Abstract
In a significant number of cases cancer therapy is followed by a resurgence of more aggressive tumors derived from immature cells. One example is acute myeloid leukemia (AML), where an accumulation of immature cells is responsible for relapse following treatment. We previously demonstrated in chronic myeloid leukemia that the bone morphogenetic proteins (BMP) pathway is involved in stem cell fate and contributes to transformation, expansion, and persistence of leukemic stem cells. Here, we have identified intrinsic and extrinsic dysregulations of the BMP pathway in AML patients at diagnosis. BMP2 and BMP4 protein concentrations are elevated within patients’ bone marrow with a BMP4-dominant availability. This overproduction likely depends on the bone marrow microenvironment, since MNCs do not overexpress BMP4 transcripts. Intrinsically, the receptor BMPR1A transcript is increased in leukemic samples with more cells presenting this receptor at the membrane. This high expression of BMPR1A is further increased upon BMP4 exposure, specifically in AML cells. Downstream analysis demonstrated that BMP4 controls the expression of the survival factor ΔNp73 through its binding to BMPR1A. At the functional level, this results in the direct induction of NANOG expression and an increase of stem-like features in leukemic cells, as shown by ALDH and functional assays. In addition, we identified for the first time a strong correlation between ΔNp73, BMPR1A and NANOG expression with patient outcome. These results highlight a new signaling cascade initiated by tumor environment alterations leading to stem-cell features and poor patients’ outcome.
Similar content being viewed by others
Introduction
The current paradigm on the initiation of leukemogenesis implies a multistep process involving different types of genetic alterations, with no obvious hierarchy and understanding of the sequential clonal selection1. Nevertheless, crosstalk between leukemic stem cells and the associated bone marrow (BM) stroma appears to be essential for leukemic progression and response to therapy2,3. More globally, understanding interactions between tumor stem cells (SCs) and their microenvironment is a challenge to develop strategies to avoid relapses after therapy. Among the main elements implicated in the crosstalk between the microenvironment and both normal and tumor SCs, we have investigated the role of bone morphogenetic proteins (BMPs), because they govern SC regulation including hematopoietic4,5, neural and epithelial systems6 by directly and indirectly affecting their niche7,8,9. Alterations of the BMP signaling pathway have been observed in numerous cancers, in some cases closely associated with cancer stem cells (CSC) properties10. According to the context, BMPs could participate in initial tumor suppression or favor CSC maintenance and metastasis8. Within the BMP family, BMP2 and BMP4 have emerged as key regulators of normal and cancer SCs11,12,13. We have previously demonstrated that alterations in the BMP pathway at intrinsic (BMP receptors and downstream partners) and extrinsic (BMP extracellular ligands) levels constitute major events in transformation, expansion and persistence of immature cells in chronic phase chronic myeloid leukemia (CML) and breast cancer, by diverting their normal functions11,12,14,15.
Acute myeloid leukemia (AML), the first tumor where CSCs were described16, is a heterogeneous disease, in which the accumulation of genetic aberrations results in the uncontrolled growth of malignant undifferentiated cells. Relapse in the first years following complete remission is prevalent and may reflect the survival of resistant immature-like tumor cells able to regenerate the entire tumor17. The BMP pathway has been implicated in adult AML. For example, the overexpression of the transcription factor MIXL1, which is sufficient to initiate AML, can be induced by BMP4. Consistent with this induction, AML cells that express MIXL1 are sensitive to type BMP type 1 receptors (BMPR1) inhibitors18. In addition, in acute megakaryoblastic leukemia, the appearance of a specific fusion protein CBFA2T3-GLIS2 leads to the overexpression of BMP2 and BMP4 by leukemic cells and is associated with colony-forming capacities, a property ascribed to immature cells19.
Here we have identified alterations of the BMP pathway and revealed their importance in immature properties exhibited by AML cells. Initially focusing on the analysis of AML patient samples collected at diagnosis and subsequently experimentally deregulating the BMP pathway, we have identified alterations in BMP ligands, receptors and target genes. Our data highlight a new signaling cascade likely involved in the cell survival and features of immature AML cells in their microenvironment.
Materials and methods
Protein quantification
Bone marrow plasma obtained from allogeneic BM healthy donors and AML patients was harvested and cleared. BMPs concentration was determined using the human BMP2-ELISA or BMP4-ELISA kits (RayBiotech) following the manufacturer’s instructions.
Primary cells, cell lines culture conditions, and treatments
Patient samples were obtained after informed consent in accordance with the Declaration of Helsinki in the hematology departments involved in this study. Mononuclear cells (MNCs) from 54 blood and BM samples were obtained from AML, excluding acute promyelocytic leukemia, patients at diagnosis. AML characteristics are presented in Table S1. Normal samples correspond to steady-state peripheral blood and BM samples from healthy donors for allogeneic BM transplantation, collected after informed consent. When necessary, primary cells were maintained in IMDM culture medium containing 10% fetal calf serum (FCS). KG1A myeloid leukemia cells were cultured in RPMI-1640 medium containing 10% FCS. BMP4 and LDN-193189 (20 nM) (Sigma-Aldrich) were added in serum-free medium as indicated18,20. Normal goat IgG control (AB-108-C) and anti-hBMPR1A (AF346) (R&D Systems) were used at 4 µg/mL.
Functional assays
Colony-forming cell (CFC) and long-term culture-initiating cell (LTC-IC) assays were performed as described20. LTC-IC number was expressed as W5-CFC/10,000 initial cells.
