Abstract
The receptor for advanced glycation end products (Rage) is involved in the development of various tumors and acts as an oncogenic protein. Rage is overexpressed in tumors including hepatocellular carcinoma (HCC). However, the molecular mechanism of Rage in HCC progression and sorafenib resistance remains unclear. In this study, enhanced Rage expression is highly associated proliferation and contributes to sorafenib resistance. Rage deficiency contributed to autophagy induction through activating AMPK/mTOR signaling pathway, which is important for sorafenib response. Moreover, the interactions between Rage and Rage ligands such as high mobility group box 1 (HMGB1) and s100a4 positively increased Rage expression. Our data indicate that Rage may be a potential target for therapeutic intervention in HCC and biomarker for sorafenib resistance.
Similar content being viewed by others
Introduction
Hepatocellular carcinoma (HCC) is the fifth common malignancy and the third main cause of cancer-associated death worldwide1. Since the high rate of vascular invasion, metastasis, and recurrence following curative liver resection, the prognosis of patients with HCC is extremely poor and the 5-year survival rate is no more than 40%2. Sorafenib has been used as the first-line systematic therapy for advanced HCC patients and improves 3 months survival3. However, the response rate of sorafenib treatment is very low and the molecular mechanism involved in sorafenib resistance has not been well elucidated. Therefore, it is imperative to find novel targets in HCC development and improve HCC response to sorafenib therapy.
Autophagy is a highly conserved catabolic process for removing and recycling damaged intracellular organelles4. Autophagy has been reported to play a critical role in many diseases including tumors5,6,7. Stress-induced autophagy such as nutrient deprivation could promote tumor cell survival, but excessive autophagy also lead to cell injury and apoptosis8. Accumulating data indicate autophagy is the leading cause for sorafenib resistance. Recently, Wu et al. reveal that the beta-2 adrenergic receptor contributes to autophagy inhibition through disrupting Beclin1/VPS34/Atg14 complex in an Akt-dependent manner, which accounts for sorafenib chemoresistance9. AMPK/mTOR signaling pathway has been demonstrated to be important in autophagy activation. Additionally, dysregulation of AMPK/mTOR is involved in sorafenib resistance10.
The receptor for advanced glycation end products (Rage) is usually expressed in diverse types of cells11. Rage has been demonstrated in the progression of many inflammatory diseases including tumors11,12,13. The interactions between Rage and its ligands, such as high mobility group box 1 (HMGB1) and s100 protein family, stimulate the activation of NF-kappaB, MAPK, and Akt/mTOR signaling pathways12,14. Recent studies indicate the relationship between Rage and autophagy15,16. However, the role of Rage in HCC development and sorafenib resistance has been rarely reported.
In our present study, Rage was highly expressed in HCC and promoted the proliferation of HCC cells. Reduced Rage expression improved sorafenib response by increasing autophagy in an AMPK/mTOR-dependent way. Moreover, interactions of Rage and Rage ligands were responsible for Rage expression, which contributed to the repression of sorafenib response. Together, we suggest Rage could be a novel target for HCC treatment and improvement of sorafenib resistance.
Results
Rage is overexpressed in HCC tissues
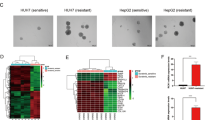
To determine the role of Rage in HCC progression, we initially evaluate Rage expression in 18 HCC specimens. We found that Rage protein expression was significantly higher in tumors compared to non-tumors (Fig. 1a, b). Moreover, we examined the expression of Rage in 68 paraffin-fixed HCC tissues (Supplementary Table S1). We observed different levels of Rage were expressed both in HCC cells and stromal cells (Fig. 1c–e). 14.9% of patients were Ragehigh in both tumor cells and stromal cells. 51.7% of patients were Ragelow in both tumor cells and stromal cells. To demonstrate the clinical relevance of Rage, Rage expression of 68 HCC tissues was quantified. Compared to non-tumor tissues, Rage expression in tumors was the highest, which further confirmed the results of protein levels (Fig. S1A–B). For further analysis, patients were seprated into two groups with high and low Rage expression groups. We found that Rage expression was statistically associated with TNM stage, which suggested that Rage played an important role in HCC progression.
