002: TTCAA CAGCT AGGTT GTCCTT; si-h-SNHG1_003: GACCU AGCUU GUUGC CAAUTT. SNHG1 overexpressed cells used pcDNA3.1 system, which was synthesized by VigeneBio co. (Shandong, China). In this study, we used lentiviral vectors (GeneChem, Shanghai, People’s Republic of China) to construct stable silencing SNHG1 PCa cell lines following to the authoritative manufacturer’s instruction. Target sequence of shRNA (short hairpin RNA) againsting SNHG1 was 5’-GACCU AGCUU GUUGC CAAU-3’. Stable SNHG1 silenced cell lines sh-SNHG1 were kept in 5 μg/mL puromycin for 10 days and then confirmed that it is validity and specificity.
Quantitative RT–PCR assay and RNA extraction
The RNAiso Plus reagent (TaKaRa) and PrimeScript RT reagent Kit (TaKaRa) were utilized to extract total RNA and generate cDNA according to the official protocols. Extraction of nuclear and cytoplasmic RNA utilized the Ambion® PARIS™ system according to the kit procedures. The quantitative reverse transcriptase–PCR (qRT–PCR) reactions was performed with the SYBR Green PCR Master Mix (TaKaRa) with ABI7500 Fast Real-Time RCR System (Applied Biosystems, USA). The Primer3 system was utilized to design the gene unique primers which were subsequently synthesized through TsingKe Biological Technology (Guangzhou, China). Every survey was conducted in three times, and GAPDH was utilized to normalize the results as internal reference. The relative quantification method which calculated by 2-ΔΔCt was used in analyzing the expression data.
CCK-8 assay
2,000 PCa cells were seeded per well in 96-well plates, and Each well contained medium containing 10% FBS with a total volume of 100 ml. According to the counting kit-8 (CK-04, Dojindo) manufacturer’s operating instructions, after the cells were adherent to the plates, the original medium was removed, RPMI1640 diluted CCK-8 reagent (10% v/v) in 100 μl was added to the cells, following 2 h of incubated. Then, the optical density (OD) at 450 nm was measured by a microplate reader (EXL800, BioTek Instruments). Experiments were performed in triplicate.
Colony formation
PCa cells were inoculated into 6 pore plates at a rate of 500 cells per well in a 2 mL medium containing 10% fetal bovine serum, and incubated for two weeks for colony formation analysis. Then, 4% paraformaldehyde were used to fix the colonies and Giemsa was applied to stain colonies. The experiment of colony formation was repeated three times with three Wells in each group.
EdU incorporation assay
Cell-Light EdU staining kit (RiboBio, Guangzhou, China) was utilized to detect cell proliferation activity according to the manufacturer’s instructions. Images were obtained by a fluorescence microscope (Olympus, Tokyo, Japan) at 200×. The proportion of EdU positively stained cells to Hoechst-stained cells (with blue fluorescence) in per well was equal to the cell proliferation rate. Experiments were performed in triplicate.
Transwell migration assay
Transwell (Costar, Corning, USA) with a membrane of numerous pole(8.0μm) was used to perform Cell migration assay. 35000 cells were hybridized in FBS-free RPMI-1640 and sowed into the upper chamber of the well. Then, 500 μL complete medium with FBS was put into the sublayer. After 24 h, 4% paraformaldehyde and Giemsa (Boster Ltd., Wuhan, China) were used to fix and stain the cells. After that, wiping out the cells on the top surface of the membrane, and the cells on the bottom surface were photographed through an inverted microscope. (Olympus DP72). Five randomly visual fields were chosen to count the cell number by using Image J software. Experiments were performed in triplicate.
Mice xenograft analysis
Xenograft models were randomly created through persistent injection of 5 × 106 DU145/sh-NC, DU145/sh-SNHG1 cells (n = 7 per group), on the axillae of nude male mice (4–5 weeks). Tumor sizes were measured twice a week and formula \(\frac{{\pi \times {\mathrm{Length}} \times {\mathrm{Width}}^2}}{6}\) was used to calculate tumor volume. Mice were raised in Specific Pathogen Free (SPF) environments for 4 weeks. Subsequently, tumor samples were carefully resected, photographed and specimens were further investigated by hematoxylin and eosin(H&E) and IHC. BALB/c nude male mice were purchased from the Animal Center of Southern Medical University, Guangzhou, China. The Animal Care and Use Committee of Southern Medical University approved all our experimental animal programs.
