Abstract
The lncRNA MALAT1 has multiple biological functions, including influencing RNA processing, miRNA sponging, and cancer development. It is acknowledged that miR663a and its targets are inflammation-related genes frequently deregulated in many cancers. The associations between MALAT1 and miR663a and their target genes remain unknown. In this study, it was found that in colon cancer (CC) cells, MALAT1 and miR663a were reciprocally repressed in cDNA array screening and qRT-PCR analysis. However, MALAT1 was significantly upregulated in CC tissues, and miR663a was significantly downregulated relative to the corresponding surgical margin (SM) tissues. An inverse relationship between MALAT1 and miR663a expression was detected among CC tissue samples (n = 172, r = −0.333, p < 0.0001). The RNA-pulldown results showed MALAT1 lncRNA–miR663a binding. The results of luciferase-reporter analysis further revealed that the MALAT1 7038–7059 nt fragment was the miR663a seed sequence. Both miR663a knockdown and MALAT1 activation alone significantly upregulated the expression levels of miR663a targets, including TGFB1, PIK3CD, P53, P21, and JUND, in the CC cell lines HCT116 and SW480. A positive relationship was also observed between the expression levels of MALAT1 and these miR663a targets in the above 172 CC samples and 160 CC samples in publicly available databases. In addition, reciprocal abolishment of the effects of miR663a overexpression and MALAT1 activation on the proliferation, migration, and invasion of cancer cells was also observed, while miR663a upregulation and MALAT1 activation alone inhibited and promoted the behaviors of these CC cell lines, respectively. All these suggested that, as a competing endogenous lncRNA, MALAT1 maybe a dominant protector for the degradation of miR663a targets. miR663a and MALAT1 may consist of a negative feedback loop to determine their roles in CC development.
Similar content being viewed by others
Background
In the genomes of mammals, less than 2% of the human genome comprises protein-coding sequences, and 98% comprises non-protein coding regions that may play roles in physiological and pathological processes1. Long non-coding RNAs (lncRNAs) are transcribed from non-protein coding regions that are longer than 200 nucleotides in length. With the discovery of thousands of lncRNAs, a few have been shown to participate in different biological processes through lncRNA–DNA, lncRNA–RNA, and lncRNA–protein interactions2. The metastasis-associated lung adenocarcinoma transcript 1 gene (MALAT1, NEAT2) is the first discovered human tumor-related lncRNA that promotes the progression and metastasis of cancers3. MALAT1 is ubiquitously expressed in normal tissues and is frequently upregulated in human cancers4,5,6,7,8. It may function as a component of RNA-splicing nuclear speckles or as a competing endogenous RNA (ceRNA)9.
miR663a is an inflammation-related miRNA that is frequently deregulated in human cancers. Although the expression level of miR663a was significantly upregulated in prostate and nasopharynx cancers, it was markedly downregulated in brain and pancreatic cancers10,11,12,13,14. It was reported that miR663a inhibited the growth of colon cancer (CC) cells15. Although several protein-coding genes have been reported to be miR663a targets, upstream networks regulating miR663a functions and the mechanisms underlying the effects of miR663a on CC development and progression are unclear.
In the present study, we found for the first time that MALAT1 and miR663a directly interacted with, and reciprocally repressed each other. Most importantly, we found that MALAT1 is a dominant inhibitor of miR663a function through preventing the degradation of most miR663a targets that are involved in CC development.
Results
MALAT1 expression is mostly decreased by miR663a in CC cells
To screen miR663a target genes, we performed cDNA microarray analyses using CC HCT116 cells at 72 h after transfection with the miR663a expression vector and its inhibitor (antisense), respectively. Using 1.5-fold change as the cutoff value, the mRNA levels of 75 different transcripts from 71 genes were decreased by miR663a overexpression and increased by the miR663a inhibitor-knockdown. Gene ontology analysis showed that these genes were related to the mitotic cell cycle, cell proliferation, apoptotic process, cell junction assembly, cell–cell adhesion, DNA repair, and oxidative stress (Fig. 1a). Notably, MALAT1 was mostly downregulated (−3.4-fold) and mostly upregulated (+2.6-fold) among the top 71 genes (Supplemental data file 1).
a Heatmap of the expression levels of MALAT1 and other genes regulated by miR663a in cDNA assay analysis and GO_names from gene ontology analysis of the protein-coding genes regulated by miR663a [Criteria for selection of these genes: absolute value of fold change >1.5 and p-value in gene ontology analysis <0.05]. b, c qRT-PCR results to detect the MALAT1 expression changes following transient transfection with the miR663a expression vector or control vector in HCT116 and SW480 cells. d, e qRT-PCR results to detect the MALAT1 expression changes following transient transfection with the miR663a mimics and inhibitor in HCT116 and SW480 cells. *p < 0.05 using Student’s t-test
The effects of miR663a expression changes on the MALAT1 expression level was further confirmed by qRT-PCR analysis. The MALAT1 level was significantly decreased after transfection with the miR663a expression vector in HCT116 and SW480 cells (Fig. 1b, c). Similarly, MALAT1 downregulation and upregulation were also observed in these cells transiently transfected with the miR663a mimics and inhibitor, respectively (Fig. 1d, e). These results suggest that MALAT1 may be a miR663a target.
