Abstract
Neat1 is widely expressed in many tissues and cells and exerts pro-proliferation effects on many cancer cells. However, little is known about the function of Neat1 in myogenesis. Here we characterized the roles of Neat1 in muscle cell formation and muscle regeneration. Gain- or loss-of-function studies in C2C12 cells demonstrated that Neat1 accelerates myoblast proliferation but suppresses myoblast differentiation and fusion. Further, knockdown of Neat1 in vivo increased the cross-sectional area of muscle fibers but impaired muscle regeneration. Mechanically, Neat1 physically interacted with Ezh2 mainly through the core binding region (1001–1540 bp) and recruited Ezh2 to target gene promoters. Neat1 promoted myoblast proliferation mainly by decreasing the expression of the cyclin-dependent kinase inhibitor P21 gene but inhibited myoblast differentiation by suppressing the transcription of myogenic marker genes, such as Myog, Myh4, and Tnni2. Altogether, we uncover a previously unknown function of Neat1 in muscle development and the molecular mechanism by which Neat1 regulates myogenesis.
Similar content being viewed by others
Introduction
Skeletal muscle is the most abundant tissue in the mammalian body and plays a pivotal role in regulating body metabolism and homeostasis1. The differentiation of skeletal muscle cells is precisely regulated by several myogenic regulatory factors (MRFs), including myogenic differentiation 1 (Myod), myogenic factor 5 (Myf5), myogenin (Myog), and Mrf42. Myod or Myf5 is necessary for skeletal muscle lineage formation and is expressed at the myoblast stage3. Myod overexpression converts fibroblasts into myoblasts and subsequent fusion into myotubes4,5. Myog and Mrf4 are expressed after Myod and Myf5 and determine terminal muscle cell differentiation. Myog knockdown reversed terminal muscle cell differentiation6. MRFs also contribute to the regeneration of injured adult muscle, as muscle regeneration demands activation of the muscle regulatory network7,8. During injury, satellite cells (SCs) are activated and undergoing proliferation, and paired box (Pax) 7 and Myod genes are upregulated at this stage. Next, SCs differentiate into myotubes, upon which Pax genes are downregulated and Myog upregulated9. Epigenetic regulation, such as DNA methylation10, histone modifications11,12, and noncoding RNA functions13,14, also play important roles in the transcriptional regulation of myogenesis and ensure the normal proliferation and differentiation of muscle progenitors15,16. Enhancer of zeste homolog 2 (Ezh2) is a subunit of the epigenetic regulator polycomb repressive complex 2 (PRC2) responsible for trimethylation of lysine 27 of histone 3 (H3k27me3), which leads to repression of gene transcription. A previous study established the important role of polycomb-mediated H3k27 methylation during myogenic differentiation17. Ezh2 overexpression suppresses myogenic differentiation by silencing muscle-specific genes18,19. Long non-coding RNAs (lncRNAs) (e.g., Linc-MD120, Lnc-mg21, LncRNA-YY122, Linc-RAM23, Myolinc24, MAR125, AK01736826, SYISL27), which are greater than 200 nucleotides in length and have no protein-coding capacity, were recently reported to play important roles in myogenesis by interacting with various proteins or acting as molecular sponges for miRNAs.
Nuclear paraspeckle assembly transcript 1 (NEAT1, known as Neat1 in mouse) is a lncRNA that is enriched in the nucleus and essential for nuclear paraspeckle formation28,29. Paraspeckles were recently identified as mammalian-specific nuclear bodies that are found in most cells cultured in vitro but are not essential in vivo30, Paraspeckles play important roles in many gene regulation processes, such as mRNA retention, A-to-I editing, and protein sequestration31,32. NEAT1 serves as a platform to recruit numerous paraspeckle proteins to maintain paraspeckle stability and integrity32,33,34. In addition, long-range interactions among NEAT1 transcripts may exert an important architectural function in paraspeckles formation35. In addition to participating in the formation of paraspeckles, NEAT1 also plays important roles in a variety of biological processes. For example, NEAT1 regulates the phenotypic switch of vascular smooth muscle cells by inhibiting SM (smooth muscle)-contractile gene expression by removing the epigenetic activator WDR5 from SM-specific gene loci36. NEAT1 is widely expressed in multiple tissues and participates in the tumorigenesis of many cancers including prostate cancer37, breast cancer38, colorectal cancer39, esophageal squamous cell carcinoma40, laryngeal squamous cell cancer41, and pancreatic cancer42. Despite the important roles of Neat1 in regulating multiple biological processes, it is unknown whether it is involved in muscle development and regeneration. In the present study, we investigated the roles of Neat1 in myogenesis and found that Neat1 regulates myoblast proliferation and differentiation by interacting with Ezh2, defining a novel function of Neat1 in muscle development and regeneration.
Materials and methods
Cell culture
Mouse C2C12 cells were cultured in DMEM (high-glucose Dulbecco’s modified Eagle’s medium) (Hyclone, USA) containing 10% fetal bovine serum (Gibco, Australia) under moist air with 5% CO2 at 37 °C for proliferation and in DMEM with 2% horse serum (Gibco, USA) at the same condition for differentiation.
Animals
C57 mice were purchased from Hubei center for disease control and housed in Huazhong Agricultural University under normal conditions with appropriate temperature and humidity and supplied with nutritional food and sufficient water. Animal feeding and tests were conducted based on the National Research Council Guide for the Care and Use of Laboratory Animals and approved by the Institutional Animal Care and Use Committee at Huazhong Agricultural University.
Plasmid construction, siRNA synthesis
The full-length sequence of Neat1, and coding sequences (CDS) of Ezh2 and P21 were amplified by polymerase chain reaction (PCR) with corresponding full-length or cds F/R primers using C2C12 cDNA as a template. The amplified sequences were cloned into pcDNA3.1 using T4 DNA ligase (Takara,Japan) to produce pcDNA3.1-Neat1, pcDNA3.1-Ezh2 and pcDNA3.1-P21. The truncated Neat1 were obtained by PCR using pcDNA3.1- Neat1 plasmid as a template and then were cloned into pcDNA3.1. The plasmids were confirmed by sequencing. The primers above were shown at Supplementary Table S1. siRNA oligos against mouse Neat1 (sense 5′- GGAGUCAUGCCUUAUACAATT-3′), Ezh2 (sense 5′- GCGCAGUAGAAUGGAGAAATT-3′) and P21 (sense 5′-UGAGCAAUGGCUGAUCCUU-3′) were designed and synthesized by GenePharma (China, Shanghai).
Transfection of plasmid, siRNA
For cell transfection, expression plasmids or siRNAs were conducted with Lipofectamine 2000 (Invitrogen, USA) as advised by the manufacturer’s protocol.
Quantitative real-time PCR
RNA samples from C2C12 cells or mice tissues were isolated using the TRIzol reagent (Invitrogen, USA). The expression of mRNA was detected by Quantitative real-time PCR (qPCR). The qPCR reaction was performed in LightCycler 480 II (Roche, Switzerland) system using SYBR®Green Real-time PCR Master Mix (Toyobo, Japan). All the experiments were designed in triplicates. The relative gene expression was calculated by the Ct (2-ΔΔCt) method according to the literature43. The sequence primers were list at Supplementary Table S2.
Cell proliferation assays
For real-time cell proliferation monitoring assay, C2C12 cells were inoculated on a 16-well E-Plate and transfected with Neat1 expression vector or siNeat1 oligos. The cell proliferation rates were recored by the RTCA xCELLigence system (Roche Applied Science, Penzberg, Upper Bavaria, Germany).
