Abstract
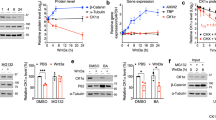
Kaposi’s sarcoma-associated herpesvirus (KSHV) causes primary effusion lymphoma (PEL). PEL cell lines require expression of the cellular FLICE inhibitory protein (cFLIP) for survival, although KSHV encodes a viral homolog of this protein (vFLIP). Cellular and viral FLIP proteins have several functions, including, most importantly, the inhibition of pro-apoptotic caspase 8 and modulation of NF-κB signaling. To investigate the essential role of cFLIP and its potential redundancy with vFLIP in PEL cells, we first performed rescue experiments with human or viral FLIP proteins known to affect FLIP target pathways differently. The long and short isoforms of cFLIP and molluscum contagiosum virus MC159L, which are all strong caspase 8 inhibitors, efficiently rescued the loss of endogenous cFLIP activity in PEL cells. KSHV vFLIP was unable to fully rescue the loss of endogenous cFLIP and is therefore functionally distinct. Next, we employed genome-wide CRISPR/Cas9 synthetic rescue screens to identify loss of function perturbations that can compensate for cFLIP knockout. Results from these screens and our validation experiments implicate the canonical cFLIP target caspase 8 and TRAIL receptor 1 (TRAIL-R1 or TNFRSF10A) in promoting constitutive death signaling in PEL cells. However, this process was independent of TRAIL receptor 2 or TRAIL, the latter of which is not detectable in PEL cell cultures. The requirement for cFLIP is also overcome by inactivation of the ER/Golgi resident chondroitin sulfate proteoglycan synthesis and UFMylation pathways, Jagunal homolog 1 (JAGN1) or CXCR4. UFMylation and JAGN1, but not chondroitin sulfate proteoglycan synthesis or CXCR4, contribute to TRAIL-R1 expression. In sum, our work shows that cFLIP is required in PEL cells to inhibit ligand-independent TRAIL-R1 cell death signaling downstream of a complex set of ER/Golgi-associated processes that have not previously been implicated in cFLIP or TRAIL-R1 function.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
CRISPR screening and RNA-seq data were deposited on GEO (accessions GSE210445, GSE210446). All cell lines and plasmids are available on request.
References
Cesarman E, Chang Y, Moore PS, Said JW, Knowles DM. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–91.
Nador RG, Cesarman E, Chadburn A, Dawson DB, Ansari MQ, Sald J, et al. Primary effusion lymphoma: a distinct clinicopathologic entity associated with the Kaposi’s sarcoma-associated herpes virus. Blood. 1996;88:645–56.
Lurain K, Polizzotto MN, Aleman K, Bhutani M, Wyvill KM, Goncalves PH, et al. Viral, immunologic, and clinical features of primary effusion lymphoma. Blood. 2019;133:1753–61.
Wies E, Mori Y, Hahn A, Kremmer E, Sturzl M, Fleckenstein B, et al. The viral interferon-regulatory factor-3 is required for the survival of KSHV-infected primary effusion lymphoma cells. Blood. 2008;111:320–7.
Godfrey A, Anderson J, Papanastasiou A, Takeuchi Y, Boshoff C. Inhibiting primary effusion lymphoma by lentiviral vectors encoding short hairpin RNA. Blood. 2005;105:2510–8.
Chaudhary PM, Jasmin A, Eby MT, Hood L. Modulation of the NF-kappa B pathway by virally encoded death effector domains-containing proteins. Oncogene. 1999;18:5738–46.
Manzano M, Patil A, Waldrop A, Dave SS, Behdad A, Gottwein E. Gene essentiality landscape and druggable oncogenic dependencies in herpesviral primary effusion lymphoma. Nat Commun. 2018;9:3263.
Hu S, Vincenz C, Buller M, Dixit VM. A novel family of viral death effector domain-containing molecules that inhibit both CD-95- and tumor necrosis factor receptor-1-induced apoptosis. J Biol Chem. 1997;272:9621–4.