MALAT1 decreases miR663a expression in CC cells
The effects of MALAT1 expression changes on miR663a expression were further studied. Because the full-length sequence of MALAT1 is >8000 bp, it is difficult to construct a full-length MALAT1 expression vector. Hence, two small activating RNAs (saR-MALAT1-1/-2) complementarily paired to the MALAT1 promoter sequence were synthesized and used to trigger endogenous MALAT1 expression in HCT116 and SW480 cells as previously reported16,17. As expected, the endogenous MALAT1 expression level was increased at 72 h following small activation RNA (saRNA) transfection (Fig. 2a). Interestingly, the miR663a expression level was consistently and significantly decreased in these cell lines following saR-MALAT1-1/-2 transfection (Fig. 2b). In contrast, knockdown of MALAT1 expression by siR-MALAT1-1/-2 remarkably induced miR663a upregulation (Fig. 2c, d). These results support that MALAT1 and miR663a maybe reciprocally repressed.
Inverse relationship between MALAT1 lncRNA and miR663a expression in CC tissues
To confirm the relationship between MALAT1 and miR663a expression in tissues, the expression levels of MALAT1 and miR663a in CC and surgical margin (SM) tissue samples from 172 patients were analyzed by qRT-PCR. It was found that, relative to SM samples, MALAT1 expression was significantly upregulated (p < 0.001; Fig. 3a) while miR663a expression was significantly downregulated in CC samples (p < 0.001; Fig. 3b). Notably, a significantly inverse relationship between these two genes was also observed in CC samples (r = −0.333; p < 0.0001; n = 172) (Fig. 3c). However, such a relationship was not detected in an equal number of SM samples (Fig. 3d). Together, the above results strongly support that the miR663a and MALAT1 expression levels were reciprocally repressed and involved in CC development.
a, b Inverse changes in MALAT1 and miR663a expression between colon cancer (CC) and paired non-cancerous surgical margin (SM) samples. p-Values were calculated using the Wilcoxon signed-rank test. c Inverse relationship between MALAT1 and miR663a expression in colon cancer tissues, as detected by qRT-PCR. The correlation coefficient and p-value were calculated using Spearman’s rank correlation analysis. d No significant relationship was found between MALAT1 and miR663a expression in the non-cancerous surgical margin (SM) samples, as detected by qRT-PCR
MALAT1 directly interacts with miR663a
MALAT1 is a well-known miRNA sponge. To investigate whether MALAT1 is also a miR663a sponge, the RNA-pulldown assay was carried out. It was found that biotin-labeled miR663a-wt pulled down MALAT1 RNA in HCT116 cells, but biotin-labeled miR663a-mut containing a mutant 5′-end did not (Fig. 4a). This indicates that miR663a may directly bind to MALAT1 RNA through its 5′ sequence.
a Binding of wildtype miR663a and its corresponding mutant to MALAT1 in the RNA pull-down assay using GAPDH as an internal reference. Sequences of the miR663a candidate target (7038–7059 nt) within MALAT1 and miR663a mutant are listed. b Influence of miR663a on the luciferase activity of various pmiR-GLO-MALAT1 fragments. c, d Binding of various wild-type MALAT1 fragments and the corresponding mutant for MALAT1-7038 to miR663a in the RNA-pulldown assay using U6 as an internal reference. Sequences of the miR663a seed site within MALAT1 and its mutant are listed. e Mutation of the 7038–7059 nt seed site within MALAT1 abolished the influence of miR663a on the pmiRGLO-MALAT1-7038 luciferase activity. f The expression status of endogenous miR663a in HCT116 and SW480 cells transfected with MALAT1-7038 and its mutant expression vector for 72 h. *p < 0.05 using Student’s t-test
In bioinformatics analysis (RNA22v2), six miR663a candidate seed sequences were found within MALAT1 RNA (Supplemental data file-2), with a high absolute value of folding energy and low p-value18. To characterize miR663a-binding sequence(s) within MALAT1 RNA, six DNA fragments (approximately 500 bp, each containing one seed sequence) were synthesized, inserted into the pmiR-GLO vector, and used in dual-luciferase reporter analysis. The results showed that miR663a only decreased the reporter activity of the MALAT1-7038 fragment (containing the 7038–7059 nt seed sequence; Fig. 4b). Similarly, miR663a was only pulled down by the biotin-labeled MALAT1-7038-wt fragment, but not by other five fragments (Fig. 4c), nor by the biotin-labeled MALAT1-7038-mut fragment containing a mutant seed sequence in RNA-pulldown assay in HCT116 cells (Fig. 4d). Mutation of the seed sequence completely abolished the influence of miR663a on the MALAT1 reporter activity (Fig. 4e). In addition, relative to the pEGFP-C1 control vector, the endogenous miR663a expression levels were significantly repressed in both HCT116 and SW480 cells transfected with the MALAT1-7038-wt vector but not with the MALAT1-7038-mut vector (Fig. 4f). The above results demonstrated that miR663a could directly bind to the MALAT1-7038 seeding site.
