Abstract
Planar cell polarity (PCP) and intercellular junctional complexes establish tissue structure and coordinated behaviors across epithelial sheets. In multiciliated ependymal cells, rotational and translational PCP coordinate cilia beating and direct cerebrospinal fluid circulation. Thus, PCP disruption results in ciliopathies and hydrocephalus. PCP establishment depends on the polarization of cytoskeleton and requires the asymmetric localization of core and global regulatory modules, including membrane proteins like Vangl1/2 or Frizzled. We analyzed the subcellular localization of select proteins that make up these modules in ependymal cells and the effect of Trp73 loss on their localization. We identify a novel function of the Trp73 tumor suppressor gene, the TAp73 isoform in particular, as an essential regulator of PCP through the modulation of actin and microtubule cytoskeleton dynamics, demonstrating that Trp73 is a key player in the organization of ependymal ciliated epithelia. Mechanistically, we show that p73 regulates translational PCP and actin dynamics through TAp73-dependent modulation of non-musclemyosin-II activity. In addition, TAp73 is required for the asymmetric localization of PCP-core and global signaling modules and regulates polarized microtubule dynamics, which in turn set up the rotational PCP. Therefore, TAp73 modulates, directly and/or indirectly, transcriptional programs regulating actin and microtubules dynamics and Golgi organization signaling pathways. These results shed light into the mechanism of ependymal cell planar polarization and reveal p73 as an epithelial architect during development regulating the cellular cytoskeleton.
Similar content being viewed by others
Introduction
The specific orientation of cells within the plane of the tissue, named planar cell polarity (PCP), is an essential feature of animal tissues1. PCP signaling is required for polarized beating of motile cilia in a variety of tissues2, including multiciliated ependymal cells (ECs), which carpet the wall of the lateral ventricles3,4. ECs, which are established perinatally from radial glial cells (RGCs) in a multistep process, display two types of PCP, rotational (rPCP) and translational (tPCP). display two types of PCP, rotational (rPCP) and translational (tPCP), which are established perinatally from radial glial cells (RGCs) in a multistep process5. rPCP is defined by the unidirectional orientation of the motile cilia within the cell and is coordinated at tissue-level. tPCP initiates in RGCs when their primary cilium is asymmetrically displaced3,6. Postnatally, immature multiciliated ECs displace their cilia clusters toward the anterior apical surface4.
PCP is regulated by asymmetric signaling through core and global regulatory modules. PCP-core module includes Frizzled (Fzd3, 6), Van Gogh-like (Vangl1/2), cadherin epidermal growth factor (EGF)-like laminin G-like seven-pass G-type receptor (Celsr1-3), Dishevelled (Dvl1-3), and Prickle (Pk1-4). In rodents, asymmetric localization of the PCP-core complexes is required for the rotational orientation of basal bodies (BBs) in multiciliated cells3,7,8,9,10,11,12. There is an initial polarization of the apical microtubule (MT) cytoskeleton, which induces the asymmetric distribution of PCP-core complexes at apical junctions13. These, in turn, will communicate polarity information to ciliary BBs distributed within the sub-apical cytoskeleton networks12,14. PCP-core signaling regulates the rotational, but not the translational, polarity in ECs15. In these cells, activation of the actin-binding protein non-muscle myosin-II (NMII) by the myosin light chain kinase (MLCK) is essential for tPCP establishment15. Thus, both actin and MT networks are key factors for setting up PCP in ECs.
The causal relationship between ciliogenesis and PCP signaling is not fully deciphered16. In this regard, the Trp73 gene is a key player in the organization of multiciliated cells17,18,19 and in EC polarity17,20. p73 belongs to the p53 family of transcription factors and generates functionally different TA and DNp73 isoforms21. Neither p73 mechanisms underlying PCP regulation nor the responsible p73-isoform, have been addressed.
In this work we analyzed the subcellular localization of PCP-regulatory proteins in ECs and the effect of Trp73 gene loss on their localization. p73 deficiency resulted in loss of PCP-core signaling complex asymmetry and lack of translational and rotational polarity in ECs. We demonstrate that p73 regulates PCP, at least in part, through TAp73-modulation of NMII activity via MLCK transcriptional regulation and regulation of MT dynamics signaling pathways.
Materials and methods
Animal care, genotyping, and isolation of wholemounts
All animal experiments were carried out in accordance with European (European Council Directive 2010/63/UE) and Spanish regulations (RD 53/2013) as well as institutional animal ethical guidelines. Mice heterozygous for Trp73 on a mixed background C57BL/6 × 129/svJae22 were backcrossed to C57BL/6, at least five times, to enrich for C57BL/6 background. Heterozygous animals were crossed to obtain the Trp73−/− mice (p73KO from now on). DNp73 and TAp73 mutant mice have been described before23,31. Male and female mice, obtained from multiple litters, were used for the experiments. Genotyping of animals was performed by PCR analysis as described before22,23,24.
For wholemount (WM) isolation, animals were euthanized and brains were dissected and placed in cold 0.1 M phosphate-buffered saline. Dissections of the lateral wall of the lateral ventricles were performed under the stereomicroscope as described before6.
Cell culture
Mouse wild type (WT) and p73KO-induced pluripotent stem cells (iPSCs) were cultured on feeder cells as described before25. For transfection assays, iPSCs were subcultured once without feeders in plates coated with Matrigel (Corning, NY, USA) and then seeded (3.7 × 104 cells/cm2) on coverslips (coated with Matrigel). Cells were transfected with the indicated plasmids using Lipofectamine™2000 Transfection Reagent (Invitrogen, Carlsbad, CA, USA) and after 24 h were fixed with 3.7% paraformaldehyde (PFA) for 15 min at room temperature (RT). For MLCK recovery assays, p73KO-iPSCs were transfected with a constitutively active MLCK expression vector (pSLIK-CA-MLCK) (Addgene plasmid # 84647). After 24 h cells were treated with doxycycline (400 ng/mL) to induce MLCK expression and were analyzed 24 h later.
HA-TAp73β-Saos-2-Tet-On cells26, (inducible TAp73-Saos-2 cells from now on) kindly provided by Dr. Karen Vousden (Cancer Research UK Beatson Institute, Glasgow), were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum (FBS), 2 mM l-glutamine, and 100 U/mL penicillin–0.1 mg/mL streptomycin. To induce TAp73 expression, cells seeded on coverslips for 24 h (3 × 104 cells/cm2) were treated with 2.5 μg/mL of doxycycline for 48 h27. Then, fixation was carried out with 3.7% PFA at RT during 15 min.
The human cell line HCT116 was cultured in RPMI supplemented with 10% FBS, 2 mM l-glutamine, and 100 U/mL penicillin–0.1 mg/mL streptomycin.
Luciferase assays
HCT116 cells were seeded in 24-well plates (3 × 105 cell/cm2) and transfected with 0.125 μg of the luciferase reporter pGL3-hMLCK(1Kb)-luc (kindly provided by Dr. Jerrold Turner, University of Chicago)28, 0.0625 μg pRLNull renilla, and 0.6 or 0.8 μg of the indicated expression vectors. Transfection was performed using X-tremeGENETM HP DNA Transfection Reagent (Roche, Basel, Switzerland).
Saos-2 were transfected with empty pcDNA3.1 or containing p73 isoforms and a Firefly luciferase reporter containing the putative WT TAp73-binding sequence (WT, Table S2) as identified by chromatin immunoprecipitation (ChIP)-seq29 or the same sequence lacking the predicted TAp73-binding motives (Mutated, Table S2). Transfection efficiency was controlled by co-transfecting a Renilla TK luciferase vector. Luciferase activity was always assayed 24 h after transfection using Dual luciferase assay. Firefly luciferase values were normalized to the corresponding Renilla luciferase levels.
