Abstract
DNA methylation and long noncoding RNAs (lncRNAs) exhibit an indispensable role in follicular development. However, the specific mechanisms regarding lncRNAs mediated by DNA methylation in follicular development remain unclearly. In this study, we found that inhibiting the expression of DNMT1 promoted granulosa cells (GCs) apoptosis to inhibit follicular development. A novel follicular development-associated lncRNA named inhibitory factor of follicular development (IFFD) was mediated by DNMT1 and showed to arrest follicular development by inhibiting GCs proliferation and estrogen (E2) secretion but promoting GCs apoptosis. Mechanistically, the deactivated Cas9-TET1 demonstrated that the hypomethylation in −1261/−1254 region of IFFD promoted the transcription of IFFD by recruiting SP1. IFFD induced the expression of GLI family zinc finger 1 through competitive binding miR-370, thereby up-regulating the expression of CASP3 to promote GCs apoptosis, as well as downregulating the expressions of PCNA and CYP19A1 to inhibit GCs proliferation and E2 secretion. Collectively, DNMT1-mediated IFFD might be a novel target for the regulation of follicular development.
Similar content being viewed by others
Introduction
In mammals, the ovarian follicles normally develop into maturations to ovulate and release a fertilizable oocyte to support generations of species, which is a complex multicellular process controlled by intrafollicular factors and peripheral hormones [1, 2]. The abnormal follicular development leads to a series of serious reproductive diseases such as polycystic ovary syndrome (PCOS) [3, 4], primary ovarian insufficiency [5], and ovarian cancer [6] in humans. Both the arrested follicular development and hyperandrogenism are commonly associated with female infertility, ovulatory dysfunction as well as depression [7]. Primary ovarian insufficiency, the result of follicular dysfunction and depletion, is characterized by the absence of regular menstrual cycles, elevated follicle-stimulating hormone (FSH) and reduced estrogen (E2) concentrations, and usually leads to sterility and reproductive diseases [8, 9]. However, no effective therapy treats and cures these devastating diseases, and the specific regulation mechanism of follicular development remains to be further explored.
As the main supporting cells in follicles, the granulosa cells (GCs) implicate in the follicular development [2, 10]. The increases in proliferation and E2 secretions of GCs significantly promote follicular development, maturation, and ovulation [11, 12], while the increase of GCs apoptosis leads to follicular atresia [13]. Studies have confirmed that DNA methylation [14, 15] and long noncoding RNAs (lncRNAs) [16] are potential therapeutic strategies to control follicular development and ovarian diseases. However, the mechanism that DNA methylation regulates follicular development remains unclear. In mice, 5-azacytidine, a DNA methyltransferase inhibitor [17], results in the obstruction of follicular development [18]. Besides, the DNA methylation of luteinizing hormone receptor gene significantly decreases by equine chorionic gonadotropin (eCG) in mice, a universal induction of follicular development [19]. Moreover, eCG treatment decreases the methylation status at promoters in approximately 40% of genes in GCs with the reduction of Dnmt1 at S phase [20]. Furthermore, we have found that the hypomethylation of RSPO2 promoter regulates the apoptosis and proliferation of GCs to control follicular development in pigs [10]. LncRNAs, defined as non-coding RNAs with transcripts over 200 nucleotides [21], have participated in numerous biological processes by acting as competing endogenous RNAs (ceRNAs) [22, 23]. In PCOS patients, lncRNA MALAT1 sponges miR-125b and miR-203a to promote GCs apoptosis [24], LINC-01572:28 blocks GCs growth by reducing the degradation of p27 [25], and lnc-MAP3K13-7:1 inhibits GCs proliferation through downregulating DNMT1 expression and the hypomethylation of CDKN1A promoter [26], thereby blocking the follicular development and ovulation. Silencing lncRNA NEAT1 causes defects in corpus luteum formation [27], and decreases the fertility in mice [28]. These results indicate that DNA methylation and lncRNAs may directly target GCs function to regulate follicular development, but the molecular mechanism has not been largely explored.
In this study, using a well-established DNA methyltransferase inhibitor 5-Aza-CdR to treat porcine GCs, we found a novel follicular development-associated lncRNA named inhibitory factor of follicular development (IFFD), which was mediated by DNMT1. Furthermore, IFFD increased the expression of GLI family zinc finger 1 (GLI1), an essential mediator of Hedgehog signaling pathway [29], by sponging miR-370 to promote GCs apoptosis, inhibit GCs proliferation and E2 secretion, and then ultimately regulate the follicular development. Our results provide new insights regarding the therapeutic benefit of DNMT1-mediated lncRNAs into follicular development.