For EdU staining, the EdU staining was performed using EDU kit (RiboBio, China) according to the manufacturer’s instructions. Images were captured with an Olympus IX51-A21PH fluorescence microscope (Olympus, Japan). Cells were further analyzed by computing the percentage of EdU+ cells.
For EdU-propidium iodide (PI) flow cytometry, Edu reagent was added to C2C12 cells at a final concentration of 50 uM and incubated for 30 min at 37℃. Then the cells were harvested and fixed in 70% ethanol at 4 ℃ overnight. The cells were further carried out with EdU staining using EdU kit (RiboBio, China) according to the manufacturer’s instructions. After that the cells were incubated in 50 mg/ml PI for 1 h at room temperature, cells were analyzed using the FACSCalibur Flow Cytometer (Becton Dickinson, Franklin Lakes, NJ, USA).
Western blotting
C2C12 cells or mouse tissues were lysed in RIPA buffer containing 1% (v/v) phenylmethylsulfonyl fluoride (PMSF) (Beyotime, Jiangsu, China). The western blotting was performed according to the previous publication44. The antibodies and their dilutions were shown as following: Myod (Santa Cruz Biotechnology, USA; sc-760; 1:1000), Myog (Santa Cruz Biotechnology, USA; sc-12732; 1:200), Myhc (Santa Cruz Biotechnology, USA; sc-376157; 1:3000), α-actin (Proteintech, China; 23660-1-AP; 1:1000), Tnni2 (Abcam, UK; ab184554; 1:1000), P21 (BOSTER, China; BM4382; 1:200), Ezh2 (Cell Signaling Technology, USA; 5246; 1:1000), Pcna (Servicebio, China; GB11010; 1:500), Ki67 (Abcam, UK; ab16667; 1:1000), β-actin (Santa Cruz Biotechnology, USA; sc-4777; 1:1000), Gapdh (BOSTER, China; BM3876; 1:200), Pax7 (Developmental Studies Hybridoma Bank; USA; 1:1000), eMyhc (Developmental Studies Hybridoma Bank, USA; BF-G6; 1:1000). The protein expression levels were normalized to corresponding β-actin or Gapdh and the western blotting bands signal intensities were quantified using ImageJ software.
Immunofluorescence
C2C12 cells were cultured in 24-well plate and differentiated for 2–3 days. The immunofluorescence staining was performed according to the previous publication44. The antibodies and their dilutions were shown as following: Myog (Santa Cruz Biotechnology, USA; sc-12732; 1:50) and Myhc (Santa Cruz Biotechnology, USA; sc-376157; 1:200), a secondary antibody (anti-mouse CY3; Beyotime Biotechnology, China). The 4′, 6-diamidino-2-phenylindole (DAPI) was used to stain the cell nuclei. The images were visualized with a fluorescence microscope (IX51-A21PH, Olympus, Japan). The cell differentiation index was calculated by the ratio of the number of nuclei in the myotubes to the total number of nuclei in one field of view. For myoblast fusion, the cells were differentiated for 5 days. Then the cells were performed with myosin (Sigma, USA; M4276; 1:1000) immunofluorescence to test myoblast fusion. The cell fusion was calculated by the number of nuclei present in one myosin-positive cell indicated.
Knockdown of Neat1 in vivo by lentivirus infection
6-week C57 male mice were injected with 100 µL final volumes of lentivirus contained small interfere Neat1 (LV3-shNeat1) or control (LV3-shNC) at 2 × 107 TU/ml into the right and left quadriceps (Qu), tibialis anterior (TA) and gastrocnemius (Gas) of the hind legs, respectively. LV3-shNeat1 or LV3-shNC was synthesized by GenePharma (China, Shanghai). LV3-shNeat1 or LV3-shNC was diluted in PBS and injected into Qu, TA and Gas of the hind legs every one week, after one month of injection, the mice were killed and the Qu, TA and Gas muscles of the hind legs were collected. For qPCR and western blotting analysis, the injection was performed in three mice. The Qu, TA and Gas muscles of the right or left hind legs of each injection mouse were collected, and used for total RNA and protein extraction. For myosin immunofluorescence, the Qu, TA and Gas muscles of the right or left hind legs of each injected mouse were fixed in 4% paraformaldehyde, respectively.
Muscle injury and regeneration
For CTX injection, 6-week male mice were injected with 100 µL final volumes of LV3-shNeat1 or LV3-shNC at 2 × 107 TU/ml into the right and left Gas muscles, respectively. One day after, the above mice were injected with 100ul of CTX at 80ug/ml into the Gas muscles at both hind legs. Mice were sacrificed and the Gas muscles were harvested at designed days, Mice were administered with 100 μg EdU (Thermo Fisher Scientific, Waltham, MA, USA) by intraperitoneal injection at 6 h before muscles harvesting. Extraction of the total RNAs and proteins were used for qPCR and western blotting analysis, respectively.
Histology staining
For immunofluorescence staining of Pax7, EdU, Myog and eMyhc, the Gas muscles were harvested at day 3. Immunofluorescence staining on frozen muscle sections was performed in accordance with previous reports21, and images were visualized using a confocal laser scanning microscope (Zeiss, LSM800, Germany). The following dilutions were used for each antibody: Pax7 (Developmental Studies Hybridoma Bank; USA; 1:20), eMyhc (Developmental Studies Hybridoma Bank, USA; BF-G6; 1:100), Myog (Santa Cruz Biotechnology, USA; sc-12732; 1:20). To detect the EdU incorporation, the sections were performed using the Life Technologies Click-iT Kit according to the manufacturer’s instructions, and images were photographed using a confocal laser scanning microscope (LSM800; Zeiss). For H&E staining, the Gas muscles were harvested at day 0, 3, 7, and 15 after CTX injection. H&E of muscle sections was performed according to previous reported methods21,45, and the cross-section area of individual myofibers was visualized using Olympus DP80 upright Metallurgical Microscope (Olympus Corporation, Japan) and qualified using ImageJ software.
RNA immunoprecipitation assay
RNA immunoprecipitation (RIP) assays were conducted using EZ-Magna RIP Kit (Millipore) according to the manufacturer’s instructions. Briefly, C2C12 cells were lysed with RIP lysis buffer, and incubated with 1 μg antibody (Ezh2, Abcam, ab3748; Suz12, Abcam, ab12073; IgG, Millipore) for RNA immunoprecipitation at 4 ℃ overnight. Then Protein A/G beads were added to the lysates to pull down antibody-protein-RNA complex. The detection of co-precipitated RNAs was performed by reverse-transcription polymerase chain reaction (RT-PCR).
RNA pulldown assays
RNA pulldown was performed as previously described46. Briefly, linearizing DNA was biotin-labeled and in vitro transcribed using the Biotin RNA Labeling Mix and T7/SP6 RNA polymerase (Roche), and purified with the RNeasy Mini Kit (QIAGEN). One milligram of protein was incubated with 3 μg of biotinylated RNA for 1 h at room temperature. After that, 40ul Streptavidin-coupled Dynabeads (Invitrogen) were added to each reaction and incubated for 1 h at room temperature. Finally, the beads were washed in RIP buffer for five times, and the pull-downed proteins were used for western blotting. For mass spectrometry, the pull-downed proteins in C2C12 cells were separated by 10% SDS-PAGE, and then performed with silver staining. The differentially expressed bands were excised and analyzed by mass spectrometry (Novogene, Beijing, China).
Chromatin immunoprecipitation assays
Chromatin immunoprecipitation (ChIP) assay was performed using ChIP Kit (Millipore, 17–371) according to the manufacturer’s instructions. Each ChIP reaction was performed using 1 μg of antibodies against Ezh2 (Abcam, ab3748), H3k27me3 (Abcam, ab6002) or IgG applied as negative control. Fold enrichment was quantified using qPCR. All promoter primers were listed in Supplementary Table S3.
