Abstract
CUL4A and CUL4B are closely related members in Cullin family and can each assemble a Cullin-RING E3 ligase complex (Cullin-RING Ligase 4A or 4B, CRL4A, or CRL4B) and participate in a variety of biological processes. Previously we showed that zebrafish cul4a, but not cul4b, is essential for cardiac and pectoral fin development. Here, we have identified cul4a as a crucial regulator of primitive erythropoiesis in zebrafish embryonic development. Depletion of cul4a resulted in a striking reduction of erythroid cells due to the inhibition of erythroid differentiation. Transcript levels for early hematopoietic regulatory genes including scl, lmo2, and gata1 are significantly reduced in cul4a-deficient embryos. Mechanistically, we demonstrated that scl and gata1, the central regulators of primitive hematopoiesis for erythroid determination, are transcriptionally upregulated by cul4a. These findings demonstrate an important role for cul4a in primitive erythropoiesis and may bear implications in regeneration medicine of anemia and related diseases.
Similar content being viewed by others
Introduction
Zebrafish (Danio rerio) has become a powerful model for investigating hematopoiesis because its transparency greatly facilitates the visualization of hematopoietic system. The Zebrafish hematopoietic system is highly analogous to that in mammals, including the phenotype of blood cells and the key hematopoietic-lineage genes1,2. Zebrafish hematopoiesis consists of two successive waves, primitive and definitive hematopoiesis3. The first wave generates primitive erythrocytes (primitive erythropoiesis) and macrophages (primitive myelopoiesis), and the second wave produces hematopoietic stem cells (HSCs)4. The primitive myelopoiesis occurs at the anterior lateral plate mesoderm, while primitive erythropoiesis takes place at the posterior lateral plate mesoderm (PLPM), which subsequently forms intermediate cell mass (ICM) at the midline5,6. A sophisticated network of many transcription factors has been described to regulate primitive and definitive hematopoiesis7. Among them, the stem cell leukemia gene (Scl/Tal1) is a central regulator of primitive hematopoiesis8,9. Gata1, a zinc finger protein, is specifically required for the maturation of proerythroblasts, and Pu.1, a transcription factor that contains an ETS domain, plays an indispensable role in primitive myelopoiesis10,11. In contrast, definitive hematopoiesis is initiated by the transcription factors of Runx1 and c-Myb6. Although the importance of these transcription factors has been demonstrated in cell-based ex vivo assays as well as in knockout mouse models, the regulation of their expression remains poorly understood.
Cullin-RING E3 ligase (CRL) complexes, which contain Cullin, RING protein, and substrate-recognition subunit as the core components, represent the largest known class of ubiquitin ligases and participate in a broad variety of physiologically and developmentally controlled processes12,13. Among Cullin family members, CUL4A and CUL4B have the highest degree of homology, with ~80% identity in protein sequences, and are believed to be derived from one common ancestor CUL414,15,16. There is only one ortholog, cul4, in lower organisms. As the scaffold protein, CUL4 assembles with damaged DNA-binding protein 1 at its N terminus and with ROC1 at its C terminus to form CRL4 complexes that target different substrates for proteosomal degradation or for protein modification17,18. Although CUL4A and CUL4B share a high degree of homology, studies have shown that the two CUL4 genes in mammals are not entirely redundant. Loss of function mutations in human CUL4B cause mental retardation, short stature, abnormal gait, impaired speech, and other abnormalities19,20,21. Cul4b null mice are embryonic lethal and tissue-specific Cul4b knockout mice exhibited a variety of developmental defects22,23,24,25,26. In contrast, no germline mutations in human CUL4A gene have been reported, and Cul4a knockout mice exhibited no apparent developmental phenotype except that the knockout males were sterile27,28. As in mammals, there are two cul4 members, cul4a and cul4b, in zebrafish. The two genes have a high degree of homology with their respective mammalian counterparts CUL4A and CUL4B29. We previously reported that cul4a, but not cul4b, is essential for zebrafish cardiac development and pectoral fin formation29. These findings indicate that the functions of cul4 paralogs are varied in different species. In this study, we determined the expression profile of cul4a during zebrafish embryogenesis and found it was localized in primary hematopoietic region. Using morpholino and recently developed four-guide Cas9 RNP targeting technology30, we demonstrated that primitive erythropoiesis was significantly reduced after cul4a ablation. Mechanistically, Cul4a was shown to be required for the expression of scl and gata1 via promoting H3K4me3. Our results revealed a crucial role for Cul4a in zebrafish primitive hematopoiesis.
Results
Knockdown of cul4a in zebrafish significantly reduces the number of erythrocytes
To assess whether cul4a is involved in the blood cell development in zebrafish, we firstly examined the expression pattern of cul4a during zebrafish embryonic development using whole-mount in situ hybridization (WISH). Maternally supplied cul4a transcripts were distributed evenly in all blastomeres through the blastula and gastrulation stages (Fig. S1a, b). Then, it was found to express elusively in two primitive hematopoietic sites, the PLPM at 12 h post fertilization (hpf) (Fig. S1c, d) and in the ICM at 24 hpf (Fig. S1e). After 48 hpf, cul4a was hardly detectable in caudal hematopoietic tissue where definitive hematopoiesis occurs (Fig. S1f). This spatiotemporal distribution pattern of cul4a expression corresponds to that of primitive hematopoiesis.