Results
DNMT1 regulates the proliferation and apoptosis of GCs
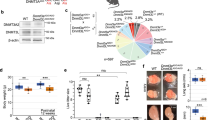
5-Aza-CdR was utilized to explore DNA methylation on the biological effects of GCs, and it was found that the viability (Fig. S1A) and number (Fig. S1B) of GCs treated with 5-Aza-CdR were notably declined in a dose-dependent manner. Also, 5-Aza-CdR significantly decreased the mRNA levels of DNMT1 and DNMT3A in GCs for 24 h (Fig. S1C) and 48 h (Fig. S1D) in a dose-dependent, but exhibited an insignificant influence on DNMT3B. To reduce the toxic effect of 5-Aza-CdR on GCs, 0.5 μM was selected for further using, and it was likely to perform the best at 24 h (Fig. S1E). We found that 5-Aza-CdR prominently declined the mRNA and protein levels of DNMT1 (Fig. 1A, B). Although the mRNA level of DNMT3A was significantly inhibited, the protein level was not markedly changed, and 5-Aza-CdR displayed an insignificant effect on DNMT3B expression (Fig. 1A, B). 5-Aza-CdR prominently declined the mRNA levels of PCNA, CDK1, CDK2, CCNA1, and CCND1, but significantly elevated the mRNA levels of CDK4, CCNA2, CCNB1, CCNB2, CCND2, CCNE2, and RRM1 of cell cycle signaling pathway (Fig. 1C). Moreover, 5-Aza-CdR significantly inhibited GCs proliferation (Fig. 1D) and reduced the number of cells in S phase (Fig. 1E). Besides, 5-Aza-CdR prominently elevated the mRNA levels of CASP3, CASP8, BAX, BAK1, BCL2L11, and BCL2, but inhibited the mRNA levels of CASP9, TP53, and MCL1 of apoptosis signaling pathway (Fig. 1F). Flow cytometry (Fig. 1G) further confirmed that 5-Aza-CdR significantly facilitated GCs apoptosis. Besides, 5-Aza-CdR notably elevated the mRNA levels of STAR, CYP11A1, and CYP17A1, but declined the mRNA levels of CYP19A1 and HSD17B1 of Estrogen signaling pathway (Fig. 1H), and significantly inhibited E2 secretion (Fig. 1I). Therefore, 5-Aza-CdR might inhibit GCs proliferation and E2 secretion, but promote GCs apoptosis by decreasing the expression of DNMT1.
Effect of 5-Aza-CdR on the mRNA (A) and protein (B) levels of DNMT1, DNMT3A and DNMT3B in porcine GCs. C Effect of 5-Aza-CdR on the mRNA levels of genes related to cell cycle in GCs. Effect of 5-Aza-CdR on GCs proliferation detected by EdU (D) and flow cytometry (E). Effect of 5-Aza-CdR on the mRNA levels of apoptosis-related genes (F) and GCs apoptosis (G). Effect of 5-Aza-CdR on the mRNA levels of E2 secretion-related genes (H) and the E2 concentration (I) in GCs. Effect of pcDNA3.1-DNMT1 (J, K) or DNMT1-siRNA (L, M) on the expression of DNMT1 in GCs. The proliferation (N) and apoptosis (O) rates of GCs after transfection with pcDNA3.1-DNMT1. The proliferation (P) and apoptosis (Q) rates of GCs after transfection with DNMT1-siRNA. * indicates P < 0.05, ** indicates P < 0.01.
To explore the role of DNMT1 in GCs, the DNMT1 overexpression plasmid (pcDNA3.1-DNMT1) and DNMT1 small interfering RNA (DNMT1-siRNA) were built. The mRNA and protein levels of DNMT1 in GCs were enhanced markedly with pcDNA3.1-DNMT1, and 500 ng of pcDNA3.1-DNMT1 showed the greatest overexpression effect (Fig. 1J, K). Three DNMT1-siRNAs (DNMT1-siRNA1, DNMT1-siRNA2, and DNMT1-siRNA3) were synthesized, and DNMT1-siRNA1 notably declined the mRNA and protein levels of DNMT1 with the strongest knockdown efficiency (Fig. 1L, M). Although the effects of pcDNA3.1-DNMT1 (Fig. S1F) and DNMT1-siRNA1 (Fig. S1G) on the DNMT1 expression were in a time-dependent manner, 24 h was selected for further using. EdU staining (Fig. 1N) and flow cytometry (Fig. 1O) showed that pcDNA3.1-DNMT1 significantly promoted GCs proliferation and inhibited GCs apoptosis. Conversely, DNMT1-siRNA significantly inhibited GCs proliferation (Fig. 1P) and promoted GCs apoptosis (Fig. 1Q). The testosterone (T) is a substrate for E2 synthesis of GCs, and FSH facilitates the proliferation of GGs [30]. To confirm the roles of 5-Aza-CdR and DNMT1 in GCs under physiological conditions, the GCs were co-cultured with FSH and T. FSH + T significantly elevated the proliferation and E2 synthesis of GCs, and 5-Aza-CdR or DNMT1-siRNA made a significant decline in the proliferation and E2 synthesis aroused by FSH + T, but pcDNA3.1-DNMT1 recovered the declined effects caused by 5-Aza-CdR or DNMT1-siRNA (Fig. S2A, B). The effects of FSH + T on apoptosis were also arrested by 5-Aza-CdR or DNMT1-siRNA in GCs (Fig. S2C). Therefore, these observations suggested that DNMT1 might promote GCs proliferation, but inhibit GCs apoptosis.
DNMT1-mediated the expression of IFFD as ceRNA for miR-370-GLI1 in GCs
The RNA-seq data of GCs treated with 5-Aza-CdR (Group A), DMSO (Group AD), DNMT1-siRNA (Group D), and siRNA-NC (Group ND) were further obtained, and 19 overlapping differential expression lncRNAs were found between 5-Aza-CdR vs. DMSO and DNMT1-siRNA vs. siRNA-NC (Fig. 2A). Among these 19 lncRNAs, MSTRG.2168.1, whom we named IFFD, upregulated both in 5-Aza-CdR and DNMT1-siRNA groups (Fig. 2A). The qRT-PCR further validated that 5-Aza-CdR (Fig. 2B) and DNMT1-siRNA (Fig. 2C) significantly upregulated the expression of IFFD. The 5′ and 3′ ends of IFFD were obtained through a rapid amplification of cDNA ends (RACE) system (Fig. 2D). We found that IFFD located at chr1:268891238-268891653 and was 395 nt in length (accession number OM105888 in NCBI), which comprised of 2 exons. The pcDNA3.1, GFP-WT, GFP-MUT, and pcDNA3.1-ORF with GFP-MUT vectors were transfected into GCs to explore the coding potential of IFFD. Compared with GFP-WT, the pcDNA3.1-ORF with GFP-MUT didn’t show GFP fluorescence (Fig. 2E), indicating that IFFD had no protein-encoding potential. Besides, IFFD was found to be predominantly expressed in the cytoplasm by using FISH (Fig. 2F) and nuclear mass separation assay (Fig. 2G). Otherwise, the oocyte-secreting factors GDF9 and BMP15 are essential for normal follicular development [20]. We found that GDF9 + BMP15 significantly declined IFFD expression (Fig. S3A) but elevated the mRNA level of DNMT1 (Fig. S3B). Moreover, FSH + T prominently declined IFFD expression (Fig. S3A).