Chromatin Isolation by RNA Purification assays
Chromatin Isolation by RNA Purification (ChIRP) assay was performed using ChIRP Kit (Millipore#17–10495) according to the manufacturer’s instructions. Biotin-labeled Neat1 probes were designed by Guangzhou Ribobio and divided into odd and even pools. C2C12 cells were cross-linked with 1% glutaraldehyde and lysed with lysis buffer before sonication for 4 h. Odd and even Neat1 probes were added into samples for incubation at 37 °C for 4.5 h with rotating. The combined chromatin fragments were enriched by C-1 streptavidin beads and purified for qPCR experiment.
Statistical analysis
All data were expressed as mean ± standard deviation (s.d.). Statistical analyses between different groups were performed using t-test. For all analyses, p < 0.05 was considered to be statistically significance.
Results
Neat1 is up-regulated during myogenic differentiation and muscle regeneration
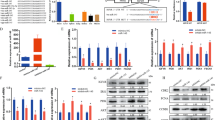
To investigate the role of Neat1 in myogenesis, we first examined the expression profiles of Neat1 as well as Myog and Myhc during myogenic differentiation. The expression levels of Neat1 and Myog were increased in C2C12 cells from day 0 (proliferating cells) to day 5 post differentiation but decreased on day 8, whereas Myhc expression gradually increased during differentiation (Fig. 1a–c). To further explore Neat1 expression during muscle regeneration, we employed an extensively used muscle regeneration model in which intramuscular injection of cardiotoxin (CTX) leads to muscle injury and induces muscle regeneration47. The expression levels of Neat1 along with Pax7 and Myod were highly increased during the early stage of regeneration and then decreased when the newly formed fibers maturation and regeneration completed. We also found that Neat1 expression peaked earlier than that of Pax7 and Myod (Fig. 1d–f). These results suggest that Neat1 is involved in myogenesis and muscle injury repair.
a qPCR results showing that Neat1 expression was upregulated in C2C12 cells from day 0 (proliferating cells) to day 5 post differentiation but downregulated on day 8. b, c qPCR results showing that Myog expression was upregulated on days 0, 2, and 5 but downregulated on day 8 (b), whereas Myhc expression was upregulated during myoblast differentiation (c). d–f Hindlimb muscles were subjected to CTX injection and harvested at 0, 1, 2, 3, 5, and 8 days post-injury for RNA analysis. qPCR results showing that the expression of Neat1 (d), Myod (e), and Pax7 (f) genes was induced at the early stage of muscle regeneration but downregulated thereafter. Relative RNA levels were normalized to those of β-actin. All values represent the mean ± standard deviation (s.d.) of three independent experiments
Neat1 promotes myoblast proliferation, but inhibits myogenic differentiation and fusion
NEAT1 promotes tumor growth in many cancer cells, including prostate cancer37, breast cancer cell lines38, colorectal cancer39. To investigate the roles of Neat1 in myoblast proliferation, we conducted Neat1 knockdown and overexpression experiments in C2C12 cells. Neat1 knockdown led to a significant reduction in Ki67 and Pcna mRNA expression and Ki67 protein expression (Fig. 2a, b), whereas Neat1 overexpression had the opposite effect (Supplementary Fig. 1a–b). RTCA xCELLigence, EdU-PI flow cytometry assays confirmed the effects of Neat1 on cell proliferation. The RTCA xCELLigence assay suggested that Neat1 knockdown significantly reduced cell growth (Fig. 2c), while Neat1 overexpression enhanced cell growth (Supplementary Fig. 1c). EdU-PI flow cytometry assays showed a significant decrease and increase in DNA replication (S-phase) after Neat1 knockdown and overexpression, respectively (Fig. 2d and Supplementary Fig. 1d-e). These observations indicate that Neat1 promotes the proliferation of C2C12 cells.
a qPCR results showing that the mRNA expression of Ki67 and Pcna was significantly decreased by Neat1 knockdown. b Western blotting analysis showing that Ki67 protein expression was significantly decreased by Neat1 knockdown. c The RTCA xCELLigence assay demonstrating that cell growth dynamics were significantly reduced by Neat1 knockdown. d The quantification of EdU-PI flow cytometry results showing that the proportion of cells in S phase was significantly reduced by Neat1 knockdown. e qPCR results showing that the mRNA expression of Myod, Myog, Myhc, α-actin, and Tnni2 was significantly increased by Neat1 knockdown in C2C12 cells on day 2 post differentiation. f Western blotting analysis showing that the protein expression of Myod, Myog, Myhc, and α-actin was significantly increased by Neat1 knockdown in C2C12 cells on days 0, 2, and 4 post differentiation. g Immunofluorescence staining of Myog showing that Myog protein expression was significantly increased by Neat1 knockdown on day 2 post-transfection. Cell nuclei were stained with DAPI. The number of Myog+ cells was quantified using ImageJ software. h Immunofluorescence staining of Myhc showing significant upregulation of C2C12 differentiation by Neat1 knockdown on day 3 post-transfection. The number of Myhc+ cells was quantified using ImageJ software. i Immunofluorescence staining of myosin in C2C12 cells differentiated for 5 days showing that knockdown of Neat1 significant enhanced the myoblast fusion. Relative RNA and protein levels were normalized to those of β-actin. All values represent the mean ± s.d. of three independent experiments. *p < 0.05, **p < 0.01, N.S. indicates not significant
To verify the functions of Neat1 during C2C12 differentiation, Neat1 was knocked down and overexpressed during C2C12 cell differentiation. Knockdown of Neat1 increased the mRNA expression of myogenic marker genes, such as Myod, Myog, Myhc, α-actin, and Tnni2 (Fig. 2e). Neat1 knockdown also enhanced myogenic differentiation, as indicated by a significant increase in the protein expression of these genes during cell differentiation (Fig. 2f and Supplementary Fig. 2a). Immunofluorescence staining of Myog and Myhc revealed an increased number of Myog+ and Myhc+ cells (Fig. 2g, h). These observations were also confirmed by Neat1 overexpression (Supplementary Fig. 2b-f). Together, these data demonstrate that Neat1 inhibits myoblast differentiation.
Finally, to investigate whether Neat1 affects myoblasts fusion, myosin immunofluorescence staining was used to analyze myoblasts fusion after Neat1 knockdown or overexpression. The results showed that Neat1 knockdown increased the ratio of myotubes with more than three nuclei, while Neat1 overexpression decreased the ratio of myotubes with more than three nuclei (Fig. 2i and Supplementary Fig. 2g). In addition, Neat1 knockdown and overexpression significantly increased and decreased the fusion marker gene Myomaker expression, respectively (Supplementary Fig. 2h–i), suggesting Neat1 inhibits myoblasts fusion.
