Abstract
Novel coronavirus disease 2019 (COVID-19) is associated with a hypercoagulable state, characterized by abnormal coagulation parameters and by increased incidence of cardiovascular complications. With this study, we aimed to investigate the activation state and the expression of transmembrane proteins in platelets of hospitalized COVID-19 patients. We investigated transmembrane proteins expression with a customized mass cytometry panel of 21 antibodies. Platelets of 8 hospitalized COVID-19 patients not requiring intensive care support and without pre-existing conditions were compared to platelets of healthy controls (11 donors) with and without in vitro stimulation with thrombin receptor-activating peptide (TRAP). Mass cytometry of non-stimulated platelets detected an increased surface expression of activation markers P-Selectin (0.67 vs. 1.87 median signal intensity for controls vs. patients, p = 0.0015) and LAMP-3 (CD63, 0.37 vs. 0.81, p = 0.0004), the GPIIb/IIIa complex (4.58 vs. 5.03, p < 0.0001) and other adhesion molecules involved in platelet activation and platelet–leukocyte interactions. Upon TRAP stimulation, mass cytometry detected a higher expression of P-selectin in COVID-19 samples compared to controls (p < 0.0001). However, we observed a significantly reduced capacity of COVID-19 platelets to increase the expression of activation markers LAMP-3 and P-Selectin upon stimulation with TRAP. We detected a hyperactivated phenotype in platelets during SARS-CoV-2 infection, consisting of highly expressed platelet activation markers, which might contribute to the hypercoagulopathy observed in COVID-19. In addition, several transmembrane proteins were more highly expressed compared to healthy controls. These findings support research projects investigating antithrombotic and antiplatelet treatment regimes in COVID-19 patients, and provide new insights on the phenotypical platelet expression during SARS-CoV-2 infection.
Similar content being viewed by others
Introduction
Despite severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)’s worldwide spread, little is known about the pathophysiological mechanisms leading to multiorgan damage in coronavirus disease 2019 (COVID-19). A hypercoagulable state with increased incidence of cardiovascular complications and venous thrombotic events has been reported in several studies1,2,3,4,5,6,7. Abnormal coagulation parameters are observed in hospitalized patients and are associated with poor prognosis8,9,10. Interestingly, two studies reported in COVID-19 alterations in platelet transcriptome and proteome, and an increased platelet reactivity11,12. A recent study described the presence microvascular thrombi in lung, heart and kidney containing neutrophil extracellular traps (NETs) in severe SARS-CoV-2 infection, as well as circulant neutrophil-platelet aggregates and immunothrombotic dysregulation, which changes with disease severity13. Recently, a computational system’s medicine platform identified as new drug target several proteins involved in the coagulation cascade14,15. However, the role of platelet activation and changes of transmembrane receptor expression in COVID-19-induced coagulopathy still needs to be further investigated.
Platelets not only play a pivotal role in vascular hemostasis but are also involved in immune response, tumor progression, and other inflammatory processes16. They are activated during sepsis and in septic shock, and antiplatelet therapy has been suggested as a novel strategy to prevent organ damage17. In fact, in the presence of severe infections or cytokine storms18,19, platelet hyperreactivity may be responsible for major cardiovascular adverse events20. Viral infections are known to be associated with coagulation disorders21. Interestingly, an increased incidence of acute coronary syndrome has been observed after influenza infection22, suggesting that viral diseases could trigger platelet activation leading to cardiovascular complications. Moreover, viral-induced coagulopathies have been already observed in SARS-CoV-1 infection including thrombocytosis, disseminated intravascular coagulation, and thromboembolism23,24. In this study, we investigated the expression of platelet transmembrane receptors and adhesion molecules at baseline level and after in vitro platelet stimulation in hospitalized COVID-19 patients without pre-existing conditions and in healthy donors using mass cytometry by time of flight (CyTOF). Here we present the largest existing CyTOF panel of platelet antibodies specifically developed to investigate platelet activation and adhesion (Table 1).
Methods
Data and code availability
All mass cytometry data have been made available at flowrepository.org and can be accessed at repository ID FR-FCM-Z2MT. The scripts used in this analysis have been deposited at github.com and can be accessed at https://github.com/biomedbigdata/SARS-CoV-2-platelets-analysis.