A Venn plots and heat maps of overlapped differentially expressed lncRNAs between 5-Aza-CdR vs. DMSO and DNMT1-siRNA vs. siRNA-NC. The expression of IFFD in GCs treated with 5-Aza-CdR (B) or DNMT1-siRNA, DNMT3A-siRNA and DNMT3B-siRNA (C). D Results of IFFD 5′ RACE and 3′ RACE. E GFP fluorescence was conducted after 48 h transfection with the plasmids. The distribution of IFFD in the cytoplasm and nuclei of GCs was determined by FISH (F) and qRT-PCR (G). U6 and Actin were used as nuclear and cytosolic controls, respectively. H The expressions of the five predicted miRNAs bound to IFFD in the IFFD pull-down complex. I Potential binding sites of miR-370 and IFFD, and dual-luciferase reporting assay after co-transfection of wild-type/mutant IFFD and miR-370 mimic/mimic NC. The overexpression (J) and interference (K) efficiency of IFFD. L Effect of pcDNA3.1-lnc IFFD or si-lnc IFFD on miR-370 expression in GCs. M Effect of 5-Aza-CdR on miR-370 expression in GCs. N Overexpression and interference efficiency of miR-370. O The potential binding sites of miR-370 and GLI1, and dual-luciferase reporting assay after co-transfection of wild-type/mutant GLI1 and miR-370 mimic/mimic NC. Effect of miR-370 mimic or miR-370 inhibitor on the mRNA (P) and protein (Q) levels of GLI1. Effect of 5-Aza-CdR on the mRNA (R) and protein (S) levels of GLI1. * indicates P < 0.05, ** indicates P < 0.01. In A, A represents 5-Aza-CdR group, AD represents DMSO group, D represents DNMT1-siRNA group, and ND represents siRNA-NC group.
Using online software, IFFD was predicted to harbor the target sites of miR-218-3p, miR-370, miR-9-1, miR-214-3p, and miR-1271-3p. Using RNA pull down, we found miR-370 expression was predominant at the IFFD probe group (Fig. 2H). Moreover, the interfering effect of miR-370 on the transcription activity of IFFD was restored by mutating the binding site of miR-370 on IFFD (Fig. 2I). Figure 2J and Fig. 2K showed that pcDNA3.1-lnc IFFD and si-lnc IFFD-1 significantly promoted and inhibited the expression of IFFD, respectively. The pcDNA3.1-lnc IFFD significantly inhibited the expression of miR-370, while si-lnc IFFD notably elevated the expression of miR-370 (Fig. 2L). Figure 2M showed that 5-Aza-CdR notably inhibited miR-370 expression. MiR-370 mimic and miR-370 inhibitor notably increased and decreased the expression of miR-370 (Fig. 2N), respectively. The interfering effect of miR-370 on the luciferase activity of GLI1 was restored by mutating the binding site of miR-370 (Fig. 2O). MiR-370 mimic prominently declined the expression of GLI1, while miR-370 inhibitor notably elevated the expression of GLI1 (Fig. 2P, Q). Alternatively, 5-Aza-CdR prominently promoted the expression of GLI1 (Fig. 2R, S). These observations showed that DNMT1-mediated the expressions of IFFD-miR370-GLI1 axis in GCs.
IFFD as ceRNA for miR-370-GLI1 regulates the proliferation and apoptosis of GCs
PcDNA3.1-lnc IFFD declined the expressions of PCNA and CCND1, and this inhibitory effect was partially weakened by miR-370 mimic (Fig. 3A, B). Similarly, si-lnc IFFD elevated the expressions of PCNA and CCND1, and this stimulative effect was partially attenuated by miR-370 inhibitor (Fig. 3C, D). The pcDNA3.1-lnc IFFD significantly reduced the proportion of GCs at S phase (Fig. 3E), inhibited GCs proliferation (Fig. 3F), and miR-370 mimic partially attenuated these effects. Conversely, si-lnc IFFD significantly increased the proportion of GCs at S phase (Fig. 3G), promoted GCs proliferation (Fig. 3H), and miR-370 inhibitor partially attenuated these effects. The pcDNA3.1-lnc IFFD declined the expression of CYP19A1 (Fig. 3I, J) as well as E2 concentration in GCs (Fig. 3K), but si-lnc IFFD exhibited the opposite effects. Besides, pcDNA3.1-lnc IFFD elevated the expressions of CASP3 and BAX, and this stimulative effect was partially weakened by miR-370 mimic (Fig. 3L, M). Si-lnc IFFD declined the expression of BAX, and miR-370 inhibitor partially attenuated this inhibitory effect (Fig. 3N, O). The pcDNA3.1-lnc IFFD significantly promoted GCs apoptosis (Fig. 3P), while si-lnc IFFD significantly inhibited GCs apoptosis, and miR-370 inhibitor partially weakened this inhibitory effect (Fig. 3Q). These results indicated that IFFD sponged miR-370 to inhibit the cell cycle, proliferation as well as E2 secretion, but promote the apoptosis of GCs.