As a master ceRNA, MALAT1 dominantly prevents the degradation of most miR663a targets
To evaluate the significance of miR663a downregulation by MALAT1, the expression changes of a set of miR663a target genes, including P5319, PIK3CD12, P2111, CXCR413, TGFB120, and JUND21, were further studied in HCT116 cells. As expected, miR663a downregulation by the inhibitor significantly increased the expression levels of these genes (Fig. 5a, #1 vs #3). Similarly, MALAT1 activation by saR-MALAT1-1 significantly increased the expression levels of most of these genes, except P21 (Fig. 5a, #1 vs #2). However, when miR663a was knocked down by the inhibitor, MALAT1 activation could no longer increase the expression levels of these genes (Fig. 5a, #3 vs #4). Similar results were also observed in SW480 cells (Fig. 5b). The results of Western blotting analysis confirmed the expression changes of these target genes in HCT116 and SW480 cells. Both MALAT1 activation and miR663a downregulation increased the protein levels of most miR663a targets, including P53, PIK3CD, P21, CXCR4, and TGFB1 (Fig. 5c, d, #1 vs #2 and #3). These results suggest that MALAT1 may dominantly prevent the degradation of miR663a target genes in a miR663a-dependent pattern.
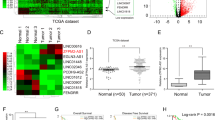
To confirm the positive relationships between the expression of MALAT1 and miR663a targets, we analyzed the correlation between the MALAT1 and miR663a target expression levels among 172 CC patients described above. As expected, the MALAT1 expression level was significantly and positively correlated with expression levels of all these genes, including P53, PIK3CD, P21, CXCR4, TGFB1, and JUND (Fig. 6a). To validate these positive relationships, the publicly available cDNA array datasets for primary colorectal cancers (n = 160; GEO GSE24551) were re-analyzed. Once again, the expression levels of almost all miR663a targets were all significantly and positively correlated with the MALAT1 RNA levels in these tissues (Fig. 6b). Collectively, the above results indicate that MALAT1 may be a master ceRNA that could greatly control the expression levels of these miR663a target genes in CC tissues.
a Colon cancer patients (n = 172) listed in Table S1. b Colon cancer patients (n = 160; from the publicly available database GSE24551). The correlation coefficient and p-value were calculated using Spearman’s rank correlation analysis
Reciprocal abolishment of the effects of miR663a and MALAT1 on the behavior of CC cells
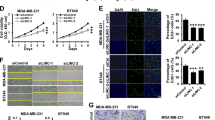
MALAT1 is the first characterized oncogenic lncRNA that promotes the progression of many cancers. To evaluate the importance of miR663a in the oncogenic effects of MALAT1 on cancer cells, we performed IncuCyte long-term dynamic proliferation/migration and Transwell invasion analysis. As expected, the downregulation of endogenous miR663a by its inhibitor promoted the proliferation and migration of HCT116 and SW480 cells (Fig. 7a–d), while the downregulation of endogenous MALAT1 by siR-MALAT1-1/2 inhibited the proliferation and migration of these cells (Fig. 7e–h). By contrast, MALAT1 upregulation by saR-MALAT1 significantly promoted the proliferation, migration, and invasion of HCT116 and SW480 cells, while miR663a overexpression decreased the proliferation, migration, and invasion of these cells (Fig. 8). Interestingly, MALAT1 upregulation combined with miR663a overexpression did not affect the proliferation, migration, and invasion of these cells, indicating reciprocal abolishment of their biological effects on these cells.
Discussion
MALAT1, as a component of nuclear paraspeckles, is a well-studied lncRNA involved in pre-mRNA splicing22. It also functions as a miRNA sponge to competitively upregulate miRNA targets23,24,25,26. Although MALAT1 is the first characterized oncogenic lncRNA, the mechanisms underlying its effects on cancer development remain unclear9. In the present study, we found, for the first time, that MALAT1 and miR663a reciprocally repressed each other through sequence-dependent binding. Moreover, MALAT1 could competitively prevent the degradation of a set of miR663a targets in vitro and in vivo, suggesting it may be a dominant regulator for miR663a functions in CC cells. MALAT1 and miR663a may consist of a negative feedback loop and are involved in CC cancer development.