Expression vectors, transfections and luciferase assay
KG1A cells were transfected with pBabe-ΔNp7321, pMXS-NANOG or empty vector (control) (Addgene) using a Neon (Thermofisher Scientific) electroporation device according to the manufacturer’s instructions. For luciferase assays, the pNANOG-Luc reporter vector was cotransfected with empty pCDNA3 or pCDNA3-ΔNp73 in KG1A cells. Luciferase assays were performed using Dual-Glo® Luciferase Assay System (Promega) following the manufacturer’s instructions. Relative activation as compared with cells transfected with the empty vector22.
RNA isolation and analysis
Quantitative RT-PCR was performed using standard protocols20. MNCs were isolated by a Ficoll gradient and total RNA was purified using TRI REAGENT™ (Sigma) and the mini-RNA extraction kit (Qiagen, Valencia, CA). For RT-qPCR, cDNA was produced using Superscript II (Invitrogen) and amplified using Sybr-green (Quantifast, Qiagen) and the Real-Time PCR system (Roche). TBP (TATA-binding protein) and HPRT (hypoxanthine-guanine phosphoribosyl transferase) genes were used for normalization. Arbitrary unit (AU) corresponds to the ratio of expression between samples and a single normal sample used as a reference in each PCR. Primer sequences are presented in the Table S2.
Flow cytometry analysis
Cells were incubated with antibodies specifically recognizing CD34, CD38 (Becton Dickinson) and/or BMPR1A (R&D System), or an irrelevant isotype-matched control antibody. MNCs separated by Ficoll gradient were subjected to sorting using BMPR1A antibody and an FACSARIA III cell sorter (Becton Dickinson). ALDH (aldehyde dehydrogenase) activity was determined using an Aldefluor kit according to the manufacturer’s recommendations (StemCell Technologies).
Western blot analysis
MNCs from AML samples were treated with BMP4 (20 ng/ml) during 24 h, then proteins were extracted. Per lane, 40 µg of proteins were loaded on SDS-PAGE and transferred onto polyvinylidene difluoride membrane (Bio-Rad). Membranes were incubated with monoclonal antibodies anti-ΔNp73 (Abcam, ab13649) or anti-βactin (Abcam, ab8226) from mouse, or anti-NANOG (Abcam, ab109250) from rabbit. Then membranes were incubated with their relevant HRP-coupled secondary antibodies (Jackson ImmunoResearch).
Datasets
Normalized Reads Per Kilobase Million (RPKM) Illumina GA-IIX RNA-seq profiles were downloaded from the TCGA AML data portal. One hundred and sixty-one patients in the TCGA AML cohort had both RNA-seq and clinical data. RPKM values were log-transformed to the base two after adding a value of 1. Expression levels were extracted from the log-transformed data. A representative probe set with the highest average intensity was selected for each gene in each dataset. The median of expression of each gene was used for patients’ stratification (high vs. low score).
Statistical analysis
Unless otherwise specified, statistical analysis was performed using the Mann−Whitney U test. Except for overall survival, the statistical analyses and graphs were performed with Graphpad prism (version 6). Significant P values are given in the text or symbolized by asterisks (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001). Overall survival (OS) curves from the date of sampling onwards are illustrated using the Kaplan−Meier method. Log-rank tests were conducted to evaluate the effect of markers on survival. Best cut-off values for expression of each gene of interest were determined with the Youden index computed on Cumulative/Dynamic time-dependent ROC curve using Inverse Probability of Censoring Weighting (IPCW) estimation between 12 and 36 months of follow-up. The best cut-off value for NANOG expression that discriminates the most death events during time was 13 at 24 months of follow-up (AUC = 0.77, sensitivity = 0.61/specificity = 0.85). The level of significance was set at 5%. OS analysis and graph were performed with the R program (version 3.2.3) using the “survival” and “ggplot2” packages.
Results
Identification of intrinsic and extrinsic alterations in the BMP pathway in AML
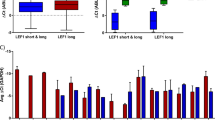
Elements of the BMP pathway were analyzed in AML samples (characteristics in Table S1) in both leukemic cells and their BM environment (Fig. S1a). We first measured the levels of soluble BMP2 and BMP4 in BM plasma from healthy donors and AML patients at diagnosis (Fig. S1b for experimental design). The levels of soluble BMP2 and BMP4 were significantly increased in AML patients compared to healthy donors by 2.9-fold (P = 0.0100) and 3.6-fold (P < 0.0001), respectively (Fig. 1a). Thus, in the leukemic microenvironment, cells are exposed to elevated concentrations of BMP2 and BMP4, the latter being almost sixfold more abundant than BMP2. In order to determine whether BM leukemic cells themselves produced these high BMP levels, the expression of the BMP2 and BMP4 genes was monitored in MNCs from both peripheral blood and BM of AML or healthy donors (Fig. 1b). BMP2 expression appeared reduced in leukemic cells compared to normal samples (P = 0.0421), while BMP4 expression was the same. Interestingly, when we analyzed BMPs transcripts in BM samples, both BMP2 and BMP4 mRNA levels were reduced in AML samples (Fig. 1c), while no significant changes were observed in circulating peripheral blood cells (Fig. S2a). These data suggest that the higher concentration of BMP2 and BMP4 in BM is not related to its autologous production by leukemic cells themselves, but is rather provided by other cells in the microenvironment.