Effects of pcDNA3.1-lnc IFFD and miR-370 mimic on the mRNA (A) and protein (B) levels of cell cycle-related genes. Effects of si-lnc IFFD and miR-370 mimic on the mRNA (C) and protein (D) levels of cell cycle-related genes. Effects of pcDNA3.1-lnc IFFD and miR-370 mimic on GCs cycle (E) and proliferation (F). Effects of si-lnc IFFD and miR-370 mimic on GCs cycle (G) and proliferation (H). Effects of IFFD and miR-370 on the mRNA (I) and protein (J) levels of E2 secretion-related genes. K Effects of IFFD and miR-370 on the E2 secretion of GCs. Effects of pcDNA3.1-lnc IFFD and miR-370 mimic on the mRNA (L) and protein (M) levels of apoptosis-related genes. Effects of si-lnc IFFD and miR-370 mimic on the mRNA (N) and protein (O) levels of apoptosis-related genes. Effects of IFFD and miR-370 on GCs apoptosis (P, Q). * indicates P < 0.05, ** indicates P < 0.01.
To investigate whether GLI1 participated in the miR-370 mediated mechanism involved in GCs, miR-370 mimic/inhibitor and GLI1-siRNA were co-transfected into GCs, respectively. Figure S4A showed that GLI1-siRNA1 performed the highest inhibitory effect on the expression of GLI1. The qRT-PCR (Fig. S4B) showed that miR-370 mimic promoted the mRNA levels of PCNA, CCNB1, and CCND1, while miR-370 inhibitor inhibited the mRNA levels of PCNA, CCNB1, and CCND1. MiR-370 mimic increased the proportion of GCs at S phase (Fig. S4C), and significantly promoted GCs proliferation (Fig. S4D). Conversely, miR-370 inhibitor reduced the proportion of GCs at S phase and significantly inhibited GCs proliferation, and GLI1-siRNA partially attenuate this effect (Fig. S4C, D). Moreover, miR-370 mimic promoted the mRNA level of HSD17B1, while miR-370 inhibitor inhibited the mRNA levels of CYP19A1 and CYP11A1 (Fig. S4E). ELISA (Fig. S4F) showed that miR-370 inhibitor significantly decreased E2 concentration in GCs, and GLI1-siRNA partially attenuated this effect. Besides, miR-370 mimic significantly decreased the mRNA level of CASP3, while miR-370 inhibitor significantly elevated the mRNA levels of CASP3, CASP8, and BAX (Fig. S4G). Flow cytometry (Fig. S4H) showed that miR-370 mimic notably inhibited GCs apoptosis, while miR-370 inhibitor significantly promoted GCs apoptosis, and GLI1-siRNA partially attenuated this effect. These results demonstrated that miR-370 targeted GLI1 to promote GCs proliferation and E2 secretion, but inhibit GCs apoptosis.
DNMT1 blocks the binding of SP1 to inhibit the transcription of IFFD
There was one CpG island (CGI) in the promoter of IFFD (named as BSP1, 319 bp in length, −1309 to −991 bp, transcription start site = +1) (Fig. 4A). To explore the DNA methylation-mediated mechanism for the transcription of IFFD, the methylation levels of BSP1, BSP2 (293 bp in length, −707 to −415 bp, non-CGI region) and BSP3 (264 bp in length, −351 to −88 bp, non-CGI region) were detected after the treatment of DNMT1-siRNA in GCs (Fig. 4A). Compared to the control, the methylation statuses of BSP1 and BSP3 regions were respectively reduced from 70 and 71.1% to 57.7 and 58.9% by DNMT1-siRNA, but it was not changed in BSP2 (Fig. 4B). Moreover, the unmethylated pGL3-BSP1 significantly promoted the dual-luciferase activity of IFFD, compared with the methylated pGL3-BSP1 (Fig. 4C). But the methylated or unmethylated of pGL3-BSP2 and pGL3-BSP3 did not show significant effect on the luciferase activity of IFFD (Fig. 4C). These observations confirmed that the methylation of BSP1 markedly regulated the transcription of IFFD.
A Schematic diagram of the CpG island (CGI) at IFFD promoter. Blue region represents CGI, and the red column represents bisulfite primers. B The methylation status of CGI (BSP1) and non-CGI (BSP2 and BSP3) at the IFFD promoter in GCs with DNMT1-siRNA. C Relative luciferase activity of IFFD promoter at different regions methylated in vitro. D The SP1-binding sites predicted in the CGI region of IFFD promoter. The mRNA (E) and protein (F) levels of SP1 in GCs transfected with pcDNA3.1-SP1. G Effect of pcDNA3.1-SP1 on the luciferase activity of CGI methylated in vitro. Effect of DNMT1-siRNA on the enrichment of SP1 at CGI was detected by ChIP-PCR (H) and ChIP-qPCR (I). J The methylation status of CGI (BSP1) in GCs transfected with dC-TET1 and gIFFD. K The expressions of IFFD, miR-370, and GLI1 in GCs transfected with dC-TET1 and gIFFD. The red boxes represent potential binding sites of SP1. * indicates P < 0.05, ** indicates P < 0.01.