Study design and participants
SARS-CoV-2-infected patients hospitalized at the Klinikum recht der Isar, Munich, Germany, between March and May 2020 with symptomatic COVID-19 not requiring intensive care unit admission and without known pre-existing conditions were recruited in our study and compared to an asymptomatic control cohort of healthy donors. Inclusion criteria for the COVID-19 group were a symptomatic (dyspnea) SARS-CoV-2 infection confirmed by a positive reverse-transcription PCR assay from any respiratory specimen or IgM antibodies in peripheral blood, age between >18 and <70 years, and written informed consent.
Exclusion criteria were known platelet dysfunctions, relevant thrombocytopenia (<100 G/l) or thrombocytosis (>500 G/l), impaired renal function (glomerular filtration rate < 60ml/min), hemoglobin < 10g/dl, leukocytes < 1 G/l, any known pre-existing condition except arterial hypertension, any medication except antihypertensive drugs, and a history of hematological neoplasia including active lymphoma, mental impairment, or pregnancy.
Blood samples were collected from patients within the first 36 h after admission. As a control group, we recruited a healthy and asymptomatic cohort of donors. All healthy donors were tested negative for SARS-CoV-2 IgG and IgM, were followed up, and did not develop any symptoms in the weeks following the recruitment. Throughout the entire study design, patients’ samples were handled together with control samples. The study complied to the Declaration of Helsinki, was approved by the local ethics committee (approval numbers 147/20 and 352/18), and all participants provided written informed consent.
Sample collection and preparation
Peripheral venous blood was collected in citrate tubes and immediately processed to produce platelet-rich plasma (PRP) as described before25,26. CyTOF staining assay was performed according to the manufacturer’s protocols. Briefly, 600 µl PRP previously inhibited by a mixture of 0.6 U Apyrase/ml, 20 mM of HEPES, and 1 mM EGTA was diluted in phosphate-buffered saline (PBS) pursuant to the gold standard protocol for mass cytometry (Fluidigm, San Francisco, CA, USA) to a final concentration of 105 platelets/µl. The PRP was stained with 5 µM Cell-IDTM Cisplatin (Fluidigm) for 5 min and then washed with 5 ml MaxPar Cell Staining Buffer (Fluidigm). After centrifugation, cells were resuspended in 50 µl Cell Staining Buffer. Two samples were prepared from each donor: one baseline sample (non-stimulated platelets) and one sample stimulated with 10 µM thrombin receptor-activating peptide (TRAP). TRAP addition was followed by a 2 min incubation at room temperature. In the same cell suspensions platelets were stained with 50 µl of the custom-made CyTOF-antibody panel in Cell Staining Buffer for 30 min (containing anti-CD3-170Er, anti-CD9-171Yb, anti-CD29-156Yb, anti-CD31-145Nd, anti-CD36-152Nd, anti-CD40-142Nd, anti-CD41-89Y, anti-CD42a-141Pr, anti-CD42b-144Nd, anti-CD47-209Bi, anti-CD61-146Nd, anti-CD62P-161Dy, anti-CD63-150Nd, anti-CD69-162Dy, anti-CD107a-151Eu, anti-CD141-166Er, anti-CD154-168Er, anti-GPVI-175Lu, anti-GPIIb/GPIIIa complex-155Gd, anti-Par1-147Sm, and anti-PEAR-147Sm; see Supplemental Material for antibody information). After washing twice with 2 ml Cell Staining Buffer at 300 g for 5 min, cells were fixed overnight at 4 °C in 1 ml of 1.6% Formaldehyde. After fixation, cells were pelleted at 800 × g for 10 min, the supernatant was aspirated and removed. Then, cells were gently vortexed and resuspended in the residual volume (~50 µl) and incubated with 125 nM Iridium in 1 ml MaxPar Fix and Perm Buffer (Fluidigm) for 1 h following the manufacturer’s protocol (Fluidigm). Afterwards, they were centrifuged at 800 × g for 5 min, then washed with 2 ml Cell Staining Buffer at 800 g for 5 min, then frozen in 10% dimethyl sulfoxide (DMSO) in fetal bovine serum until acquisition27. After thawing the samples, they were washed twice with Cell Staining Buffer and once with water at 800 × g for 5 min to eliminate DMSO remnants. Cells were then handled according to the manufacturer’s protocol. Prior to measurement, cells were diluted to a final concentration of 103 platelets/µl before addition of EQ calibration beads. Cells were measured using a Helios mass cytometer (Fluidigm). Throughout the study, patients’ samples were measured with at least one control sample to reduce batch effect. In total, 476,756 ± 151,746 events were acquired at a rate of 300–500 events per second. Experiments were carried out by the same scientist and antibodies were from the same lot. See Supplemental Material for a complete reagent list.