It was reported that the 6918–8441 nt fragment within the MALAT1 RNA plays a pivotal role in the biological processes of cell proliferation, migration, and invasion of CC cells27. We found that the 7038–7059 nt sequence within the 6918–8441 nt fragment is essential for the miR663a–MALAT1 RNA–RNA binding. Mutation of this miR663a binding site within MALAT1 RNA could completely abolish the miR663a–MALAT1 interaction in both the RNA-pulldown and dual-fluorescence reporter assays. Interaction of miR663a with other MALAT1 fragments was not observed. These results demonstrated that the 7038–7059 nt sequence is the only miR663a binding site within MALAT1 RNA and may be an essential point in the feedback loop.
It is well recognized that miRNA promotes the degradation of mRNA through interacting with mRNA 3UTR. Although it is not necessary to expect a ceRNA to control the stability of its interacting molecules, however, the present study indicates that a ceRNA MALAT1 could promote the endogenous miR663a degradation and vice versa. This suggests important effects of the miR663a–MALAT1 loop on the reciprocal regulation of their biological functions.
MALAT1 may be sheared by endogenous RNase P and RNase Z into a longer segment [1–7072 nt] and a shorter tRNA-like RNA (~60 nt) from the 3′ terminus that would be transported into the cytoplasm28. The miR663a-binding 7038–7059 nt sequence is located within the longer MALAT1 segment. Although the longer MALAT1 segment stays in the nucleus, many reports have shown that MALAT1 could function as a sponge for many miRNAs29,30,31. In proliferating cells from prophase to anaphase in the cell cycle, disruption of the nuclear membrane leads to merging of the cytoplasm with the nucleus. If nuclear lncRNAs indeed interact with cytoplasmic miRNAs in cells at these mitotic stages, these interactions should be cell proliferation-dependent. This might account for the significantly inverse miR663a–MALAT1 relationship observed in CC tissues containing a high proportion of proliferating cancer cells but not in an equal number of SM samples containing a few proliferating tissue stem cells. It is an important issue to address the possible roles of the disruption of the nuclear membrane in actively proliferating cells in occurrence of interactions between nuclear components and cytoplasm components.
Most importantly, our results revealed that miR663a overexpression accompanied with MALAT1 upregulation did not affect the proliferation, migration, and invasion of CC cells, whereas miR663a downregulation alone or MALAT1 upregulation alone promoted the proliferation, migration, and invasion of CC cells. The miR663a gene is a primate-specific gene that is absent in the mouse and rat genomes. To study whether MALAT1 may affect the proliferation, migration, and invasion of CC cells in a miR663a-dependent manner, other animal models should be used.
Chronic inflammation may play a causal role in CC development. It was reported that MALAT1 expression is increased32,33,34 and miR663a expression is decreased in inflammatory tissues35,36. We found obvious MALAT1 overexpression and miR663a downregulation in CC tissues. The long-term administration of aspirin, as an antioxidant, decreases CC risk in the population. Antioxidant resveratrol treatment not only could prevent inflammation but could also restore the expression status of MALAT1 and miR663a to normal levels20,21,37. It is unknown whether aspirin treatment has similar effects. The effects of re-balancing MALAT1 and miR663a expression by antioxidants on CC development should be studied in the future.
We found that the expression levels of a set of miR663a targets were positively and significantly associated with MALAT1 expression in both 172 CC samples analyzed in the present study and 160 CC samples from public databases and that both miR663a knockdown and MALAT1 activation increased the expression levels of these target genes. These phenomena indicated that MALAT1 may be a dominant regulator for miR663a targets.
In conclusion, we found that MALAT1 was significantly upregulated and miR663a was significantly downregulated in CC tissues. MALAT1 and miR663a could reciprocally repress each other through direct RNA–RNA binding. MALAT1 might be a dominant regulator for miR663a targets through competitive interactions with miR663a. MALAT1 and miR663a could reciprocally abolish their biological functions in CC development, which might be a useful intervention target for CC prevention.
Materials and methods
Synthesis of RNA nucleotides and plasmids
For MALAT1 upregulation, saRNA sequences were designed to target the MALAT1 promoter, including saR-MALAT1-1 (574–592 nt; forward: 5′-cgaga auucu agacu aguatt-3′; reverse: 5′-uacua gucua gaauu cucgtt-3′) and saR-MALAT1-2 (330–348 nt; forward: 5′-gcaga guagc gaccg agaatt-3′; reverse: 5′-uucuc ggucg cuacu cugctt-3′)16,17. For MALAT1 knockdown, siRNA sequences were siR-MALAT1-1 (5098–5116 nt; forward: 5′-gcaaa ugaaa gcuac caau-3′; reverse: 5′-auugg uagcu uucau uugctt-3′) and siR-MALAT1-2 (6392–6410 nt; forward: 5′-gcaga ggcau uucau ccuu-3′; reverse: 5′-aagga ugaaa ugccu cugctt-3′).