Knockdown of Neat1 promotes postnatal muscle growth in vivo
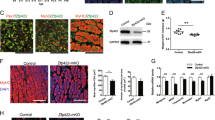
To investigate the effects of Neat1 on muscle growth in vivo, 6-week-old C57 mice were injected with LV3-shNeat1 or LV3-shNC particles into the right and left hindlimbs, respectively. The injection scheme is shown in Fig. 3a. The injection of LV3-shNeat1 particles led to a significant reduction of Neat1 expression (Fig. 3b). The mRNA and protein levels of Myog, Myhc, Tnni2, and α-actin genes were significantly increased after Neat1 knockdown (Fig. 3b, c). Further, the volume and weight of the Qu, TA, and Gas muscles in Neat1 knockdown groups were larger than those of the controls (Fig. 3d, e). Immunofluorescence staining for myosin showed that the cross-sectional areas of the Qu, TA, and Gas muscles injected with LV3-shNeat1 particles were dramatically larger than those injected with LV3-shNC particles (Fig. 3f). These results indicate that Neat1 knockdown enhances muscle growth.
a The injection scheme for LV3-shNeat1 or LV3-shNC particles into the right or left hindlimb muscles of C57 mice. The injection was given every one week for one month. b qPCR results showing that Neat1 expression was reduced after LV3-shNeat1 particle injection, while the mRNA expression of Myog, Myhc, α-actin, and Tnni2 was significantly increased after LV3-shNeat1 particle injection. c Western blotting analysis showing that the protein expression of Myog, Myhc, α-actin, and Tnni2 was increased after LV3-shNeat1 particle injection. The protein levels of these genes were quantified using ImageJ software. d Representative photograph of the three muscles from the right or left hindlimbs showing that the volume of the Qu, TA, and Gas muscles of the right hindlimb were larger than those of the left hindlimb. e Quantification of the weight of three muscles from the right or left hindlimbs of 12 injected mice showing that the weight of the Qu, TA, and Gas muscles of the right hindlimb were higher than those of the left hindlimb. f Representative photograph of myosin immunofluorescence staining in Qu, TA, and Gas muscles from the right or left hindlimb following injection with LV3-shNeat1 or LV3-shNC particles. Compared with LV3-shNC particle injection, LV3-shNeat1 particle injection increased the average cross-sectional areas of the indicated muscles. The fiber sizes of the Qu, TA, and Gas muscles were quantified using ImageJ. Relative RNA and protein levels were normalized to those of Gapdh. All values represent the mean ± s.d. of three independent experiments. *p < 0.05, **p < 0.01
Neat1 knockdown delays muscle regeneration after CTX injection in vivo
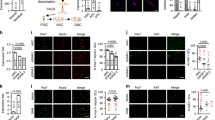
To determine whether Neat1 regulates CTX-induced muscle regeneration, 6-week-old C57 mice were injected with LV3-shNeat1 or LV3-shNC particles into the Gas muscles of the right and left hindlimbs, respectively, followed by CTX injection (Fig. 4a). The muscle regeneration phenotype was evaluated by H&E staining of muscle sections on days 0, 3, 7, and 15 after CTX treatment. On day 7 post-CTX treatment, the LV3-shNeat1 injection group displayed more inflammatory cells and fewer newly formed myofibers. On day 15 post-CTX treatment, the newly formed myofibers with the central nucleus were smaller in the LV3-shNeat1 injection group than those of the LV3-shNC injection group (Fig. 4b), and the percentage of fibers with the central nucleus was higher in LV3-shNeat1 group than that in LV3-shNC groups (Fig. 4c). Knockdown of Neat1 also led to a decrease in both the mRNA and protein levels of Pax7, Myod, Myog, and eMyhc genes, as well as the Ki67 mRNA level, on day 7 post-CTX injection (Fig. 4d, e). Immunofluorescence staining in muscle sections showed that the percentage of both Pax7+ SCs and proliferating Pax7 (Pax7+/EdU+) SCs were reduced in the LV3-shNeat1 injection group compared with the controls on day 3 post-CTX injection (Fig. 4f). Immunofluorescence staining in muscle sections also showed that knockdown of Neat1 decreased the number of cells with positive staining of Myog and eMyhc on day 3 post-CTX injection (Fig. 4g, h). Reduced Myog and eMyhc expression in the LV3-shNeat1 injection groups is likely the consequence of impaired SC proliferation rather than impaired differentiation, as Neat1 knockdown promoted Myog and Myhc expression. Together, these results suggest that Neat1 knockdown delays muscle regeneration following CTX treatment.
a The scheme for LV3-shNeat1 or LV3-shNC particle injection and CTX injection into the Gas muscles and harvesting time points for subsequent analysis. b H&E staining results showing that LV3-shNeat1 particle injection significantly delayed muscle regeneration compared with LV3-shNC particle injection. The injected muscles were harvested on days 0, 3, 7, and 15 post-CTX injections and used for H&E staining. c The quantification of the percentage of fibers with central localized nuclei (CLN) at 15 days after CTX injection. The results showed that the percentage of fibers with CLN were significantly higher in LV3-shNeat1 groups than that in LV3-shNC groups. d qPCR results showing that the mRNA expression levels of Neat1, Pax7, Myod, Myog, eMyhc, and Ki67 genes were remarkably reduced after LV3-shNeat1 particle injection. The injected muscles were harvested on day 7 post-CTX injection. e Western blotting analysis showing that the protein expression levels of Pax7, Myod, Myog, and eMyhc were reduced after LV3-shNeat1 particle injection. The protein levels of these genes were quantified using ImageJ software. The injected muscles were harvested on day 7 post-CTX injection. f Immunofluorescence staining of Pax7, EdU and DAPI in muscle sections on day 3 post-CTX injection showing that Pax7+ SCs and proliferating SCs were reduced after LV3-shNeat1 particle injection. The percentage of Pax7+ SCs were indicated for the proportion of the number of Pax7+ nuclei in total number of DAPI, the percentage of proliferating satellite cells were indicated for the proportion of the number of Pax7+/EdU+ nuclei in total number of Pax7+ nuclei. g, h Immunofluorescence staining of Myog (g) and eMyhc (h) in muscle sections on day 3 post-CTX showing significantly reduced expression of these proteins after LV3-shNeat1 particle injection. Relative RNA and protein levels were normalized to those of Gapdh. All values represent the mean ± s.d. of three independent experiments. *p < 0.05, **p < 0.01, N.S. indicates not significant
Neat1 physically interacts with Ezh2
As a well-known nuclear lncRNA, Neat1 may play a role at the transcriptional level. A recent study revealed that NEAT1 interacts with EZH2 in human glioblastoma cells48. Our previous studies showed that Ezh2 plays important roles in myoblast proliferation and differentiation by increasing the level of H3k27me3 binding at gene promoters27; therefore, we inferred that Neat1 may also regulate myoblast proliferation and myogenic differentiation by interacting with Ezh2. First, we performed RNA immunoprecipitation (RIP) assays to confirm the interaction between Neat1 and Ezh2, and as expected, Neat1 was significantly enriched on the Ezh2 antibody compared with the IgG antibody (negative control) (Fig. 5a). To confirm the interaction, biotin-labeled full-length Neat1 was used to pull down target proteins. RNA pull-down assays revealed that Neat1 transcripts pulled down endogenous Ezh2 but not Myod or Pcna (Fig. 5b), and there was no mutual effect between Neat1 and Ezh2 expression (Supplementary Fig. 3a–c), suggesting that Neat1 physically binds to Ezh2. As PRC2 contains several members, such as Ezh2 and Suz12, we also evaluated the interaction between Neat1 and Suz12. As expected, Neat1 was also pulled down by the Suz12 antibody (Supplementary Fig. 3d). Next, a series of deletions in full-length Neat1 was performed to determine the core binding domain (Fig. 5c). Interestingly, the third fragment (1001–1540, F3) was efficiently pulled down Ezh2, whereas fragments F1, F2, and F4 rarely pulled down Ezh2 (Fig. 5c). To further verify the core functional domain of Neat1, F1, F2, F3, F4, and full-length Neat1 were overexpressed in C2C12 cells. EdU staining showed that the overexpression of F3 and full-length Neat1 significantly increased EdU incorporation compared with the control (Fig. 5d and Supplementary Fig. 3e). Immunofluorescence staining also revealed a significantly decreased number of Myog+ and Myhc+ cells when F3 and full-length Neat1 were overexpressed (Fig. 5e, f and Supplementary Fig. 3f–g). These results indicate that the F3 fragment of Neat1 is required for its recruitment of Ezh2 in myogenesis. Besides, we also identified 122 Neat1-binding proteins in C2C12 cells by mass spectrometry including some known interacting paraspecle proteins (Sfpq, Hnrnpr, Hnrnpm and Matr3) and some proteins involved in muscle development and disease (Eef2, Dnm2, Trim28, Trpv2, Wfs1, Pml, Vcp, Lmna and Matr3) (Supplementary Fig. 3h and Supplementary Table 4).