Mass cytometry
CyTOF allows multidimensional relative protein quantification for single-cell datasets and we adapted it for platelets using a customized mass cytometry panel of 21 antibodies (Table 1). For custom-made antibody conjugations, 100 mg of carrier-free antibody was coupled to metal-labeled X8 polymer according to the manufacturer’s instructions (Fluidigm). Briefly, using the MaxPAR antibody conjugation kit (Fluidigm) following the manufacturer’s recommended protocol, six antibodies were conjugated to isotopically enriched lanthanide metals. After labeling, the antibodies were stored in an antibody stabilization buffer (Boca Scientific, Westwood, MA, USA) at 4 °C. The other antibodies were pre-conjugated, CyTOF-ready, and commercially available (Fluidigm Sciences). Please see the Supplemental Materials for the reagent list. All custom-conjugated antibodies were validated with calibration beads. In detail, 0.5 µl of the conjugated antibody was added to one drop of beads and incubated for 15 min. After two washing steps with 1.5 ml PBS at 300 × g for 10 min, the mixture was washed twice with de-ionized water at 300 × g for 10 min, and resuspended in 200 µl water until acquisition.
CyTOF processing
After acquisition, samples were cleaned up according to the latest standard of data pre-gating (Fluidigm) using the Cytobank™ software (www.cytobank.org, Beckman-Coulter, Brea, CA, USA)28. To avoid leukocyte contamination, we gated the acquired events for platelet-specific markers: only CD41 (GPIIb)- and CD61-(GPIIIa) positive events were selected for further analysis and defined as platelets (Supplemental Fig. I). CD3 marker was included in the panel as an additional negative control (Supplemental Fig. II).
Computational analysis
All models were built and assessed using the statsmodels v0.11.1 python package29. CyTOF data were processed using Cytobank and analyzed using R 4.0 (R Development Core Team, 2005) and Python 3.6 (Python 3 Reference Manual, 2009). For visualization of activation markers in reduced dimensions, we performed uniform manifold approximation and projection based on 16 markers (excluding the activation markers) using the CATALYST v1.12.1 R package. To account for differences in coverage between samples, we randomly sampled the minimum number of events acquired (41,525 events per sample, Fig. 1). Following standard practice for differential marker expression in CyTOF30, we built mixed-effect linear models for the TRAP-stimulated and non-stimulated sample groups, respectively. We considered the sample-wise median signal intensity as dependent variable, disease status as fixed and patient IDs as random effect, i.e., each patient has a different intercept. Furthermore, we built a linear model for all samples (TRAP-stimulated and non-stimulated) with an interaction term to assess whether activation is significantly affected by disease status. P-values of model coefficients were corrected for multiple hypothesis testing using the Benjamini–Hochberg method (false discovery rate < 0.05)31.
Differential analysis of overall marker expression
Statistical significance is evaluated based on regression analysis. To estimate whether the sample-wise median expression of a marker is significantly different between COVID-19 patients and healthy controls, the following linear mixed-effect model was used:
where Yij is the median expression of the j-th marker for i-th patient, xci is a binary variable indicating if a patient i belongs to case or control group, and \(\gamma _i \sim N\left( {0,\sigma _i^2} \right)\) is a random intercept for each patient. The latter allows us to disentangle within-sample and within-group variance.
Slope coefficients \(\beta _{cj}\) were tested for significance of the linear relationship between the independent variable xc and the dependent variable Y:
Difference in TRAP stimulation effect for COVID-19 patients
To analyze if a higher expression of activation markers in COVID-19 quiescent platelets is coupled with a reduced capacity to react upon activation stimuli, we compared slope coefficients for the covariate that corresponds to activation for cases and controls separately. We used the following model for healthy controls:
where h belongs to a set of indices for all healthy controls and xah is a binary variable that indicates if a sample h was TRAP-stimulated or not. We computed a set of slopes \(\beta _{aj}\) that show an average linear increase in expression after activation for every marker j.