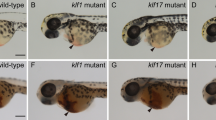
Our previous study showed that embryos injected with cul4a-MO (a splicing blocking morpholino) or cul4a-MO1 (a translational-blocking morpholino) exhibited an array of morphological defects, including tail curling, hindbrain edema, stunted or completely absent pectoral fins and pericardial edema that was accompanied by heart looping impairment29. Moreover, blood cells were markedly reduced or absent in the circulation of cul4a morphants. We therefore here focused on the hematopoiesis in the embryos injected with cul4a-MO, or cul4a-MO1 or control MO (CoMO) as described in our previous study29. Indeed, the cul4a morphants had significantly decreased number of red blood cells as shown by o-dianisidine staining (Fig. 1a). To rule out that the reduction in erythrocytes in cul4a morphants was due to heart defects, we performed WISH and quantitative real-time PCR (qRT-PCR) assays for globin transcripts in cul4a morphants at 24 hpf, when heart tubes in cul4a morphants were indistinguishable from those in controls. As shown in Fig. 1b, c, the expression of hbbe3 was significantly downregulated in cul4a morphants compared to controls. To determine whether the hematopoietic defects observed in cul4a morphants resulted from a nonspecific morpholino effect, we performed rescue experiments using a mismatch cul4a mRNA to avoid targeting by cul4a-MO. After coinjection of cul4a-MO together with cul4a mRNA, the decreased erythrocytes in cul4a morphants could be efficiently rescued (Fig. 1a–c). Furthermore, embryos injected with cul4a-MO1 exhibited similar defects compared with the control embryos (Fig. 1d, e). To further confirm these phenotypes, we generate cul4a−/−, cul4b−/−, or double knockout mutants using an optimized four-guide Cas9 RNP targeting system30. Sequencing analysis showed that four-guide sgRNAs (a, b, d, and f for cul4a, a′, b′, d′, and e′ for cul4b in Table S1) could efficiently delete cul4a and cul4b gene, respectively (Fig. S1g, h). As shown in Fig. 1f–h, depletion of cul4a, but not cul4b, resulted in a significant reduction in the number of erythrocytes as demonstrated by o-dianisidine staining or hbbe3 expression. Moreover, double knockout mutants exhibited similar phenotypes to cul4a knockout mutants. These results indicated that cul4a is required for primitive hematopoiesis.
a o-dianisidine staining showed depletion of erythrocytes in cul4a-morphant embryos, compared with controls. Zebrafish cul4a mRNA rescued primitive erythropoiesis in cul4a-morphant embryos. b Expression of the embryonic hemoglobin, hbbe3, was analyzed by WISH at 24 hpf in embryos injected with CoMO or cul4a-MO, or MO coinjected with zebrafish cul4a mRNA. Lateral views are shown with anterior to the left. c Relative mRNA level of hbbe3 was assayed by qRT-PCR in embryos injected with CoMO or cul4a-MO, or MO coinjected with cul4a mRNA at 24 hpf. d WISH was performed with hbbe3 probes in embryos at 24 hpf injected with CoMO or cul4a-MO1. e Relative mRNA level of hbbe3 was measured by qRT-PCR in embryos injected with CoMO or cul4a-MO1. f o-dianisidine staining showed decreased erythrocytes in cul4a−/− and double knockout, but not in cul4b−/− knockout embryos. Embryos shown are lateral views with anterior to the left. g The images of WISH with hbbe3 mRNA probes in control (cas9-tail injected), cul4a −/−, cul4b −/−, and double knockout mutants at 24 hpf. h Relative mRNA level of hbbe3 was assayed by qRT-PCR in control (cas9-tail injected), cul4a−/−, cul4b−/−, and double knockout mutants at 24 hpf. The number in the top right-hand corner indicates the phenotypic embryos/total embryos. qRT-PCR experiments were performed in triplicate. ***p < 0.001. All scale bars represent 250 μm
Zebrafish cul4a regulates the primitive erythropoiesis, but not primitive myelopoiesis or definitive hematopoiesis
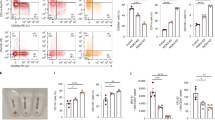
Hematopoiesis occurs in two successive waves and is regulated by lineage-specific genes. Gata1 and pu.1 encode two transcription factors that regulate primitive erythropoiesis and primitive myelopoiesis, respectively3,37,38. To assess the role of cul4a in primitive hematopoiesis, we performed WISH in cul4a morphants for gata1, a marker for erythroid progenitors. Consistent with the scarcity of erythrocytes in cul4a morphants, the expression level of gata1 was dramatically downregulated by cul4a knockdown (Fig. 2a). Coinjection of in vitro synthesized cul4a mRNA rescued the decreased expression of gata1 in cul4a morphants (Fig. 2a). qRT-PCR assays further confirmed the reduction in gata1 expression in cul4a morphants and its rescue by cul4a mRNA (Fig. 2b). Consistent with unaltered number of erythrocytes in cul4b-deficient embryos, gata1 expression was not affected by cul4b knockdown (Fig. 2a, b). To further confirm this finding, we injected cul4a-CoMO or cul4a-MO into zebrafish embryos of the transgenic line Tg (gata1: EGFP) in which the differentiated erythroid cells are labeled by EGFP. We found that the EGFP-positive population was substantially reduced in cul4a morphants at 24 hpf, and the reduction in EGFP-positive cells was efficiently reversed by coinjection of cul4a mRNA (Fig. 2c). To determine whether decreased number of erythrocytes was mediated by downregulation of gata1 expression, we performed rescue experiments using in vitro synthesized gata1 mRNA. As shown in Fig. 2d–f, coinjection of in vitro synthesized gata1 mRNA could efficiently block the reduction in the number of erythrocytes caused by cul4a depletion. These results indicated that cul4a promotes primitive erythropoiesis by regulating gata1 expression.