Western blot analysis
Western blotting was performed as previously described30 and is detailed in the Supplementary Experimental Procedures.
Immunostaining and image analysis
Immunostaining protocols are described in the Supplementary Experimental Procedures. WM samples were analyzed using the Zeiss LSM800 and Olympus FluoView FV10i confocal laser scanning microscopes. iPSCs and inducible-TAp73-Saos-2 samples were analyzed using a Zeiss LSM800 Confocal Laser Scanning Microscope and a Nikon Eclipse TE2000 Confocal Microscope. Confocal z-stack images were taken in all cases. Images were processed using ZEN blue software, FV10-ASW 2.1 viewer software, EZ-C1 software, and ImageJ software.
Morphometric analysis
For the quantification of BB displacement in RGCs (P1) and in immature ECs (P15 or P21), at least three independent mice from each genotype were considered, counting at least three non-overlapping fields (50 μm × 50 μm) of each animal. Image analysis was performed using ImageJ software, quantifying the displacement by measuring a vector from the cell center to the BB (P1) or to the patch centroid (P15/P21).
For the quantification of number and intensity of p-MLC2 membrane dots, five independent mice from each genotype were considered, counting six non-overlapping fields (1 cell/field; ±20 μm × 20 μm) of each animal. Single plane images were obtained from z-stacks. Image analysis was performed using ZEN blue software, analyzing number and intensity of p-MLC dots in membrane region.
For the quantification of BB docking, three independent adult mice from each genotype were considered, counting two non-overlapping fields of each animal (50 μm × 50 μm; ±40 cells per animal). Orthogonal view of z-stack image analysis was performed with ZEN blue software, counting the number of cells with BBs aligned with the plasma membrane and cells with BBs scattered within the cytoplasm.
For the quantification of rotational PCP, a combined staining of FGFR1 Oncogene Partner (FOP) and γ-tubulin was used to delineate cilia polarity3. The FOP signal is adjacent to that of γ-tubulin at the side opposite to the basal foot. A vector that represents the direction of the individual cilia beat was defined by the direction from FOP (blue) to γ-tubulin (red) dots, using ImageJ software and all data were transformed to positive angles. The angle of polarity formed by this vector with respect to a horizontal axis was drawn. For each cell, a mean rotational angle and the circular standard deviation (CSD) were calculated with Oriana software. Five independent mice from each genotype were considered, counting three non-overlapping fields of each animal (50 μm × 50 μm; ±21 cells per animal).
For the quantification of Vangl2 and Frizzled localization at the plasma membrane, confocal z-stack images were taken before and after transiently transfecting the indicated expression vectors into p73KO-iPSCs. Single-plane images were obtained from z-stacks. Gaussian Blur filter (σ = 10) was applied and then images were binarized using automatic threshold. The midline along the membrane was created using Skeletonize Option. This network was dilated (15 iterations) to create the plasma membrane selection. which was overlapped with Vangl2 and Frizzled3 staining and the mean fluorescence intensity was measured.
RNA isolation and real-time quantitative reverse transcription-PCR analysis
Total RNA from cultured cells or dissected WMs was isolated using TRI reagent (Ambion, TX, USA). Kidney, bladder, and retina total RNA were isolated using RNeasy mini Kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. cDNA was synthesized using up to 2 μg of total RNA and the High Capacity RNA-to-cDNA kit (Applied Biosystems, Carlsbad, CA, USA). Gene expression was detected by real-time quantitative reverse transcription-PCR (qRT-PCR) in a StepOnePlus Real-Time PCR System (Applied Biosystems, Carlsbad, CA, USA) using FastStart Universal SYBR Green Master (Roche, Basel, Switzerland). qRT-PCR conditions were described before31. mRNA expression levels were expressed as 2ΔCt (ΔCt = Ct internal reference − Ct gene), normalized to 18S mRNA expression. Primer sequences were as follows: 18S mouse gene32: F 5′-AGTTCCAGCACATTTTGCGAG-3′ and R 5′-TCATCCTCCGTGAGTTCTCCA-3′; Mlck mouse gene (Primer Bank Database ID: 29650205a1): F 5′-TGGGGGACGTGAAACTGTTTG-3′ and R 5′-GGGGCAGAATGAAAGCTGG-3′; TAp73 mouse gene F 5′-GCACCTACTTTGACCTCCCC-3′ and R 5′-GCACTGCTGAGCAAATTGAAC-3′, and Eb3 mouse gene F 5′-ACTAGAACACGAGTACATCCACA-3′ and R 5′-CCATCATAGTTTGCGTCAAAGA-3′. 18S human gene: F 5′-GGCGCCCCCTCGATGCTCTTA-3′ and R 5′-GCTCGGGCCTGCTTTGAACAC-3′; MLCK human gene: F 5′-CACCGTCCATGAAAAGAAGAGTAG-3′ and R 5′-GAGAGGCCCTGCAGGAAGATGG-3′; TAp73 human gene: F 5′-GCACCTACTTCGACCTTCCC-3′ and R 5′-GTGCTGCTCAGCAGATTGAAC-3′; EB3 human gene: F 5′-GCCAATGATGCCTGAGATAACAG-3′ and R 5′-GCAGAGGGTTGTAATCCTTTCCA-3′.
Sequence analysis and ChIP
In silico prediction of putative p73-response elements within the mouse Mlck gene was performed using the open-access database for eukaryotic transcription factor-binding profiles JASPAR33. Matrix models MA0861.1 and MA0525.1 were applied for the mouse Mlck gene and for the analysis of the ChIP-seq peak detected in the human MLCK gene, respectively. Only the sites with a score over 9.0 were selected for further in vitro analysis.
ChIP analyses were carried out as previously described25 and are detailed in the Supplementary Experimental Procedures.
Gene ontology analysis
Gene ontology (GO) analysis was carried out using selected genes from GSE15780. DAVID Bioinformatics Resources 6.834,35 and PANTHER™ GO slim (version 13.1, released 2018-02-03)36 were used for the analysis. The results were plotted using the ggplot2 library from R (3.5.1) Core Team (2014), a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL http://www.R-project.org/.
Statistical analysis
Statistical analysis and Figure generation were performed with GraphPad Prism 7.04 software (Figs. 1–3, 6, 7) and Oriana 4 Software (Fig. 4). Data are expressed as means ± standard error (S.E.) of the mean. Differences were considered significant when p < 0.05 (*p < 0.05, **p < 0.01, ***p < 0.001). To test for normal distribution D’Agostino and Pearson tests were performed and non-parametric assumptions were made when necessary. In that case, the means of two experimental groups were compared with Mann-Whitney’s test; otherwise, unpaired two-tailed Student’s t-test was applied. In case of multiple comparisons, statistical differences were evaluated using either analysis of variance or Kruskal-Wallis test with Dunn’s multiple comparisons. To compare docking between WT and p73KO-ECs, a contingency analysis (Fisher’s exact test) was performed. Data in Fig. 4 are graphically represented using Oriana software (Kovach Computing Services, Anglesey, Wales) and statically compared by circular analysis (Watson’s U2 test).