a RNA immunoprecipitation (RIP) assays performed in C2C12 cells using Ezh2 antibodies. The retrieved Neat1 transcripts were assessed by PCR. b Biotin-labeled full-length Neat1 was used to pull down Ezh2. Western blotting analysis was performed to detect the precipitated Ezh2 protein. Myod and Pcna were used as negative controls. c The interactions between a series of Neat1 fragments (FL, F1, F2, F3, F4) and Ezh2 assessed by RNA pull-down assays. d Transfection of a series of Neat1 fragments (FL, F1, F2, F3, and F4) into C2C12 cells, followed by EdU staining in the cells on day 2 after transfection. The quantification of EdU staining results showed that the overexpression of F3, but not the other fragments, and full-length Neat1 significantly promoted C2C12 cell proliferation. e, f Transfection of a series of Neat1 fragments (FL, F1, F2, F3, and F4) into C2C12 cells, followed by immunofluorescence staining of Myog and Myhc at 2 days after differentiation. The quantification of immunofluorescence staining results showed that the overexpression of F3, but not the other fragments, and full-length Neat1 significantly inhibited Myog protein expression (e) and C2C12 cell differentiation (f). U1 was used as controls for the RIP assays. Myod and Pcna were used as controls for RNA pull-down assays. All values represent the mean ± s.d. of three independent experiments. *p < 0.05, **p < 0.01, N.S. indicates not significant
Neat1 enhances the proliferation of C2C12 cells via Ezh2-mediated H3k27me3 enrichment at the P21 promoter
Previous studies have shown that lncRNAs such as SNHG2049, Xist50 and SYSIL27 repress the cyclin-dependent kinase inhibitor 1 A (P21) by interacting with Ezh2, as well as promote cell proliferation. Our above mentioned results showed that Neat1 promotes proliferation and interacts with Ezh2 in C2C12 cells (Fig. 2a–d and Supplementary Fig. 1 and Fig. 5). Further, the overexpression of P21 significantly reduced the number of EdU+ cells, whereas Ezh2 overexpression increased the number of EdU+ cells (Supplementary Fig. 4a-b), demonstrating that P21 and Ezh2 have the opposite effects on C2C12 cell proliferation. Thus, we inferred that Neat1 facilitates C2C12 cell proliferation by Ezh2-mediated inhibition of P21 expression. First, we explored whether Neat1 regulates P21 expression and found that knockdown of Neat1 inhibited cell cycle-dependent kinase 4 (Cdk4) and enhanced P21 protein expression, indicating that Neat1 inhibits P21 expression in C2C12 cells (Fig. 6a). A previous study showed that Ezh2 inhibits P21 expression by directly binding to its promoter27. Therefore, we performed ChIP-qPCR assays to explore whether Neat1 affects the Ezh2- and H3k27me3-binding capacities at the P21 promoter. Knockdown and overexpression of Neat1 decreased and increased the enrichment of Ezh2 and H3k27me3 at the P21 promoter, respectively (Fig. 6b–e). To further elucidate whether Neat1 affected P21 expression through Ezh2, Ezh2 siRNA fragment and Neat1 expression vector were co-transfected into C2C12 cells. Immunofluorescence staining of P21 revealed that Neat1 overexpression inhibited P21 expression, but had no effect when co-transfected with Ezh2 siRNA fragments (Fig. 6f and Supplementary Fig. 4c). We also co-transfected Neat1 siRNA fragments with Ezh2 expression vector in C2C12 cells and assessed cell proliferation by EdU staining. Knockdown of Neat1 significantly reduced the percentage of EdU+ cells. After transfection with Ezh2 expression vector, Neat1 knockdown did not reduce the number of EdU+ cells (Fig. 6g and Supplementary Fig. 4d), indicating that Neat1 regulation of myoblast proliferation is dependent on Ezh2. To further confirm whether Neat1 promoted cell proliferation through P21 pathway, Neat1 and P21 siRNA fragments were co-transfected into C2C12 cells, and then the cell proliferation ability was detected by EdU staining. The results showed that Neat1 knockdown significantly reduced the percentage of EdU+ cells, but had no effect after co-transfection with P21 siRNA fragment (Fig. 6h and Supplementary Fig. 4e). Together, these results suggest that Neat1 inhibits P21 expression by increasing the Ezh2-binding capacities at its promoter, thereby promoting C2C12 cell proliferation.
a Western blotting analysis showing that Neat1 knockdown enhanced P21 protein expression but decreased Cdk4 protein expression in C2C12 cells. The relative protein levels of P21 and Cdk4 were quantified using ImageJ software. b, c ChIP-qPCR results revealed that the enrichments of Ezh2 (b) and H3k27me3 (c) at the P21 promoter were significantly decreased after Neat1 knockdown. d, e ChIP-qPCR results revealed that the enrichments of Ezh2 (d) and H3k27me3 (e) at the P21 promoter were significantly increased after Neat1 overexpression. f Co-transfection of Ezh2 siRNA fragment and Neat1 expression vector in C2C12 cells for 2 days. Immunofluorescence staining of P21 was performed, and P21 expression was quantified by ImageJ. The quantification of P21 immunofluorescence staining results showed that the overexpression of Neat1 inhibited P21 protein expression, but had no significant effect on P21 expression after co-transfection with Ezh2 siRNA fragment. g Neat1 siRNA fragment and Ezh2 expression vector were co-transfected into C2C12 cells and the cells were performed with EdU staining at 2 days after transfection. The percentage of EdU+ cells was quantified. The quantification of EdU staining results showed that Neat1 knockdown inhibited myoblast proliferation. After co-transfected with Ezh2 expression vector, Neat1 knockdown can not inhibit myoblast proliferation. h Neat1 and P21 siRNA fragments were co-transfected into C2C12 cells and the cells were performed with EdU staining at 2 days after transfection. The quantification of EdU staining results showed that Neat1 knockdown significantly reduced the percentage of EdU+ cells, but did not reduce the number of EdU+ cells after co-transfection with P21 siRNA fragment. Protein levels were normalized to those of β-actin. All values represent the mean ± s.d. of three independent experiments. *p < 0.05, **p < 0.01, N.S. indicates not significant
Neat1 inhibits myogenic differentiation by epigenetically silencing the expression of myogenic markers
Previous studies have shown that Ezh2 suppresses myogenic differentiation by increasing levels of the epigenetic silencing marker H3k27me3 to repress the transcription of myogenic markers such as Myog18,51, myh418,51,52, and Tnni252. Therefore, we performed ChIP assays to determine whether Neat1 affects the inhibitory effects of Ezh2 on the expression of these myogenic genes. ChIP-qPCR assays suggested that knockdown of Neat1 decreased the binding of Ezh2 and H3k27me3 at the Myog, Myh4, and Tnni2 gene promoters (Fig. 7a, b), which was confirmed by Neat1 overexpression (Fig. 7c, d). To further elucidate whether Neat1 affects these target genes via Ezh2, Neat1 expression vector and Ezh2 siRNA fragment were co-transfected into C2C12 cells. As expected, the overexpression of Neat1 remarkably reduced the expression of Myog, Myhc, and Tnni2, but not when co-transfected with the Ezh2 siRNA fragments (Fig. 7e and Supplementary Fig. 5a–b). Immunofluorescence staining of Myhc also showed that Neat1 knockdown enhanced Myhc expression, but not when co-transfected with Ezh2 expression vector (Fig. 7f and Supplementary Fig. 5c).