a Immunoblot analysis shows that Rage expression is higher in tumors than non-tumors. b Protein levels of Rage were normalized to GAPDH and analyzed. c IHC of Rage in HCC samples. Representative images show different intensities of staining of Rage in tumor cells and stromal cells. Scale bars, 100 μm. d, e Different intensities of staining of Rage in tumor cells and stromal cells were analyzed. Data are mean ± SEM, *p < 0.05, **p < 0.01, ***p < 0.001 by unpaired Student’s t-test
High expression of Rage promotes HCC proliferation
We evaluated Rage expression in four HCC cell lines, finding different protein levels of Rage in various cell lines (Fig. 2a, Fig. S2). To explore the role of Rage in HCC development, Rage siRNA was well designed and Rage-deficient cells (Bel7402 siR and HCCLM3 siR) were established (Fig. 2b). We found that, compared to normal cells, the proliferation of Bel7402 siR and HCCLM3 siR was significantly decreased (Fig. 2c). Moreover, edu assays and colony formation experiments were performed and results were similar, which enhanced the role of Rage in proliferation of HCC cells. Proliferation-related proteins, such as p53, Cyclin D, and Bcl-2 were all downregulated in Rage ablation cells. To further investigate the role of Rage in proliferation, immunohistochemistry staining Ki67 were conducted in HCC tissues and analyzed. We demonstrated that there was a positive correlation between Rage and Ki67. These finding indicated that high expression of Rage promoted the proliferation of HCC cells.
a Immunoblot analysis of different expression of Rage in four HCC cell lines. b Immunoblot analysis confirmed the downregulation of Rage in cells treated with Rage siRNA. c Rage ablation inhibited proliferation of HCC cells. d Edu assays were performed and analyzed. e Colony formations were performed and analyzed. f, g Rage ablation contributed to reduction of cyclin D, p53, and Bcl-2. Protein levels of cyclin D, p53, and Bcl-2 were normalized to GAPDH and analyzed. h Rage expression was positively associated with Ki67 in HCC samples. Data are mean ± SEM, *p < 0.05, **p < 0.01, ***p < 0.001 by unpaired Student’s t-test
Autophagy is involved in Rage-mediated proliferation
Recent studies reported that Rage promoted tumor cells survival via sustaining autophagy15. Autophagy has been demonstrated to be involved in tumor progression7. Stress-induced autophagy could promote cell survival, but excessive autophagy would contribute to apoptosis8,17. To visualize the change of autophagy, immunofluorescence staining lc3b were performed. Accumulating of lc3 puncta was observed in Rage ablation cells, which suggested that Rage deficiency could increase the level of autophagy (Fig. 3a). In addition, the ratio of lc3b/lc3a in Rage knockdown cells was higher than normal cells, which confirmed the findings that autophagy was induced in Rage-deficient cells (Fig. 3b). Using transmission electron microscope (TEM), we observed more autophagesomes characterized by double-membrane vehicles engulfing cellular organelles in Rage ablation cells (Fig. 3c). To determine the relationship between autophagy and impaired proliferation both caused by Rage ablation, Bel7402 siR and HCCLM3 siR were treated with chloroquine (CQ), an autophagy inhibitor, for 24 h. With the treatment of CQ, we found that the impairment of proliferation in Rage ablation cells was partly rescued, which indicated that autophagy induction was responsible for proliferation repression (Fig. 2d). Similar results were got from edu assays (Fig. 2e). Moreover, we found repression autophagy in Rage ablation cells via CQ re-expressed the expressions of p53 and Cyclin D (Fig. 3f, g). These data suggested that autophagy induction caused by Rage deficiency accounted for proliferation inhibition.