a–g Wholemounts (WMs) from WT, p73KO (a–e) or TAp73KO (f, g) LW were stained with the indicated antibodies and confocal z-stack images were generated. WMs were stained with anti-γ-tubulin (red, basal bodies), p-MLC (green), and β-catenin (blue, cell contour, red in g). (a, a′) Comparative quantification of BB displacement in P1-RGCs (a) showing 10–30 mm2 apical surface or P15-ECs (a′) using ImageJ software. Scale bar: 5 µm. A vector of displacement was drawn from the cell center to the BB-cluster centroid. Bars represent mean values ± S.E. (RGCs: n = 24 cells from three mice per genotype; ECs: n = 15 cells from five mice per genotype; ***p < 0.001). b–e WMs from P1-RGCs (b) or P15-ECs (c–e) were stained with anti-p-MLC to detect activated NMII. p-MLC and primary cilia BB colocalization are indicated by arrows. Magnification insert at the right most panel. Scale bar: 5 µm. c BB-clusters crescents and orientation are indicated by arrows. p-MLC polarized membrane localization is marked by a dotted box. Scale bar: 5 µm. d Orthogonal projections show p-MLC signal (green arrows) adjacent to BB (red arrows). Scale bar: 2 µm. e Quantification of p-MLC dot number (upper panel) and average dot intensity (lower panel) at the plasma membrane. Scale bar: 5 µm. Right graphs represent the frequency distribution of those values in the cellular population. Bars represent mean values ± SE (n = 15 cells from five mice per genotype). f, g Analysis and quantification of BB-clusters displacement (f) and p-MLC localization (g) in P21 TAp73KO-ECs. f BB-cluster displacement in WT cells is depicted by white arrows and lack of coordination in BB-cluster displacement in p73KO cells by yellow arrows. Scale bar: 5 µm. g Scale bar: 5 µm
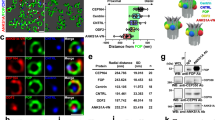
a TAp73 ChIP-seq genome tracks of the human MLCK locus (NG_029111.1). The transcriptional direction is marked with an horizontal arrow. TAp73 peak matches to intronic sequence upstream the exon that contains the ATG (region 70,706–70,883 bp from NCBI Reference Sequence: NG_029111.1). Localization of the identified response elements (RE) is indicated by an orange arrow (p53RE) and a pink arrow (p73RE enhancer) in the human MLCK gene, and by a green arrow (p73RE) in the mouse Mlck gene. b Luciferase reporter assays were performed with the indicated expression vectors together with the reporter constructs pGL3-hMLCK(1Kb)-luc, containing the human MLCK regulatory region with the p53RE (left panel), or p73RE-enh-MLCK-luc, containing a minimal promoter and the putative p73 enhancer element (p73RE, right panel). Mutated control reporter contains the same DNA region but lacks the TP73-binding motifs. Three independent experiments per reporter were performed in in triplicate p53-deficient cell lines: HCT116 (p53RE-MLCK-luc) and Saos-2 (p73RE-enh-MLCK-luc). Fold increase activity was calculated relative to control vector. Error bars represent standard error of the mean. Differences were considered significant when p < 0.05. c, d Direct binding of ectopic (c) or endogenous (d) TAp73 analyzed by ChIP assays that were carried out in inducible-TAp73-Saos2 cells (c, n = 3) or in mouse brain tissue (d, n = 3). The p53RE (c, left panel) and the p73RE enhancer element (c, right panel) of the hMLCK promoter region, or the p73RE of the proximal mouse Mlck promoter (d), were amplified by q-PCR. The data were normalized to input chromatin samples for each case and to IgG values = 1. Dox: doxycycline e, f Quantitative RT-PCR analysis of Mlck expression levels in WT and p73KO samples, from either ventricle LW of mice brains (e, n = 9 WT brains and n = 4 p73KO brains) or iPSC (e, n = 3 clones per genotype). f MLCK protein expression analysis in iPSCs by western blot. g, h Induction of Mlck analyzed by qRT-PCR (g) or western blot (h) in p73KO-iPSCs after ectopic expression of TAp73 or empty vector. Two independent transfection experiments were performed with two clones per genotype. qRT-PCR assays were repeated four times by duplicate. Bars represent mean values ± S.E. *p < 0.05, **p < 0.01, ***p < 0.001, n.s: non-significant
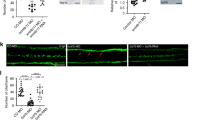
a, b Detection of activated NMII and actin cytoskeleton in WT vs. p73KO-iPSC colonies (a), or after ectopic TAp73 expression in p73KO-iPSCs (b). a Orthogonal projections from WT and p73KO-iPSC where arrows indicate F-actin and p-MLC colocalization at intercellular junctions. Scale bar: 10 µm. b p-MLC colocalization with cortical actin bundles is recovered (arrows) upon TAp73 transfection, but not vector control. Scale bar: 10 µm. c–f Confocal images of WT (c, e) and p73KO (d, f) P15 WMs displaying as indicated: actin cytoskeleton (Phalloidin), p-MLC (green), and BBs alone (γ-tubulin, red) or with the cell membrane (β-catenin and γ-tubulin, blue). The white arrows point to cortical actin; the white arrowheads indicate apical actin lattices and blue arrowheads mark the sub-apical actin filaments. Scale bar: 5 µm. e, f p-MLC puncta associated with actin lattice around BBs or with plasma membrane are marked by white arrowheads and white arrows, respectively. f Yellow arrowheads and yellow arrows indicate the lack of such p-MLC association. Scale bar: 5 µm. (g, g′) BB docking was analyzed in WT and p73KO (g) or TAp73KO (g′) ECs. Lateral views of WM confocal images labeled with anti-γ-tubulin (red) and β-catenin (blue). Scale bar: 5 µm. Graphs represent the percentage of cells with correct docking (green bars) versus cells with incomplete alignment (purple bars) within the indicated genotypes. Contingency analysis (Fisher’s exact test) was performed to evaluate statistical differences (g: n = 125 WT cells and 140 p73KO cells; g': n = 117 WT cells and 127 TAp73KO cells; images from three mice per genotype were analyzed ; ***p < 0.001)
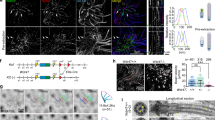
a–i Representative confocal images of WMs from WT, p73KO (a, b, c–f, h), TAp73KO (b′, g, i) and DNp73KO (b″) mice stained with the indicated antibodies. b, b″ Immunostaining against α-tubulin (red) was used to visualize the cilia axoneme. Scale bar: 10 µm. Parallel and organized bundles of cilia in WT and DNp73, but disorganized in p73KO and TAp73KO-ECs are marked by yellow arrows. White arrows indicate cells lacking cilia axoneme. c, d To delineate cilia polarity and quantified rPCP, combined staining of FGFR1 Oncogene Partner (FOP, blue) and γ-tubulin (red) on WT and p73KO WMs was performed. Scale bar: 5 µm. The direction of each BBs was drawn by a vector (white arrows) from FOP-dot to γ-tubulin-dot within a BB. An angle of polarity, named rotational angle (RA), was drawn with respect to the horizontal axis for each BB and the mean RA for each cell was determined. d Mean vectors and circular standard deviation (CSD) of RAs around the mean in WT and p73KO were calculated (n = 105 cells from five mice per genotype). Data were graphically represented using Oriana software and statistically compared by circular analysis (Watson’s U2 test). e–g Representative confocal images of WMs from the indicated genotype stained for α-tubulin to mark MTs (e, green) and for EB3 to determine MT polarization (e′, green; f, g, red). e Arrowheads indicate the MT-meshwork underlying BB-clusters. Scale bar: 5 µm. e′–g White arrows show a second polarized MT network located at anterior intercellular junctions illustrating their coordinated orientation. Scale bar: 5 µm. h, i WMs from WT and p73KO (h) or TAp73KO (i) mice were stained against Fat4 (green), γ-tubulin (red), and β-catenin (blue). Scale bar: 5 µm. h Fat4 polarized distribution was indicated by dashed arrows. Orthogonal projections show the close association of Fat4 and the BBs indicated by a white arrow