Furthermore, the PROMO website predicted that there was a potential binding site of SP1 in the −1261 to −1254 region (Fig. 4D). To determine the role of SP1 on the transcription of IFFD, the overexpression plasmid of SP1 (pcDNA3.1-SP1) was built, and pcDNA3.1-SP1 notably elevated the expression of SP1 (Fig. 4E, F). Compared with the control group, pcDNA3.1-SP1 significantly induced the dual-luciferase activity of unmethylated pGL3-BSP1 by 3.8 times, while only increasing the activity of methylated pGL3-BSP1 by 1.4 times (Fig. 4G). Results of ChIP and ChIP-qPCR showed that DNMT1-siRNA obviously promoted SP1 binding in the −1261 to −1254 region (Fig. 4H, I). Moreover, compared to the control, the deactivated Cas9-TET1 and gIFFD vector obviously reduced the methylation status of the SP1-binding site on BSP1 (Fig. 4J), but significantly enhanced the expressions of IFFD and GLI1, while decreasing miR-370 expression (Fig. 4K). These results suggested that DNMT1 induced hypermethylation of IFFD and then inhibited its transcription by blocking the binding of SP1.
IFFD as ceRNA for miR-370-GLI1 regulates follicular development in pigs
To explore the function of IFFD on follicular development, LV-lnc IFFD or sh-lnc IFFD was infected into porcine follicles in vitro. Two days after transfection, the expressions of IFFD, miR-370, GLI1, PCNA, CCNB1, CCND1, CASP3, CASP8, BAX, CYP19A1, and HSD17B1 were measured. LV-lnc IFFD significantly induced the mRNA expressions of IFFD and GLI1, but significantly inhibited the expression of miR-370 (Fig. 5A). Meanwhile, sh-lnc IFFD significantly declined the mRNA expressions of IFFD and GLI1, but significantly elevated the expression of miR-370 (Fig. 5A). In addition, the expression of CASP3 significantly increased in follicles infected LV-lnc IFFD, but significantly decreased in follicles infected sh-lnc IFFD (Fig. 5B, C). The expressions of PCNA and CYP19A1 significantly reduced in follicles infected LV-lnc IFFD, but significantly elevated in follicles infected sh-lnc IFFD (Fig. 5B, C). Moreover, LV-lnc IFFD promoted the turbidity of follicular fluid, the degeneration of blood vessels (Fig. 5D), and the apoptosis of GCs (Fig. 5E), while sh-lnc IFFD significantly inhibited GCs apoptosis (Fig. 5E).
The mRNA (A, B) and protein (C) levels of IFFD, miR-370, GLI1, PCNA, CCND1, CASP3, BAX, and CYP19A1 in the porcine follicles transfected with LV-lnc IFFD or sh-lnc IFFD. D The appearance of porcine follicles after transfected with LV-lnc IFFD or sh-lnc IFFD on the 1st, 3rd, and 5th day. E The apoptosis of follicular GCs after transfecting with LV-lnc IFFD or sh-lnc IFFD on the 3rd day. F The methylation level of CGI (BSP1) in small, medium, and large follicles detected by bisulfite sequencing PCR. The expressions of IFFD (G), DNMT1 (H), miR-370 (I) and GLI1 (J, K) in small, medium, and large follicles. * indicates P < 0.05, ** indicates P < 0.01.
Furthermore, in the normal follicles of pigs, the DNA methylation level of BSP1 increased gradually with follicular development (Fig. 5F), the mRNA level of IFFD significantly decreased (Fig. 5G), and the mRNA level of DNMT1 significantly increased (Fig. 5H). The expression of miR-370 notably elevated in large follicles (Fig. 5I), and the expression of GLI1 prominently declined in large follicles compared with small and medium follicles (Fig. 5J, K). These results suggested that the hypomethylation of IFFD might promote GCs apoptosis to inhibit the development of porcine follicles.
IFFD as ceRNA for miR-370-Gli1 controls follicular development in mice
Using FISH, we found that IFFD was widely expressed in different follicular stages in blank mice (Fig. 6A). To further explore the biological function of IFFD on follicular development in vivo, LV-lnc IFFD or sh-lnc IFFD was infected into mouse ovaries. We found that LV-lnc IFFD notably elevated the expressions of IFFD and Gli1, and significantly inhibited the expression of miR-370 (Fig. 6B). However, sh-lnc IFFD notably declined the expressions of IFFD and Gli1, and prominently elevated the expression of miR-370 (Fig. 6B). The expression of Casp3 significantly increased in ovaries infected LV-lnc IFFD, but significantly decreased in ovaries infected sh-lnc IFFD (Fig. 6C, D). The expressions of Pcna and Cyp19a1 significantly reduced in ovaries infected LV-lnc IFFD, but significantly elevated in ovaries infected sh-lnc IFFD (Fig. 6C, D). Moreover, LV-lnc IFFD significantly delayed the age of pubertal initiation compared to LV-NC (48.8 ± 2.68 d vs. 38.4 ± 2.07 d, P < 0.01, n = 5), while sh-lnc IFFD markedly shortened the age of pubertal initiation, compared to sh-NC (35 ± 3.08 d, vs. 40.4 ± 1.14 d, P < 0.01, n = 5) (Fig. 6E). Furthermore, LV-lnc IFFD significantly decreased E2 concentration, while sh-lnc IFFD significantly increased E2 concentration (Fig. 6F). The LV-lnc IFFD significantly increased the number of pre-antrum follicles and antrum follicles, but significantly decreased the number of corpus luteum, and the sh-lnc IFFD significantly increased the number of corpus luteum (Fig. 6G). Furthermore, TUNEL determined that LV-lnc IFFD obviously increased GCs apoptosis, but sh-lnc IFFD inhibited GCs apoptosis (Fig. 6H and Fig. S5).