a Gata1 was analyzed by WISH in embryos at 6 somite injected with CoMO, cul4a-MO, cul4b-MO, cul4a/cul4b-MO, or cul4a-MO coinjected with zebrafish cul4a mRNA. b Relative gata1 expression was analyzed by qRT-PCR in embryos injected with CoMO, cul4a-MO, cul4b-MO, cul4a/cul4b-MO, or MO coinjected with cul4a mRNA, respectively. c cul4a-MO was injected into Tg (gata1: EGFP) transgenic embryos and decreased EGFP-positive cells were observed. Coinjection of cul4a mRNA rescued the reduction in gata1+ cells caused by cul4a MOs. d–f Effects of gata1 mRNA in rescuing hematopoietic defects in cul4a-morphant embryos as demonstrated by staining of o-dianisidine (d), WISH (e) and qRT-PCR (f) of hbbe3 probes. The number in the top right-hand corner indicates the phenotypic embryos/total embryos. qRT-PCR experiments were performed in triplicate. ***p < 0.001. All scale bars represent 250 μm
We then examined the role of cul4a in the development of myeloid cells during primitive hematopoiesis. The expression levels of pu.1, a marker for myeloid progenitors, were comparable between cul4a morphants and CoMO-injected embryos, as indicated by WISH and qRT-PCR (Fig. 3a, b). To confirm these results, we examined the pu.1 expression in cul4a−/−, cul4b−/−, and double knockout mutants. As shown in Fig. 3c, d, depletion of cul4a or cul4b or both did not alter the expression of pu.1. Similarly, the expression of mpo, a marker for granulocytes, was not affected by cul4a MOs (Fig. 3e, f). These results suggest that both cul4a and cul4b are dispensable for the development of myeloid cells during primitive hematopoiesis.
a Pu.1 was analyzed by WISH at 24 hpf in embryos injected with CoMO or cul4a-MO. b Relative expression of pu.1 mRNA was analyzed by qRT-PCR in embryos injected with CoMO or cul4a-MO. c, d Expression of pu.1 in control and cul4a−/−, cul4b−/−, and double knockout mutants was assayed by WISH (c) and by qRT-PCR (d). e, f Mpo was analyzed by WISH (e) and qRT-PCR (f) at 24 hpf in embryos injected with CoMO or cul4a-MO. g–j runx1 and c-myb expression were analyzed by WISH (g, i) or qRT-PCR (h, j) in embryos injected with CoMO or cul4a-MO. k, l Expression of runx1 in control and cul4a−/−, cul4b −/−, and double knockout embryos. The number in the top right-hand corner indicates the phenotypic embryos/total embryos. qRT-PCR experiments were performed in triplicate. All scale bars represent 250 μm
We also assessed whether Cul4a affected definitive hematopoiesis. WISH and qRT-PCR were performed with markers for definitive hematopoiesis, runx1 and c-myb. The expression of neither runx1 nor c-myb in cul4a morphants appeared to be affected by cul4a morpholinos (Fig. 3g–j). Similarly, the expression of runx1 was not affected by depletion of cul4a, cul4b, or both cul4a and cul4b (Fig. 3k, l). These data indicated that lack of cul4a and cul4b has no impact on the emergence of primitive myeloid progenitors or definitive stem cells.
The expression of the early hematopoietic markers scl and lmo2 is downregulated in cul4a morphants
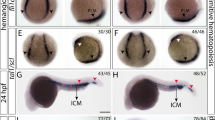
To investigate the molecular mechanism by which Cul4a regulates primitive erythropoiesis, we examined the expression of early hematopoietic progenitor markers scl and lmo2, which are required for the generation of hematopoietic progenitors within PLPM. The expression levels of these two genes were remarkably reduced in cul4a-MO-injected embryos compared with those in embryos injected with CoMO (Fig. 4a, b). The downregulation of scl and lmo2 in cul4a morphants was confirmed by qRT-PCR assays (Fig. 4c, d). Importantly, coinjection of cul4a mRNA significantly blocked the reduction in the expression levels of scl and lmo2, both confirmed by WISH and qRT-PCR (Fig. 4a–d). We further confirmed that scl and lmo2 expression levels were uniquely decreased in cul4a−/− mutants rather than cul4b−/− or double knockout mutants (Fig. 4e–h).
a, b WISH was performed with scl (a) or lmo2 (b) probes in embryos at 6 somite and 24 hpf, respectively. Embryos at 6-somite stage were in poster order view with anterior to the top. The ICM regions of embryos were lateral views with anterior to the left. c, d qRT-PCR analyzed the expression of scl and lmo2 in embryos injected with CoMO or cul4a-MO, or MO coinjected with zebrafish cul4a mRNA at 6 somite and 24 hpf. e, f The images of WISH with scl or lmo2 mRNA probes in control (cas9-tail injected), cul4a −/−, cul4b −/−, and double knockout mutants at 24 hpf. g, h Relative expression of scl and lmo2 mRNA were analyzed by qRT-PCR in control and cul4a −/−, cul4b −/−, and double knockout mutants. The number in the top right-hand corner indicates the phenotypic embryos/total embryos. qRT-PCR experiments were performed in triplicate. ***p < 0.001. All scale bars represent 250 μm
Impaired primitive erythropoiesis caused by lack of cul4a is mediated by downregulated expression of scl-α
It has been shown that zebrafish produces two scl isoforms, full-length scl-α and a shorter isoform scl-β, through alternative promoter sites. While scl-α and scl–β are redundant for the initiation of primitive hematopoiesis, scl-β, but not scl-α, is required for specification of definitive HSCs39. The fact that lack of cul4a impaired primitive erythropoiesis, but not definitive hematopoiesis, suggests that Cul4a regulates the expression of scl-α, but not that of scl-β. To confirm this notion, we used specific primer pairs to distinguish two isoforms. As expected, knockdown of cul4a in zebrafish resulted in a downregulation of scl-α, but had no effect on the expression of scl-β (Fig. 5a). To determine whether the impaired erythropoiesis caused by cul4a depletion is also medicated by the downregulation of scl, we performed rescue experiments with in vitro synthesized scl mRNA. We found that coinjection of in vitro synthesized scl-α mRNA rather than scl-β rescued the decreased primitive erythropoiesis in cul4a morphants (Fig. 5b). Consistently, the reduced expression of lineage-specific genes gata1 and hbbe3 was rescued by coinjection with scl-α mRNA as indicated by WISH and qRT-PCR (Fig. 5c–f). Interestingly, coinjection of scl-α mRNA also blocked the reduction in the expression of lmo2 (Fig. 5g, h), suggesting that the reduction of lmo2 is caused by the downregulation of scl-α. Taken together, these observations indicate that the impaired erythropoiesis due to cul4a knockdown is mediated by the downregulation of scl-α expression.