a, b ChIP-qPCR results revealing that Neat1 knockdown significantly decreased the enrichments of Ezh2 (a) and H3k27me3 (b) at the Myog, Myh4, and Tnni2 promoters. c, d ChIP-qPCR results indicating Neat1 overexpression significantly increased the enrichments of Ezh2 (c) and H3k27me3 (d) at the Myog, Myh4, and Tnni2 promoters. e Neat1 expression vector and Ezh2 siRNA fragment were co-transfected into C2C12 cells. The indicated genes expression were measured by qPCR after Neat1 expression vector and Ezh2 siRNA fragment were co-transfected 3 days post differentiation. The results showed that overexpression of Neat1 inhibits the expression of Myog, Myhc and Tnni2, but had no significant effect when co-transfected with Ezh2 siRNA fragment. f Neat1 siRNA fragment was con-transfected with Ezh2 expression vector into C2C12 cells at 5 days post differentiation, the cells were performed with Myhc immunofluorescence staining. The quantification of Myhc immunofluorescence staining results showed Neat1 knockdown alone increased Myhc protein expression but not when co-transfected with Ezh2 expression vector. g, h ChIRP-qPCR results revealed that Neat1 binds directly to the Myog, Myh4, and Tnni2 promoters but not to the Myod promoter in C2C12 cells (g) and primary myoblasts (h). Relative RNA levels were normalized to those of β-actin. All values represent the mean ± s.d. of three independent experiments. *p < 0.05, **p < 0.01, N.S. indicates not significant
Lastly, we performed chromatin isolation by RNA purification (ChIRP) to confirm that Neat1 binds directly to the Myog, Myh4, Tnni2, and P21 gene promoters but not to the Myod promoter in C2C12 cells and mouse primary myoblasts (Fig. 7g, h), consistent with the pattern of Ezh2 occupancy at its target genes (Fig. 6b–d and Fig. 7a, c and Supplementary Fig. 5d), indicating Neat1 regulated Myod expression independent of Ezh2. Altogether, these results suggest that Neat1 suppresses C2C12 myogenic differentiation mainly by increasing Ezh2 enrichment at the promoters of target genes.
Discussion
NEAT1 is involved in multiple biological processes in vitro and in vivo. In vitro, NEAT1 affects the proliferation, migration, invasion, and apoptosis of multiple cancer cells53; for example, NEAT1 promotes the proliferation and invasion of colorectal cancer cells54. Knockdown of NEAT1 suppresses the migration and invasion of glioma cells55. NEAT1 is regulated by c-myc and inhibits imatinib-induced apoptosis of chronic myeloid leukemia cells56. Besides, NEAT1 also plays an important role in vascular smooth muscle cell phenotypic switching36. In vivo, NEAT1 is overexpressed in many solid tumors, including small cell lung cancer57 and hepatocellular carcinoma58,59. Neat1 knockout mice display impaired corpus luteum differentiation60. The genetic ablation of Neat1 leads to abnormal mammary gland morphogenesis and lactation defects61. Neat1 exerts anti-apoptotic and anti-inflammatory functions in C57BL/6 mice after traumatic brain injury62. The functions of Neat1 in myogenesis and skeletal muscle development remain unexplored. Here, we demonstrated that Neat1 promotes myoblast proliferation and inhibits myogenic differentiation. Moreover, knockdown of Neat1 improved the cross-sectional area of muscle fibers, mainly by increasing the expression of myogenic genes, and delaying muscle regeneration, primarily via a reduction in the number of Pax7+ cells. In general, our study found a previously unidentified function of Neat1 in regulating muscle development and regeneration.
LncRNAs regulate gene expression at the transcriptional and post-transcriptional levels or by chromatin modifications63,64,65. As a well-known nuclear lncRNA, NEAT1 functions mainly as a transcriptional regulator. Capture hybridization analysis of RNA targets (CHART) analysis revealed that NEAT1 binds directly to both the transcriptional start sites and transcriptional termination sites of target genes. CHART-mass spectrometry assays identified a large number of NEAT1-interacting proteins66, suggesting that the function of NEAT1 in transcriptional regulation may be mediated by many proteins. In the present study, we found that Neat1 promoted myoblast proliferation and inhibited myogenic differentiation by guiding Ezh2 to target gene promoters, such as Myog, Myh4, Tnni2, and P21, and repressed their transcription. Ezh2 is an important epigenetic inhibitory factor involved in many biological processes, including myogenesis. Ezh2 suppresses myogenic differentiation mainly by inhibiting the expression of myogenic marker genes, such as Myog, myh4, and Mck18,51,52. A previous study showed that the conditional knockout of Ezh2 in SCs resulted in decreased muscle regeneration and number of Pax7+ cells67, consistent with the phenotype of Neat1 knockdown. Moreover, Neat1 knockdown in vivo increased the expression of myogenic genes and the myofiber cross-sectional area, which may be also mediated by Ezh2, because Ezh2 muscle-specific knockout SCs also enhanced myogenic differentiation reflected by increased Myog expression67. Therefore, we conclude that the Neat1 is an important regulator of Ezh2-mediated epigenetic regulation in myogenesis and muscle regeneration. In addition to interacting with Ezh2, Neat1 may regulate myogenesis via other proteins or signaling pathways, because Neat1 also inhibited Myod expression in an Ezh2-independent manner. Further endeavors will be devoted to determining the other mechanisms by which Neat1 affects myogenesis.
LncRNAs are highly conserved in terms of their position within the genome, and conserved lncRNAs may play similar functions among species. For example, linc-YY1 and lncMyoD are both conserved in their genomic positions and are involved in myogenesis in both mouse and human68,69. Compared with protein-coding genes, lncRNAs have low sequence conservation. However, some lncRNAs that possess “ultraconserved” regions also have conserved functions70,71. For example, the lncRNA THOR exerts a carcinogenic role by interacting with IGF2BP1 via an ultraconserved region in human, mouse, and zebrafish72. In the current study, we observed that the F3 region (1001–1540 bp) of Neat1 encompasses the functional domain due to its interaction with Ezh2 and regulation of myogenesis, and further sequence alignment analysis demonstrated that this 1001–1540 bp region is also conserved between mouse and human (Supplementary Fig. 6), indicating a potentially ultraconserved region. In addition, a previous study showed that lncRNA higher-order structures are highly conserved, and these conserved secondary and tertiary structures are related to their biological functions and protein-binding potential73,74. Therefore, we speculated that this 1001–1540 bp region of Neat1 may also contribute to the formation of Neat1 higher-order structures to allow binding to its interacting proteins, and Neat1 may have a conserved function in myogenesis between human and mouse.
In conclusion, our findings provide a novel function of Neat1 in muscle development and regeneration. We demonstrated that Neat1 promoted myoblast proliferation and repressed myogenic differentiation, and knockdown of Neat1 promoted muscle growth but impaired muscle regeneration. Mechanistically, Neat1 recruited Ezh2 to increase the level of H3k27me3 binding at the P21 promoter, leading to repression of P21 expression and promotion of myoblast proliferation. Meanwhile, Neat1 inhibited the expression of muscle-specific genes, such as Myog, Myh4, and Tnni2, by recruiting Ezh2 to target gene promoters and thereby suppressing myogenic differentiation (Fig. 8).