Irmler M, Thome M, Hahne M, Schneider P, Hofmann K, Steiner V, et al. Inhibition of death receptor signals by cellular FLIP. Nature. 1997;388:190–5.
Thome M, Schneider P, Hofmann K, Fickenscher H, Meinl E, Neipel F, et al. Viral FLICE-inhibitory proteins (FLIPs) prevent apoptosis induced by death receptors. Nature. 1997;386:517–21.
Krueger A, Schmitz I, Baumann S, Krammer PH, Kirchhoff S. Cellular FLICE-inhibitory protein splice variants inhibit different steps of caspase-8 activation at the CD95 death-inducing signaling complex. J Biol Chem. 2001;276:20633–40.
Kataoka T, Budd RC, Holler N, Thome M, Martinon F, Irmler M, et al. The caspase-8 inhibitor FLIP promotes activation of NF-kappaB and Erk signaling pathways. Curr Biol. 2000;10:640–8.
Chugh P, Matta H, Schamus S, Zachariah S, Kumar A, Richardson JA, et al. Constitutive NF-κB activation, normal Fas-induced apoptosis, and increased incidence of lymphoma in human herpes virus 8 K13 transgenic mice. Proc Natl Acad Sci USA. 2005;102:12885–90.
Bélanger C, Gravel A, Tomoiu A, Janelle ME, Gosselin J, Tremblay MJ, et al. Human herpesvirus 8 viral FLICE-inhibitory protein inhibits Fas-mediated apoptosis through binding and prevention of procaspase-8 maturation. J Hum Virol. 2001;4:62–73.
Tolani B, Matta H, Gopalakrishnan R, Punj V, Chaudhary PM. NEMO is essential for Kaposi’s sarcoma-associated herpesvirus-encoded vFLIP K13-induced gene expression and protection against death receptor-induced cell death, and its N-terminal 251 residues are sufficient for this process. J Virol. 2014;88:6345–54.
Galluzzi L, Vitale I, Aaronson SA, Abrams JM, Adam D, Agostinis P, et al. Molecular mechanisms of cell death: recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 2018;25:486–541.
Fritsch M, Gunther SD, Schwarzer R, Albert MC, Schorn F, Werthenbach JP, et al. Caspase-8 is the molecular switch for apoptosis, necroptosis and pyroptosis. Nature. 2019;575:683–7.
Newton K, Wickliffe KE, Maltzman A, Dugger DL, Reja R, Zhang Y, et al. Activity of caspase-8 determines plasticity between cell death pathways. Nature. 2019;575:679–82.
Yu L, Alva A, Su H, Dutt P, Freundt E, Welsh S, et al. Regulation of an ATG7-beclin 1 program of autophagic cell death by caspase-8. Science. 2004;304:1500–2.
Bell BD, Leverrier S, Weist BM, Newton RH, Arechiga AF, Luhrs KA, et al. FADD and caspase-8 control the outcome of autophagic signaling in proliferating T cells. Proc Natl Acad Sci USA. 2008;105:16677–82.
Jimbo A, Fujita E, Kouroku Y, Ohnishi J, Inohara N, Kuida K, et al. ER stress induces caspase-8 activation, stimulating cytochrome c release and caspase-9 activation. Exp Cell Res. 2003;283:156–66.
Munoz-Pinedo C, Lopez-Rivas A. A role for caspase-8 and TRAIL-R2/DR5 in ER-stress-induced apoptosis. Cell Death Differ. 2018;25:226.
Grundhoff A, Ganem D. Mechanisms governing expression of the v-FLIP gene of Kaposi’s sarcoma-associated herpesvirus. J Virol. 2001;75:1857–63.
Liu L, Eby MT, Rathore N, Sinha SK, Kumar A, Chaudhary PM. The human herpes virus 8-encoded viral FLICE inhibitory protein physically associates with and persistently activates the Ikappa B kinase complex. J Biol Chem. 2002;277:13745–51.
Field N, Low W, Daniels M, Howell S, Daviet L, Boshoff C, et al. KSHV vFLIP binds to IKK-gamma to activate IKK. J Cell Sci. 2003;116(Pt 18):3721–8.