a The relative expression of the two isoforms of scl, scl-α and scl-β, were measured by qRT-PCR in embryos injected with CoMO or cul4a-MO. b o-dianisidine staining showed depletion of erythroid cells in cul4a-morphant embryos compared with controls. Zebrafish scl-α, but not scl-β, mRNA rescued the reduction in primitive erythropoiesis of cul4a-morphant embryos. c, d WISH was performed with gata1 or hbbe3 probes in 24 hpf embryos injected with CoMO or cul4a-MO, or MO coinjected with zebrafish scl-α or scl-β mRNA. e, f Relative expression of gata1 or hbbe3 in embryos injected with CoMO or cul4a-MO, or MO coinjected with zebrafish scl-α or scl-β mRNA at 24 hpf. g WISH was performed with lmo2 probes in embryos at 6 somite and 24 hpf, respectively. Reduced expression of lmo2 in cul4a-morphant embryos were rescued by coinjection of zebrafish scl-α, but not scl-β mRNA. h Relative expression of lmo2 was measured by qRT-PCR in embryos injected with CoMO or cul4a-MO, or MO coinjected with zebrafish scl-α or scl-β mRNA at 6 somite and 24 hpf. The number in the top right-hand corner indicates the phenotypic embryos/total embryos. qRT-PCR experiments were performed in triplicate. ***p < 0.001. All scale bars represent 250 μm
Cul4a activates scl-α and gata1 transcription by an epigenetic mechanism
To further characterize whether cul4a regulates scl-α expression directly by binding to its promoter, we next performed a whole-embryo quantitative chromatin immunoprecipitation (E-qChIP) analysis to test whether Cul4a binds to scl-α promoter using six pairs of primers covering a region approximately ranging from −2000 bp to +200 bp of scl-α transcription start site. The E-qChIP assay revealed that the Cul4a occupancy peaked in the region at around −1643 bp to −1143 bp of the scl promoter (Fig. 6a). Notably, H3K4me3, a histone marker for transcription activation, was also enriched in the same region (Fig. 6b). We next examined the effect of cul4a knockdown on the enrichment of these proteins on the scl–α promoter and found that accompanying a marked reduction in the enrichment of cul4a bound to the scl promoter, the level of H3K4me3 at the scl promoter was also markedly decreased when cul4a was knocked-down (Fig. 6c, d). These results show that Cul4a can bind to scl-α promoter and thereafter promotes H3K4 trimethylation en route to activate scl-α transcription.
a, b E-qChIP assays of wild-type zebrafish at 24 hpf with antibodies against Cul4a (a) and H3K4me3 (b). The presence of scl promoter sequences in the input DNA and antibody-bound chromatin segments was analyzed by qPCR and agarose gel electrophoresis. c, d E-qChIP assays of scl at 24 hpf embryos injected with CoMO or cul4a-MO. e, f The presence of gata1 promoter sequences in the input DNA and antibody-bound chromatin segments was analyzed by qPCR and agarose gel electrophoresis. g, h E-qChIP assays of gata1 at 24 hpf embryos injected with CoMO or cul4a-MO. Experiments were performed in triplicate. ***p < 0.001, **p < 0.01
The facts that depletion of cul4a downregulated the expression of gata1 and coinjection of in vitro synthesized gata1 mRNA could efficiently block the reduction of erythrocytes in cul4a morphants suggests that Cul4a might directly regulate the transcription of gata1 gene. To test this hypothesis, we used the ChIP assay to examine whether gata1 gene is bound by Cul4a using seven pairs of primers specific for a region located approximately −2000 bp to +200 bp of gata1 transcription start site. The ChIP assay showed that Cul4a could directly bind to the region at −1059 bp to −538 bp of gata1 promoter. H3K4me3 was also bound to the same region. Moreover, the quantitative ChIP assay showed that knockdown of cul4a significantly decreased the binding of cul4a on the gata1 promoter (Fig. 6g). Consistently, the level of H3K4me3 at the gata1 promoter was also markedly decreased when cul4a was depleted (Fig. 6h). Together, these data show that cul4a can activate gata1 transcription by promoting H3K4 trimethylation.