Antibodies and western blot analyses
Related cell lysates were run on 4–12% SDS PAGEs to conduct Western blot analyses. RIPA lysis buffer containing protease inhibitors (#KGP250, KeyGEN BioTECH, Nanjing, China) was used to extracted PCa cells protein following to the operation protocol. Then, equal amounts of 30 μg proteins were separated by SDS/PAGEs and transferred onto PVDF membranes (Millipore, Billerica, MA, USA) electrically. Then, membranes were blocked with TBS(Tris/saline solution with 0.1% Tween-20) including 5% milk without fat for 1 h and incubated overnight at 4 °C covering by unique antibodies: rabbit anti-β-actin (#4970, CST), rabbit anti-Slug (#9585), rabbit anti-ZEB1(#3396, CST), rabbit anti-E-Cadherin (#3195, CST) and rabbit anti-hnRNPL (4D11, #ab6106, Abcam). All membranes were subsequently incubated at room temperature for 1 h with horseradish peroxidase-linked secondary anti-rabbit or anti-mouse IgG antibodies (Cell Signaling Technology). Final bands were visualized by ECL kit (Pierce Biotechnology, Rockford, IL, USA). Image J was applied to quantify the intensity of the band. Experiments were performed in triplicate.
Plasmid transfection
The full-length wild-type human SNHG1 and 401-607 bp truncated mutant SNHG1-mut were inserted into pcDNA3.1 expression vector acquired from Shangdong Vigene Biosciences Co. All constructs were confirmed by sequencing. Referring to the Lipofectamine™ 3000 Transfection Reagent instructions (L3000015, Thermo Fisher Scientific, USA), DU145/sh-SNHG1 cell line was transiently transfected with SNHG1-mut or SNHG1, DU145 and C4-2 cells were transfected with SNHG1 or meaningless vector. Expression of Plasmid was confirmed by qRT-PCR.
Immunohistochemical analysis and assessment
Prostate tissue IHC were employed to investigate protein expression in tissues as described previously. Relative proteins expression was investigated in Prostate tissue IHC as previously referred to32. In IHC, we incubated the specimens with 1: 500 anti-Ki-67 antibodies. IHC results were evaluated by calculating the percentage of positive nuclei, the percentage of positive cells and the staining concentration. Every assessment was carried out by three separated advanced pathologists utilizing the parallel microscope. Experiments were performed in triplicate.
RNA-seq processing
RNA sequencing and sequence quality control was performed by BGISEQ platform. Human genome reference was established from UCSC version GRCh38/hg38 chromosomes 1-22, X, Y and mitochondrial DNA. The further analysis including heatmap, gene set enrichment analysis, and gene annotation of sequencing data was completed by BGI Dr. Tom system.
RNA-protein pulldown and RNA immunoprecipitation (RIP)
In vitro biotin-labeled RNAs (SNHG1, AR-3’UTR and Ploy (A)25) and RNA-protein pull-down assay were done with Pierce™ Magnetic RNA-Protein Pull-Down Kit (#20164, Thermo Scientific™). Pull-down protein samples were ran on SDS-PAGE gels, following silver staining to Identify the difference bands, then the difference bands were analyzed by mass spectrometry. RIP assays were performed by utilizing EZ-Magna RIP™ Kit (#17-701, Merck Millipore) following the protocol.
In silico binding prediction
To obtain potential SNHG1 and hnRNPL binding sites, we first predicted the interaction probability between SNHG1 and hnRNPL sequences in RNA-Protein Interaction Prediction(RPISeq). Furthermore, catRAPID was used to predict the binding site of hnRNPL on SNHG1. The highest-raking site at RNA position 401-677 bp includes CA-rich motifs and thus used for subsequent analysis.
Statistical analysis
Statistical analyses were carried out with GraphPad Prism8 (GraphPad Software, La Jolla, USA). Student’s t test or chi-square test was applied to determine the statistical significance of differences between distinct groups, if proper. All results were expressed as mean ± standard deviation (SD), and the two-tailed p value of 0.05 was considered statistically significant.
Data availability statement
All data generated or analyzed during this study are included in this published article.
Change history
14 March 2024
A Correction to this paper has been published: https://doi.org/10.1038/s41419-024-06561-5
References
Siegel R. L., Miller K. D., Jemal A. Cancer statistics, 2020. CA Cancer J. Clin. 2020.