a BMP2 and BMP4 concentrations in bone marrow (BM) supernatants from AML patients (n = 17) and healthy donors (n = 19) evaluated by ELISA. b BMP2 and BMP4 mRNA level in blood and bone marrow MNCs from healthy donors and AML patients’ samples at diagnosis. c BMP2 and BMP4 mRNA level in bone marrow MNCs only, from healthy donors and AML patients’ samples at diagnosis. d Expression of BMP receptors: BMPR1A, BMPR1B and BMPR2, and BMP target genes: Id1 and RunX1 evaluated in normal (n = 14–25) and AML (n = 34–51) grouped BM and blood samples by RT-qPCR. Arbitrary unit (AU): relative expression compared to a healthy sample used as a reference for each PCR experiment. P values were determined using the Mann−Whitney U test. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001
We next analyzed gene expression profiles of intrinsic BMP pathway components in AML MNCs, with a focus on BMP membrane receptors and target genes. We observed significantly (P = 0.0005) higher levels of BMPR1A transcripts, but not of BMPR1B and BMPR2, as compared to MNCs from healthy donors (Fig. 1d). With respect to the BMP-target genes, Id1 and RUNX1 expression was significantly upregulated compared to the levels found in samples from healthy donors (P < 0.0001 and P = 0.0001; respectively) in leukemic cells (Fig. 1d), whereas Id3 and RUNX2 expression remained unchanged (Fig. S2b). The observed increases in expression were detected both in BM and peripheral blood MNCs from AML patients (Fig. S2c). These results identify intrinsic dysregulations of the BMP pathway at different levels of the signaling cascade in addition to extrinsic alterations in human primary adult AML samples at diagnosis.
BMP4 and BMPR1A increases are associated with stemness features
In order to identify BMP-responsive cells in BM, we assessed whether a correlation existed between deregulated BMP elements and the presence of BMPR1A at the cell surface. By flow cytometry analysis, we found a significantly higher number of primary MNCs overexpressing membrane-BMPR1A (BMPR1A+; P = 0.0024) in AML than in normal samples (Fig. 2a). We also observed that the expression of BMPR1A was significantly correlated with that of Id1 in AML samples (P = 0.0017) (Fig. 2b) and that after sorting MNCs according to their membrane-BMPR1A content (Fig. S3a), we observed a higher Id1 expression in BMPR1A+ cells (Fig. S3b). Furthermore, when we exposed healthy and AML MNCs to BMP4 for 24 h, we could observe an increase in BMPR1A expression at the transcript level in only AML MNCs (P = 0.0195; Fig. 2c). This was confirmed at the protein level by the increase of BMPR1A-expressing cell numbers in AML samples following BMP4 treatment (P = 0.0313; Fig. 2d and illustrated in Fig. S3c).
a Percentage of mononuclear cells (MNCs) of healthy donors (n = 8) and AML (n = 11) samples expressing BMPR1A at their membrane determined by flow cytometry. b Correlation between the expressions of BMPR1A and Id1 in AML samples at diagnosis (n = 50). c Relative expression of BMPR1A in MNCs from healthy donors (n = 6) and AML patients (n = 9), exposed or not, to BMP4 (10 ng/mL) for 24 h. d Effect of BMP4 (10 ng/mL) exposure on the proportion of BMPR1A-positive cells at the surface in healthy and AML MNC samples (n = 6 patients for each cohort). e Effect of BMP4 (10 ng/mL) exposure for 24 h on the proportion of very immature leukemic cells in AML mononuclear cells (MNCs), evaluated by long-term culture-initiating cells (LTC-IC) assay (n = 3). f Flow cytometric analysis of the expression of BMPR1A at the cell surface according to CD34 and CD38 status of MNCs from an AML patient. g Number of colony-forming cells (CFC) and LTC-IC among peripheral blood MNCs from AML patients according to their low (n = 8) or high (n = 6) BMPR1A mRNA level. h AML samples with low (n = 6) or high (n = 7) BMPR1A mRNA levels were analyzed for ALDH activity by flow cytometry. NT non-treated, NBM normal bone marrow. Statistical analysis: P values were determined using Spearman’s nonparametric test (b); Wilcoxon matched-pairs signed rank test (c, d); *P < 0.05; **P < 0.01
Since BMP4 is linked to the regulation of SCs10,11,12,20,23, we evaluated its involvement in immature features observed in leukemic cells. We first analyzed the effect of BMP4 exposure of AML cells on the number of long-term culture-initiating cells (LTC-IC) as a read-out of SCs properties maintenance through time (8 weeks assay). As previously reported for normal hematopoietic cells20,24, BMP4 treatment of AML MNCs induced a reproducible increase in the number of LTC-IC (Fig. 2e). This indicates that exposure to BMP4 favors the survival of very immature AML cells. Through cytometry analysis, we observed that BMPR1A+ AML cells were specifically detected in the CD34+CD38− and CD34+CD38+ BM fractions, respectively enriched in stem and progenitor cells (Fig. 2f). We then divided at-diagnosis AML MNC samples into two groups according to their BMPR1A transcript expression. The “BMPR1A-low” group represented AML samples that expressed BMPR1A mRNA levels similar to healthy donors, while the “BMPR1A-high” group displayed BMPR1A levels exceeding the maximum value measured in healthy donors’ cells. Functional assays confirmed that AML samples expressing high levels of BMPR1A contain 15-fold more immature cells able to generate LTC-IC (P = 0.0426) but as many more mature progenitors identified as CFCs as BMPR1Alow AML samples (Fig. 2g). This was confirmed by the observation that such samples are also enriched in ALDH-positive cells (P = 0.0394), recognized as an SC marker in several cancer types including AML (Fig. 2h)25,26. Taken together, these findings indicate that leukemic cells harboring high levels of membrane BMPR1A receptors preferentially display immature features, this latter being increased after BMP4 exogenous exposure.