In proliferating myoblasts, Neat1 guides Ezh2 to the P21 promoter and inhibits P21 expression, leading to the promotion of myoblast proliferation. Upon differentiation, Neat1 recruits Ezh2 to inhibit the expression of muscle-specific genes, such as Myog, Myh4, and Tnni2, and suppresses myogenic differentiation
References
Li, Y., Chen, X., Sun, H. & Wang, H. Long non-coding RNAs in the regulation of skeletal myogenesis and muscle diseases. Cancer Lett. 417, 58–64 (2018).
Sabourin, L. A. & Rudnicki, M. A. The molecular regulation of myogenesis. Clin.Genet. 57, 16–25 (2000).
Rudnicki, M. A. et al. MyoD or Myf-5 is required for the formation of skeletal muscle. Cell 75, 1351–1359 (1993).
Tapscott, S. J. et al. MyoD1: a nuclear phosphoprotein requiring a Myc homology region to convert fibroblasts to myoblasts. Science 242, 405–411 (1988).
Choi, J. et al. MyoD converts primary dermal fibroblasts, chondroblasts, smooth muscle, and retinal pigmented epithelial cells into striated mononucleated myoblasts and multinucleated myotubes. Proc. Natl Acad. Sci. USA 87, 7988–7992 (1990).
Mastroyiannopoulos, N. P., Nicolaou, P., Anayasa, M., Uney, J. B. & Phylactou, L. A. Down-regulation of myogenin can reverse terminal muscle cell differentiation. PloS ONE 7, e29896 (2012).
Charge, S. B. & Rudnicki, M. A. Cellular and molecular regulation of muscle regeneration. Physiol. Rev. 84, 209–238 (2004).
Tosic, M. et al. Lsd1 regulates skeletal muscle regeneration and directs the fate of satellite cells. Nat. Commun. 9, 366 (2018).
Dhawan, J. & Rando, T. A. Stem cells in postnatal myogenesis: molecular mechanisms of satellite cell quiescence, activation and replenishment. Trends Cell Biol. 15, 666–673 (2005).
Reik, W., Dean, W. & Walter, J. Epigenetic reprogramming in mammalian development. Science 293, 1089–1093 (2001).
Jenuwein, T. & Allis, C. D. Translating the histone code. Science 293, 1074–1080 (2001).
Meissner, A. Epigenetic modifications in pluripotent and differentiated cells. Nature Biotechnol. 28, 1079–1088 (2010).
Bernstein, E. & Allis, C. D. RNA meets chromatin. Genes Dev. 19, 1635–1655 (2005).
Pauli, A., Rinn, J. L. & Schier, A. F. Non-coding RNAs as regulators of embryogenesis. Nat. Rev. Genet. 12, 136–149 (2011).
Perdiguero, E., Sousa-Victor, P., Ballestar, E. & Munoz-Canoves, P. Epigenetic regulation of myogenesis. Epigenetics 4, 541–550 (2009).
Saccone, V. & Puri, P. L. Epigenetic regulation of skeletal myogenesis. Organogenesis 6, 48–53 (2010).
Asp, P. et al. Genome-wide remodeling of the epigenetic landscape during myogenic differentiation. Proc. Natl Acad. Sci. USA 108, E149–E158 (2011).
Caretti, G., Di Padova, M., Micales, B., Lyons, G. E. & Sartorelli, V. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 18, 2627–2638 (2004).
Stojic, L. et al. Chromatin regulated interchange between polycomb repressive complex 2 (PRC2)-Ezh2 and PRC2-Ezh1 complexes controls myogenin activation in skeletal muscle cells. Epigenet. Chromatin 4, 16 (2011).
Cesana, M. et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147, 358–369 (2011).
Zhu, M. et al. Lnc-mg is a long non-coding RNA that promotes myogenesis. Nat. Commun. 8, 14718 (2017).
Zhou, L. et al. Linc-YY1 promotes myogenic differentiation and muscle regeneration through an interaction with the transcription factor YY1. Nat. Commun. 6, 10026 (2015).
Yu, X. et al. Long non-coding RNA Linc-RAM enhances myogenic differentiation by interacting with MyoD. Nat. Commun. 8, 14016 (2017).
Militello, G. et al. A novel long non-coding RNA Myolinc regulates myogenesis through TDP-43 and Filip1. J. Mol. Cell Biol. 10, 102–117 (2018).
Zhang, Z. K. et al. A newly identified lncRNA MAR1 acts as a miR-487b sponge to promote skeletal muscle differentiation and regeneration. J. Cachexia, Sarcopenia Muscle 9, 613–626 (2018).
Liang, T. et al. lncRNA AK017368 promotes proliferation and suppresses differentiation of myoblasts in skeletal muscle development by attenuating the function of miR-30c. FASEB 32, 377–389 (2018).
Jin, J. J. et al. Long noncoding RNA SYISL regulates myogenesis by interacting with polycomb repressive complex 2. Proc. Natl Acad. Sci. USA 115, E9802–E9811 (2018).
Sunwoo, H. et al. MEN ε/β nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 19, 347–359 (2009).
Souquere, S., Beauclair, G., Harper, F., Fox, A. & Pierron, G. Highly ordered spatial organization of the structural long noncoding NEAT1 RNAs within paraspeckle nuclear bodies. Mol. Biol. Cell 21, 4020–4027 (2010).
Nakagawa, S., Naganuma, T., Shioi, G. & Hirose, T. Paraspeckles are subpopulation-specific nuclear bodies that are not essential in mice. J Cell Biol. 193, 31–39 (2011).
Bond, C. S. & Fox, A. H. Paraspeckles: nuclear bodies built on long noncoding RNA. J. Cell Biol. 186, 637–644 (2009).
Hirose, T. et al. NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Mol. Biol. Cell 25, 169–183 (2014).
Fox, A. H. & Lamond, A. I. Paraspeckles. Cold Spring Harbor Perspect. Biol. 2, a000687 (2010).
Imamura, K. et al. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol. Cell 53, 393–406 (2014).
Lin, Y., Schmidt, B. F., Bruchez, M. P. & McManus, C. J. Structural analyses of NEAT1 lncRNAs suggest long-range RNA interactions that may contribute to paraspeckle architecture. Nucleic Acids Res. 46, 3742–3752 (2018).
Ahmed, A. S. I. et al. Long noncoding RNA NEAT1 (nuclear paraspeckle assembly transcript 1) is critical for phenotypic switching of vascular smooth muscle cells. Proc. Natl Acad. Sci. USA 115, E8660–E8667 (2018).
Chakravarty, D. et al. The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat. Commun. 5, 5383 (2014).
Choudhry, H. et al. Tumor hypoxia induces nuclear paraspeckle formation through HIF-2alpha dependent transcriptional activation of NEAT1 leading to cancer cell survival. Oncogene 34, 4546 (2015).
Wu, Y. et al. Nuclear-enriched abundant transcript 1 as a diagnostic and prognostic biomarker in colorectal cancer. Mol. Cancer 14, 191 (2015).
Chen, X., Kong, J., Ma, Z., Gao, S. & Feng, X. Up regulation of the long non-coding RNA NEAT1 promotes esophageal squamous cell carcinoma cell progression and correlates with poor prognosis. Am. J. Cancer Res. 5, 2808–2815 (2015).
Wang, P. et al. Long noncoding RNA NEAT1 promotes laryngeal squamous cell cancer through regulating miR-107/CDK6 pathway. J. Exp. Clin. Cancer Res. 35, 22 (2016).
Mello, S. S. et al. Neat1 is a p53-inducible lincRNA essential for transformation suppression. Genes Dev. 31, 1095–1108 (2017).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408 (2001).