a Immunofluorescence staining lc3b were performed. b Immunoblot analysis of lc3b and Rage. c Autophagesomes were shown by transmission electron microscopy. d, e Autophagy induction caused by Rage deficiency was responsible for proliferation inhibition. CCK8 and edu assays were performed and analyzed. CQ (50 μM) was used to inhibit autophagy induction. f, gImmunoblot analysis shows that autophagy induction caused by Rage deficiency accounts for decrease of cyclin D and p53. Protein levels of Cyclin D and p53 were normalized to GAPDH and analyzed. CQ (50 μM) was used to inhibit autophagy induction. Data are mean ± SEM, *p < 0.05, **p < 0.01, ***p < 0.001 by unpaired Student’s t-test
AMPK/mTOR is essential for autophagy induction in Rage-deficient cells
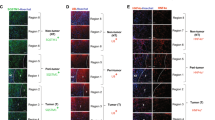
Many signaling pathways have been reported to be involved in autophagy induction, including PI3K/AKT/mTOR and AMPK/mTOR18,19,20,21. mTOR has been demonstrated to inhibit the ULK1complex, which was essential for autophagy induction at an early stage22. To examine the relationship between Rage and mTOR, immunohistochemistry staining p-mTOR and Rage were performed. We found that patients with high expression of Rage are characterized with high expression of p-mTOR (Fig. S2A). Moreover, in vitro experiments, we observed that PI3K/AKT and NF-κB signaling pathway were repressed, but AMPK signaling was activated (Fig. S2B). Both inactivation of PI3K/AKT and activation of AMPK are responsible for p-mTOR inhibition. To test the role of AMPK in Rage ablation cells, Bel7402 siR and HCCLM3 siR cells were treated with AMPK siRNA to silence AMPK expression. With the silence of AMPK, the decrease of p-mTOR and p-AKT was partly rescued, indicating that AMPK signaling played a more important role in Rage-deficient cells (Fig. S3C). Moreover, we found that silencing AMPK could repress autophagy induction caused by Rage ablation (Fig. 4a, b). Following the decrease level of autophagy, cell proliferation abilities of Rage-deficient cells were partly recovered (Fig. 4c). To further determine the relationship between Rage and AMPK/mTOR signaling, we performed immunohistochemistry staining relevant proteins and demonstrated the inverse correlation of Rage and AMPK/mTOR activation (Fig. 4d). These findings suggested that Rage deficiency-mediated autophagy induction was dependent on AMPK/mTOR signaling pathway.
a, b Autophagy induction caused by Rage ablation was dependent on AMPK/mTOR signaling pathway. c Proliferation repression caused by Rage ablation was dependent on AMPK/mTOR signaling pathway. CCK8 assays were conducted and analyzed. d IHC of Rage, p-AMPK, beclin-1, lc3b, and p62 in HCC samples. Representative images show different intensities of staining of Rage, p-AMPK, beclin-1, lc3b, and p62. Scale bars, 100 μm. Data are mean ± SEM from three independent experiments, *p < 0.05, **p < 0.01, ***p < 0.001 by unpaired Student’s t-test
Rage induces sorafenib resistance in HCC cells by modulating AMPK/mTOR signaling pathway
As a multi-kinase inhibitor, sorafenib has been proven to an effectively systematic treatment for advanced HCC. However, sorafenib treatment has demonstrated to be with a low response rate and acquired resistance of sorafenib obviously limited survival benefits. Recent studies showed that alternative activation of mTOR signaling was responsible for sorafenib resistance10,15. As reportedly, we found AMPK activation and mTOR inhibition in three HCC cell lines treated with sorafenib (Fig. S4A). To determine the role of AMPK/mTOR in sorafenib response, AMPK siRNA was used to repress AMPK activation in sorafenib-treated cells. With the inactivation of AMPK, we demonstrated that sorafenib-induced apoptosis was suppressed, which suggested that activation of AMPK was essential for sorafenib response (Fig. S4B–C). Moreover, Rapamycin or Metformin treatment both targeting inhibition of mTOR could significantly enhance sorafenib response (Fig. S4D).
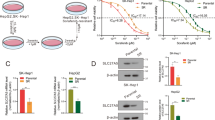
Former studies confirmed that Rage promoted HCC proliferation via regulating AMPK/mTOR pathway. To explore whether Rage was involved in sorafenib response, HCC cells were cultured with varied concentration of sorafenib. Interestingly, we found that sorafenib treatment contributed to Rage decrease and activation of AMPK/mTOR in a dose-dependent manner (Fig. 5a). Moreover, compared to normal cells, IC50 of sorafenib treatment in Rage-deficient cells was markedly decreased (Fig. 5b). Rage inhibition not only strengthened sorafenib causing apoptosis but also proliferation repression (Fig. 5c–f). Moreover, AMPK silencing partly recovered impaired proliferation caused by sorafenib treatment in Rage ablation cells (Fig. 5e, f). To unveil the underlying molecular mechanism of Rage in sorafenib response, western blot was performed. We demonstrated that Rage ablation elevated autophagy levels in HCC cells treated by sorafenib through further activating AMPK/mTOR signaling and silencing AMPK could inhibited autophagy process and then partly rescued proliferation repression as well as apoptosis induction caused by sorafenib (Fig. 5g, h). These findings suggested Rage participated in sorafenib response by regulating autophagy level, which was dependent on AMPK/mTOR pathway.