Sun Q, Matta H, Chaudhary PM. The human herpes virus 8-encoded viral FLICE inhibitory protein protects against growth factor withdrawal-induced apoptosis via NF-kappa B activation. Blood. 2003;101:1956–61.
Bagneris C, Rogala KB, Baratchian M, Zamfir V, Kunze MB, Dagless S, et al. Probing the solution structure of IkappaB Kinase (IKK) subunit gamma and its interaction with Kaposi sarcoma-associated herpes virus flice-interacting protein and IKK subunit beta by EPR spectroscopy. J Biol Chem. 2015;290:16539–49.
Hu WH, Johnson H, Shu HB. Activation of NF-kappaB by FADD, Casper, and caspase-8. J Biol Chem. 2000;275:10838–44.
Kataoka T, Tschopp J. N-terminal fragment of c-FLIP(L) processed by caspase 8 specifically interacts with TRAF2 and induces activation of the NF-kappaB signaling pathway. Mol Cell Biol. 2004;24:2627–36.
Baratchian M, Davis CA, Shimizu A, Escors D, Bagneris C, Barrett T, et al. Distinct activation mechanisms of NF-kappaB regulator inhibitor of NF-kappaB kinase (IKK) by isoforms of the cell death regulator cellular FLICE-like inhibitory protein (cFLIP). J Biol Chem. 2016;291:7608–20.
Shisler JL, Moss B. Molluscum contagiosum virus inhibitors of apoptosis: the MC159 v-FLIP protein blocks Fas-induced activation of procaspases and degradation of the related MC160 protein. Virology. 2001;282:14–25.
Nichols DB, Shisler JL. The MC160 protein expressed by the dermatotropic poxvirus molluscum contagiosum virus prevents tumor necrosis factor alpha-induced NF-kappaB activation via inhibition of I kappa kinase complex formation. J Virol. 2006;80:578–86.
Biswas S, Smith GL, Roy EJ, Ward B, Shisler JL. A comparison of the effect of molluscum contagiosum virus MC159 and MC160 proteins on vaccinia virus virulence in intranasal and intradermal infection routes. J Gen Virol. 2018;99:246–52.
Ma Y, Walsh MJ, Bernhardt K, Ashbaugh CW, Trudeau SJ, Ashbaugh IY, et al. CRISPR/Cas9 screens reveal Epstein-Barr virus-transformed B cell host dependency factors. Cell Host Microbe. 2017;21:580–91.e587.
Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34:184–91.
Sherman BT, Hao M, Qiu J, Jiao X, Baseler MW, Lane HC, et al. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022;50(W1):W216–21.
Komatsu M, Chiba T, Tatsumi K, Iemura S, Tanida I, Okazaki N, et al. A novel protein-conjugating system for Ufm1, a ubiquitin-fold modifier. EMBO J. 2004;23:1977–86.
Kang SH, Kim GR, Seong M, Baek SH, Seol JH, Bang OS, et al. Two novel ubiquitin-fold modifier 1 (Ufm1)-specific proteases, UfSP1 and UfSP2. J Biol Chem. 2007;282:5256–62.
Tatsumi K, Sou YS, Tada N, Nakamura E, Iemura S, Natsume T, et al. A novel type of E3 ligase for the Ufm1 conjugation system. J Biol Chem. 2010;285:5417–27.
Lemaire K, Moura RF, Granvik M, Igoillo-Esteve M, Hohmeier HE, Hendrickx N, et al. Ubiquitin fold modifier 1 (UFM1) and its target UFBP1 protect pancreatic beta cells from ER stress-induced apoptosis. PLoS One. 2011;6:e18517.
Chen C, Itakura E, Weber KP, Hegde RS, de Bono M. An ER complex of ODR-4 and ODR-8/Ufm1 specific protease 2 promotes GPCR maturation by a Ufm1-independent mechanism. PLoS Genet. 2014;10:e1004082.
Cai Y, Pi W, Sivaprakasam S, Zhu X, Zhang M, Chen J, et al. UFBP1, a key component of the Ufm1 conjugation system, is essential for ufmylation-mediated regulation of erythroid development. PLoS Genet. 2015;11:e1005643.