Shen, M. M. & Abate-Shen, C. Molecular genetics of prostate cancer: new prospects for old challenges. Genes Dev. 24, 1967–2000 (2010).
Kalluri, R. & Weinberg, R. A. The basics of epithelial-mesenchymal transition. J. Clin. Investig. 119, 1420–1428 (2009).
Mittal, V. Epithelial mesenchymal transition in tumor metastasis. Annu. Rev. Pathol. 13, 395–412 (2018).
Brabletz, T., Kalluri, R., Nieto, M. A. & Weinberg, R. A. EMT in cancer. Nat. Rev. Cancer 18, 128–134 (2018).
Body, J. J., Casimiro, S. & Costa, L. Targeting bone metastases in prostate cancer: improving clinical outcome. Nat. Rev. Urol. 12, 340–356 (2015).
Osielska, M. A. & Jagodzinski, P. P. Long non-coding RNA as potential biomarkers in non-small-cell lung cancer: What do we know so far? Biomedicine Pharmacother. 101, 322–333 (2018).
Alahari, S. V., Eastlack, S. C. & Alahari, S. K. Role of long noncoding RNAs in neoplasia: special emphasis on prostate cancer. Int Rev. Cell Mol. Biol. 324, 229–254 (2016).
Prensner, J. R. & Chinnaiyan, A. M. The emergence of lncRNAs in cancer biology. Cancer Disco. 1, 391–407 (2011).
Zhao, Z., Liang, S. & Sun, F. LncRNA DLX6-AS1 promotes malignant phenotype and lymph node metastasis in prostate cancer by inducing LARGE Methylation. Front Oncol. 10, 1172 (2020).
Cui, R. et al. LncRNA AC245100.4 binds HSP90 to promote the proliferation of prostate cancer. Epigenomics 12, 1257–1271 (2020).
Thin, K. Z., Tu, J. C. & Raveendran, S. Long non-coding SNHG1 in cancer. Clin. Chim. Acta 494, 38–47 (2019).
Li, J. et al. SNHG1 lncRNA negatively regulates miR-199a-3p to enhance CDK7 expression and promote cell proliferation in prostate cancer. Biochem Biophys. Res. Commun. 487, 146–152 (2017).
Xie, M., Zhang, Z. Y. & Cui, Y. P. Long noncoding RNA SNHG1 contributes to the promotion of prostate cancer cells through regulating miR-377-3p/AKT2 axis. Cancer Biother Radio 35, 109–119 (2020).
Siegel, R. L., Miller, K. D. & Jemal, A. Cancer statistics, 2018. CA Cancer J. Clin. 68, 7–30 (2018).
Lughezzani, G. et al. Multicenter European external validation of a prostate health index-based nomogram for predicting prostate cancer at extended biopsy. Eur. Urol. 66, 906–912 (2014).
Yang, G. D., Lu, X. Z. & Yuan, L. J. LncRNA: a link between RNA and cancer. Bba-Gene Regul. Mech. 1839, 1097–1109 (2014).
Soares, J. C. et al. Detection of the prostate cancer biomarker PCA3 with electrochemical and impedance-based biosensors. ACS Appl Mater. Interfaces 11, 46645–46650 (2019).
Han, D. et al. Long noncoding RNAs: novel players in colorectal cancer. Cancer Lett. 361, 13–21 (2015).
Tay, Y., Rinn, J. & Pandolfi, P. P. The multilayered complexity of ceRNA crosstalk and competition. Nature 505, 344–352 (2014).
Peng, W. et al. Lnc-FAM84B-4 acts as an oncogenic lncRNA by interacting with protein hnRNPK to restrain MAPK phosphatases-DUSP1 expression. Cancer Lett. 494, 94–106 (2020).
Klingenberg, M. et al. The long noncoding RNA cancer susceptibility 9 and RNA binding protein heterogeneous nuclear ribonucleoprotein L form a complex and coregulate genes linked to AKT signaling. Hepatology 68, 1817–1832 (2018).
Zhou, X. et al. HnRNP-L promotes prostate cancer progression by enhancing cell cycling and inhibiting apoptosis. Oncotarget 8, 19342–19353 (2017).