a Immunoblot analysis reveals that sorafenib inhibited Rage expression and activated AMPK/mTOR pathway. HCCLM3 cells were treated with different concentration of soarfenib for 24 h. Autophagy activity was measured by analyzing the fold change of Lc3II/Lc3I. b Rage ablation enhanced sorafenib response. CCK8 assays were performed and analyzed. c, d Apoptosis assays were conducted and analyzed. Bel7402 and HCCLM3 cells were cultured with sorafenib (10 μM) for 24 h. e, f Colony formation experiments were conducted and analyzed. Bel7402 and HCCLM3 cells were cultured with sorafenib (10 μM) for 24 h. g Immunoblot analysis indicates that targeting Rage strengthened sorafenib response, which was dependent on AMPK/mTOR pathway. Bel7402 and HCCLM3 cells were cultured with sorafenib (10 μM) for 24 h. Autophagy activity was measured by analyzing the fold change of Lc3II/Lc3I. Data are mean ± SEM from three independent experiments, *p < 0.05, **p < 0.01, ***p < 0.001 by unpaired Student’s t-test
Rage expression is enhanced by its ligands, such as HMGB1 and s100a4
The interactions between Rage and its ligands, such as HMGB1 and s100a4, play a critical role in tumor progression through activating MAPK, NF-κB, and producing inflammatory cytokines12. Accumulating data demonstrated that HMGB1 and s100a4 were both involved in inflammatory diseases including tumors and inflammatory factors could contributed to the production of HMGB1 and s100a423,24,25,26. Through analyzing expression of HMGB1, s100a4, and Rage in TCGA data, we found there was a positive correlation between Rage and its ligands (Fig. 6a). To determine the role of Rage ligands in Rage expression, HCC cells were treated with rhHMGB1 and rhs100a4. Data suggested that the interactions of Rage and its ligands resulted in Rage increase (Fig. 6b, c). Moreover, we demonstrated that Rage ligand HMGB1 could promote proliferation and repress sorafenib response in a Rage-dependent way (Fig. 6d). To further exploring the relationship between Rage and HMGB1, immunohistochemistry staining HMGB1, Rage, and Ki67 were performed and quantified. We observed HMGB1 was positively associated with Rage and proliferation (Fig. 6e, f). These observations indicated the important role of Rage ligand HMGB1 in Rage expression.
a Rage expression was positively associated with Rage ligands, sush as HMGB1 and s100a4. Data were extracted from TCGA database (https://xenabrowser.net/heatmap/). b HMGB1 and s100a4 promoted Rage expression and activated AMPK/mTOR pathway. c Rage expression was analyzed through immunofluorescence. Bel7402 cells were cultured with HMGB1 (1 μg/mL) for 24 h. d HMGB1 repressed sorafenib response, which was dependent on Rage. CCK8 assays were conducted and analyzed. e IHC of Rage, HMGB1, and ki67 in HCC samples. Representative images show different intensities of staining of Rage, HMGB1, and ki67. Scale bars, 100 μm. f HMGB1 expression was positively associated with Rage expression. IHC of Rage, HMGB1, and ki67 in HCC samples were analyzed. Data are mean ± SEM from three independent experiments, *p < 0.05, **p < 0.01, ***p < 0.001 by unpaired Student’s t-test
Rage induces sorafenib resistance in vivo
To further confirm the crucial role of Rage in sorafenib resistance, HCCLM3-shRage cells were well established and tumor xenografts and orthotopic models in nude mice were performed. Compared to mice bearing HCCLM3 cells, tumor growth inhibition caused by sorafenib treatment in those bearing HCCLM3-shRage cells was more significant (Fig. 7a, b, Fig. S5). Taken together, our study provided evidence that Rage ablation inhibited HCC proliferation via regulating AMPK/mTOR pathway, which was responsible for autophagy induction. Moreover, targeting Rage could enhance sorafenib response by increasing autophagy level. Lastly, Rage ligands were important for Rage expression and alleviate sorafenib response in a Rage-dependent manner.