Discussion
Many transcription factors regulating primitive and definitive hematopoiesis have been identified and mechanistically characterized. However, the regulation of their expression is less understood. In this study we showed that Cul4a is essential for zebrafish primitive erythropoiesis. Depletion of cul4a resulted in a severe reduction of the number of erythrocytes. We obtained several lines of evidences that support the conclusion that zebrafish Cul4a is directly involved in embryonic erythrocyte development. First, the reduction in the number of red blood cells was observed as early as 24 hpf, when heart tubes in cul4a morphants were indistinguishable from those in controls. Second, depletion of cul4a results in the downregulation of gata1, a gene essential of primitive erythroid-lineage development, which was efficiently restored by coinjection of cul4a mRNA. Third, knockdown of cul4a leads to significant downregulation of scl, a key transcription factor in primitive hematopoiesis. Furthermore, coinjection of zebrafish scl-α mRNA is able to restore erythropoiesis as well as gata1 expression. Finally, the ChIP assay shows that Cul4a directly binds to both scl-α and gata1 promoter, and promotes H3K4 trimethylation, a histone mark of active transcription. Together, these results indicate that zebrafish cul4a regulates primitive erythropoiesis by promoting scl-α and gata1 transcription.
SCL was originally identified as a proto-oncogene through the study of T-cell acute lymphoblastic lecukemia patients with a chromosomal translocation, and encodes a basic helix-loop-helix transcription factor8,9,40. The importance of SCL in the initiation of primitive and definitive hematopoiesis, erythrocyte and megakaryocyte differentiation, angiogenesis, and astrocyte development has been demonstrated in cell-based ex vivo assays as well as in knockout mouse models41,42,43,44. To our surprise, while knockdown of cul4a led to significant downregulation of gata1, one of the downstream genes of scl and lmo2, the markers for primitive myelopoiesis as well as definitive hematopoiesis were not found to be reduced, which is not due to the redundant role of Cul4b, since neither cul4b−/− nor double knockout mutants exhibited a reduction in the expression of these markers. Previous study has elucidated that zebrafish produces two scl isoforms, scl-α and scl-β, which exert distinct functions in the regulation of primitive erythroid differentiation and definitive hematopoietic stem cell specification39,45. We thus examined the effect of cul4a knockdown on the expression of two scl isoforms, and revealed that knockdown of cul4a downregulated scl-α, but not scl-β. Further studies are required to elucidate how Cul4a complex regulates the temporal and spatial patterns of expression of the different isoforms. On the other hand, it has been identified that Lmo2 works together with Scl to regulate both primitive and definitive hematopoiesis46. The decreased scl levels led by cul4a depletion could attenuate the effect of downregulated lmo2 on hematopoiesis. Intriguingly, Cul4a also directly regulates the expression of gata1, supporting that Cul4a plays more important roles in regulating gata1 expression than that in regulating scl expression.
Epigenetic modifications have essential roles in cellular development and differentiation. For example, a recent study revealed that zebrafish TET2 plays an essential role in hematopoiesis by activating lineage-specific genes, scl, gata1, and c-myb, via DNA oxidative demethylation47. CUL4B complex was found to function as a transcriptional repressor by catalyzing histone H2A monoubiquitination at lysine 119, which facilitates the recruitment of repressive complexes PRC2, HDAC, and DNMT to chromatin and in turn catalyzes trimethylation of histone H3K27, histone deacetylation, and DNA methylation, respectively48,49,50. In contrast, CUL4A complex has been found to function as transcriptional activator. For example, Cul4A was reported to activate ZEB1 in tumor progression and tbx5 expression in zebrafish development by promoting H3K4 methylation29,51. In this study, we showed that Cul4a could bind to scl-α and gata1 promoters, and promote H3K4 trimethylation. Future studies need to determine how CUL4A complex functions as a transcriptional activator for scl-α and gata expression. Cul4a knockout mice do not show obvious developmental or health defects except for male infertile28. Cul4a zebrafish morphants, on the other hand, failed to undergo heart looping29 and primitive erythropoiesis. While loss of function mutation of human CUL4B leads to an array of developmental and behavioral abnormalities20,21, and constitutive deletion of Cul4b in mouse is incompatible with embryonic development22,23, cul4b zebrafish morphants exhibit no remarkable phenotypes29. These results suggest that CUL4A and CUL4B may each have species-specific functions.
In summary, this study unveiled a function of Cul4a in primitive erythropoiesis in zebrafish. Cul4a binds to scl and gata1 promoters and promotes their transcription during primitive erythropoiesis. Our study provides new insight into erythropoiesis, which may have general implications in regeneration medicine of anemia and related diseases.
Materials and methods
Zebrafish maintenance
The Tübingen strain and Tg transgene zebrafish were maintained in a circulating water system with a 14-h light/10-h dark cycle at 28.5 °C. The Tg transgene zebrafish was kindly provided by Professor Yiyue Zhang (South China University of Technology). Embryos were collected from adult fish after natural mating and raised at 28.5 °C in N-Phenylthiourea (PTU, Sigma Aldrich, St. Louis, MO, USA, P7629)/Holtfreter solution (0.003%, final) to prevent pigmentation implemented with 280 μg/L methylene blue (Sigma Aldrich, M4159). Four nanograms of Morpholinos (Gene Tools LLC, Philomath, OR, USA) was injected at the 1- to 2-cell stage of embryos with splice-blocking (MO: 5′-CTTGGGTCTGTCTGTAACACACAGA-3′), translation-blocking (MO1: 5′-GCTGGTGCTGAACATCTTCTGCCAT-3′), or control MO (CoMO: 5′-CTAGCGTCTCTCTCTAACACACACA-3′) that was used as the negative control as described previously29. For the rescue experiments, 80–100 pg mRNA was injected.