A The expression and localization of IFFD in ovaries detected by FISH. The mRNA (B, C) and protein (D) levels of IFFD, miR-370, Gli1, Pcna, Ccnd1, Casp3, Bax, and Cyp19a1 in mouse ovary transfected with LV-lnc IFFD or sh-lnc IFFD. E The age of pubertal initiation in mice after ovarian transfection with LV-lnc IFFD and sh-lnc IFFD. The E2 concentration in serum (F), follicular statistics (G), and GCs apoptosis (H) of 50-day-old mice after ovarian transfection with LV-lnc IFFD and sh-lnc IFFD. I The mechanistic scheme of DNMT1-mediated DNA hypomethylation of IFFD to block follicular maturation. * indicates P < 0.05, ** indicates P < 0.01.
Discussion
DNA methylation has been reported to participate in follicular development. Lomniczi et al. used 5-azacytidine to inhibit DNA methylation in rats and found that the ovaries of 5-azacytidine-treated rats had antral follicles, but no corpora lutea, and the rats treated with 5-azacytidine had failed to ovulation [18]. However, Lomniczi et al. [18] focused on the regulations of DNA methylation in the hypothalamus. It is not clear how DNA methylation disturbed the follicular development. In our study, 5-Aza-CdR was found to inhibit GCs proliferation (Fig. 1D) and E2 secretion (Fig. 1I), and promote GCs apoptosis in pigs (Fig. 1G). Moreover, DNMT1 was found to increase GCs proliferation (Fig. 1N) and decrease GCs apoptosis (Fig. 1O). In line with previous studies that 5-Aza-CdR may inhibit proliferation and facilitate apoptosis by reducing DNMT1 expression in colon cancer cell lines [31], as well as that DNMT1 downregulation inhibits GCs proliferation and growth by increasing cyclin-dependent kinase inhibitor 1A expression [26].
To explore how DNA methylation involved in follicular development, we analyzed the transcriptomes of porcine GCs treated with 5-Aza-CdR and DNMT1-siRNA (Fig. 2A), and found that both 5-Aza-CdR (Fig. 2B) and DNMT1-siRNA (Fig. 2C) significantly increased the expression of IFFD. In addition, GDF9 + BMP15 prominently declined the expression of IFFD (Fig. S3A) but elevated the mRNA level of DNMT1 (Fig. S3B), and FSH + T prominently declined the expression of IFFD (Fig. S3A), indicating the potential roles of DNMT1 and IFFD in the regulation of follicular development. Moreover, we also found a negative correlation between IFFD (Fig. 5G) and DNMT1 (Fig. 5H) expression during normal follicular development in pigs. Therefore, we hypothesized that DNMT1 might be involved in follicular development by regulating IFFD expression.
Similar to protein-coding genes, the expressions of lncRNAs are also regulated by epigenetics [32, 33]. To determine the mechanism of IFFD during follicular development, we analyzed the promoter of IFFD and found that the DNA hypermethylation of its CGI resulted in the decreased expression of IFFD (Fig. 5F). Transcription factors have been verified to be closely related to the transcription of lncRNAs [34, 35]. We found that IFFD was activated by SP1 (Fig. 4G), which is a ubiquitous regulator of basal promoter activity [36]. SP1 mediates FSH induced the epithelial regulatory protein expression in GCs [37]. MiR-375 needs to bind with SP1 to regulate the synthesis of E2 in porcine ovaries [38]. In addition, we innovatively demonstrated that DNA methylation mediated the binding of SP1 to IFFD promoter using the CRISPR-dCas9. The deactivated Cas9-TET1 and gIFFD vector obviously reduced the methylation status of SP1-binding site on BSP1 (Fig. 4J), but significantly elevated the expression of IFFD (Fig. 4K). Moreover, the hypomethylation made IFFD promoter more likely to be bound by SP1 (Fig. 4I).
The role of lncRNAs mainly relates to its subcellular localization [39]. We found that IFFD was expressed in both cytoplasm and nucleus, but was highly expressed in cytoplasm (Fig. 2G). Compared to normal ovarian GCs, miR-370 expression was declined in GCs from PCOS and correlated with the number of dominant follicles [40]. Also, miR-370 has been shown to mediate FSHR expression and participate in follicular formation and oocyte maturation [40, 41]. GLI1, a key molecule of the Hedgehog signaling pathway, is closely related to the proliferation and apoptosis of many cells and follicular growth [42,43,44]. In mouse follicles, Gli1 is involved in follicular development through mediating interactions between GCs and theca cells [29, 45]. In our study, we found that IFFD inhibited GCs proliferation (Fig. 3F) and E2 secretion (Fig. 3K) and promoted GCs apoptosis (Fig. 3P) by sponging miR-370, leading to the increased expression of GLI1. A previous study has indicated that lncRNA Neat1 knockout leads to ovarian defects by impairing luteum formation and decreasing fertility [28]. To further characterize the function of IFFD, we built the lentiviral for IFFD overexpression or knockdown, and found that IFFD promoted the loss of follicular blood vessels, follicular fluid turbidity (Fig. 5D), and GCs apoptosis (Fig. 5E) in pigs. Also, IFFD promoted GCs apoptosis (Fig. 6H), impeded E2 secretion (Fig. 6F) and follicular development (Fig. 6G), or delayed pubertal initiation in mice (Fig. 6E).