Wu J, Lei G, Mei M, Tang Y, Li H. A novel C53/LZAP-interacting protein regulates stability of C53/LZAP and DDRGK domain-containing Protein 1 (DDRGK1) and modulates NF-kappaB signaling. J Biol Chem. 2010;285:15126–36.
Jiang Y, Cheng Z, Mandon EC, Gilmore R. An interaction between the SRP receptor and the translocon is critical during cotranslational protein translocation. J Cell Biol. 2008;180:1149–61.
Song W, Raden D, Mandon EC, Gilmore R. Role of Sec61alpha in the regulated transfer of the ribosome-nascent chain complex from the signal recognition particle to the translocation channel. Cell. 2000;100:333–43.
Mason N, Ciufo LF, Brown JD. Elongation arrest is a physiologically important function of signal recognition particle. EMBO J. 2000;19:4164–74.
Walter P, Blobel G. Translocation of proteins across the endoplasmic reticulum III. Signal recognition protein (SRP) causes signal sequence-dependent and site-specific arrest of chain elongation that is released by microsomal membranes. J Cell Biol. 1981;91(2 Pt 1):557–61.
Walczak CP, Leto DE, Zhang L, Riepe C, Muller RY, DaRosa PA, et al. Ribosomal protein RPL26 is the principal target of UFMylation. Proc Natl Acad Sci USA. 2019;116:1299–308.
Wang L, Xu Y, Rogers H, Saidi L, Noguchi CT, Li H, et al. UFMylation of RPL26 links translocation-associated quality control to endoplasmic reticulum protein homeostasis. Cell Res. 2020;30:5–20.
García-Suárez O, García B, Fernández-Vega I, Astudillo A, Quirós LM. Neuroendocrine tumors show altered expression of chondroitin sulfate, glypican 1, glypican 5, and syndecan 2 depending on their differentiation grade. Front Oncol. 2014;4:15.
Boztug K, Järvinen PM, Salzer E, Racek T, Mönch S, Garncarz W, et al. JAGN1 deficiency causes aberrant myeloid cell homeostasis and congenital neutropenia. Nat Genet. 2014;46:1021–7.
Hwang S, Kim S, Kim K, Yeom J, Park S, Kim I. Euchromatin histone methyltransferase II (EHMT2) regulates the expression of ras-related GTP binding C (RRAGC) protein. BMB Rep. 2020;53:576–81.
Xu L, Gao X, Yang P, Sang W, Jiao J, Niu M, et al. EHMT2 inhibitor BIX-01294 induces endoplasmic reticulum stress mediated apoptosis and autophagy in diffuse large B-cell lymphoma cells. J Cancer. 2021;12:1011–22.
Sheard MA, Asgharzadeh S, Liu Y, Lin TY, Wu HW, Ji L, et al. Membrane-bound TRAIL supplements natural killer cell cytotoxicity against neuroblastoma cells. J Immunother. 2013;36:319–29.
Mirandola P, Ponti C, Gobbi G, Sponzilli I, Vaccarezza M, Cocco L, et al. Activated human NK and CD8+ T cells express both TNF-related apoptosis-inducing ligand (TRAIL) and TRAIL receptors but are resistant to TRAIL-mediated cytotoxicity. Blood. 2004;104:2418–24.
Chen JJ, Knudsen S, Mazin W, Dahlgaard J, Zhang B. A 71-gene signature of TRAIL sensitivity in cancer cells. Mol Cancer Ther. 2012;11:34–44.
Sullivan GP, O’Connor H, Henry CM, Davidovich P, Clancy DM, Albert ML, et al. TRAIL receptors serve as stress-associated molecular patterns to promote ER-stress-induced inflammation. Dev Cell. 2020;52:714–30.e715.
Dufour F, Rattier T, Constantinescu AA, Zischler L, Morle A, Ben Mabrouk H, et al. TRAIL receptor gene editing unveils TRAIL-R1 as a master player of apoptosis induced by TRAIL and ER stress. Oncotarget. 2017;8:9974–85.