Generation of cul4-depleted zebrafish embryos by CRISPR/Cas9 system
Six site-specific CRISPR-Cas9 sgRNAs for the zebrafish cul4a or cul4b gene were designed using the online software CRISPR design tool (http://www.crisprscan.org) CRISPRscan, and the primer sequences are listed in Table S1. We chose four highly efficient sgRNAs for cul4a and cul4b, respectively. To produce sgRNA templates for in vitro transcription, cul4a and cul4b sgRNAs were produced by in vitro transcription using the MEGAshortscript T7 Transcription Kit (Ambion Life Technologies, CA, USA) as described previously31. Zebrafish embryos were injected with 1 nL mixed solution containing 150 pg of sgRNA, 120 pg of cas9 protein (Script, Z03389-50, New-Jersey, USA), and 1.5 ng of phenol red (Sigma-Aldrich, P0290) tracer. After 24 hpf, 10 of the injected embryos were assayed for sgRNA activity by DNA extraction, PCR amplification, restriction digestion, and DNA sequencing.
The screening primers were designed around the cul4a (Exon15, 13, 11, 5, and 2) or cul4b (Exon1, 12, and 17) sgRNA target sites. The primer sequences were listed in Table S2. The fragments were cloned using the TA Cloning Kit (TAKARA, 6028, Japan) and then were sequenced.
Whole-mount in situ hybridization (WISH)
WISH was performed using single-stranded RNA probes labeled with digoxigenin-uridine 5-triphosphate (Roche, Mannheim, Germany, 11277073910) essentially as described with minor changes32,33. Briefly, embryos were manually dechorionated, fixed in 4% PFA at 4 °C overnight. Embryos over 24 hpf were permeabilized using 10 μg/ml Proteinase K (Ambion, Austin, TX, USA, AM2546) in PBST (1 x PBS + 0.1% Tween-20) for 5 min and post-fixed in 4% PFA for 20 min at the room temperature. Hybridization was performed using 100 ng Digoxigenin-labeled cul4a, scl, lmo2, gata1, hbbe3, pu.1, mpo, and runx1, and c-myb probes synthetized using the T7 RNA polymerase (Promega, Fitchburg, WI, USA, P2075). The probes were detected by incubating the embryos with an anti-Digoxigenin antibody coupled to alkaline phosphase (Roche, 11093274910) diluted 1:2500 in blocking solution (Roche, 11921673001). The staining reaction was performed using NBT/BCIP (Sigma, N6639, B-8503) in staining buffer (100 mM Tris-HCl pH 9.5, 50 mM MgCl2, 100 mM NaCl, 0.1% Tween 20). All in situ probes were synthesized as described previously34. Plasmid clones for the generation of probes of zebrafish c-myb and runx1, were kindly provided by Professor Yiyue Zhang (South China University of Technology) and Professor Weijun Pan (Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences). Plasmid clones of zebrafish gata1 and pu.1 were provided by Professor Yiyue Zhang. Plasmid clones of zebrafish mpo were provided by Travis Maslow in Harvard university. Other plasmid clones for generating probes of zebrafish lmo2, hbbe3, and full-lengh scl were kindly provided by Professor Anming Meng (Institute of Zoology, Chinese Academy of Sciences). Images were acquired using the Olympus SZX16 system (Olympus, Tokyo, Japan), and analyzed with Image-pro plus software.
Quantitative real-time PCR(qRT-PCR)
Total mRNA was extracted with TRIzol (Ambion, 175803) from 24 hpf embryos (at least 100 embryos). RNA was used as template for cDNAs synthesis, which was performed using the RevertAid Reverse transcriptase (ThermoFisher, Rockford, IL, USA, EP0441). qRT-PCR was performed on a LightCycler 480 using SYBR Green (Roche, 04887352001). The sequence-specific primers were designed using Primer 5 software and shown in supplementary Table S3.
o-dianisidine Staining
For detection of hemoglobin in red blood cells of zebrafish embryos, the staining was performed as described previously35.
Whole-embryo quantitative chromatin immunoprecipitation(E-qChIP)
E-qChIP was performed as previously described with modifications36. Briefly, 24 hpf zebrafish embryos (at least 1000 per sample) were hand-dechorionated and fixed in 4% formaldehyde (final concentration) for 20 min at room temperature, followed by glycine (0.125 M) treatment for 5 min. The embryos were then homogenized in cell lysis buffer (10 mM Tris-HCl pH 7.5, 10 mM NaCl, 0.5% NP-40, and protease inhibitors), and incubated for 10 min on ice. Nuclei were collected by centrifugation, resuspended in nuclei lysis buffer (50 mM Tris-HCl pH 7.5, 10 mM EDTA, 1% SDS, and protease inhibitors), and incubated for 10 min. Chromatin was sonicated by Diagenode Bioruptor to yield fragments of 300–500 bp. Chromatin was pre-cleared with protein G agarose beads (Invitrogen, Carlsbad, CA, USA, 00507406), then incubated with antibodies with rotation overnight at 4 °C. Antibodies used were Cul4a (Abcam, ab72548), and H3K4me3 (Chemicon/Millpore, Billerica, MA, USA, 07-473). Beads were washed five times with RIPA wash buffer (50 mM HEPES pH 7.6, 1 mM EDTA, 0.7% Deoxycholic acid sodium, 1% NP-40, and 0.5% LiCl) and once with 1X Triton wash buffer (50 mM HEPES pH 7.6, 150 mM NaCl, and 1% Triton X-100) at 4 °C. Bound complexes were eluted from the beads at 65 °C with vortexing in elution buffer (1 M NaHCO3, 1% SDS). Cross-links were reversed for 6–12 h at 65 °C and the chromatin purified the DNA Clean-up Kit (CWBIO, Beijing, China, CW2301M). The primers used in qPCR were shown in supplementary Table S4.