A similar model was used for COVID-19 patients:
where p belongs to a set of indices for all COVID-19 patients. Although we can compare \(\beta _a\) coefficients from the model (3) and model (4) directly, we cannot conclude if the difference between slope coefficients is statistically significant. To evaluate if there is a statistically significant difference in the reaction to TRAP stimulation between patients and controls, we used a single model with an interaction effect term. Significance of the Interaction effect means that activation status and patient condition (disease or control) combined have a significantly larger effect on median signal intensity as compared to the sum of the individual factors alone. Formally, this results in the following model:
where xint is an interaction term defined as xa × xc. The slope coefficient for the interaction \({\beta}_{int}\) was then tested for statistical significance as shown in Eq. (2).
Clustering analysis: FlowSOM algorithm
Automated clustering analysis was done using the FlowSOM algorithm32. After gating (Supplemental Fig. I), data were compensated, transformed with an estimated logical transformation, and scaled. Cells were assigned to a 10 × 10 self-organizing map and then metaclustering in 12 clusters was performed. The number of clusters was selected based on relative change in area under the cumulative distribution function curve that indicated that cells stratification in more than 12 clusters cannot improve the clustering results. For each cluster-marker pair, a p-value was computed using a linear mixed-effect model:
where Yij is the median expression of the j-th marker for i-th cluster, xci is a binary variable indicating if cell population Yij belongs to case or control group, and \(\gamma _i \sim N\left( {0,\sigma _i^2} \right)\) is a random intercept for each cell subpopulation. Slope coefficients \({\beta}_{{{cj}}}\)were tested for significance of the linear relationship between the independent variable xc and the dependent variable Y.
Results
Study population characteristics
Eight hospitalized symptomatic COVID-19 patients without pre-existing conditions requiring oxygen support were recruited and compared to a cohort of 11 asymptomatic healthy donors, tested negative for SARS-CoV-2 (mean age COVID-19: 51.4 ± 11.7, controls: 44.7 ± 13.0, p = 0.27; male COVID-19 62.5%, controls: 45% p = 0.49). Seven patients showed typical COVID-19 pulmonary lesions in chest computed tomography. Patients were admitted through the emergency department and moved to normal wards due to dyspnea. During the hospitalization, one patient was transferred to an intermediated care unit (2 days after blood collection) for a few hours, for the purpose of monitoring due to respiratory deterioration. No patient required assisted ventilation and all were discharged in good condition (average hospitalization 9.5 ± 6.3 days). No major adverse events (bleeding and thromboembolic events) were reported. All admitted patients were not under regular medication, except one with two antihypertensive medications: amlodipine and valsartan. For a detailed description of the study population, see the Supplementary Table 1.
Platelet surface receptor and adhesion molecule expression in non-stimulated platelets
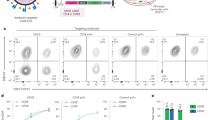
Compared to healthy controls, non-stimulated platelets of COVID-19 patients showed a significantly higher spontaneous expression of specific platelet activation markers (Fig. 1), such as P-Selectin (0.67 vs. 1.87 median signal intensity for healthy donors vs. patients, p = 0.0015) and LAMP-3 (0.37 vs. 0.81 median signal intensity, p = 0.0004, Fig. 2a), as well as the the GPIIb/IIIa complex (4.58 vs. 5.03 median signal intensity, p < 0.0001). In addition, we detected a higher spontaneous expression of some constitutive receptors and adhesion molecules involved in platelet activation and aggregation in COVID-19 platelets, such as the transmembrane integrins GPIIb (p = 0.0001) and GPIIIa (p < 0.0001), as well as the glycoproteins GPIbα (p = 0.0086) and GPIX (p = 0.0126, Fig. 2b). The expression level of all other activation markers and adhesion molecules are shown in Supplemental Fig. II.