Lv, D. et al. HnRNP-L mediates bladder cancer progression by inhibiting apoptotic signaling and enhancing MAPK signaling pathways. Oncotarget 8, 13586–13599 (2017).
Fei, T. et al. Genome-wide CRISPR screen identifies HNRNPL as a prostate cancer dependency regulating RNA splicing. Proc. Natl Acad. Sci. USA 114, E5207–E5215 (2017).
Li, Z. et al. The long noncoding RNA THRIL regulates TNFα expression through its interaction with hnRNPL. Proc. Natl Acad. Sci. USA 111, 1002–1007 (2014).
Nieto, M. A., Huang, R. Y.-J., Jackson, R. A. & Thiery, J. P. EMT: 2016. Cell 166, 21–45 (2016).
Skrypek, N., Goossens, S., De Smedt, E., Vandamme, N. & Berx, G. Epithelial-to-mesenchymal transition: epigenetic reprogramming driving cellular plasticity. Trends Genet. 33, 943–959 (2017).
Sun, L. & Fang, J. Epigenetic regulation of epithelial-mesenchymal transition. Cell Mol. Life Sci. 73, 4493–4515 (2016).
Serrano-Gomez, S. J., Maziveyi, M. & Alahari, S. K. Regulation of epithelial-mesenchymal transition through epigenetic and post-translational modifications. Mol. Cancer 15, 18 (2016).
Raj, A., van den Bogaard, P., Rifkin, S. A., van Oudenaarden, A. & Tyagi, S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat. methods 5, 877–879 (2008).
Qiao, H. et al. Phosphoribosyl pyrophosphate synthetases 2 knockdown inhibits prostate cancer progression by suppressing cell cycle and inducing cell apoptosis. J. Cancer 11, 1027–1037 (2020).
Funding statement
This work was financed by grants from the National Natural Science Foundation of China (No.81773277), Science and Technology Program of Guangzhou (No.201803010014), China Postdoctoral Science Foundation funded project (No.2018M643126 and 2019M662865), the Guangdong Basic and Applied Basic Research Foundation (No.2019A1515110033); Distinguished Young Talents in Higher Education Foundation of Guangdong Province (NO.2019KQNCX115); Achievement cultivation and clinical transformation application cultivation projects of the First Affiliated Hospital of Guangzhou Medical University (NO.ZH201908).
Author information
Authors and Affiliations
Contributions
M.X.M, S. F. P, and L.C.H conceived and designed the study. T.X, C.W.B, L.D.J carried out the experiments. Y. T. W, W. K.H, and Z.L.B, collected and analyzed the clinical data. L.J.Q and Z.X.M analyzed and interpreted the data. C.W.B and L.D.J wrote the manuscript. M.X.M, S. F. P, and L.D.J executed the funding acquisition. All authors have reviewed and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval and consent to participate
All experiment protocols were approved by the Medical Ethics Committee of the Zhujiang Hospital Affiliated to Southern Medical University and performed following the World Medical Association Declaration of Helsinki. All procedures were approved by the Medical Ethics Committee of the Zhujiang Hospital Affiliated to Southern Medical University, and performed according to the Guide for the Care and Use of Laboratory Animals.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by G. Calin
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Tan, X., Chen, Wb., Lv, Dj. et al. LncRNA SNHG1 and RNA binding protein hnRNPL form a complex and coregulate CDH1 to boost the growth and metastasis of prostate cancer. Cell Death Dis 12, 138 (2021). https://doi.org/10.1038/s41419-021-03413-4
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41419-021-03413-4
This article is cited by
-
Long noncoding RNA SNHG1 promotes breast cancer progression by regulating the miR-641/RRS1 axis
Scientific Reports (2024)
-
LncRNA SNHG1 Accelerates Cell Proliferation, Migration, and Invasion of Hepatoblastoma Through Mediating miR-6838-5p/PIM3/RhoA Axis
Biochemical Genetics (2024)
-
LncRNA SNHG1: role in tumorigenesis of multiple human cancers
Cancer Cell International (2023)
-
Long Non-coding RNA SNHG1 Suppresses the Osteogenic Differentiation of Bone Marrow Mesenchymal Stem Cells by Binding with HMGB1
Biochemical Genetics (2023)
-
Molecular Landscape of LncRNAs in Prostate Cancer: A focus on pathways and therapeutic targets for intervention
Journal of Experimental & Clinical Cancer Research (2022)