a 5 × 106 HCCLM3-con, HCCLM3-shRage cells were subcutaneously injected into nude mice and those mice were treated with or without sorafenib (50 mg/kg/d). Four weeks later, tumors derived from indicated cells were removed and shown, n = 4. b The tumor growth curves for three groups as described in a were measured. c Our study suggested that Rage was elevated in tumors and regulates proliferation via modulating AMPK/mTOR pathway. Autophagy induction caused by Rage ablation was dependent on AMPK/mTOR signaling. Moreover, soarfenib response was enhanced by downregulation of Rage whose repression activated AMPK/mTOR pathway. Lastly, we found Rage ligands, especially HMGB1, promoted Rage expression and decreased sorafenib response through inactivating AMPK/mTOR pathway
Discussion
Accumulating data demonstrate that Rage is overexpressed in various types of tumors and has a vital role tumor progression12,14. Recent studies reported that Rage promotes proliferation via activating NF-κB/cyclin D pathway13. Moreover, targeting Rage could enhance chemotherapeutic effects and promote tumor cell survival by reducing autophagy16. Our study suggested that Rage was expressed in both tumor and stromal cells and confirmed that Rage expression was highly elevated in tumors. We observed that targeting Rage indeed contributed to autophagy induction in HCC cell lines. Mounting evidence has been reported to show the paradox role of autophagy in pro-survival and pro-apoptosis27. Our study demonstrated that autophagy induction caused by Rage deficiency was responsible for proliferation impairment in HCC cells. Recent studies indicate that Rage ablation results in increase of phosphorylation of mTOR in pancreatic tumor cells, which is responsible for Rage-sustained autophagy15,16,28. In our study, we confirmed that activation of AMPK/mTOR pathway in Rage ablation cells accounted for autophagy increase and proliferation impairment, which might explain the different relationship between autophagy and Rage in varied tumor cells. More efforts are needed to be performed to explore the underlying mechanisms of Rage and AMPK/mTOR pathway.
The detailed molecular basis for acquired sorafenib resistance remains complicated. Many cellular signaling pathways have been proven to be involved in sorafenib-resistant cells. Emerging evidence demonstrated that alternative activation of mTOR accounts for sorafenib resistance in varied cells, including AML and HCC9,10. Inhibition of mTOR is a crucial step for autophagy process, which promotes the formation of ULK complex18,29. Moreover, the role of autophagy in sorafenib therapy is still controversial30. A lot of studies indicate that sorafenib-induced autophagy was responsible for tumor survival, which inhibited the cytotoxic effects of sorafenib treatment. Combining autophagy inhibitors and sorafenib could enhance sorafenib response27,31,32,33. However, contrary results were also reported. Sorafenib was demonstrated to promote autophagic death of HCC34. Additionally, activation of ARDB2 was confirmed to promote sorafenib resistance by inhibiting autophagy, which suggested that autophagy inhibition might be a potential therapy for enhancing sorafenib response9. Our study showed Rage ablation elevated autophagy levels through activating AMPK/mTOR signaling pathway. We firstly reported that Rage decrease and AMPK/mTOR activation in sorafenib-treated HCC cells. We found inhibition of AMPK impaired sorafenib response and combination of Rapamycin or Metformin, both targeting repression of mTOR, strengthened it35. Moreover, Rage ablation increased the efficiency of sorafenib-induced apoptosis in a AMPK manner, which suggested that Rage was involved in sorafenib response through regulating AMPK activity.
The interactions of Rage and its ligands have been reported to promote tumor progression by activating MAPK, NF-κB, PI3K/AKT, and cdc42 pathways14. Our study demonstrated that Rage ligands including HMGB1 and s100a4 significantly increased Rage expression. Additionally, HMGB1 treatment inhibited sorafenib response in a Rage-dependent way. Accumulating data demonstrated that inflammatory microenvironment of HCC played an important role in sorafenib response36. Recent studies indicated that patients with less inflammation benefited more from sorafenib therapy36. Rage ligands and Rage were tightly associated with inflammation, which indicated that inflammatory microenvironment of HCC might repress sorafenib response and resulted in sorafenib resistance through regulating Rage expression and stimulating the interactions between Rage and its ligands.
In conclusion, we demonstrated the critical role of Rage in HCC proliferation. Autophagy induction caused by Rage ablation via activating AMPK/mTOR pathway was responsible for impaired proliferation. Moreover, Rage was involved in sorafenib response through modulating AMPK activity and Rage ablation significantly increased sorafenib-induced apoptosis. Lastly, we observed that HMGB1 treatment, as a Rage ligand, not only enhanced Rage expression but contributed to repression of sorafenib repsonse. Consequently, our study indicated targeting Rage would be a potential therapy for HCC and sorafenib resistance.