a–c WMs from WT, p73KO (a), or TAp73KO (a′) were stained for PCP-core proteins Vangl2 (a, a′, b, green), Celsr1 (b, blue), and Prickle (c, green). Scale bar: 10 µm. d–f ECs were stained with anti-γ-tubulin (d, e, red) and α-tubulin (f, red) to visualize the BBs and axoneme, respectively. PCP-effectors were marked by anti-Dvl2 (d, green) and anti-Rac1 (e, green; f, blue). Orthogonal projections (d, e) show Dvl2 and Rac1 localization, indicated either by white arrows (WT) or yellow arrows (p73KO). Scale bar: 5 µm
a–e Representative confocal images of WT and p73KO-iPSC, transfected with the indicated expression vectors (b–e). PCP-core proteins were visualized by staining with anti-Vangl2 (a, b, e, green), anti-Celsr1 (a, e, blue), anti-Fzd3 (a, c, green), and anti-Prickle2 (a, green), and plasma membrane by β-catenin (a,c, red; b, blue). b, c Transfected TAp73 and DNp73 (b, light blue; c, red) were visualized using specific antibodies. Scale bar: 10 µm. d Vangl2 and Fdz3 localization at the plasma membrane was quantified by ImageJ software. Graphs represent the mean florescence intensity (MFI). Two independent transfection experiments were performed with two clones per genotype. Five pictures from each experiment were randomly selected and quantified (n = 20 pictures per set). Bars represent mean values ± S.E. Kruskal-Wallis test together with Dunn’s multiple comparisons test was performed to evaluate statistical differences; **p < 0.01, ***p < 0.001, n.s: non-significant. e Ectopic expression of TAp73 restores Celsr1 (blue) and Vangl2 (green) colocalization. Scale bar: 10 µm
a WT and p73KO-iPSCs were treated with MLCK inhibitor, ML7, and p-MLC (a) or Vangl2 (b) localization was analyzed by confocal microscopy. a' (bold font) The effect of ML7 treatment over p-MLC levels was confirmed by western blot b' Experiments to quantify Vangl2 localization at the plasma membrane were performed as in Figure 6.b. c, d p73KO-iPSCs were transfected with inducible constitutively active MLCK (CA-MLCK); VENUS reporter expression shown in light blue) and upon MLCK induction, p-MLC (c) and Vangl2 (d) colocalization with F-actin at the cellular cortex was analyzed by confocal microscopy. Scale bar: 20 µm
Results
p73 coordinates tPCP establishment through the regulation of polarized NMII activity
To address tPCP in transforming RGCs3, we quantified primary cilia displacement (CD) from the cell center in WMs from P1 WT and p73KO lateral wall (LW) of the lateral ventricle. (Fig. 1a, white arrows). We found reduced CD in p73KO-RGCs. Thus, in the absence of p73, tPCP establishment at single-cell level is either delayed or defective. We next analyzed the effect of p73 deficiency on NMII activation and localization, both essential for tPCP establishment15. In WT-RGCs, activated NMII (p-MLC) was located in distinct puncta at the plasma membrane (Fig. 1b), colocalizing with the primary cilia BB (inserts, white arrows). At the same stage, p73KO-RGCs, with defective CD, have lower levels of p-MLC, especially at the plasma membrane (Fig. 1b), altogether confirming that NMII activation is associated with tPCP establishment.
In immature ECs, BBs packed tightly during migration and get arranged into clusters, termed “crescents”, coordinately displaced toward the anterior apical surface (Fig. 1c, arrows). p-MLC was closely associated to BB within crescents (Fig. 1c, dotted box, Fig. 1d, arrows), advocating the idea that NMII activation plays a fundamental role in BB polarized migration15. p73KO-ECs had less CD, crescents were never detected (Fig. 1a, c), and their BBs were scattered throughout the apical surface (Fig. 1c, solid arrows) with little p-MLC associated to them (Fig. 1d, green and red arrows and 1e, arrowheads). p-MLC also localize in puncta at membrane junctions, polarized in the same direction of the BB-cluster (Fig. 1c, dotted box), which are reduced in p73-deficient cells (Fig. 1e), highlighting the correlation between p73 expression and NMII activation and localization.
Analysis of immature ECs from TAp73KO brains, lacking only TAp73 isoforms31, revealed a significant decrease in BB-cluster displacement (Fig. 1f) in consonance with their low p-MLC levels (Fig. 1g). Moreover, CD in neighboring TAp73KO-ECs followed different directions, confirming the lack of tissular coordination (Fig. 1f, yellow arrows). Analysis of DNp73KO-ECs, lacking DNp73 isoforms23, did not reveal defects in BB-cluster displacement and orientation, or NMII activation and localization (Supplementary 1a,b), indicating that TAp73, but not DNp73, plays a key role in tPCP establishment.
TAp73 could regulate tPCP, at least in part, by controlling NMII activity via the regulatory kinase MLCK15. A proximal p53 response element (p53RE) had been previously reported within the human MLCK promoter37 (Fig. 2a). Moreover, ChIP-seq analysis of TAp73-transfected Saos-2 cells29 revealed a TAp73 peak, which could correspond to a putative enhancer element (Fig. 2a, p73RE-enh). We performed luciferase and ChIP assays with the identified p53 and p73REs (Fig. 2b, c). Transcriptional analysis in HCT116 cells with the p53RE-MLCK-luc reporter28 (Fig. 2b, left panel), and in Saos-2 cells with the p73RE-enh-MLCK-luc (right panel), demonstrated that TAp73 can significantly activate both reporters. Under the same conditions, DNp73 had a weak transactivating activity on the proximal p53RE but none over the enhancer. In inducible-TAp73-Saos-2 cells26 TAp73 induction correlated with increased MLCK expression (Supplementary 2a) and ChIP assays revealed a significant TAp73-interaction with both sites (Fig. 2c). To demonstrate the relevance of this interaction in a physiological context, we searched for p73/p53REs (JASPAR Core Vertebrata Database) in the murine Mlck gene and predicted a strong p73RE within the proximal promoter (−1741 bp from Exon1*38, Fig. 2a). ChIP assays showed a significant interaction of endogenous TAp73 with this site in WT, but not in p73KO brain tissue (Fig. 2d). p73-deficiency correlated with lower Mlck levels in brain (Fig. 2e and Supplementary 2b), in iPSCs25 (Fig. 2f) and in retina and kidney, but not in bladder (Supplementary 2c–e). Moreover, TAp73 overexpression in p73KO-iPSCs significantly induced Mlck levels (Fig. 2g, h). These data, as a whole, uncover MLCK as a direct TAp73-transcriptional target.
MLCK influences cortical localization of NMII, which functions as a cortical organizer to concentrate E-cadherin to zonula adherens39. Thus, p73-deficiency might affect the establishment of epithelial cell junctions. To address this, we used iPSCs, a model with epithelial-like cell junctions25,40. In WT-iPSCs, p-MLC is strongly expressed at intercellular junctions localizing with F-actin (Fig. 3a, arrows). p73KO-iPSCs, with an impaired epithelial phenotype25, had less and mostly cytoplasmic p-MLC (Fig. 3a and Supplementary 3a), not colocalizing with F-actin. TAp73 overexpression induced NMII phosphorylation (Supplementary 3b), restoring p-MLC levels and localization at plasma membrane in p73KO-iPSCs (Fig. 3b, arrows). In agreement, upon TAp73 induction, p-MLC increases and colocalizes with cortical actin bundles41 (Supplementary 3c, arrows), highlighting the functional interaction of TAp73, NMII activity, and the actin cytoskeleton.