Median signal intensity of activation markers (A) and relevant transmembrane proteins (B) in non-stimulated platelets. COVID-19 patients are plotted in red, whereas controls are plotted in blue. The horizontal line within the box plot represents the median, the top and bottom the interquartile range(Q1–Q3), whisker bars indicate the largest observation that is less than or equal to the upper inner fence (UIF = Q3 + 1.5 × IQR) or the smallest observation that is greater than or equal to the lower inner fence (LIF = Q1–1.5 × IQR) and the circles indicate outliers, if present; *P < 0.01. N = 8 COVID-19 patients, 11 healthy donors.
Diseased platelet reactivity after TRAP stimulation
To further investigate platelet reactivity, we stimulated the collected platelets with 10 µM TRAP. Upon TRAP stimulation, mass cytometry also detected a significantly higher expression of the platelet activation marker P-selectin in samples of COVID-19 patients compared to healthy controls (p = 0.0176), but LAMP-3 did not show significant differences (p = 0.40, Fig. 3a). Interestingly, the GPIIb/GPIIIa complex remained upregulated in COVID-19 patients after TRAP stimulation (p < 0.0001). Similar to non-stimulated platelets, we also observed a higher expression level for the integrins GPIIb (p < 0.0001), GPIIIa (p = 0.0009), as well as for the glycoproteins GPIbα in TRAP-stimulated platelets compared to healthy controls (p < 0.0001, Fig. 3b). In Table 2, we provide a complete result list of markers tested.
Median signal intensity of activation markers (A) and relevant transmembrane proteins (B) in TRAP-stimulated platelets (10 µM TRAP). COVID-19 patients are plotted in red, whereas controls are plotted in blue. The horizontal line within the box plot represents the median, the top and bottom the interquartile range (Q1–Q3), whisker bars indicate the largest observation that is less than or equal to the upper inner fence (UIF = Q3 + 1.5 × IQR) or the smallest observation that is greater than or equal to the lower inner fence (LIF = Q1–1.5 × IQR) and the circles indicate outliers, if present; *P < 0.01. N = 8 COVID-19 patients, 11 healthy donors.
To assess the reaction capacity of platelets upon stimulation, we compared the expression of activation markers before and after stimulation with TRAP. Interestingly, we observed a significantly reduced capacity of COVID-19 platelets to increase the expression of the activation markers LAMP-3 and P-Selectin (p = 0.04 and p = 0.04, respectively) upon stimulation (Fig. 4).
Median signal intensity increase of activation marker expression after TRAP stimulation (10 µM) compared to non-stimulated platelets (baseline). Linear model analysis detected a reduced capacity of COVID-19 patients to increase expression of activation markers LAMP-3 and P-Selectin (p = 0.04 and p = 0.04) after TRAP stimulation. A P-Selectin, B LAMP-3, C LAMP-1. α: signal increment slope coefficient (for details see Methods); ns: nonsignificant. N = 8 COVID-19 patients, 11 healthy donors.
Discussion
We analyzed the expression of activation markers and transmembrane receptors in platelets of hospitalized stable COVID-19 patients without pre-existing conditions and without anticoagulants or antiplatelet drugs (except prophylactic low-molecular-weight heparin during hospitalization). As a major result, we detected significant higher levels of the platelet activation markers P-Selectin and LAMP-3 compared to controls, as well as significantly higher levels of the transmembrane proteins GPIIb/GPIIIa complex, GPIbα, GPIX, CD9, and CD40. After TRAP stimulation, platelets of COVID-19 patients showed significantly higher levels of the collagen receptor GPVI, whereas the receptor PEAR1 showed lower levels in COVID-19. These findings indicate the presence of a hyperactivated phenotype of platelets during SARS-CoV-2 infection, which might contribute to the hypercoagulopathy observed in COVID-19 and might influence disease progression. The adhesion protein P-Selectin translocates to the plasma membrane upon activation and regulates platelet–leukocyte interactions resulting in activation of neutrophil integrins and inducing NETs formation33,34. Moreover, platelet–leukocyte interaction may trigger the tissue factor expression as recently described in severe COVID-1935. P-Selectin expression together with the upregulation of the integrins GPIIb (CD41) and GPIIIa (CD61), and the subunits of the von Willebrand receptor GPIbα and GPIX, known to regulate platelet–leukocyte interactions, may contribute to the COVID-19 inflammatory response33,36. Consistent with our data, Manne et al.11 recently reported a higher surface expression of P-Selectin and higher levels of circulating platelet–leukocyte aggregates in COVID-19 patents. Moreover, the study showed a faster platelet aggregation and increased spreading on fibrinogen and collagen in COVID-19 patients compared to controls. The higher surface expression of integrins and adhesion protein detected in our study may provide a first mechanistic explanation to these findings.