The wildtype MALAT1 fragments (approximately 500 bp; Supplemental data file-2) and its miR663a-seeding-site mutant were synthesized and inserted into the pmiR-GLO vector. To construct the MALAT1-7038 fragment and its mutant expression vectors, the PCR products were amplified from pmiR-GLO-MALAT1 and pmiR-GLO-7038-mutant vectors using a primer set (forward: 5′-ggaaa tttct gcagt tttaa-3′; reverse: 5′-ttcac ctgtt ttcct cattt-3′) and were inserted into the pEGFP-C1 vector.
The sequences of the miR663a mimics were as follows: forward: 5′-aggcg gggcg ccgcg ggaccgc-3′; reverse: 5′-ggucc cgcgg cgccc cgccuuu-3′. A scrambled siRNA set (NC, forward 5′-uucuc cgaac guguc acgutt-3′ and reverse 5′-acgug acacg uucgg agaatt-3′) was used as the saRNA/siRNA/mimics negative control. The sequence of the miR663a inhibitor (antisense) was 5′-gcggu cccgc ggcgc cccgc cu-3′. A scrambled RNA inhibitor (Inhibitor NC, 5′-cagua cuuuu gugua guacaa-3′) was used as the inhibitor negative control. All these synthesized RNA products were purchased from Genepharma (Shanghai, China). The miR663a expression vector pcDNA3.1b-pri-miR663a was constructed using 93 bp PCR products amplified from the genomic DNA of A549 cells using HiFi DNA polymerase (Transgen Biotech, Beijing, China) and the primer set (forward: 5′-ccttc cggcg tccca ggcg-3′; reverse: 5′-catgg ccggg ccacc aggaaa-3′).
Cells culture and tissue samples
Human CC cell lines HCT116 and SW480 were purchased from the American Type Culture Collection (ATCC, Manassas, USA). These cell lines were cultured in RPMI 1640 medium or DMEM medium containing 10% FBS and 100 U/mL penicillin/streptomycin (Invitrogen, CA, USA) at 37 °C in a humidified incubator with 5% CO2. X-tremeGENE siRNA Transfection Reagent or X-tremeGENE HP DNA Transfection Reagent (Roche, Mannheim, Germany) were used in the cell transfection of miRNAs (final concentration, 100 nM) or plasmids (2 μg/well in 6-well plates) following the manufacturer’s instructions. The overexpression or knockdown efficiency was determined after transfection by reverse transcription-PCR (RT-PCR) and Western blotting at the indicated time points.
The CC tissues and paired non-cancerous SM samples were collected from 172 patients (average age, 61.64 years; 101 males and 71 females; 89 CCs at pTNM stage I–II and 83 CCs at the stage III–IV) at the Peking University Cancer Hospital and Institute from 2004 to 2011 (Table S1). The Peking University Cancer Hospital and Institute Review Boards approved this study. All patients provided written informed consent to participate in the study.
cDNA array and bioinformatics analysis
HCT116 cells were harvested at 72 h after transient transfection of the pcDNA3.1b-pri-miR663a vector, antisense/inhibitor, and corresponding negative controls. Total RNA was extracted with TRIzol reagent (Life Technologies, CA, USA), reverse transcribed to cDNA, and used for Affymetrix U133Plus 2.0 array analysis. The gene expression levels were assessed by the fluorescence signal attached to the probe and then to template strand in PCR. After log transformation, the fold changes were calculated according to the miR663a expression vector group vs pcDNA3.1b vector control group, or antisense/inhibition group vs inhibitor NC group. The gene expression levels changed inversely in the above two comparisons were selected as miR663a candidate targets (Supplemental data file-1).
Quantitative RT-PCR
The quality and concentration of RNA samples were monitored using the NanoDrop 2000 system (Thermo Fisher Scientific, Waltham, MA, USA). Qualified RNA samples were used to synthesize cDNA using the TransScript First-Strand cDNA Synthesis SuperMix (TransGen Biotech, Beijing, China). Because of the absence of introns within MALAT1 RNA, to exclude false MALAT1-amplification from DNA templates, the total RNA was pre-digested with DNase before cDNA synthesis. Quantitative RT-PCR (qRT-PCR) was performed using a StepOne Real-time PCR System (Applied Biosystems, Foster City, CA, USA) and SYBR Green PCR master mix reagents (FastStart Universal SYBR Green Master, Roche, Mannheim, Germany). MALAT1 and the protein-coding gene expression levels were normalized to those of GAPDH (for cells) and Alu (for tissues). The relative mRNA level was calculated using the classical delta-delta-Ct method. The Bulge-Loop™ miRNA qRT-PCR starter kit (RuiBO, Guangzhou, China) was used to determine the miR663a levels, and the U6 transcription level was used as a reference. The sequences of primers used in these PCR assays are listed in Table S2. Each sample was determined in triplicate.