BMP4 induces ΔNp73, a p53 family member associated with stemness features
BMP4, BMPR1A, and Id1 are commonly associated with cell survival probably by controlling cell proliferation and differentiation, as reported in normal and leukemic contexts4,12,20. In order to identify new BMP4/BMPR1A downstream partners, we evaluated the expression of other genes involved in cell survival, such as the p53 family members27. We focused on ΔNp73, the antiapoptotic isoform produced by the TP73 gene27,28, which has been found to be overexpressed in AML patients29,30. We confirmed this finding in our samples where a highly significant (P < 0.0001) overexpression of ΔNp73 (Fig. 3a, right panel) at diagnosis, but not of the proapoptotic TAp73 isoform (Fig. 3a, left panel). Interestingly, ΔNp73 expression was strongly correlated with that of BMPR1A (P < 10−5) (Fig. 3b). This correlation was further confirmed by cell sorting MNCs from AML BM samples with an increase in ΔNp73 expression within the BMPR1A+ cells subfraction being found (Fig. S4a). Furthermore, treatment of AML or normal primary MNCs with exogenous soluble BMP4 for 24 h resulted in a significant increase in ΔNp73 transcripts only in AML samples (P = 0.0059; Fig. 3c). This increase was also observed at the protein level (Fig. 3d). A similar ΔNp73 upregulation after exposure to BMP4 was observed in the KG1A cell line, a model of immature AML31 (P = 0.0078; Fig. 3e). To confirm the direct link between BMP4, BMPR1A and the increase of ΔNp73 expression, KG1A cells were treated with BMP4 in the presence of either an anti-BMPR1A blocking antibody or the chemical BMPR1 inhibitor, LDN-193189. BMP4-mediated induction of ΔNp73 was abrogated by both the BMPR1A blocking antibody (Fig. 3f) and the LDN 193189 (Fig. S4b). Quantification of CFC and LTC-IC in two groups of samples based on their ΔNp73 expression revealed a dramatic and significant increase in LTC-IC number in the ΔNp73-high group (P = 0.0356) (Fig. 3g, right panel), whereas CFC remained equal (Fig. 3g, left panel). Next, we determined the number of ALDH+ cells in ΔNp73-low and -high expression groups and observed a significant correlation between ΔNp73 expression and ALDH positivity (P = 0.0350; Fig. 3h). These data show that deregulation of the BMP pathway contributes to the overexpression of the survival factor ΔNp73 in AML cells through the binding of BMP4 to BMPR1A-overexpressing leukemic cells. As this upregulation is associated with immature cell features, we next investigated potential mechanism through which ΔNp73 could participate in SC regulation.
a Expression of TAp73 and ΔNp73 in mononuclear cell (MNC) samples from healthy donors (n = 19–29) and AML at diagnosis (n = 46–54) quantified by RT-qPCR. Arbitrary Unit (AU), relative expression compared to a healthy sample used as a reference. b Correlations between the expressions of ΔNp73 and BMPR1A (n = 51). P values were determined using Spearman’s nonparametric test. c Relative expression of ΔNp73 in MNCs from healthy donors (n = 5) and AML patients (n = 7), exposed or not, to BMP4 (10 ng/mL) for 24 h; NT not treated. d Western blot analysis of AML cells exposed or not, to BMP4 (20 ng/mL) for 24 h (n = 2); N not treated. e Relative expression of ΔNp73 in KG1A cells exposed to BMP4 (10 ng/mL) for 24 h with (n = 8). f Expression of ΔNp73 in KG1A cells exposed to BMP4, in presence of either an antibody specific for BMPR1A or a control antibody, measured by RT-qPCR. Ratio was calculated according to untreated KG1A cells. g CFC and LTC-IC values in AML samples according to low (<8 AU, n = 6) or high (>8 AU, n = 9) ΔNp73 levels. h Flow cytometry analysis of ALDH activity in AML samples with low (n = 7) or high (n = 9) ΔNp73 expressions levels. Statistical analysis: Wilcoxon matched-pairs signed rank test was used for (c, e, f); *P < 0.05; **P < 0.01; ***P < 0.001
ΔNp73 activates NANOG expression in BMP4-responsive cells
Since ΔNp73 has been described as a regulator of NANOG32,33, we evaluated the potential link between ΔNp73 and genes known to drive SC features in AML samples. For this, we analyzed the expression of the three self-renewal regulators NANOG, SOX2, and OCT433,34,35. Although no difference was observed as compared with normal MNCs for OCT4 (P = 0.5500), a significantly higher expression of NANOG (P = 0.0025) and SOX2 (P = 0.0359) was measured in AML MNCs (Fig. 4a). In addition, the expression of ΔNp73 and NANOG and ΔNp73 and SOX2 were strongly correlated (Fig. 4b). Using KG1A cells, we observed that ectopic ΔNp73 induced NANOG expression (P = 0.0286; Fig. 4c), whereas ectopic NANOG did not affect ΔNp73 expression (Fig. 4c). In addition, the reduction of ΔNp73 expression by RNA interference consistently decreased NANOG expression (P = 0.0286; Fig. 4d). Finally, using a luciferase reporter assay, we showed that ΔNp73 is able to activate the NANOG promoter (Fig. 4e), as previously reported in a reprogramming iPS fibroblast model32.