To further investigate platelet reactivity in COVID-19, we induced platelet activation with TRAP, which activates platelets by thrombin signaling. After activation, we detected significantly higher levels of platelet activation markers P-Selectin and GPIIb/GPIIIa complex but not LAMP-3 in COVID-19 patients compared to healthy controls. Interestingly, we observed a decreased activation capacity in platelets of COVID-19 patients compared to controls, suggesting that the chronic platelet activation during SARS-CoV-2 correlates with an altered reactivity upon stimuli, which is possibly due to an higher activation level at rest in COVID-19 (Fig. 4)8,33. Of note, CD40 ligand (CD154) expression did not provide informative data: signal increased after TRAP stimulation but we did not detected any differences among groups (Supplemental Fig. II).
Subgroups investigations using FlowSOM analysis detected some differences in platelet activation patterns between healthy donors and COVID-19 patients (Supplemental Fig. III). However, as shown in Fig. 1, we did not find any defined and distinct subgroups, highlighting the lower heterogeneity of platelets compared to other cells in peripheral blood. Nonetheless, the FlowSOM trees shown in Supplemental Fig. III report a different activation pattern in COVID-19 patients compared to controls involving different platelet subgroups. Further studies are needed, to dissect the role of platelet heterogeneity in COVID-19 platelet activation.
Although the pathophysiological mechanisms behind the high incidence of thromboembolic events in hospitalized COVID-19 patients remain unclear, our data describe with high resolution the presence of activated platelets, which may provide one explanation for COVID-19 coagulopathy and suggests platelet inhibition as a possible therapeutic option in COVID-19 patients. Our data are consistent with previous studies reporting an immunothrombotic dysregulation as a typical marker of SARS-CoV-2 infection11,13. However, the key drivers behind platelet activation in COVID-19 remain to be determined. SARS-CoV-2 tropism for thrombocytes has not been proven yet and platelet activation may be induced by infected endothelium as well as by the cytokine storm occurring during SARS-CoV-2 infection37. Clinical trials investigating empirically different anticoagulation schemes and antiplatelet therapies are ongoing worldwide, and may provide more insights concerning the clinical relevance of antithrombotic regimes for COVID-19 patients38.
A strength of our analysis is the simultaneous measurement in a healthy control group, minimizing the risk that the observed higher platelet activation in COVID-19 was due to procedural biases. In addition, we restricted our measurements to stable COVID-19 patients not requiring supported respiration or extracorporeal perfusion, which may induce non-disease associated platelet activation. A further strength of our study consists in the high-resolution achieved by our measurements using mass cytometry, avoiding the spectral limitation of flow cytometry and allowing the measurements of 21 markers at single-cell level with virtually no overlapping.
Limitations of this study consist in the limited number of patients and in its ex vivo observational nature: our research was limited to the phenotypical observation of platelet surface receptor expressions and we did not assess the pathophysiological mechanisms triggering platelet activation. In fact, other pathways including the cytokine storm and the pro-inflammatory state during SARS-CoV-2 infection may play a relevant role in COVID-19 coagulopathy. Moreover, we did not include patients with non-COVID-19 inflammation and/or other types of viremia (e.g. influenza or other respiratory viruses) as an additional control group. Thus, we cannot quantify the severity of platelet activation in COVID-19 comparing it with other pathological settings. Nevertheless, here we provide the first mass cytometric analysis of platelets in COVID-19 and our results provide the basis for further research regarding pathways of platelet activation in COVID-19 patients as well as for further investigations of platelet biology in other pathological settings.
In conclusion, mass cytometry of COVID-19 patients revealed higher expression levels of platelet activation markers and adhesion proteins compared to healthy controls. These findings provide new insights into COVID-19 coagulopathy and support research projects investigating antithrombotic and antiplatelet treatment regimes in COVID-19.