Zhang, Y. et al. FHL3 differentially regulates the expression of MyHC isoforms through interactions with MyoD and pCREB. Cell. Signal. 28, 60–73 (2016).
Wang, Z. et al. The long noncoding RNA Chaer defines an epigenetic checkpoint in cardiac hypertrophy. Nat. Med. 22, 1131–1139 (2016).
Rinn, J. L. et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129, 1311–1323 (2007).
Wang, L. et al. LncRNA Dum interacts with Dnmts to regulate Dppa2 expression during myogenic differentiation and muscle regeneration. Cell Res. 25, 335–350 (2015).
Chen, Q. et al. Long Noncoding RNA NEAT1, Regulated by the EGFR pathway, contributes to glioblastoma progression through the WNT/beta-catenin pathway by scaffolding EZH2. Clin. Cancer Res.: An Off. J. Am. Assoc. Cancer Res. https://doi.org/10.1158/1078-0432.CCR-17-0605 (2017).
Chen, Z. et al. Long non-coding RNA SNHG20 promotes non-small cell lung cancer cell proliferation and migration by epigenetically silencing of p21 expression. Cell Death Dis. 8, e3092 (2017).
Xu, T., Jiang, W., Fan, L., Gao, Q. & Li, G. Upregulation of long noncoding RNA Xist promotes proliferation of osteosarcoma by epigenetic silencing of p21. Oncotarget 8, 101406–101417 (2017).
Hernandez-Hernandez, J. M., Mallappa, C., Nasipak, B. T., Oesterreich, S. & Imbalzano, A. N. The Scaffold attachment factor b1 (Safb1) regulates myogenic differentiation by facilitating the transition of myogenic gene chromatin from a repressed to an activated state. Nucleic Acids Res. 41, 5704–5716 (2013).
Zhou, L. et al. A novel target of microRNA-29, Ring1 and YY1-binding protein (Rybp), negatively regulates skeletal myogenesis. J. Biol. Chem. 287, 25255–25265 (2012).
Dong, P. et al. Long Non-coding RNA NEAT1: A Novel Target for Diagnosis and Therapy in Human Tumors. Front. Genet. 9, 471 (2018).
Luo, Y. et al. Long non-coding RNA NEAT1 promotes colorectal cancer progression by competitively binding miR-34a with SIRT1 and enhancing the Wnt/beta-catenin signaling pathway. Cancer Lett. 440 – 441, 11–22 (2019).
Zhou, K. et al. Knockdown of long non-coding RNA NEAT1 inhibits glioma cell migration and invasion via modulation of SOX2 targeted by miR-132. Mol. Cancer 17, 105 (2018).
Zeng, C. et al. The c-Myc-regulated lncRNA NEAT1 and paraspeckles modulate imatinib-induced apoptosis in CML cells. Mol. Cancer 17, 130 (2018).
Pan, L. J. et al. Upregulation and clinicopathological significance of long non-coding NEAT1 RNA in NSCLC tissues. Asian Pac. J. Cancer Prevention: APJCP 16, 2851–2855 (2015).
Guo, S. et al. Clinical implication of long non-coding RNA NEAT1 expression in hepatocellular carcinoma patients. Intl. J. Clin. Exp. Pathol. 8, 5395–5402 (2015).
Liu, Z. et al. Long non-coding RNA NEAT1 overexpression is associated with unfavorable prognosis in patients with hepatocellular carcinoma after hepatectomy: A Chinese population-based study. Eur. J Surg. Oncol.: J. Eur. Soc. Surg. Oncol. British Assoc. Surg. Oncol. 43, 1697–1703 (2017).
Nakagawa, S. et al. The lncRNA Neat1 is required for corpus luteum formation and the establishment of pregnancy in a subpopulation of mice. Development 141, 4618–4627 (2014).
Standaert, L. et al. The long noncoding RNA Neat1 is required for mammary gland development and lactation. Rna 20, 1844–1849 (2014).
Zhong, J. et al. The long non-coding RNA Neat1 is an important mediator of the therapeutic effect of bexarotene on traumatic brain injury in mice. Brain, Behavior, Immun. 65, 183–194 (2017).
Mercer, T. R., Dinger, M. E. & Mattick, J. S. Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 10, 155–159 (2009).
Kung, J. T., Colognori, D. & Lee, J. T. Long noncoding RNAs: past, present, and future. Genetics 193, 651–669 (2013).
Cheetham, S. W., Gruhl, F., Mattick, J. S. & Dinger, M. E. Long noncoding RNAs and the genetics of cancer. British J. Cancer 108, 2419–2425 (2013).
West, J. A. et al. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol. Cell 55, 791–802 (2014).
Juan, A. H. et al. Polycomb EZH2 controls self-renewal and safeguards the transcriptional identity of skeletal muscle stem cells. Genes Dev. 25, 789–794 (2011).
Sun, K., Zhou, L., Zhao, Y., Wang, H. & Sun, H. Genome-wide RNA-seq and ChIP-seq reveal Linc-YY1 function in regulating YY1/PRC2 activity during skeletal myogenesis. Genom. Data 7, 247–249 (2016).
Gong, C. et al. A long non-coding RNA, LncMyoD, regulates skeletal muscle differentiation by blocking IMP2-mediated mRNA translation. Dev. Cell 34, 181–191 (2015).
Ulitsky, I., Shkumatava, A., Jan, C. H., Sive, H. & Bartel, D. P. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell 147, 1537–1550 (2011).
Calin, G. A. et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell 12, 215–229 (2007).
Hosono, Y. et al. Oncogenic Role of THOR, a Conserved Cancer/Testis Long Non-coding RNA. Cell 171, 1559–1572 e1520 (2017).
Johnsson, P., Lipovich, L., Grander, D. & Morris, K. V. Evolutionary conservation of long non-coding RNAs; sequence, structure, function. Biochim. Biophys. Acta 1840, 1063–1071 (2014).
Li, R., Zhu, H. & Luo, Y. Understanding the functions of long non-coding rnas through their higher-order structures. Intl J. Mol. Sci. 17, https://doi.org/10.3390/ijms17050702 (2016).
Acknowledgements
This work was supported by National Project for Breeding of Transgenic Animal Grants 2018ZX0801012B and 2016ZX08006-002, Fundamental Research Funds for Central Universities Grant 2662018PY045, and the Agricultural Innovation Fund of Hubei Province (2016-620-000-001-043).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by E. Baehrecke
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, S., Zuo, H., Jin, J. et al. Long noncoding RNA Neat1 modulates myogenesis by recruiting Ezh2. Cell Death Dis 10, 505 (2019). https://doi.org/10.1038/s41419-019-1742-7
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41419-019-1742-7
This article is cited by
-
MAL expression downregulation through suppressive H3K27me3 marks at the promoter in HPV16-related cervical cancers is prognostically relevant and manifested by the interplay of novel MAL antisense long noncoding RNA AC103563.8, E7 oncoprotein and EZH2
Clinical Epigenetics (2024)
-
Prediction of lncRNA functions using deep neural networks based on multiple networks
BMC Genomics (2023)
-
LncRNA-TBP mediates TATA-binding protein recruitment to regulate myogenesis and induce slow-twitch myofibers
Cell Communication and Signaling (2023)
-
A bioinformatics approach to identify a disulfidptosis-related gene signature for prognostic implication in colon adenocarcinoma
Scientific Reports (2023)
-
Long noncoding RNA NEAT1 promotes cardiac fibrosis in heart failure through increased recruitment of EZH2 to the Smad7 promoter region
Journal of Translational Medicine (2022)