Lu M, Lawrence DA, Marsters S, Acosta-Alvear D, Kimmig P, Mendez AS, et al. Opposing unfolded-protein-response signals converge on death receptor 5 to control apoptosis. Science. 2014;345:98–101.
Iurlaro R, Puschel F, Leon-Annicchiarico CL, O’Connor H, Martin SJ, Palou-Gramon D, et al. Glucose deprivation induces ATF4-mediated apoptosis through TRAIL death receptors. Mol Cell Biol. 2017;37:e00479–16.
van Raam BJ, Lacina T, Lindemann RK, Reiling JH. Secretory stressors induce intracellular death receptor accumulation to control apoptosis. Cell Death Dis. 2017;8:e3069.
Lam M, Marsters SA, Ashkenazi A, Walter P. Misfolded proteins bind and activate death receptor 5 to trigger apoptosis during unresolved endoplasmic reticulum stress. Elife. 2020;9:e52291.
Ramkumar P, Abarientos AB, Tian R, Seyler M, Leong JT, Chen M, et al. CRISPR-based screens uncover determinants of immunotherapy response in multiple myeloma. Blood Adv. 2020;4:2899–911.
Manzano M, Gunther T, Ju H, Nicholas J, Bartom ET, Grundhoff A, et al. Kaposi’s sarcoma-associated herpesvirus drives a super-enhancer-mediated survival gene expression program in primary effusion lymphoma. mBio. 2020;11:e01457–20.
De Leo A, Chen HS, Hu CC, Lieberman PM. Deregulation of KSHV latency conformation by ER-stress and caspase-dependent RAD21-cleavage. PLoS Pathog. 2017;13:e1006596.
Johnston BP, Pringle ES, McCormick C. KSHV activates unfolded protein response sensors but suppresses downstream transcriptional responses to support lytic replication. PLoS Pathog. 2019;15:e1008185.
Azfer A, Niu J, Rogers LM, Adamski FM, Kolattukudy PE. Activation of endoplasmic reticulum stress response during the development of ischemic heart disease. Am J Physiol Heart Circ Physiol. 2006;291:H1411–20.
Zhang Y, Zhang M, Wu J, Lei G, Li H. Transcriptional regulation of the Ufm1 conjugation system in response to disturbance of the endoplasmic reticulum homeostasis and inhibition of vesicle trafficking. PLoS One. 2012;7:e48587.
Liu J, Wang Y, Song L, Zeng L, Yi W, Liu T, et al. A critical role of DDRGK1 in endoplasmic reticulum homoeostasis via regulation of IRE1α stability. Nat Commun. 2017;8:14186.
Hu X, Pang Q, Shen Q, Liu H, He J, Wang J, et al. Ubiquitin-fold modifier 1 inhibits apoptosis by suppressing the endoplasmic reticulum stress response in Raw264.7 cells. Int J Mol Med. 2014;33:1539–46.
Tsherniak A, Vazquez F, Montgomery PG, Weir BA, Kryukov G, Cowley GS, et al. Defining a cancer dependency map. Cell. 2017;170:564–76.e516.
Zhu H, Bhatt B, Sivaprakasam S, Cai Y, Liu S, Kodeboyina SK, et al. Ufbp1 promotes plasma cell development and ER expansion by modulating distinct branches of UPR. Nat Commun. 2019;10:1084.
Hagelkruys A, Wirnsberger G, Stadlmann J, Wohner M, Horrer M, Vilagos B, et al. A crucial role for Jagunal homolog 1 in humoral immunity and antibody glycosylation in mice and humans. J Exp Med. 2021;218:e20200559.
Nosak C, Silva PN, Sollazzo P, Moon KM, Odisho T, Foster LJ, et al. Jagn1 is induced in response to ER stress and regulates proinsulin biosynthesis. PLoS One. 2016;11:e0149177.
Charnaux N, Brule S, Hamon M, Chaigneau T, Saffar L, Prost C, et al. Syndecan-4 is a signaling molecule for stromal cell-derived factor-1 (SDF-1)/ CXCL12. FEBS J. 2005;272:1937–51.