References
Wichmann, D. et al. Autopsy findings and venous thromboembolism in patients with COVID-19: a prospective cohort study. Ann. Intern. Med. 173, 268–277 (2020).
Poissy, J. et al. Pulmonary embolism in COVID-19 patients: awareness of an increased prevalence. Circulation 142, 184–186 (2020).
Helms et al. High risk of thrombosis in patients with severe SARS-CoV-2 infection: a multicenter prospective cohort study. Intens. Care Med. 46, 1089–1098 (2020).
Llitjos, J. et al. High incidence of venous thromboembolic events in anticoagulated severe COVID‐19 patients. J. Thromb. Haemost. 18, 1743–1746 (2020).
Oxley, T. J. et al. Large-vessel stroke as a presenting feature of Covid-19 in the young. N. Engl. J. Med. 382, e60 (2020).
Klok, F. A. et al. Incidence of thrombotic complications in critically ill ICU patients with COVID-19. Thromb. Res. 191, 145–147 (2020).
Bangalore, S. et al. ST-segment elevation in patients with Covid-19 — a case series. N. Engl. J. Med. 382, 2478–2480 (2020).
Tang, N., Li, D., Wang, X. & Sun, Z. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J. Thromb. Haemost. 18, 844–847 (2020).
Zhou, F. et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet 395, 1054–1062 (2020).
Bowles, L. et al. Lupus anticoagulant and abnormal coagulation tests in patients with Covid-19. N. Engl. J. Med. 383, 288–290 (2020).
Manne, B. K. et al. Platelet gene expression and function in COVID-19 patients. Blood https://doi.org/10.1182/blood.2020007214 (2020).
Zaid, Y. et al. Platelets can associate with SARS-CoV-2 RNA and are hyperactivated in COVID-19. Circ. Res. 127, 1404–1418 (2020).
Nicolai, L. et al. Immunothrombotic dysregulation in COVID-19 pneumonia is associated with respiratory failure and coagulopathy. Circulation https://doi.org/10.1161/circulationaha.120.048488 (2020).
Sadegh, S. et al. Exploring the SARS-CoV-2 virus-host-drug interactome for drug repurposing. Nat. Commun. 11, 3518 (2020).
Matschinske, J. et al. Individuating possibly repurposable drugs and drug targets for COVID-19 treatment through hypothesis-driven systems medicine using CoVex. Assay. Drug Dev. Technol. 18, 348–355 (2020).
Semple, J. W., Italiano, J. E. & Freedman, J. Platelets and the immune continuum. Nat. Rev. Immunol. 11, 264–274 (2011).
Wang, Y., Ouyang, Y., Liu, B., Ma, X. & Ding, R. Platelet activation and antiplatelet therapy in sepsis: a narrative review. Thromb. Res. 166, 28–36 (2018).
Yang, Y. & Tang, H. Aberrant coagulation causes a hyper-inflammatory response in severe influenza pneumonia. Cell Mol. Immunol. 13, 432–442 (2016).
Qu, R. et al. Platelet‐to‐lymphocyte ratio is associated with prognosis in patients with coronavirus disease‐19. J. Med. Virol. 92, 1533–1541 (2020).
Stoppelaar, Sde, Veer, Cvan’t & Poll, Tvander The role of platelets in sepsis. Thromb. Haemost. 112, 666–677 (2014).
Goeijenbier, M. et al. Review: viral infections and mechanisms of thrombosis and bleeding. J. Med. Virol. 84, 1680–1696 (2012).
Kwong, J. C. et al. Acute myocardial infarction after laboratory-confirmed influenza infection. N. Engl. J. Med. 378, 345–353 (2018).
Umapathi, T. et al. Large artery ischaemic stroke in severe acute respiratory syndrome (SARS). J. Neurol. 251, 1227–1231 (2004).
Wong, R. S. M. et al. Haematological manifestations in patients with severe acute respiratory syndrome: retrospective analysis. Bmj 326, 1358 (2003).
Bernlochner, I. et al. Sorting and magnetic-based isolation of reticulated platelets from peripheral blood. Platelets 1–7 https://doi.org/10.1080/09537104.2020.1724923 (2020).