Distribution of normal mononuclear cells (MNCs) (n = 14–27) and AML samples from patients (n = 31–54) according to a NANOG, OCT4, and SOX2 expression. b Correlation between NANOG (n = 52), SOX2 (n = 41), OCT4 (n = 41) and ΔNp73 mRNA levels in AML samples. Statistical analysis: Spearman’s nonparametric test. c Expression of NANOG and ΔNp73 in KG1A cells 48 h after transfection with pBabe-ΔNp73, pMXS-NANOG or empty vector (control). Expression ratio is the relative expression after transfection with the indicated vector as compared to transfection with the empty vector (n = 6). d Expression of NANOG and ΔNp73 48 h after KG1A cells transfection with an anti-ΔNp73-specific shRNA (n = 6). e Luciferase assay using the pNANOG-Luc, as reporter, cotransfected with a vector expressing ΔNp73 or an empty vector in KG1A cells (n = 6). f Correlation between BMPR1A and NANOG mRNA levels (n = 51). Statistical test: Spearman’s nonparametric test. g NANOG mRNA levels in AML cells sorted according to their surface expression BMPR1A (n = 3). RT-qPCR was used for mRNA quantification. Arbitrary Unit (AU), relative expression compared to a healthy sample used as a reference. h Western blot analysis of primary AML cells exposed or not, to BMP4 (20 ng/mL) for 24 h. Statistical analysis: P values were determined using Wilcoxon signed rank test for c, d; Wilcoxon matched-pairs signed rank test was used for e; *P < 0.05; **P < 0.01; ***P < 0.001
A significant correlation between BMPR1A and NANOG expression in AML samples was also observed (P < 10−5; Fig. 4f). This link was further confirmed in BM cells isolated from three different AML samples in which we detected higher levels of NANOG transcripts in total MNC cells sorted only for the BMPR1A cell membrane expression (Fig. 4g). Furthermore and in agreement with the robust correlation between BMPR1A, ΔNp73, and NANOG expression, we observed the induction of the NANOG protein after BMP4 treatment (Fig. 4h). Lastly, treating primary AML cells by BMP4 allows correlation in the same cells of the BMP4-increased expression of BMPR1A, ΔNp73, and NANOG with the amplification of cells with stemness features identified by the LTC-IC assay (Supplemental Table S4). Altogether, our data indicate that BMP4 induces the expression of ΔNp73, which in turn, activates NANOG transcription.
The combined BMPR1A/ΔNp73/NANOG overexpression at diagnosis identifies AML patients with a higher risk of early relapse
Finally, we analyzed the significance of BMPR1A, ΔNp73, and NANOG transcript levels in AML patients at diagnosis with respect to clinical outcome at 3 years post-diagnosis. We observed that levels of the three markers at diagnosis did not predict the initial complete remission status (Table S3). Conversely, high expression of either BMPR1A, ΔNp73 or NANOG at diagnosis was associated with an increased rate of relapse within 3 years (Fig. 5a). Combining the three markers led to an increase in the clinical predictability for AML patient outcome at diagnosis, by identifying patients with a higher risk of relapse (from 33 to 86% risk of relapse; Fig. 5b). In order to evaluate the importance of NANOG, we performed a Kaplan−Meyer analysis on a larger number of patients and observed that high NANOG transcript levels are associated with poor survival (Fig. 5c). We performed a multivariate parameter analysis and, as expected, identified a significant correlation only between NANOG and ΔNp73 or Sox2 or Oct4 expression. No correlation with any other parameters appeared significant, as indicated in Supplemental Table S5. Lastly, a similar analysis using the independent TCGA dataset36, this time restricted to patients within the intermediate risk group (n = 92), showed again the association between high expression level of NANOG and shorter survival (P = 0,054). Altogether, it suggests that NANOG could be considered as an independent risk factor and of high interest for patient within the intermediate risk group.
Percentage of AML patients in remission or relapse 3 years after diagnosis, according to BMPR1A, ΔNp73 or NANOG expression levels separately (n = 19) (a) or in combination (n = 13) (b). c Overall survival (OS) curve of AML patients from the date of sampling onwards, by using the Kaplan−Meier method. The level of significance was determined by the log-rank test (n = 52; 23 high and 29 low NANOG level). d Overall survival (OS) curve of AML patients from the date of sampling onwards, by using the Kaplan−Meier method. TCGA dataset from AML patients of the intermediate group. e Schematic representation of the role played by BMPs on homeostasis under healthy conditions (top) and the cascade of dysregulations associated with increased concentration of BMP4 in the microenvironment of leukemic immature cells (bottom)
These results demonstrate the importance of measuring levels of BMPR1A, ΔNp73 and even more so of NANOG, as they could contribute to predicting at diagnosis the risk of relapse of AML patients.
Discussion
Resistance to treatment is often associated with the persistence of cancer SCs within their microenvironment. In AML, owing to the poor clinical efficacy of current treatments, deciphering mechanisms through which the microenvironment can sustain and promote the survival of immature-like cancer cells remains an important issue for patient outcome. Among the mechanisms through which the microenvironment can affect leukemic SCs’ survival, cytokine production by stromal cells is of major importance. We focused on the BMP pathway as BMP2 and BMP4 regulate SC fate, maintenance and differentiation processes20,24,37 and contribute to cancer SC emergence11,14, maintenance and expansion15. Here, we found an abnormally elevated concentration of BMP4, and to a lesser extent of BMP2 in the BM plasma of AML patients. These cytokines are likely to be produced by the BM microenvironment of leukemic cells, since we did not observe any increase in BMP2 and BMP4 transcripts levels in primitive leukemic cells themselves as compared to normal hematopoietic cells. This appears to be a general mechanism as we have found similar profiles in CML at the time of diagnosis12 and also in luminal breast cancer11. Conversely, we observed an increased expression of intrinsic actors of the BMP pathway in MNCs of AML patients. In particular, we identified a highly significant increase in the expression of BMPR1A both at the mRNA and protein levels, in agreement with the important role of BMPR1 in the early steps of the AML transforming process, as previously described18. Interestingly, unlike the chronic phase of CML, in which we identified BMP2 and BMPR1B as driving the deregulation of the BMP pathway and SC/progenitor maintenance and expansion12, in AML BM cells, BMP4 and BMPR1A alterations were already detected at diagnosis. Interestingly, with CML progression towards more advanced phases and the acquisition of resistance to treatment, the BMP4 signal becomes predominant38. This suggests that the BMP4 signal is related to more aggressive or advanced disease as identified in cancers of other origins such as liver (HCC)39, thyroid (PTC)40 and bladder41. In malignant glioma, while BMP4 appears to induce differentiation of glioma cancer stem-like cells42,43, an epigenetic repression of the BMPR1B receptors contributes to the CSC phenotype in these tumors10.