Material and methods
Patients and specimens
Samples were achieved from 68 HCC patients who had undergone curative resection between 2014 and 2017 and were pathologically confirmed HCC at Medical School of Nanjing University Affiliated Drum Tower Hospital. Informed consent was obtained from each recruited patient, and the protocol was approved by the Institutional Research Ethics Committee. The clinical signatures of all patients are summarized in Supplementary Table 1.
Animals and chemical reagents
Male BALB/c nu/nu mice (6–8 weeks old, Shanghai Institute of Material Medicine, Chinese Academy of Science) were housed in specific pathogen-free conditions. All animals received humane care according to the criteria outlined in the “Guide for the Care and Use of Laboratory Animals” prepared by the National Academy of Sciences and published by the National Institutes of Health (NIH publication 86–23 revised 1985). Sorafenib (No. S7397), Rapamycin (No. S1039), and Metformin (No. S1950) were purchased from Selleck Chemicals (Houston, TX, USA). The PI (propidiumiodide)/Annexin V-FITC apoptosis detection kit was from BD Biosciences (San Jose, CA, USA). The Cell Counting Kit-8 (CCK-8) kit was purchased from Dojindo Laboratories (Kumamoto, Japan). Edu assays (KGA331-100) were purchased from KeyGENE BioTECH (Nanjing, China).
Antibodies
The antibody catalog numbers were provided in Supplementary Table 2.
Cell culture
The human HCC cell line HepG2, HCCLM3, Huh7, Bel7402 was achieved from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Four HCC cells were al cultured in 4.5 g/L glucose DMEM containing 10% FBS (Gbico, USA) and 10% FBS, penicillin (100 U/mL), and streptomycin (100 μg/mL). LO2 cells were cultured in RPMI1640 medium containing 10% FBS, penicillin (100 U/mL), and streptomycin (100 μg/mL). All cells were incubated at 37 °C in humidified air with 5% CO2.
Immunoblot analysis
Total protein was extracted by lysing cells in RIPA buffer containing protease inhibitor cocktail. Protein samples boiled with 1× loading buffer were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred onto polyvinylidene fluoride membranes. After blocking with 5% BSA in TBS-T, membranes were incubated with the primary antibody at 4 °C overnight. Goat-anti-rabbit or mouse IgG conjugated to horseradish peroxidase was used as the secondary antibody. Protein was imaged by Tanon System.
Cell transfection
For stably knockdown of Rage with lenti-virus shRNA, 2 × 105 cells were planted onto 6-well plates. After 24 h, the liquid containing shRNA was added to cultural medium according protocol. To select stable transfectants, cells were cultured in complete DMEM with 10 μg/mL puromycin (Sigma-Aldrich, USA) for some weeks. AMPK siRNA, Rage siRNA, and control siRNA (RiboBio, China) were transfected into cells using lipofectin 2000 according to the manufacturer’s instructions. At the end of the siRNA treatment (48–72 h), the cells were collected for western blot and q-PCR.
Immunofluorescence
Immunofluorescence analysis was performed according to protocols. Cells were implanted in 24-well dishes and fixed by 4% paraformaldehyde 24 h later. Fixed cells were stained with autophagy-related proteins (Cell Signaling Technology, USA), Rage (Cell Signaling Technology), followed by FITC-conjugated anti-mouse IgG and Cy3-conjugated anti-rabbit IgG (Abcam). Representative images were detected by fluorescent microscopy (Leica, German) and data were processed via ImagePro Plus.
Immunohistochemistry staining
Immunohistochemistry of HCC samples were performed as previously described. Briefly, after incubation with HMGB1 (Abcam), Rage (Abcam), autophagy-related markers (Cell Signaling Technology, USA), p-AMPK (Cell Signaling Technology, USA), mTOR (Cell Signaling Technology, USA), the sections were stained in an Envision System (Dako Cytomation, USA). IHC results were scored according to 0, <25%; 1, <50%; 2, <75%; 3, >75% by two experienced pathologists. Data are shown as mean ± SEM.