To address the functional linkage between p73 and the actin cytoskeleton in vivo, we examined F-actin distribution in ECs. These cells displayed cortical F-actin staining3 (Fig. 3c, white arrow), a polarized and organized actin apical lattice between the BBs (orthogonal projections, white arrowheads), and a set of sub-apical actin filaments underneath the BBs (blue arrowheads). p-MLC puncta appeared to be associated to the actin lattices within the BBs, as well as with the cortical actin anchored to the plasma membrane (Fig. 3e, white arrowheads and arrows, respectively), suggesting that activated NMII have a key role orienting and keeping together the actin meshwork and, concomitantly, the BBs clusters. Consistent with this hypothesis, p73-deficient cells, with less p-MLC, lacked the organized apical actin lattices associated with BBs and the sub-apical actin filaments (Fig. 3d). Moreover, p-MLC puncta were not associated to the BB cluster or polarized at the plasma membrane (Fig. 3f, yellow arrowheads and arrows, respectively).
Thus, p73-deficiency results in blunted NMII activation leading to a failure in the actin-network assembly, implicated in BB apical migration, docking42, and spacing43. In agreement, BBs were clearly identified and aligned with the plasma membrane in WT-ECs (Fig. 3g, g'), but p73KO-ECs (Fig. 3g) or TAp73KO-ECs (Fig. 3g′) had BBs scattered and deep within the cytoplasm, demonstrating TAp73 essential role in sub-apical cytoskeleton dynamics and BB docking.
TAp73 regulates the polarized junctional MT assembly promoting PCP-core complex asymmetric localization
p73 deficiency not only affected BB-cluster displacement, docking, and spacing, but also BB orientation (Fig. 4a). WT-ECs displayed well-organized and straight parallel bundles of cilia, while p73KO (Fig. 4b) and TAp73KO-ECs (Fig. 4b′) had disorganized cilia with a “disheveled” appearance (yellow arrows). Similar cilia alterations have been attributed to BBs’ rotational alignment defects9,11. Thus, we measured rotational polarity of individual BBs by calculating an angle of polarity formed by a vector (marking the orientation of each BB) and the horizontal axis3 (Fig. 4c). Next, we determined the mean vector for each EC and calculated the CSD (Fig. 4d). This value was significantly larger in p73KO than in WT cells (p < 0.001), demonstrating p73 requirement for rPCP. Only few TAp73KO-ECs lacked cilia axoneme (Fig. 4b′, white arrows), but we did not detect ciliary axoneme defects in DNp73KO-ECs, which displayed well-organized and parallel bundles of cilia and well-oriented BBs (Fig. 4b″ and Supplementary 1b).
p73 regulation of actin dynamics, by itself, could not fully explain the generation of refined rPCP40. Thus, we expanded our analysis to the MT cytoskeleton, since polarized MT dynamics is fundamental for rPCP3,12,13. α-tubulin staining revealed a fine and polarized MT-meshwork (Fig. 4e, arrowhead), with similar disposition to the actin lattice within BB (Fig. 3c). In p73KO-ECs this meshwork was lax and disorganized (Fig. 4e, arrowhead). To address MT-growth orientation, we analyzed EB3, a plus-end MT protein, which marks newly synthesized MTs. In WT cells, a second MT network grew asymmetrically from the center of the cell toward the anterior region of the apical cell cortex contacting the plasma membrane (Fig. 4e′, arrow). These MT-anchoring points3,12,44 were polarized at tissue-level (Fig. 4f, g, arrows). Interestingly, p73 deficiency resulted in lower and diffuse levels of EB3 staining around the scattered BBs, and a total absence of polarized MT-anchoring points at cell junctions (Fig. 4e–g), demonstrating TAp73 requirement for polarized MT network formation in developing ECs.
The protocadherins Fat (Fat4 in mammals4) and its ligand Dachsous (Ds) conform the global-PCP module, known to control the polarized junctional alignment and asymmetry of MT13,45. Confocal images of WT-ECs revealed Fat4 polarized distribution toward the anterior apical cell surface (Fig. 4h, dashed arrows). Orthogonal planes revealed that Fat4 was closely associated with the BBs (white arrow). p73KO-ECs had weak Fat4 staining, scattered throughout the cytoplasm without association with the BBs, lacking any asymmetric distribution (Fig. 4h, i).
Polarized MTs are required for targeting PCP proteins to the proper asymmetric membrane domains12,46. PCP-core molecular complexes formed by Vangl2, Celsr1, and Pk2 displayed asymmetric distribution in WT-ECs (Fig. 5a–c), with Vangl2 exhibiting the characteristic zigzag pattern3 indicative of tissular coordination of polarity. This asymmetric distribution was completely lost in p73KO-ECs (Fig. 5a–c) and Vangl2 had cytoplasmic localization (Fig. 5a, a′) suggesting a defect in Vangl2-vesicle trafficking.
PCP-core proteins transduce the polarity signals through the adaptor protein Dvl29,15 and its effector Rac147. Orthogonal projections demonstrated that Dvl2 was located at the base of the individual BBs (Fig. 5d, white arrows) clustered at the anterior cell surface, establishing a polarized Dvl2-distribution. As expected, Rac1 was localized on top of the BBs (Fig. 5e, white arrows) at the base of the axoneme (Fig. 5f, white arrows). In p73 absence, Dvl2 did not associate with BBs (Fig. 5d, yellow arrows), and Rac1 was found scattered throughout the disheveled axonemes (Fig. 5e, f, yellow arrow), all in consonance with a lack of the polarized anchoring structures required to scaffold and orient the PCP signaling machinery.
TAp73 induces PCP-core protein membrane localization and modulates MT dynamics
TAp73 acts as a transcriptional integrator of multiciliogenesis18,19 and regulator of EC differentiation17. Therefore, it was possible that like in airway epithelial cells16, the defects observed in p73KO-ECs were a consequence of their defective differentiation and ciliogenesis impairment. To rule this out, we analyzed PCP-core protein localization in a ciliogenesis-independent model, the iPSCs. In WT-iPSCs, Vangl2, Celsr1, and Fzd3 localized at the membrane, with intense staining at the lateral junctions (Fig. 6a, white arrows). Prickle2 also had cortical localization, close to Celsr1 (Fig. 6a, yellow arrows). Lack of p73 resulted in cytoplasmic Vangl2, which had lost most of its colocalization with Celsr1, confirming p73 requirement for Vangl2 membrane localization. In p73KO-iPSCs Fzd3 had faint and diffuse cytoplasmic staining (Fig. 6a, arrowheads), while little Prickle2 localized at the cell cortex associated with Celsr1 (Fig. 6a, yellow arrowheads). To test whether p73 was sufficient for PCP-core protein membrane localization, we expressed TAp73 or DNp73 in p73KO-iPSCs and analyzed the restoration of Vangl2 and Fzd3 at the plasma membrane and Celsr1 colocalization. Confocal microscopy (Fig. 6b, c, e) and image quantification (Fig. 6d) revealed that TAp73, but not DNp73, could rescue the p73KO phenotype and demonstrated that TAp73 is necessary and sufficient for membrane localization of PCP-core proteins, independently of its function as multiciliogenesis regulator.
MLCK is required for the establishment of epithelial cell junctions, which are necessary for polarized transport. Moreover, in some systems MLCK mediates intracellular trafficking over MTs48. Therefore, we asked whether MLCK could be involved in PCP-core protein membrane transport. We treated iPSCs with the MLCK inhibitor ML715 and confirmed that upon treatment p-MLC went down, disappearing from intercellular junctions (Fig. 7a, a′). Concomitantly, Vangl2 membrane localization decreased (Fig. 7b, b′), partially phenocopying p73KO-iPSC. Furthermore, overexpression of constitutively active MLCK (CA-MLCK49) in p73KO-iPSCs resulted in an enhancement of p-MLC levels (Fig. 7c) and a partial restoration of PCP-core proteins membrane localization (Fig. 7d), indicating that MLCK/NMII signaling pathway is required for PCP-core proteins membrane delivery, but other factors are also involved.