Bongiovanni, D. et al. Transcriptome analysis of reticulated platelets reveals a prothrombotic profile. Thromb. Haemost. 119, 1795–1806 (2019).
Sumatoh, H. R., Teng, K. W. W., Cheng, Y. & Newell, E. W. Optimization of mass cytometry sample cryopreservation after staining. Cytom. Part A 91, 48–61 (2017).
Kotecha, N., Krutzik, P. O. & Irish, J. M. Web‐based analysis and publication of flow cytometry experiments. Curr. Protoc. Cytom. 53, 10.17.1–10.17.24 (2010).
Seabold, S. & Perktold, J. Statsmodels: Econometric and Statistical Modeling with Python 92–96 https://doi.org/10.25080/majora-92bf1922-011 (2010).
Nowicka, M. et al. CyTOF workflow: differential discovery in high-throughput high-dimensional cytometry datasets. F1000research 6, 748 (2017).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 57, 289–300 (1995).
Gassen, S. V. et al. FlowSOM: Using self‐organizing maps for visualization and interpretation of cytometry data. Cytom. Part A 87, 636–645 (2015).
Blair, T. A., Frelinger, A. L. & Michelson, A. D. 35 flow cytometry. Platelets 627–651 https://doi.org/10.1016/b978-0-12-813456-6.00035-7 (2019).
Etulain, J. et al. P-selectin promotes neutrophil extracellular trap formation in mice. Blood 126, 242–246 (2015).
Hottz, E. D. et al. Platelet activation and platelet-monocyte aggregates formation trigger tissue factor expression in severe COVID-19 patients. Blood https://doi.org/10.1182/blood.2020007252 (2020).
Rayes, J., Bourne, J. H., Brill, A. & Watson, S. P. The dual role of platelet‐innate immune cell interactions in thrombo‐inflammation. Res. Pr. Thrombosis Haemost. 4, 23–35 (2020).
Conti, P. et al. IL-1 induces throboxane-A2 (TxA2) in COVID-19 causing inflammation and micro-thrombi: inhibitory effect of the IL-1 receptor antagonist (IL-1Ra). J. Biol. Reg. Homeos. Ag. 34, 1623–1627 (2020).
Bikdeli, B. et al. Pharmacological agents targeting thromboinflammation in COVID-19: review and implications for future research. Thromb. Haemost. 120, 1004–1024 (2020).
Acknowledgements
We thank all the health workers of the Internal Medicine Department I and II of University hospital rechts der Isar for their support regardless of extremely challenging working conditions due to the SARS-CoV-2 outbreak. We thank Mayur Bakshi, Qianjun Zhang, and Dries van Hemelen for excellent technical assistance. This work was supported by the German Center for Cardiovascular Research (DZHK) grant number 81X3600606 to D.B. J.B. and M.L. are grateful for financial support from BMBF grant Sys_CARE [grant number01ZX1908A] of the Federal German Ministry of Research and Education. The work of O.L. was funded by the Bavarian State Ministry of Science and the Arts as part of the Bavarian Research Institute for Digital Transformation (bidt). Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by A. Stephanou
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bongiovanni, D., Klug, M., Lazareva, O. et al. SARS-CoV-2 infection is associated with a pro-thrombotic platelet phenotype. Cell Death Dis 12, 50 (2021). https://doi.org/10.1038/s41419-020-03333-9
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41419-020-03333-9
This article is cited by
-
The efficacy of aspirin to inhibit platelet aggregation in patients hospitalised with a severe infection: a multicentre, open-label, randomised controlled trial
Clinical and Experimental Medicine (2023)
-
Differential platelet activation through an interaction with spike proteins of different SARS-CoV-2 variants
Journal of Thrombosis and Thrombolysis (2023)
-
Variant-derived SARS-CoV-2 spike protein does not directly cause platelet activation or hypercoagulability
Clinical and Experimental Medicine (2023)
-
Neutrophil Activation and Immune Thrombosis Profiles Persist in Convalescent COVID-19
Journal of Clinical Immunology (2023)
-
Role of aging in Blood–Brain Barrier dysfunction and susceptibility to SARS-CoV-2 infection: impacts on neurological symptoms of COVID-19
Fluids and Barriers of the CNS (2022)