Cell proliferation
Cell proliferation was tested by Cell Counting Kit-8 and edu assays. According to manufacturer’s instructions, cells were seeded in 96-well plates at 5 × 103 per well. Cells were cultured with different treatments for 24, 48, and 72 h. 10 μL CCK-8 solutions was added into each well, and after incubation for 2–4 h at 37 °C, the absorbance at 450 nm for each well was measured to estimate the number of viable cells. For edu assays, all performances were according to manufacturer’s instructions.
Transmission electron microscopy
Cells seeded onto 6-well plate were fixed with fixative buffer containing 2% paraformaldehyde and 2.5% glutaraldehyde in 0.1 M PBS. After embedded, samples were cut into 0.12-μm sections and stained with 0.2% lead citrate and 1% uranyl acetate. The images were detected by a JEOL TEM-2000 EX II (JEOL, Tokyo, Japan).
Statistical analysis
Fisher’s exact tests and χ2 tests were used to determine clinicopathological correlations. The association between Rage, autophagy markers, and p-mTOR in HCC tissues was evaluated by Spearman’s correlation. The Student’s t-test was used for comparison between variables. GraphPad Prism 6 was used for all statistical analyses. P < 0.05 was considered statistically significant.
References
Han, Z. G. Functional genomic studies: insights into the pathogenesis of liver cancer. Annu. Rev. Genom. Human Genet. 13, 171–205 (2012).
Portolani, N. et al. Early and late recurrence after liver resection for hepatocellular carcinoma: prognostic and therapeutic implications. Ann. Surg. 243, 229–235 (2006).
Llovet, J. M. et al. Sorafenib in advanced hepatocellular carcinoma. N. Engl. J. Med. 359, 378–390 (2008).
Mizushima, N. & Komatsu, M. Autophagy: renovation of cells and tissues. Cell 147, 728–741 (2011).
Czaja, M. J. Functions of autophagy in hepatic and pancreatic physiology and disease. Gastroenterology 140, 1895–1908 (2011).
Rautou, P. E. et al. Autophagy in liver diseases. J. Hepatol. 53, 1123–1134 (2010).
Lee, Y. J. & Jang, B. K. The role of autophagy in hepatocellular carcinoma. Int. J. Mol. Sci. 16, 26629–26643 (2015).
Kroemer, G., Marino, G. & Levine, B. Autophagy and the integrated stress response. Mol. Cell 40, 280–293 (2010).
Wu, F. Q. et al. ADRB2 signaling promotes HCC progression and sorafenib resistance by inhibiting autophagic degradation of HIF1alpha. J. Hepatol. 65, 314–324 (2016).
Masuda, M. et al. Alternative mammalian target of rapamycin (mTOR) signal activation in sorafenib-resistant hepatocellular carcinoma cells revealed by array-based pathway profiling. Mol. Cell. Proteomics 13, 1429–1438 (2014).
Bhawal, U. K. et al. Association of expression of receptor for advanced glycation end products and invasive activity of oral squamous cell carcinoma. Oncology 69, 246–255 (2005).
Yan, S. F., Ramasamy, R. & Schmidt, A. M. Receptor for AGE (RAGE) and its ligands-cast into leading roles in diabetes and the inflammatory response. J. Mol. Med. 87, 235–247 (2009).
Yaser, A. M. et al. The role of receptor for advanced glycation end products (RAGE) in the proliferation of hepatocellular carcinoma. Int. J. Mol. Sci. 13, 5982–5997 (2012).
Logsdon, C. D., Fuentes, M. K., Huang, E. H. & Arumugam, T. RAGE and RAGE ligands in cancer. Curr. Mol. Med. 7, 777–789 (2007).
Kang, R. et al. The receptor for advanced glycation end products (RAGE) sustains autophagy and limits apoptosis, promoting pancreatic tumor cell survival. Cell Death Differ. 17, 666–676 (2010).
Zhao, Z. et al. Receptor for advanced glycation end product blockade enhances the chemotherapeutic effect of cisplatin in tongue squamous cell carcinoma by reducing autophagy and modulating the Wnt pathway. Anticancer Drugs 28, 187–196 (2017).
Tian, Y. et al. Autophagy inhibits oxidative stress and tumor suppressors to exert its dual effect on hepatocarcinogenesis. Cell Death Differ. 22, 1025–1034 (2015).
Yu, L. et al. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature 465, 942–946 (2010).
Kim, J., Kundu, M., Viollet, B. & Guan, K. L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 13, 132–141 (2011).