To determine if a defect in MT dynamics was causing the observed p73KO-iPSCs defects, we asked whether Fat4, which promotes the formation of polarized MT networks, could rescue the phenotype, similarly to TAp73 overexpression13,50. p73KO cells displayed a loosely formed MT meshwork (Fig. 8a, white arrowheads); however, upon Fat4 or TAp73 expression, the MT cytoskeleton formed a tight cortical network at the proximity of the cell periphery (where PCP-core proteins were localized), resembling WT phenotype (Fig. 8a, yellow arrowheads). Concurrently, Vangl2 and Fzd3 levels at the plasma membrane significantly increased, approaching those of WT-iPSCs (Fig. 8a′). Thus, Fat4 was sufficient to rescue the MT disposition and restore PCP-core proteins membrane localization in p73KO-iPSCs, confirming that defective MT dynamics were an underlying cause of p73KO phenotype.
a Analysis of Vangl2 and Fzd3 membrane localization by confocal microscopy on WT and p73KO-iPSC, transfected with the indicated expression vector. Plasma membrane was visualized by β-catenin staining (red), MT by α-tubulin (red), Vangl2 (blue), and Fzd3 (green). The lax meshwork of MTs in p73KO is marked by white arrowheads while tight cortical networks are marked by yellow arrowheads. a′ ImageJ quantification of the amount of Vangl2 and Fdz3 localized at the plasma membrane. Graphs represent the mean florescence intensity (MFI). Two independent transfection experiments were performed with two clones per genotype. Five pictures from each experiment were randomly selected and quantified (n = 20 pictures per set). Bars represent mean values ± S.E. Kruskal-Wallis test together with Dunn’s multiple comparisons test was performed to evaluate statistical differences; *p < 0.05, ***p < 0.001, n.s: non-significant. b The Golgi apparatus was marked by GM130 (red) and in the magnification panel the contour of a single cell is delineated by a white line. c–g Analysis of MT cytoskeleton changes induced upon TAp73 expression (+Dox) in TAp73-Saos-2 cells stained with specific antibodies against TAp73 (c, red), Vangl2 (d–f, green), α-tubulin (d, g, blue), acetylated α-tubulin (e, red), EB3 (g, red), and Phalloidin (g, green). Nuclei are visualized by Dapi (c, e, f). a, b Scale bar: 10 µm; c–f Scale bar: 20 µm. h Plot representing GO PANTHER enrichment analysis of TAp73 peak-containing differentially expressed genes from Saos-2 cells with ectopic expression of TAp73 (GSE15780). GO Biological process complete was used as annotation data set. Proposed model of TAp73 regulation of PCP in ependymal cells through the regulation of microtubules and actin organization signaling pathways
MTs play a central role in the Golgi complex integrity and agents that alter MT distribution induce Golgi fragmentation51,52. As expected, cis-Golgi fragmentation was detected in p73KO-iPSCs (Fig. 8b). Next, we analyzed MT cytoskeleton and Vangl2 localization in inducible-TAp73-Saos-2 cells. Untreated cells, with very low levels of p73 (Fig. 8c), displayed a lax MT disposition and diffuse cytoplasmic Vangl2 staining (Fig. 8c–e). Upon TAp73 induction, changes in Vangl2 expression pattern, concurrently with changes in MTs disposition, were observed. These adopted a radial structure that radiated from a juxtanuclear cellular compartment where Vangl2 was enriched (Fig. 8d, arrowheads). This subcellular compartment accumulated acetylated-tubulin, which colocalized with Vangl2 (Fig. 8e, arrowheads) and Golgi marker GM130 (Fig. 8f). Therefore, it was consistent with Golgi-derived MTs, reported to preferentially accumulate acetylated-tubulin53 and indispensable for cell polarity54. Upon TAp73 induction we detected an increase on EB3 levels (Fig. 8g), confirmed by RT-PCR in inducible-TAp73-Saos-2 cells and p73KO-iPSCs (Supplementary Fig. 4a, b), all demonstrating that p73 could induce changes in MT dynamics. These changes were accompanied by a dramatic alteration in actin cytoskeleton and peripheral stress fiber architecture, readouts of MLCK enhancement55.
To determine whether TAp73 could induce transcriptional programs involved in these processes, we analyzed previously published TAp73β-ChIP-seq/RNA-seq data from Saos-2 cells (GSE15780)29. We used differentially expressed genes (DEGs) upon TAp73β expression that contained TAp73 peaks either at the promoter and/or at regulatory regions (within 25 kb). GO analysis identified terms like “Regulation of MT based processes”, “Regulation of MT polymerization”, “Post-Golgi vesicle–mediated transport”, and “Regulation of Actin cytoskeleton organization” as highly enriched biological processes (Fig. 8h and Supplementary 4c,d). These DEGs included key regulators of Golgi organization and MT dynamics like AKAP9 (AKAP 450), PDE4DIP, GOLGA1, or CLIP1 also required for cell polarity56,57. Among actin cytoskeleton regulators, we confirmed MYLK (MLCK) as a TAp73β-DEG. These results support that TAp73 modulates, directly and/or indirectly, transcriptional programs regulating actin and MT dynamics and Golgi organization.
Discussion
Polarity establishment relies on polarized actin and MT cytoskeletal rearrangements, but knowledge of the signals that organize these processes remains incomplete43. This work demonstrates an essential role of TAp73 in PCP through the regulation of both, actin and MT-polarized network assembly, providing a model to the mechanism of planar polarization regulated by TAp73 (Fig. 8h).
We show that TAp73, but not DNp73, is necessary for the efficient tPCP establishment in RGCs, suggesting that the initial mechanoreceptors signaling, which instruct primary CD, cannot work properly in the absence of p73. While TAp73KO-ECs show PCP- and BB-docking defects, cilia axoneme alterations are milder than in the total p73KO mice, suggesting that DNp73 could have a compensatory role. However, consistent with the lack of DNp73 expression in ECs23, we did not detect any cilia or PCP defects in DNp73KO-ECs. These data suggest that compensatory and redundant ciliary programs are induced in the absence of TAp73 when DNp73 is present, but not in total p73 deficiency. Nevertheless, these signaling programs do not compensate for PCP establishment in the absence of TAp73, uncoupling ciliogenesis, and PCP signaling in ECs.
Actin remodeling and the actin-based motor protein NMII are essential to establish tPCP15. Our results support a model in which NMII activation and membrane localization correlates with anterior migration of the BB crescent. Mechanistically, we identify TAp73 isoform as an upstream modulator of NMII through the transcriptional regulation of MLCK in vivo and in vitro. This directly links p73 with actin microfilament dynamics, essential for proper cilia docking and spacing, and for the establishment of global coordination of cilia polarity43,58.
PCP regulation in epithelial cells relies on the PCP-core and the Fat/Ds modules to generate morphological asymmetries59. PCP-core complexes transduce polarity signals through the adaptor protein Dvl29,15 and its effectors aPKC, Rac1, and RhoA, leading to reorganization of the actin cytoskeleton9. We show that Rac1 is localized at the base of the axoneme in a region corresponding to the ciliary pocket, a proposed interface with the actin cytoskeleton and a platform for vesicle trafficking60. These molecular complexes are not established in the absence of p73. We report for the first time in ECs4, the expression of the Fat/Ds-module components13,45 at the base of the cilia closely associated with the BBs, supporting its role in ciliary functions. Interestingly, p73-deficient cells displayed a defective global-PCP signaling that could be underlying, at least in part, the lack of polarized MT-junctional structures in p73KO cells.