RNA-pulldown assay
In total, 3 × 106 HCT116 cells were seeded on the 10-cm plate for 24 h. Next, these cells were transfected with biotin-labeled NC (forward: 5′-uucuc cgaac guguc acgutt-3′; reverse: 5′-acgug acacg uucgg agaatt-3′), biotin-labeled miR663a-wt (forward: 5′-aggcg gggcg ccgcg ggacc gc-3′; reverse 5′-gcggu cccgc ggcgc cccgc cu-3′), or biotin-labeled miR663a-mut (forward: 5′-cuuac auucg ccgcg ggacc gc-3′; reverse: 5′-gcggu cccgc ggcga augua ag-3′) at a final concentration of 100 nM. The cells were harvested at 48 h post-transfection. Activated Streptavidin-Dyna beads (Dyna beads M-280 Streptavidin, #11205D, Invitrogen) were coated with 10 μL per sample yeast tRNA (10 mg/mL stock; Ambion, Austin, USA) and 10 μL BSA (10 ng/mL stock) and were incubated in the lysis buffer (480 μL) with rotation at 4 °C for 0.5 h. The beads were then washed, and the sample lysates (600 μL) were mixed with pre-coated beads (50 μL per sample) and incubated at 4 °C for 4 h on a rotator. The beads were then pelleted down the next day to remove unbound materials at 4 °C for 2 min, 500g, and were washed six times with 500 μL of ice cold lysis buffer. The MALAT1 levels in the pulldown samples were detected by qRT-PCR and normalized using GAPDH as an internal reference.
In the MALAT1-pulldown assay, various wild-type MALAT1 fragments and MALAT1-7038 mutant control were amplified from the corresponding pmiR-GLO-MALAT1 vector using the corresponding primer set (Table S2), digested with restriction enzyme SalI, inserted into the pGEM-T vector, and transcribed by T7 RNA Polymerase using Riboprobe in vitro Transcription Systems (P1460, Promega, Madison, WI, USA). The RNA was labeled with PierceTM RNA 3′ End Desthiobiotinylation Kit (20163, Thermo Scientific, Rockford, IL, USA). The MALAT1 binding RNA was captured by magnetic beads and was used to pull down MALAT1-interacting RNA as described above. The miR663a levels in the pulldown samples were detected by qRT-PCR, and normalized using U6 as an internal reference.
Western blotting
The primary monoclonal antibodies used in Western blot analyses were sc-126 for P53, sc-55589 for PIK3CD, sc-53534 for CXCR-4 (Santa Cruz Biotechnology, Inc, Santa Cruz, CA, USA), CST-2947 for P21 (Cell Signaling Technology, Danvers, MA, USA), ab-25121 for TGFB1 (Abcam, Cambridge, UK), QJ221464 for JUND (Thermo Fisher Scientific, Waltham, MA, USA), and 60004-1-Ig for GAPDH (Proteintech, Rosemont, IL, USA). The signals were visualized using the enhanced chemiluminescence kit (Pierce, Rockford, IL, USA).
Dual-luciferase reporter assay
The pmiR-GLO-MALAT1 and pmiR-GLO-7038-mutant vectors were used to transfect HCT116 cells in 24-well plate. MiR663a mimics or NC were transfected on the second day (3 wells/group). At 72 h post-transfection, the activities of both Renilla and firefly luciferases were measured using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA). The results were presented after normalization with the measured values of the firefly luciferase.
Cell proliferation and migration assays using IncuCyte
HCT116 and SW480 cells were seeded into 96-well plates (3000 cells/well, 5 wells/group), and were cultured for at least 96 h to determine the proliferation curves. The cells were photographed every 12 h in the long-term dynamic observation platform (IncuCyte, Essen, MI, USA). The cell confluence was analyzed using IncuCyte ZOOM software (Essen, Ann Arbor, MI, USA). For continuous observation for cell migration, the cells were seeded into 96-well plates at a density of 10,000 cells/well and were cultured for 24 h. After a wound was scratched, the cells were washed three times with PBS. The cells were regularly cultured and photographed every 12 h for at least 96 h. Both the relative wound density and width were calculated using the same software.
Transwell assays
Twenty-four-well plates of Transwell permeable, which supports with a 6.5-mm insert and an 8.0-μm polycarbonate membrane (Corning 3422, Kennebunk, ME, USA), were used in the cell invasion assay. The upper chamber was pre-coated with 100 μL of the BD Matrigel mixture (BD Biosciences, San Jose, CA, USA) and 1640 medium (Gibco, Waltham, MA, USA, 1:5, v/v). To the lower chamber was added 800 μL of 1640 with 10% FBS. The HCT116 cells were seeded into the upper chamber (30,000 cells/well, 3 wells/group) with 1640 medium without FBS. After 24 or 36 h of incubation, the 6.5-mm insert was removed from the plate and fixed in 4% paraformaldehyde, followed by staining with crystal violet staining solution. Non-migrated/noninvasive cells on the upper surface of the insert were wiped with a cotton swab. The migrated/invaded cell number was manually counted in four randomly selected fields under a light microscope. All experiments were performed at least three times.