Statistical analysis
All experiments were repeated at least three times. Quantitative data are expressed as mean ± SD. Statistical significance was determined by the Student’s t-test. P value of less than 0.05 was considered statistically significant.
References
Ellett, F. & Lieschke, G. J. Zebrafish as a model for vertebrate hematopoiesis. Curr. Opin. Pharmacol. 10, 563–570 (2010).
Jing, L. & Zon, L. I. Zebrafish as a model for normal and malignant hematopoiesis. Dis. Models. Mech. 4, 433–438 (2011).
Paik, E. J. & Zon, L. I. Hematopoietic development in the zebrafish. Int. J. Dev. Biol. 54, 1127–1137 (2010).
Gore, A. V., Pillay, L. M., Venero Galanternik, M. & Weinstein, B. M. The zebrafish: a fintastic model for hematopoietic development and disease. Wiley Interdiscip. Rev. Dev. Biol. 7, e312 (2018).
Davidson, A. J. & Zon, L. I. The ‘definitive’ (and ‘primitive’) guide to zebrafish hematopoiesis. Oncogene 23, 7233–7246 (2004).
de Jong, J. L. & Zon, L. I. Use of the zebrafish system to study primitive and definitive hematopoiesis. Annu. Rev. Genet. 39, 481–501 (2005).
Orkin, S. H. & Zon, L. I. Hematopoiesis: an evolving paradigm for stem cell biology. Cell 132, 631–644 (2008).
Chen, Q. et al. The tal gene undergoes chromosome translocation in T cell leukemia and potentially encodes a helix-loop-helix protein. EMBO J. 9, 415–424 (1990).
Hoang, T., Lambert, J. A. & Martin, R. SCL/TAL1 in hematopoiesis and cellular reprogramming. Curr. Top. Dev. Biol. 118, 163–204 (2016).
Migliaccio, A. R., Rana, R. A., Vannucchi, A. M. & Manzoli, F. A. Role of GATA-1 in normal and neoplastic hemopoiesis. Ann. N. Y. Acad. Sci. 1044, 142–158 (2005).
Koschmieder, S., Rosenbauer, F., Steidl, U., Owens, B. M. & Tenen, D. G. Role of transcription factors C/EBPalpha and PU.1 in normal hematopoiesis and leukemia. Int. J. Hematol. 81, 368–377 (2005).
Petroski, M. D. & Deshaies, R. J. Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6, 9–20 (2005).
Bosu, D. R. & Kipreos, E. T. Cullin-RING ubiquitin ligases: global regulation and activation cycles. Cell Div. 3, 7 (2008).
Higa, L. A. & Zhang, H. Stealing the spotlight: CUL4-DDB1 ubiquitin ligase docks WD40-repeat proteins to destroy. Cell Div. 2, 5 (2007).
Jackson, S. & Xiong, Y. CRL4s: the CUL4-RING E3 ubiquitin ligases. Trends Biochem. Sci. 34, 562–570 (2009).
Hannah, J. & Zhou, P. Distinct and overlapping functions of the cullin E3 ligase scaffolding proteins CUL4A and CUL4B. Gene 573, 33–45 (2015).
He, Y. J., McCall, C. M., Hu, J., Zeng, Y. & Xiong, Y. DDB1 functions as a linker to recruit receptor WD40 proteins to CUL4-ROC1 ubiquitin ligases. Genes Dev. 20, 2949–2954 (2006).
Lee, J. & Zhou, P. Pathogenic role of the CRL4 ubiquitin ligase in human disease. Front Oncol. 2, 21 (2012).
Tarpey, P. S. et al. Mutations in CUL4B, which encodes a ubiquitin E3 ligase subunit, cause an X-linked mental retardation syndrome associated with aggressive outbursts, seizures, relative macrocephaly, central obesity, hypogonadism, pes cavus, and tremor. Am. J. Hum. Genet. 80, 345–352 (2007).
Zou, Y. et al. Mutation in CUL4B, which encodes a member of cullin-RING ubiquitin ligase complex, causes X-linked mental retardation. Am. J. Hum. Genet. 80, 561–566 (2007).
Vulto-van Silfhout, A. T. et al. Variants in CUL4B are associated with cerebral malformations. Hum. Mutat. 36, 106–117 (2015).
Liu, L. et al. Essential role of the CUL4B ubiquitin ligase in extra-embryonic tissue development during mouse embryogenesis. Cell Res. 22, 1258–1269 (2012).
Jiang, B. et al. Lack of Cul4b, an E3 ubiquitin ligase component, leads to embryonic lethality and abnormal placental development. PloS ONE 7, e37070 (2012).
Chun-Yu, C. et al. Rescue of the genetically engineered Cul4b mutant mouse as a potential model for human X-linked mental retardation. Hum Mol. Genet. 21, 4270–4285 (2012).
Qian, Y. et al. The CUL4B/AKT/beta-catenin axis restricts the accumulation of myeloid-derived suppressor cells to prohibit the establishment of a tumor-permissive microenvironment. Cancer Res. 75, 5070–5083 (2015).
Li, P. et al. Lack of CUL4B in adipocytes promotes PPARgamma-mediated adipose tissue expansion and insulin sensitivity. Diabetes 66, 300–313 (2017).
Kopanja, D. et al. Cul4A is essential for spermatogenesis and male fertility. Dev. Biol. 352, 278–287 (2011).