Polarized MTs are an integral component of early steps in cell polarity driving asymmetric accumulation of PCP-core proteins45,61. The cytoplasmic distribution of Vangl2 in p73-deficient ECs suggests a possible defect in Vangl2-vesicle trafficking and points toward a possible role of p73 in MT dynamics. Moreover, disassembling of MTs by nocodazole treatment of brain ventricles impairs the trafficking and distribution of PCP-core proteins in ECs3, phenocopying the observed p73KO phenotype. Lack of p73 affected both sets of MT described in ECs. Such defects could impair PCP-core proteins traffic to the plasma membrane at the MT-anchoring points and the assembly of PCP-asymmetric complexes.
We demonstrate that TAp73 is necessary and sufficient for membrane localization of PCP-core proteins independently of multiciliogenesis. Moreover, MLCK/NMII signaling pathway is necessary for PCP protein plasma membrane delivery, but not sufficient to completely rescue p73KO phenotype. Since the reciprocal interaction between MTs and the actin cytoskeleton is necessary for intracellular transport62, it is possible that similar functional interactions are triggered by TAp73 in this system.
We found that p73-deficient ECs displayed lower levels of EB3, a TIP+-binding protein required to modulate MT dynamics and the Golgi apparatus organization63. Interestingly, EB1/3 disruption is known to perturb non-centrosomal MT organization57. Congruently, TAp73 overexpression induces enhanced EB3 staining and dramatic changes in the dynamics of non-centrosomal Golgi-derived MTs, indispensable for post-Golgi trafficking and cell polarity54.
GO analysis of DEGs containing TAp73 peaks showed enrichment in Golgi organization and MT dynamics, as well as actin cytoskeleton organization GO-terms, confirming TAp73 role as modulator of actin and MT dynamics signaling programs. However, further studies will be required to unravel how p73, directly or indirectly, regulates these modules.
Altogether, our data demonstrate that TAp73 not only regulates ECs development and ciliogenesis but also is an essential regulator of PCP establishment. This novel p73 function as an epithelial architect is not only fundamental during development but could also play a relevant role in TAp73 tumor suppression.
References
Butler, M. T. & Wallingford, J. B. Planar cell polarity in development and disease. Nat. Rev. Mol. Cell Biol. 18, 375–388 (2017).
Wallingford, J. B. & Mitchell, B. Strange as it may seem: the many links between Wnt signaling, planar cell polarity, and cilia. Genes Dev. 25, 201–213 (2011).
Boutin, C. et al. A dual role for planar cell polarity genes in ciliated cells. Proc. Natl Acad. Sci. USA 111, E3129–E3138 (2014).
Ohata, S. & Alvarez-Buylla, A. Planar organization of multiciliated ependymal (E1) cells in the brain ventricular epithelium. Trends Neurosci. 39, 543–551 (2016).
Spassky, N. et al. Adult ependymal cells are postmitotic and are derived from radial glial cells during embryogenesis. J. Neurosci. 25, 10–18 (2005).
Mirzadeh, Z., Han, Y. G., Soriano-Navarro, M., Garcia-Verdugo, J. M. & Alvarez-Buylla, A. Cilia organize ependymal planar polarity. J. Neurosci. 30, 2600–2610 (2010).
Liu, T., Jin, X., Prasad, R. M., Sari, Y. & Nauli, S. M. Three types of ependymal cells with intracellular calcium oscillation are characterized by distinct cilia beating properties. J. Neurosci. Res. 92, 1199–1204 (2014).
Montcouquiol, M. et al. Identification of Vangl2 and Scrb1 as planar polarity genes in mammals. Nature 423, 173–177 (2003).
Ohata, S. et al. Loss of Dishevelleds disrupts planar polarity in ependymal motile cilia and results in hydrocephalus. Neuron 83, 558–571 (2014).
Shi, D. et al. Celsr1 is required for the generation of polarity at multiple levels of the mouse oviduct. Development 141, 4558–4568 (2014).
Tissir, F. et al. Lack of cadherins Celsr2 and Celsr3 impairs ependymal ciliogenesis, leading to fatal hydrocephalus. Nat. Neurosci. 13, 700–707 (2010).
Vladar, E. K., Bayly, R. D., Sangoram, A. M., Scott, M. P. & Axelrod, J. D. Microtubules enable the planar cell polarity of airway cilia. Curr. Biol. 22, 2203–2212 (2012).
Matis, M., Russler-Germain, D. A., Hu, Q., Tomlin, C. J. & Axelrod, J. D. Microtubules provide directional information for core PCP function. eLife 3, e02893 (2014).
Kunimoto, K. et al. Coordinated ciliary beating requires Odf2-mediated polarization of basal bodies via basal feet. Cell 148, 189–200 (2012).
Hirota, Y. et al. Planar polarity of multiciliated ependymal cells involves the anterior migration of basal bodies regulated by non-muscle myosin II. Development 137, 3037–3046 (2010).
Vladar, E. K., Nayak, J. V., Milla, C. E. & Axelrod, J. D. Airway epithelial homeostasis and planar cell polarity signaling depend on multiciliated cell differentiation. JCI Insight 1, pii: e88027 (2016).
Gonzalez-Cano, L. et al. p73 is required for ependymal cell maturation and neurogenic SVZ cytoarchitecture. Dev. Neurobiol. 76, 730–747 (2016).
Marshall, C. B. et al. p73 is required for multiciliogenesis and regulates the Foxj1-associated genenetwork. Cell Rep. 14, 2289–2300 (2016).
Nemajerova, A. et al. TAp73 is a central transcriptional regulator of airway multiciliogenesis. Genes Dev. 30, 1300–1312 (2016).
Fujitani, M., Sato, R. & Yamashita, T. Loss of p73 in ependymal cells during the perinatal period leads to aqueductal stenosis. Sci. Rep. 7, 12007 (2017).
Candi, E., Agostini, M., Melino, G. & Bernassola, F. How the TP53 family proteins TP63 and TP73 contribute to tumorigenesis: regulators and effectors. Hum. Mutat. 35, 702–714 (2014).
Yang, A. et al. p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature 404, 99–103 (2000).
Tissir, F. et al. DeltaNp73 regulates neuronal survival in vivo. Proc. Natl Acad. Sci. USA 106, 16871–16876 (2009).
Flores, E. R. et al. Tumor predisposition in mice mutant for p63 and p73: evidence for broader tumor suppressor functions for the p53 family. Cancer Cell 7, 363–373 (2005).
Martin-Lopez, M. et al. p73 is required for appropriate BMP-induced mesenchymal-to-epithelial transition during somatic cell reprogramming. Cell Death Dis. 8, e3034 (2017).
Balint, E., Phillips, A. C., Kozlov, S., Stewart, C. L. & Vousden, K. H. Induction ofp57(KIP2) expression by p73beta. Proc. Natl Acad. Sci. USA 99, 3529–3534 (2002).
Melino, G. et al. p73 Induces apoptosis via PUMA transactivation and Bax mitochondrial translocation. J. Biol. Chem. 279, 8076–8083 (2004).
Graham, W. V. et al. Tumor necrosis factor-induced long myosin light chain kinase transcription is regulated by differentiation-dependent signaling events. Characterization of the human long myosin light chain kinase promoter. J. Biol. Chem. 281, 26205–26215 (2006).
Koeppel, M. et al. Crosstalk between c-Jun and TAp73alpha/beta contributes to the apoptosis-survival balance. Nucleic Acids Res. 39, 6069–6085 (2011).
Fernandez-Garcia, B. et al. p73 cooperates with Ras in the activation of MAP kinase signaling cascade. Cell Death Differ. 14, 254–265 (2007).
Tomasini, R. et al. TAp73 knockout shows genomic instability with infertility and tumor suppressor functions. Genes Dev. 22, 2677–2691 (2008).