Statistical analyses
All statistical analyses were performed using SPSS 18.0 software. The Kolmogorov–Smirnov test was used to estimate the normality of distributions. The Mann–Whitney U-test was conducted for non-normally distributed data. Student’s t-test was conducted for normally distributed data. The relationship between the levels of MALAT1 and miR663a target genes was measured using the nonparametric correlation test and curvilinear regression model. Statistical significance was assigned at p < 0.05 (*) or p < 0.01 (**). All experiments were performed at least three times with triplicate samples.
References
Birney, E. et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project ENCODE Project Consortium. Nature 447, 799–816 (2007).
Lin, C. & Yang, L. Long noncoding RNA in cancer: wiring signaling circuitry. Trends Cell Biol. 28, 287–301 (2018).
Ji, P. et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 22, 8031–8041 (2003).
Zidan, H. E., Karam, R. A., El-Seifi, O. S. & Abd Elrahman, T. M. Circulating long non-coding RNA MALAT1 expression as molecular biomarker in Egyptian patients with breast cancer. Cancer Genet. 220, 32–37 (2018).
Li, C. et al. High expression of long noncoding RNA MALAT1 indicates a poor prognosis and promotes clinical progression and metastasis in bladder cancer. Clin. Genitourin. Cancer 15, 570–576 (2017).
Droop, J. et al. Diagnostic and prognostic value of long noncoding RNAs as biomarkers in urothelial carcinoma. PLoS One 12, e0176287 (2017).
Li, C. et al. The role of lncRNA MALAT1 in the regulation of hepatocyte proliferation during liver regeneration. Int. J. Mol. Med. 39, 347–356 (2017).
Wang, X. et al. LncRNA MALAT1 promotes development of mantle cell lymphoma by associating with EZH2. J. Transl. Med. 14, 346 (2016).
Zhang, X., Hamblin, M. H. & Yin, K. J. The long noncoding RNA Malat1: its physiological and pathophysiological functions. RNA Biol. 14, 1705–1714 (2017).
Wang, S., Liu, J., Li, C. & Yang, X. Research of the effect of miR-663 on the proliferation of prostate cancer cells, and the correlations of miR-663 with pathological grade and clinical stage. J. BUON 22, 1011–1016 (2017).
Yi, C. et al. MiR-663, a microRNA targeting p21(WAF1/CIP1), promotes the proliferation and tumorigenesis of nasopharyngeal carcinoma. Oncogene 31, 4421–4433 (2012).
Shi, Y. et al. Primate-specific miR-663 functions as a tumor suppressor by targeting PIK3CD and predicts the prognosis of human glioblastoma. Clin. Cancer Res. 20, 1803–1813 (2014).
Shi, Y. et al. miR-663 suppresses oncogenic function of CXCR4 in glioblastoma. Clin. Cancer Res. 21, 4004–4013 (2015).
Zang, W. et al. miR-663 attenuates tumor growth and invasiveness by targeting eEF1A2 in pancreatic cancer. Mol. Cancer 14, 37 (2015).
Kuroda, K. et al. miR-663a regulates growth of colon cancer cells, after administration of antimicrobial peptides, by targeting CXCR4-p21 pathway. BMC Cancer 17, 33 (2017).
Yang, M. H. et al. MALAT1 promotes colorectal cancer cell proliferation/migration/invasion via PRKA kinase anchor protein 9. Biochim. Biophys. Acta 1852, 166–174 (2015).
Hu Z. Y., et al. Long non-coding RNA MALAT1 increases AKAP-9 expression by promoting SRPK1-catalyzed SRSF1 phosphorylation in colorectal cancer cells. Oncotarget. 7, 11733–11743 (2016).
Miranda, K. C. et al. A pattern-based method for the identification of microRNA binding sites and their corresponding heteroduplexes. Cell 126, 1203–1217 (2006).
Cho, J. G. et al. ZNF224, Krüppel like zinc finger protein, induces cell growth and apoptosis-resistance by down-regulation of p21 and p53 via miR-663a. Oncotarget 7, 31177–31190 (2016).
Tili, E. et al. Resveratrol modulates the levels of microRNAs targeting genes encoding tumor-suppressors and effectors of TGFβ signaling pathway in SW480 cells. Biochem. Pharmacol. 80, 2057–2065 (2010).
Tili, E. et al. Resveratrol decreases the levels of miR-155 by upregulating miR-663, a microRNA targeting JunB and JunD. Carcinogenesis 31, 1561–1566 (2010).