Wang, D. et al. The role of AKT1 and autophagy in the protective effect of hydrogen sulphide against hepatic ischemia/reperfusion injury in mice. Autophagy 8, 954–962 (2012).
Lee, J. W., Park, S., Takahashi, Y. & Wang, H. G. The association of AMPK with ULK1 regulates autophagy. PLoS ONE 5, e15394 (2010).
Klionsky, D. J. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat. Rev. Mol. Cell Biol. 8, 931–937 (2007).
Wang, X. et al. The role of HMGB1 signaling pathway in the development and progression of hepatocellular carcinoma: a review. Int. J. Mol. Sci. 16, 22527–22540 (2015).
Chen, M. et al. High-mobility group box 1 promotes hepatocellular carcinoma progression through miR-21-mediated matrix metalloproteinase activity. Cancer Res. 75, 1645–1656 (2015).
Naglova, H. & Bucova, M. HMGB1 and its physiological and pathological roles. Bratisl. Lek. Listy 113, 163–171 (2012).
Tang, D. et al. Quercetin prevents LPS-induced high-mobility group box 1 release and proinflammatory function. Am. J. Respir. Cell Mol. Biol. 41, 651–660 (2009).
Pan, H. et al. Autophagy inhibition sensitizes hepatocellular carcinoma to the multikinase inhibitor linifanib. Sci. Rep. 4, 6683 (2014).
Chen, J. et al. Paeoniflorin ameliorates AGEs-induced mesangial cell injury through inhibiting RAGE/mTOR/autophagy pathway. Biomed. Pharmacother. 89, 1362–1369 (2017).
Jung, C. H., Ro, S. H., Cao, J., Otto, N. M. & Kim, D. H. mTOR regulation of autophagy. FEBS Lett. 584, 1287–1295 (2010).
Prieto-Dominguez, N. et al. Modulation of autophagy by sorafenib: effects on treatment response. Front. Pharmacol. 7, 151 (2016).
Zhai, B. et al. Inhibition of Akt reverses the acquired resistance to sorafenib by switching protective autophagy to autophagic cell death in hepatocellular carcinoma. Mol. Cancer Ther. 13, 1589–1598 (2014).
Shi, Y. H. et al. Targeting autophagy enhances sorafenib lethality for hepatocellular carcinoma via ER stress-related apoptosis. Autophagy 7, 1159–1172 (2011).
Shimizu, S. et al. Inhibition of autophagy potentiates the antitumor effect of the multikinase inhibitor sorafenib in hepatocellular carcinoma. Int. J. Cancer 131, 548–557 (2012).
Tai, W. T. et al. Mcl-1-dependent activation of Beclin 1 mediates autophagic cell death induced by sorafenib and SC-59 in hepatocellular carcinoma cells. Cell Death Dis. 4, e485 (2013).
Ling, S. et al. Combination of metformin and sorafenib suppresses proliferation and induces autophagy of hepatocellular carcinoma via targeting the mTOR pathway. Int. J. Oncol. 50, 297–309 (2017).
Bruix, J. et al. Prognostic factors and predictors of sorafenib benefit in patients with hepatocellular carcinoma: analysis of two phase III studies. J. Hepatol. 67, 999–1008 (2017).
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (Grant No. 81670566), Jiangsu Province’s Key Provincial Talents Program (Grant No. ZDRCA2016066).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, J., Wu, PW., Zhou, Y. et al. Rage induces hepatocellular carcinoma proliferation and sorafenib resistance by modulating autophagy. Cell Death Dis 9, 225 (2018). https://doi.org/10.1038/s41419-018-0329-z
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41419-018-0329-z
This article is cited by
-
Targeting and regulation of autophagy in hepatocellular carcinoma: revisiting the molecular interactions and mechanisms for new therapy approaches
Cell Communication and Signaling (2023)
-
Cholesterol sensor SCAP contributes to sorafenib resistance by regulating autophagy in hepatocellular carcinoma
Journal of Experimental & Clinical Cancer Research (2022)
-
Data Mining and Systems Pharmacology to Elucidate Effectiveness and Mechanisms of Chinese Medicine in Treating Primary Liver Cancer
Chinese Journal of Integrative Medicine (2022)
-
Autophagy, an accomplice or antagonist of drug resistance in HCC?
Cell Death & Disease (2021)
-
AGE-RAGE synergy influences programmed cell death signaling to promote cancer
Molecular and Cellular Biochemistry (2021)