Grespi, F. & Melino, G. P73 and age-related diseases: is there any link with Parkinson disease? Aging 4, 923–931 (2012).
Mathelier, A. et al. JASPAR 2016: a major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 44, D110–D115 (2016).
Huang da, W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009).
Huang, D. W., Sherman, B. T. & Lempicki, R. A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 (2009).
Mi, H. et al. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 45, D183–D189 (2017).
Ye, D. & Ma, T. Y. Cellular and molecular mechanisms that mediate basal and tumour necrosis factor-alpha-induced regulation of myosin light chain kinase gene activity. J. Cell. Mol. Med. 12, 1331–1346 (2008).
Herring, B. P., El-Mounayri, O., Gallagher, P. J., Yin, F. & Zhou, J. Regulation of myosin light chain kinase and telokin expression in smooth muscle tissues. Am. J. Physiol. Cell. Physiol. 291, C817–C827 (2006).
Smutny, M. et al. Myosin II isoforms identify distinct functional modules that support integrity of the epithelial zonula adherens. Nat. Cell Biol. 12, 696–702 (2010).
Krtolica, A. et al. Disruption of apical-basal polarity of human embryonic stem cells enhances hematoendothelial differentiation. Stem Cells 25, 2215–2223 (2007).
Totsukawa, G. et al. Distinct roles of ROCK (Rho-kinase) and MLCK in spatial regulation of MLC phosphorylation for assembly of stress fibers and focal adhesions in 3T3 fibroblasts. J. Cell Biol. 150, 797–806 (2000).
Vladar, E. K. & Axelrod, J. D. Dishevelled links basal body docking and orientation in ciliated epithelial cells. Trends Cell Biol. 18, 517–520 (2008).
Werner, M. E. et al. Actin and microtubules drive differential aspects of planar cell polarity in multiciliated cells. J. Cell Biol. 195, 19–26 (2011).
Takagishi, M. et al. Daple coordinates planar polarized microtubule dynamics in ependymal cells and contributes to hydrocephalus. Cell Rep. 20, 960–972 (2017).
Harumoto, T. et al. Atypical cadherins Dachsous and Fat control dynamics of noncentrosomal microtubules in planar cell polarity. Dev. Cell 19, 389–401 (2010).
Axelrod, J. D. Progress and challenges in understanding planar cell polarity signaling. Semin. Cell Dev. Biol. 20, 964–971 (2009).
Schlessinger, K., Hall, A. & Tolwinski, N. Wnt signaling pathways meet Rho GTPases. Genes Dev. 23, 265–277 (2009).
Dewerchin, H. L., Desmarets, L. M., Noppe, Y. & Nauwynck, H. J. Myosins 1 and 6, myosin light chain kinase, actin and microtubules cooperate during antibody-mediated internalisation and trafficking of membrane-expressed viral antigens in feline infectious peritonitis virus infected monocytes. Vet. Res. 45, 17–17 (2014).
Wong, S. Y. et al. Constitutive activation of myosin-dependent contractility sensitizes glioma tumor-initiating cells to mechanical inputs and reduces tissue invasion. Cancer Res. 75, 1113–1122 (2015).
Aw, W. Y. & Devenport, D. Planar cell polarity: global inputs establishing cellular asymmetry. Curr. Opin. Cell Biol. 44, 110–116 (2017).
Ho, W. C., Allan, V. J., van Meer, G., Berger, E. G. & Kreis, T. E. Reclustering of scattered Golgi elements occurs along microtubules. Eur. J. Cell Biol. 48, 250–263 (1989).
Yang, W. & Storrie, B. Scattered Golgi elements during microtubule disruption are initially enriched in trans-Golgi proteins. Mol. Biol. Cell 9, 191–207 (1998).
Chabin-Brion, K. et al. The Golgi complex is a microtubule-organizing organelle. Mol. Biol. Cell 12, 2047–2060 (2001).
Zhu, X. & Kaverina, I. Golgi as an MTOC: making microtubules for its own good. Histochem. Cell. Biol. 140, 361–367 (2013).
Kassianidou, E., Hughes, J. H. & Kumar, S. Activation of ROCK and MLCK tunes regional stress fiber formation and mechanics via preferential myosin light chain phosphorylation. Mol. Biol. Cell 28, 3832–3843 (2017).
Wu, J. et al. Molecular pathway of microtubule organization at the Golgi apparatus. Dev. Cell 39, 44–60 (2016).
Yang, C. et al. EB1 and EB3 regulate microtubule minus end organization and Golgi morphology. J. Cell Biol. 216, 3179–3198 (2017).
Rauzi, M., Lenne, P. F. & Lecuit, T. Planar polarized actomyosin contractile flows control epithelial junction remodelling. Nature 468, 1110–1114 (2010).
Devenport, D. The cell biology of planar cell polarity. J. Cell Biol. 207, 171–179 (2014).
Molla-Herman, A. et al. The ciliary pocket: an endocytic membrane domain at the base of primary and motile cilia. J. Cell Sci. 123(Pt 10), 1785–1795 (2010).
Shimada, Y., Yonemura, S., Ohkura, H., Strutt, D. & Uemura, T. Polarized transport of Frizzled along the planar microtubule arrays in Drosophila wing epithelium. Dev. Cell 10, 209–222 (2006).
Akhmanova, A. & Steinmetz, M. O. Control of microtubule organization and dynamics: two ends in the limelight. Nat. Rev. Mol. Cell Biol. 16, 711–726 (2015).
Zhang, R., Alushin, G. M., Brown, A. & Nogales, E. Mechanistic origin of microtubule dynamic instability and its modulation by EB proteins. Cell 162, 849–859 (2015).
Acknowledgements
This work was supported by Grants SAF2015-71381-R from Spanish Ministerio de Economía y Competitividad co-financed by FEDER funds (to M.C.M.) and LE021P17 from Junta de Castilla y Leon, and from the Queen Elisabeth Medical Foundation to F.T. J.V.-F. and S.F.-A. are holders of predoctoral fellowships from the Junta de Castilla y León. L.M.-A. is supported by a predoctoral scholarship from the Asociación Española contra el Cáncer (AECC). F.T. is a Research Director of the FNRS. M.W. and M.L. are funded by the Deutsche Forschungsgemeinschaft (DFG) under grant number LI 2405/2.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by G. Melino
Electronic supplementary material
41419_2018_1205_MOESM3_ESM.pdf
Supplementary Figure 1. Lack of DNp73 has no effect on PCP establishment or pMLC localization or rotational PCP Basal Bodies organization
41419_2018_1205_MOESM6_ESM.pdf
Supplementary Figure 4. TAp73 overexpression results in changes in “Golgi and MT organization” signaling pathways as well as Actin dynamics
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Fuertes-Alvarez, S., Maeso-Alonso, L., Villoch-Fernandez, J. et al. p73 regulates ependymal planar cell polarity by modulating actin and microtubule cytoskeleton. Cell Death Dis 9, 1183 (2018). https://doi.org/10.1038/s41419-018-1205-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41419-018-1205-6
This article is cited by
-
Dual-color live imaging unveils stepwise organization of multiple basal body arrays by cytoskeletons
EMBO Reports (2024)
-
A Review of Cerebrospinal Fluid Circulation and the Pathogenesis of Congenital Hydrocephalus
Neurochemical Research (2024)
-
p73 is required for vessel integrity controlling endothelial junctional dynamics through Angiomotin
Cellular and Molecular Life Sciences (2022)
-
Tissue-specific expression of p73 and p63 isoforms in human tissues
Cell Death & Disease (2021)
-
Modeling CADASIL vascular pathologies with patient-derived induced pluripotent stem cells
Protein & Cell (2019)