In conclusion, we found that DNMT1 downregulation induces the hypomethylation of IFFD and then promotes its transcription by recruiting the binding of SP1. IFFD increases GLI1 expression by sponging miR-370, and promotes GCs apoptosis, inhibits GCs proliferation, E2 secretion, and ultimately blocks follicular development (Fig. 6I). These results indicate that DNMT1-mediated IFFD may act as a potential therapeutic strategy for regulation of the follicular development.
Materials and methods
Animals
Three-week-old female C57BL/6J mice (n = 20) obtained from Guangdong medical laboratory animal center (China) were assigned into LV-NC (n = 5), LV-lnc IFFD (n = 5), sh-NC (n = 5), and sh-lnc IFFD (n = 5) groups, randomly. The lentiviral vector that overexpression or knockdown of lnc IFFD (LV-lnc IFFD or sh-lnc IFFD) was synthesized by Dongze Biotechnology Co., LTD (Guangzhou, China). The lentivirus was injected in the same way as our previous study [10]. Briefly, 1 × 107 TU of lentivirus was infected into mice via intraperitoneal injection and was given once a week for 3 weeks. In this study, the pubertal initiation of mice was characterized by the opening of vaginal orifice.
Isolation and culture of GCs and follicles
More than 500 ovaries of prepubertal gilts were obtained from a large slaughterhouse, washed with PBS containing streptomycin/penicillin, and transported to the cell chamber. GCs collected from 3–5 mm follicles through syringes were cultured with DMEM containing 10–12% FBS and incubated at 37 °C and 5% CO2. Plasmids were transfected into GCs through LipofectamineTM 3000 (Thermo Scientific, USA). 3–5 mm antral follicles separated from ovaries were cultured with serum-free DMEM/F12 in 24-well plate and incubated at 38.5 °C and 5% CO2. 100 ng/mL GDF9 (MCE, USA), 50 ng/mL BMP15 (MCE, USA), 100 ng/mL FSH (MCE, USA), 10 ng/mL testosterone (MCE, USA), and 0.5 μM 5-Aza-CdR were used to treat porcine GCs for 24 h.
RNA sequencing and data analysis
The porcine GCs treated with 5-Aza-CdR (named as group A), DMSO (named as group AD), DNMT1-siRNA (named as group D), and siRNA-NC (named as group ND) were used for RNA-seq. The quality and quantity of total RNA extracted by TRIzol were detected by Nanodrop 2000 spectrophotometer and gel electrophoresis. RNA-seq was then conducted with the Illumina Hiseq 2500. The adapter sequences paired reads in which the content of N in paired-end sequencing reads exceeds 3% of the read length and low-quality reads were removed. The cleaned reads were mapped to the Sus scrofa11.1. reference genome using HISAT2 [46], and the mapped reads were assembled and transcripts utilizing Stringtie [47]. CPC2 [48], CNCI [49], and FEELnc [50] software were used for the prediction of coding ability. Finally, the transcripts whose coding ability was not predicted by all three were selected, and the ones whose RNA length > 200 were judged to be lncRNAs. The screening criteria for differentially expressed lncRNAs is based on the FPKM value. DESeq2 [51] of R software was used to differential screening, and |log2FC| > 1 and FDR < 0.05 as the cut-off.
Real-time quantitative PCR
The cDNA of samples was obtained through the RevertAid First Strand cDNA Synthesis Kit (Thermo). Quantitative PCR was conducted using SYBR Green qRT-PCR Mix (2x) (Thermo) and detected by the CFX96 Touch PCR instrument (Bio-Rad, Berkeley, USA). Glyceraldehyde phosphate dehydrogenase (GAPDH) or U6 was selected as endogenous control, and the 2−ΔΔct method was applied for the analysis of expression level. The primer sequences were shown in Tables 1 and 2.
EdU assay
EdU assay was performed based on instruction of the Cell-LightTM Edu Apollo Kit (RiboBio, China). The GCs transfected with plasmids in 48-well plates after 24 h were treated with Triton X-100 for 10–15 min, EdU staining buffer for 30–45 min and Hoechst for 10–15 min. GCs were counted in three random fields per well under an inverted fluorescence microscope. The proliferation rate of GCs in each group was the ratio of EdU-labeled proliferating cells to all Hoechst-labeled cells.
Flow cytometry
Based on the instruction of the Annexin V-FITC Apoptosis Detection Kit (BioVision, USA), the GCs transfected with plasmids in 6-well plates for 24 h were resuspended with 1× Annexin V buffer, and then treated with Annexin V-FITC and PI in darkness. GCs apoptosis was analyzed through a flow cytometry (guava easyCyte, Luminex, USA), and counted as the sum of early and late apoptosis.
ELISA
ELISA was conducted according to the instruction of the mouse estrogen ELISA Kit (Enzyme-linked Biotechnology, ml063198, Shanghai, China) and porcine estrogen ELISA Kit (ml002366). Standard in different concentrations and samples were incubated with 100 μl of horse radish peroxidase for 1 h at 37 °C, and 50 μl each of substrate A and B for 15 min. The termination solution was then added to obtain OD value at 450 nm. The standard curve was drawn based on the concentration of the standard and the corresponding OD value.
Immunofluorescence, HE staining, and TUNEL assay
The prepared paraffin sections of ovaries were placed in dewaxing solution for 10–15 min and anhydrous ethanol for 15 min. The washed ovarian sections were incubated with EDTA antigen retrieval buffer and repaired in a microwave oven. A histochemical pen was used to draw a circle around the tissue of the slices, and BSA was dropped and incubated for 30 min, following incubating primary antibodies overnight and secondary antibodies for 50 min. DAPI dye solution was incubated at 37 °C in darkness for 10 min, and autofluorescence quench was treated for 5 min. The tablets were sealed with anti-fluorescence quench sealing agent. The dewaxed sections were incubated with hematoxylin for 5 min and eosin dye solution for 3 min, then dehydrated and sealed. Finally, the sections were photographed under a fluorescence microscope.