We identify here that the compartment of cells expressing surface BMPR1A is enriched in very immature AML cells as demonstrated by functional and ALDH assays. In addition, an increase in LTC-IC after exposure to BMP4 confirmed the involvement of this cytokine provided by the tumor niche and of its receptor in promoting AML stem-like cells. The BM-niche was previously reported to promote survival of leukemic cells in AML by activating different cytokine-related pathways, such as SDF1/CXCR4, Wnt/β-catenin, integrins2. Here, the elevated concentrations of both BMP2 and BMP4 in BM of AML patients at diagnosis also support their involvement in leukemic cells survival, as described in CML12. These results, obtained in the context of AML, highlight the importance under pathological conditions of this pathway already found to be clearly involved in SC regulation in neural44, epithelial11,14 and hematopoietic systems20,24 under physiological conditions.
We investigated the interplay between BMP4/BMPR1A signaling and the ΔNp73 isoform, as this member of the p53 family has been shown to be overexpressed in AML and to regulate CSCs23,30. Crosstalk between the p53 family and the BMP pathway has already been reported during breast45 and epidermis46 development, as well as in tumorigenesis47, though no evidence for a link between BMPs and the ΔNp73 isoform has so far been reported. Here, we clearly demonstrate a functional link between BMP4 and ΔNp73. First, we identified a specific ΔNp73 upregulation already present at the time of diagnosis in MNCs of AML patients, while no such effect is seen for TAp73. Subsequently, we showed that this expression was higher within the BMPR1A+ subpopulation and after exposure to BMP4. Finally, we established the link between both the BMP pathway and ΔNp73 with immature cell features. Indeed, AML MNCs expressing high levels of BMPR1A and ΔNp73 were mainly ALDH positive, contained more LTC-IC and, after BMP4 treatment, exhibited an increased capacity to form colonies in an in vitro human functional SC assay.
Our results suggest that extrinsic (BMP4) and intrinsic (BMPR1A) alterations of the BMP pathway, associated with the induction of ΔNp73 expression, contribute to the increase in the number of immature cells in AML BM. The molecular mechanisms underlying such a phenomenon were evaluated focusing on the expression of some genes associated with immaturity, in particular NANOG, a known ΔNp73-target gene32,33. We observed a significant correlation between the expression of ΔNp73 and NANOG and also of BMPR1A and NANOG in AML patients. These results highlight for the first time a cascade of events in AML, initiated by the binding of BMP4 to its type 1A receptor that leads to ΔNp73 expression, which in turn induces NANOG by promoter transactivation. This activation is p53-independent, since it was confirmed using the KG1A AML cell line, which does not express functional p5331.
The increased expression of three genes involved in self-renewal and in cell reprogramming, ΔNp7332 NANOG and SOX232,48, in AML patients and the induction of ΔNp73 and NANOG upon BMP4 treatment supports the hypothesis that BMP4 could promote the reprogramming of cells towards immature leukemic cells. This is consistent with a recent study that identified BMP4 and its signaling pathway as a driving element of adult cell reprogramming, including fibroblasts towards functional hematopoietic SCs49.
Resistance to treatment and relapse are often associated with the persistence of cancer SCs within their microenvironment. Indeed, BMP4 involvement in resistance to treatment has been demonstrated in several tumor types, including HCC50 and ovarian cancer51. BMP type 1 receptors have also been associated with resistance in CML38 and AML18, as has ΔNp7352. These latter, as well as NANOG, are also associated with resistance and progression in other cancers53,54,55.
In conclusion, our study deciphers a new signaling cascade through which alterations of BMPs secretion in the microenvironment can sustain and promote resistant immature-like leukemic cells (Fig. 5e). Activation of this signaling cascade is associated with a poor prognosis for AML patients. Thus, measuring transcript levels of the three markers BMPR1A, ΔNp73, and NANOG is likely to increase the ability to predict patient outcome at diagnosis. Since involvement of CSC in clinical outcome may be a common feature of relapsing cancers, it will be worthwhile to investigate the potential involvement of this novel cascade in other cancer types.
References
Hirsch, P. et al. Genetic hierarchy and temporal variegation in the clonal history of acute myeloid leukaemia. Nat. Commun. 7, 12475 (2016).
Zhou, H. S., Carter, B. Z. & Andreeff, M. Bone marrow niche-mediated survival of leukemia stem cells in acute myeloid leukemia: Yin and Yang. Cancer Biol. Med. 13, 248–259 (2016).
Shafat, M. S. et al. Leukemic blasts program bone marrow adipocytes to generate a protumoral microenvironment. Blood 129, 1320–1332 (2017).
Toofan, P. & Wheadon, H. Role of the bone morphogenic protein pathway in developmental haemopoiesis and leukaemogenesis. Biochem. Soc. Trans. 44, 1455–1463 (2016).