Yin, Y. et al. The E3 ubiquitin ligase Cullin 4A regulates meiotic progression in mouse spermatogenesis. Dev. Biol. 356, 51–62 (2011).
Zhao, X. et al. Zebrafish cul4a, but not cul4b, modulates cardiac and forelimb development by upregulating tbx5a expression. Hum. Mol. Genet. 24, 853–864 (2015).
Wu, R. S. et al. A rapid method for directed gene knockout for screening in G0 zebrafish. Dev. Cell 46, 112–125 e114 (2018).
Varshney, G. K. et al. High-throughput gene targeting and phenotyping in zebrafish using CRISPR/Cas9. Genome Res. 25, 1030–1042 (2015).
Jowett, T. & Yan, Y. L. Double fluorescent in situ hybridization to zebrafish embryos. Trends Genet. 12, 387–389 (1996).
Thisse, B. & Thisse, C. In situ hybridization on whole-mount zebrafish embryos and young larvae. Methods Mol. Biol. 1211, 53–67 (2014).
McReynolds, L. J., Gupta, S., Figueroa, M. E., Mullins, M. C. & Evans, T. Smad1 and Smad5 differentially regulate embryonic hematopoiesis. Blood 110, 3881–3890 (2007).
Danilova, N., Sakamoto, K. M. & Lin, S. Ribosomal protein L11 mutation in zebrafish leads to haematopoietic and metabolic defects. Br. J. Haematol. 152, 217–228 (2011).
Wardle, F. C. et al. Zebrafish promoter microarrays identify actively transcribed embryonic genes. Genome Biol. 7, R71 (2006).
Bennett, C. M. et al. Myelopoiesis in the zebrafish, Danio rerio. Blood 98, 643–651 (2001).
Arinobu, Y. et al. Reciprocal activation of GATA-1 and PU.1 marks initial specification of hematopoietic stem cells into myeloerythroid and myelolymphoid lineages. Cell Stem Cell 1, 416–427 (2007).
Qian, F. et al. Distinct functions for different scl isoforms in zebrafish primitive and definitive hematopoiesis. PLoS Biol. 5, e132 (2007).
Lecuyer, E. & Hoang, T. SCL: from the origin of hematopoiesis to stem cells and leukemia. Exp. Hematol. 32, 11–24 (2004).
Porcher, C., Chagraoui, H. & Kristiansen, M. S. SCL/TAL1: a multifaceted regulator from blood development to disease. Blood 129, 2051–2060 (2017).
Vagapova, E. R., Spirin, P. V., Lebedev, T. D. & Prassolov, V. S. The Role of TAL1 in hematopoiesis and leukemogenesis. Acta Nat. 10, 15–23 (2018).
Deleuze, V. et al. TAL-1/SCL and its partners E47 and LMO2 up-regulate VE-cadherin expression in endothelial cells. Mol. Cell. Biol. 27, 2687–2697 (2007).
Muroyama, Y., Fujiwara, Y., Orkin, S. H. & Rowitch, D. H. Specification of astrocytes by bHLH protein SCL in a restricted region of the neural tube. Nature 438, 360–363 (2005).
Jin, S. et al. Splicing factor SF3B1K700E mutant dysregulates erythroid differentiation via aberrant alternative splicing of transcription factor TAL1. PloS ONE 12, e0175523 (2017).
Patterson, L. J. et al. The transcription factors Scl and Lmo2 act together during development of the hemangioblast in zebrafish. Blood 109, 2389–2398 (2007).
Ge, L. et al. TET2 plays an essential role in erythropoiesis by regulating lineage-specific genes via DNA oxidative demethylation in a zebrafish model. Mol. Cell. Biol. 34, 989–1002 (2014).
Hu, H. et al. CRL4B catalyzes H2AK119 monoubiquitination and coordinates with PRC2 to promote tumorigenesis. Cancer Cell 22, 781–795 (2012).
Ji, Q. et al. CRL4B interacts with and coordinates the SIN3A-HDAC complex to repress CDKN1A and drive cell cycle progression. J. Cell Sci. 127, 4679–4691 (2014).
Yang, Y. et al. CRL4B promotes tumorigenesis by coordinating with SUV39H1/HP1/DNMT3A in DNA methylation-based epigenetic silencing. Oncogene 34, 104–118 (2015).
Wang, Y. et al. CUL4A induces epithelial-mesenchymal transition and promotes cancer metastasis by regulating ZEB1 expression. Cancer Res. 74, 520–531 (2014).
Acknowledgements
We thank all members of Dr M.S. lab for helpful advice on WISH and ChIP in zebrafish. We sincerely acknowledge Prof. Yiyue Zhang to provide Gata1-EGFP transgenetic zebrafish and c-myb, runx1, gata1, and pu.1 clones, Prof. Weijun Pan for kindly providing gata1, pu.1, runx1, and c-myb expression clones, Prof. Anming Meng for scl, lmo2, and hbbe3 clones, and Dr Kanki and Travis Maslow for mpo clone. This projected was supported by the Natural Science Foundation of Shandong Province (ZR2016HZ01 to Y.G.); the Key Research and Development Program of Shandong Province (2016ZDJS07A08 to Y.G.); the Foundation for Outstanding Young Scientist in Shandong Province (BS2014SW004), and China Postdoctoral Science Foundation to H.H.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by D. Guardavaccaro
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yang, F., Hu, H., Liu, Y. et al. Cul4a promotes zebrafish primitive erythropoiesis via upregulating scl and gata1 expression. Cell Death Dis 10, 388 (2019). https://doi.org/10.1038/s41419-019-1629-7
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41419-019-1629-7