Farzan M, Babcock GJ, Vasilieva N, Wright PL, Kiprilov E, Mirzabekov T, et al. The role of post-translational modifications of the CXCR4 amino terminus in stromal-derived factor 1α association and HIV-1 entry. J Biol Chem. 2002;277:29484–9.
Kremer KN, Peterson KL, Schneider PA, Meng XW, Dai H, Hess AD, et al. CXCR4 chemokine receptor signaling induces apoptosis in acute myeloid leukemia cells via regulation of the Bcl-2 family members Bcl-XL, Noxa, and Bak. J Biol Chem. 2013;288:22899–914.
Pham H, Kearns NA, Maehr R. Transcriptional regulation with CRISPR/Cas9 effectors in mammalian cells. Methods Mol Biol. 2016;1358:43–57.
Pelossof R, Fairchild L, Huang CH, Widmer C, Sreedharan VT, Sinha N, et al. Prediction of potent shRNAs with a sequential classification algorithm. Nat Biotechnol. 2017;35:350–3.
Patil A, Manzano M, Gottwein E. CK1alpha and IRF4 are essential and independent effectors of immunomodulatory drugs in primary effusion lymphoma. Blood. 2018;132:577–86.
Bogerd HP, Whisnant AW, Kennedy EM, Flores O, Cullen BR. Derivation and characterization of Dicer- and microRNA-deficient human cells. RNA. 2014;20:923–37.
Martin M. CUTADAPT removes adapter sequences from high-throughput sequencing reads. EMBnetjournal. 2011;17.
Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25.
Li W, Xu H, Xiao T, Cong L, Love MI, Zhang F, et al. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 2014;15:554.
Cock PJA, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A, et al. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009;25:1422–3.
Clement K, Rees H, Canver MC, Gehrke JM, Farouni R, Hsu JY, et al. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat Biotechnol. 2019;37:224–6.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–8.
Virtanen P, Gommers R, Oliphant TE, Haberland M, Reddy T, Cournapeau D, et al. Author Correction: SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat Methods. 2020;17:352–352.
Waskom M. seaborn: statistical data visualization. JOSS. 2021;6:3021.
Zhou F, Shimoda M, Olney L, Lyu Y, Tran K, Jiang G, et al. Oncolytic reactivation of KSHV as a therapeutic approach for primary effusion lymphoma. Mol Cancer Ther. 2017;16:2627–38.
Acknowledgements
We would like to thank Dr Joanna Shisler for sharing MCV FLIP protein expression vectors and Dr Ethel Cesarman for sharing a control aliquot of BC-3 cells. Imaging work was performed at the Northwestern University Center for Advanced Microscopy generously supported by NCI CCSG P30 CA060553 awarded to the Robert H Lurie Comprehensive Cancer Center. This work was supported by the Northwestern University Interdepartmental Immunobiology Flow Cytometry, NuSeq, and Sanger Sequencing Core Facilities and the University of Chicago Genomics Facility.
Funding
This work was supported by NCI R01 CA247619 and R01 CA247619-01A1S1 to EG; MM was supported by K22 CA241355.
Author information
Authors and Affiliations
Contributions
Study design: NK, SMO, MM and EG. MM performed exploratory resistance screens in BC-3, which were not included in the final manuscript due to very low cFLIP dependency of this cell line. NK performed most experiments. EG performed shRNA cloning, pLX and pCW cell line construction, and shRNA resistance screens until the harvest of cell pellets. SMO performed all immunofluorescence staining and analyses. ZL performed pLX and pCW vector cloning. Computational and statistical analyses were performed by NK. NK and SMO put together figures, which were edited by EG. NK, SMO, and EG wrote the manuscript. EG supervised all aspects of the work. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kuehnle, N., Osborne, S.M., Liang, Z. et al. CRISPR screens identify novel regulators of cFLIP dependency and ligand-independent, TRAIL-R1-mediated cell death. Cell Death Differ 30, 1221–1234 (2023). https://doi.org/10.1038/s41418-023-01133-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-023-01133-0