Clemson, C. M. et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell 33, 717–726 (2009).
Fu, Z. et al. Malat1 activates autophagy and promotes cell proliferation by sponging miR-101 and upregulating STMN1, RAB5A and ATG4D expression in glioma. Biochem. Biophys. Res. Commun. 492, 480–486 (2017).
Shi, B., Wang, Y. & Yin, F. MALAT1/miR-124/Capn4 axis regulates proliferation, invasion and EMT in nasopharyngeal carcinoma cells. Cancer Biol. Ther. 18, 792–800 (2017).
Hou, Z. et al. The long non-coding RNA MALAT1 promotes the migration and invasion of hepatocellular carcinoma by sponging miR-204 and releasing SIRT1. Tumour Biol. 39, https://doi.org/10.1177/1010428317718135 (2017).
Chen, L., Yao, H., Wang, K. & Liu, X. Long non-coding RNA MALAT1 regulates ZEB1 expression by sponging miR-143-3p and promotes hepatocellular carcinoma progression. J. Cell. Biochem. 118, 4836–4843 (2017).
Xu, C., Yang, M., Tian, J., Wang, X. & Li, Z. MALAT-1: a long non-coding RNA and its important 3′ end functional motif in colorectal cancer metastasis. Int. J. Oncol. 39, 169–175 (2011).
Jeremy, E., Freier, S. M. & Spector, D. L. 3′ End processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell 135, 919–932 (2008).
Leucci, E. et al. microRNA-9 targets the long non-coding RNA MALAT1 for degradation in the nucleus. Sci. Rep. 3, 2535 (2013).
Li, J. et al. LncRNA MALAT1 exerts oncogenic functions in lung adenocarcinoma by targeting miR-204. Am. J. Cancer Res. 6, 1099–1107 (2016).
Liu, W. et al. Long non-coding RNA MALAT1 contributes to cell apoptosis by sponging miR-124 in Parkinson disease. Cell Biosci. 7, 19 (2017).
Zhang, J. et al. Genome-wide analysis of long noncoding RNA profiling in PRRSV-infected PAM cells by RNA sequencing. Oncol. Rep. 35, 3371–3386 (2016).
Yang, S., Yao, H., Li, M., Li, H. & Wang, F. Long non-coding RNA MALAT1 mediates transforming growth factor beta1-induced epithelial–mesenchymal transition of retinal pigment epithelial cells. PLoS ONE 11, e0152687 (2016).
Puthanveetil, P., Chen, S., Feng, B., Gautam, A. & Chakrabarti, S. Long non-coding RNA MALAT1 regulates hyperglycaemia induced inflammatory process in the endothelial cells. J. Cell. Mol. Med. 19, 1418–1425 (2015).
Dalal, S. R. & Kwon, J. H. The role of microRNA in inflammatory bowel disease. Gastroenterol. Hepatol. 6, 714–722 (2010).
Tili, E. & Michaille, J. J. Resveratrol, microRNAs, inflammation, and cancer. J. Nucleic Acids 2011, 102431 (2011).
Ji, Q. et al. Resveratrol inhibits invasion and metastasis of colorectal cancer cells via MALAT1 mediated Wnt/b-catenin signal pathway. PLoS ONE 8, e78700 (2013).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 81372592) to D.D. We thank Dr. Jian-Ping Guo from the Dana Farber Cancer Institute for guidance in the RNA pull-down assay. We also thank Dr. Jin-Bing Bai from Emory University for revising the paper.
Author information
Authors and Affiliations
Contributions
D.D. designed the research; W.T., Y.D., Y.M., L.G., and J.Z. performed the research; W.T., Y.D., and D.D. analyzed data and wrote the paper; and D.D. conceived the idea, obtained the financial support, and supervised the entire project. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by G. Ciliberto
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Tian, W., Du, Y., Ma, Y. et al. MALAT1–miR663a negative feedback loop in colon cancer cell functions through direct miRNA–lncRNA binding. Cell Death Dis 9, 857 (2018). https://doi.org/10.1038/s41419-018-0925-y
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41419-018-0925-y
This article is cited by
-
LncRNA miR663AHG represses the development of colon cancer in a miR663a-dependent manner
Cell Death Discovery (2023)
-
TTC22 promotes m6A-mediated WTAP expression and colon cancer metastasis in an RPL4 binding-dependent pattern
Oncogene (2022)
-
Modulation of macrophage inflammatory function through selective inhibition of the epigenetic reader protein SP140
BMC Biology (2022)
-
Knockdown of LINC00467 contributed to Axitinib sensitivity in hepatocellular carcinoma through miR-509-3p/PDGFRA axis
Gene Therapy (2021)
-
Long noncoding RNAs: functions and mechanisms in colon cancer
Molecular Cancer (2020)