Kirmizitas, A., Meiklejohn, S., Ciau-Uitz, A., Stephenson, R. & Patient, R. Dissecting BMP signaling input into the gene regulatory networks driving specification of the blood stem cell lineage. Proc. Natl Acad. Sci. USA 114, 5814–5821 (2017).
Rendl, M., Polak, L. & Fuchs, E. BMP signaling in dermal papilla cells is required for their hair follicle-inductive properties. Genes Dev. 22, 543–557 (2008).
Zhang, J. et al. Identification of the haematopoietic stem cell niche and control of the niche size. Nature 425, 836–841 (2003).
Pickup, M. W., Owens, P. & Moses, H. L. TGF-beta, bone morphogenetic protein, and activin signaling and the tumor microenvironment. Cold Spring Harb. Perspect. Biol. 9, https://doi.org/10.1101/cshperspect.a022285 (2017).
Katagiri, T. & Watabe, T. Bone morphogenetic proteins. Cold Spring Harb. Perspect. Biol. 8, https://doi.org/10.1101/cshperspect.a021899 (2016).
Lee, J. et al. Epigenetic-mediated dysfunction of the bone morphogenetic protein pathway inhibits differentiation of glioblastoma-initiating cells. Cancer Cell 13, 69–80 (2008).
Chapellier, M. et al. Disequilibrium of BMP2 levels in the breast stem cell niche launches epithelial transformation by overamplifying BMPR1B cell response. Stem Cell Rep. 4, 239–254 (2015).
Laperrousaz, B. et al. Primitive CML cell expansion relies on abnormal levels of BMPs provided by the niche and on BMPRIb overexpression. Blood 122, 3767–3777 (2013).
Sagorny, K., Chapellier, M., Laperrousaz, B. & Maguer-Satta, V. [BMP and cancer: the Yin and Yang of stem cells]. Med. Sci. (Paris) 28, 416–422 (2012).
Clement, F. et al. Long-term exposure to bisphenol A or benzo(a)pyrene alters the fate of human mammary epithelial stem cells in response to BMP2 and BMP4, by pre-activating BMP signaling. Cell Death Differ. 24, 155–166 (2017).
Zylbersztejn, F. et al. The BMP pathway: a unique tool to decode the origin and progression of leukemia. Exp. Hematol. https://doi.org/10.1016/j.exphem.2018.02.005 (2018).
Bonnet, D. & Dick, J. E. Human acute myeloid leukemia is organized as a hierarchythat originates from a primitive hematopoietic cell. Nat. Med. 3, 730–737 (1997).
Shlush, L. I. et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature 506, 328–333 (2014).
Raymond, A. et al. A role for BMP-induced homeobox gene MIXL1 in acute myelogenous leukemia and identification of type I BMP receptor as a potential target for therapy. Oncotarget 5, 12675–12693 (2014).
Gruber, T. A. et al. An Inv(16)(p13.3q24.3)-encoded CBFA2T3-GLIS2 fusion protein defines an aggressive subtype of pediatric acute megakaryoblastic leukemia. Cancer Cell 22, 683–697 (2012).
Jeanpierre, S. et al. BMP4 regulation of human megakaryocytic differentiation is involved in thrombopoietin signaling. Blood 112, 3154–3163 (2008).
Accardi, R. et al. Skin human papillomavirus type 38 alters p53 functions by accumulation of deltaNp73. EMBO Rep. 7, 334–340 (2006).
Rodda, D. J. et al. Transcriptional regulation of nanog by OCT4 and SOX2. J. Biol. Chem. 280, 24731–24737 (2005).
Basu, S. & Murphy, M. E. p53 family members regulate cancer stem cells. Cell Cycle 15, 1403–1404 (2016).
Khurana, S. et al. A novel role of BMP4 in adult hematopoietic stem and progenitor cell homing via Smad independent regulation of integrin-alpha4 expression. Blood 121, 781–790 (2013).
Cheung, A. M. et al. Aldehyde dehydrogenase activity in leukemic blasts defines a subgroup of acute myeloid leukemia with adverse prognosis and superior NOD/SCID engrafting potential. Leukemia 21, 1423–1430 (2007).
Ran, D. et al. Aldehyde dehydrogenase activity among primary leukemia cells is associated with stem cell features and correlates with adverse clinical outcomes. Exp. Hematol. 37, 1423–1434 (2009).
Buhlmann, S. & Putzer, B. M. DNp73 a matter of cancer: mechanisms and clinical implications. Biochim. Biophys. Acta 1785, 207–216 (2008).
Ferraiuolo, M., Di Agostino, S., Blandino, G. & Strano, S. Oncogenic intra-p53 family member interactions in human cancers. Front. Oncol. 6, 77 (2016).
Alexandrova, E. M. & Moll, U. M. Role of p53 family members p73 and p63 in human hematological malignancies. Leuk. Lymphoma 53, 2116–2129 (2012).
Rizzo, M. G. et al. Analysis of p73 expression pattern in acute myeloid leukemias: lack of DeltaN-p73 expression is a frequent feature of acute promyelocytic leukemia. Leukemia 18, 1804–1809 (2004).
Furley, A. J. et al. Divergent molecular phenotypes of KG1 and KG1a myeloid cell lines. Blood 68, 1101–1107 (1986).
Lin, Y., Cheng, Z., Yang, Z., Zheng, J. & Lin, T. DNp73 improves generation efficiency of human induced pluripotent stem cells. Bmc Cell Biol. 13, 9 (2012).
Meier, C., Hardtstock, P., Joost, S., Alla, V. & Putzer, B. M. p73 and IGF1R regulate emergence of aggressive cancer stem-like features via miR-885-5p control. Cancer Res. 76, 197–205 (2016).
Lagadec, C., Vlashi, E., Della, D. L., Dekmezian, C. & Pajonk, F. Radiation-induced reprogramming of breast cancer cells. Stem Cells 30, 833–844 (2012).
Mathieu, J. et al. HIF induces human embryonic stem cell markers in cancer cells. Cancer Res. 71, 4640–4652 (2011).
Cancer Genome Atlas Research, N. et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 45, 1113–1120 (2013).
Maguer-Satta, V. et al. Regulation of human erythropoiesis by activin A, BMP2, and BMP4, members of the TGFbeta family. Exp. Cell Res. 282, 110–120 (2003).
Grockowiak, E. et al. Immature CML cells implement a BMP autocrine loop to escape TKI treatment. Blood 130, 2860–2871 (2017).
Zeng, S. et al. BMP4 promotes metastasis of hepatocellular carcinoma by an induction of epithelial-mesenchymal transition via upregulating ID2. Cancer Lett. 390, 67–76 (2017).
Meng, X. et al. Expression of BMP-4 in papillary thyroid carcinoma and its correlation with tumor invasion and progression. Pathol. Res. Pract. 213, 359–363 (2017).
Martinez, V. G. et al. BMP4 Induces M2 Macrophage Polarization and Favors Tumor Progression in Bladder Cancer. Clin. Cancer Res. 23, 7388–7399 (2017).
Raja, E. et al. Bone morphogenetic protein signaling mediated by ALK-2 and DLX2 regulates apoptosis in glioma-initiating cells. Oncogene 36, 4963–4974 (2017).
Xi, G., Best, B., Mania-Farnell, B., James, C. D. & Tomita, T. Therapeutic potential for bone morphogenetic protein 4 in human malignant glioma. Neoplasia 19, 261–270 (2017).
Mira, H. et al. Signaling through BMPR-IA regulates quiescence and long-term activity of neural stem cells in the adult hippocampus. Cell Stem Cell 7, 78–89 (2010).
Balboni, A. L. et al. DeltaNp63alpha-mediated activation of bone morphogenetic protein signaling governs stem cell activity and plasticity in normal and malignant mammary epithelial cells. Cancer Res. 73, 1020–1030 (2013).
Aberdam, D. et al. Key role of p63 in BMP-4-induced epidermal commitment of embryonic stem cells. Cell Cycle 6, 291–294 (2007).
Balboni, A. L. et al. p53 and DeltaNp63alpha coregulate the transcriptional and cellular response to TGFbeta and BMP signals. Mol. Cancer Res. 13, 732–742 (2015).
Takahashi, K. & Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 (2006).
Cheng, H. et al. Reprogramming mouse fibroblasts into engraftable myeloerythroid and lymphoid progenitors. Nat. Commun. 7, 13396 (2016).
Ma, J. et al. BMP4 promotes oxaliplatin resistance by an induction of epithelial-mesenchymal transition via MEK1/ERK/ELK1 signaling in hepatocellular carcinoma. Cancer Lett. 411, 117–129 (2017).
Coffman, L. G. et al. Human carcinoma-associated mesenchymal stem cells promote ovarian cancer chemotherapy resistance via a BMP4/HH signaling loop. Oncotarget 7, 6916–6932 (2016).
Lucena-Araujo, A. R. et al. The expression of DeltaNTP73, TATP73 and TP53 genes in acute myeloid leukaemia is associated with recurrent cytogenetic abnormalities and in vitro susceptibility to cytarabine cytotoxicity. Br. J. Haematol. 142, 74–78 (2008).
Lucena-Araujo, A. R. et al. High DeltaNp73/TAp73 ratio is associated with poor prognosis in acute promyelocytic leukemia. Blood 126, 2302–2306 (2015).
Engelmann, D., Meier, C., Alla, V. & Putzer, B. M. A balancing act: orchestrating amino-truncated and full-length p73 variants as decisive factors in cancer progression. Oncogene 34, 4287–4299 (2015).
Jeter, C. R., Yang, T., Wang, J., Chao, H. P. & Tang, D. G. Concise review: NANOG in cancer stem cells and tumor development: an update and outstanding questions. Stem Cells 33, 2381–2390 (2015).
Acknowledgements
We thank I. Durand from the CRCL cytometry platform, G. Clapisson and S. Tabone-Eglinger, managers of Plateforme de Gestion des Echantillons Biologiques of Centre Léon Bérard, Prof. E. Delabesse (Institut de Cancérologie de Toulouse, France) for access to samples from HIMIP collection and Prof. O. Herault for helpful discussions. We greatly thank Mr. S. Morisset for expert statistical analyses, Dr. Janet Hall for improving the quality of the manuscript and Dr. S. Gobert for administrative support. We thank the association Anim’ Montbernier, Ruy Montceau and its president Mr. Armand Glasson for support. This work was supported by the French Ligue Nationale Contre le Cancer: Comité de la Loire and Comité de Saône et Loire, Fondation de France (20140047501), Association Laurette Fugain (ALF2014/03) and the association “Cent pour Sang la Vie”. M.F.-V. and F.Z. respectively received Conacyt and ARC PhD grants.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by D. Aberdam
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Voeltzel, T., Flores-Violante, M., Zylbersztejn, F. et al. A new signaling cascade linking BMP4, BMPR1A, ΔNp73 and NANOG impacts on stem-like human cell properties and patient outcome. Cell Death Dis 9, 1011 (2018). https://doi.org/10.1038/s41419-018-1042-7
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41419-018-1042-7