TUNEL assay was performed based on instruction of the TUNEL Apoptosis Assay Kit (Beyotime, China). Follicles which cut into paraffin sections were incubated with xylene for 10 min, ethanol for 5–10 min, and protease K for 15 min, respectively. The washed sections were incubated with TUNEL reagent for 1 h in darkness and observed under Nikon ECLIPSE Ti2 fluorescence microscope.
Bisulfite sequencing PCR (BSP)
BSP was conducted on the grounds of instruction of the EZ DNA Methylation-GoldTM Kit (ZYMO, USA). Using the purified DNA treated with CT conversion reagent as template, the corresponding fragments were amplified by BSP primer. QUMA (http://quma.cdb.riken.jp/) was used to compare the sequencing results with the corresponding original sequences and produce dot plots. Ten clones in each group were used to obtain the methylation rate. The primers were listed in Table 3.
The pdCas9-TET1-CD vector (dC-TET1, 83340, ADDGENE, USA) and sgRNA targeted to the −1261/−1254 region of IFFD promoter (gIFFD) were used to specific demethylation of the SP1-binding site on CGI. The sgRNA was synthesized by Guangzhou Dongze Biotechnology Co., LTD (China). The target sequence of sgRNA vector was GCTGGGATCCATGAGGATGC.
Chromatin Immunoprecipitation assay (ChIP)
Based on the instruction of the PierceTM ChIP kit (Thermo), the GCs treated with formaldehyde after 10–15 min were quenched by glycine. The chromatin fragments solution obtained from automatic ultrasonic crushing machine was then incubated with SP1 antibody (21962-1-AP, proteintech, Wuhan, China) and IgG antibody (12–370, Millipore) at 4 °C overnight. The purified DNA was used for real-time quantitative PCR and the primers were listed in Table 3.
FISH assay
The RiboTM Fluorescent In Situ Hybridization Kit (RiboBio) was selected for FISH assay. The GCs cultured at 24-well plates after 24 h were treated with paraformaldehyde and Triton X-100 for 15 min, prehybridization solution for 30 min, and then incubated with IFFD probe mix overnight. The washed GCs were treated with DAPI for 10 min and observed with confocal microscope (FV3000, OLYMPUS, JAPAN).
Dual-luciferase reporter gene assay
The RNA22 website (https://cm.jefferson.edu/rna22/) was used to predict the potential binding relation of IFFD and miR-370. The sequence of IFFD 3′ UTR containing the binding sites of miR-370 (lnc IFFD-WT) and the mutated binding site of miR-370 (lnc IFFD-MUT) were cloned into pmirGLO-reporter (Promega, Madison, USA) by the endonuclease sites Sac1 and Sall. The GCs at 70% fusion were transfected with lnc IFFD-WT and lnc IFFD-MUT mixed with mimics NC and miR-370 mimic for 48 h, respectively. The luciferase activity reporter assay was conducted by using the Dual-Luciferase Reporter Assay System (Promega, USA) and detected by Synergy Neo2 Reader (BioTek, USA).
The potential binding relation of miR-370 and GLI1 3′ UTR was predicted at the website (http://www.targetscan.org/). The sequence of GLI1 3′ UTR containing the binding sites of miR-370 (GLI1-WT) and the mutated binding site of miR-370 (GLI1-MUT) were synthesized and used for plasmid construction. The GCs at 70% fusion were transfected with GLI1-WT and GLI1-MUT mixed with mimics NC and miR-370 mimic for 48 h, respectively.
Western blot analysis
The concentration of extracted total protein was determined through the BCA Protein Assay Kit (Beyotime, China). The equal proteins of each sample were electrophoresed by SDS-PAGE gels and transferred to polyvinylidene difluoride (PVDF) membranes. The PVDF membranes treated with 5% skimmed milk at 37 °C for 1 h were incubated with DNMT1 (ab188453, abcam, 1:1000), DNMT3A (32578, CST, 1:1000), DNMT3B (57868, CST, 1:1000), SP1 (21962-1-AP, proteintech, 1:5000), PCNA (10205-2-AP, proteintech, 1:2000), CCND1 (26939-1-AP, proteintech, 1:1000), CASP3 (19677-1-AP, proteintech, 1:1000), BAX (50599-2-Ig, proteintech, 1:5000), CYP19A1 (bs-0114R, bioss, 1:2000), CYP11A1 (bs-3608R, bioss, 1:2000), GLI1 (66905-1-Ig, proteintech, 1:3000) and Tubulin (ab7291, Abcam, 1:10,000) at 4 °C overnight. The washed membranes were then incubated with goat anti-mouse (ab6789, Abcam, 1:5000) or goat anti-rabbit IgG H&L (ab205718, Abcam, 1:10000) at 37 °C for 2 h. The Tanon 5200 Muti (Shanghai, China) and ImageJ were used to observe and measure the blots, respectively.
RNA pull down
According to the instruction of PierceTM Magnetic RNA-Protein Pull-Down Kit (Thermo), the GCs were lysed using standard lysate buffers and the 3 ‘of the target lncRNA labeled with biotin. The labeled RNA bound to the streptavidin magnetic beads was treated with equal amount of 1× RNA capture buffer and 100 μl 1× protein-RNA-binding solution. The RNA-bound magnetic beads were treated with 100 μl Master Mix for 30–60 min, and then the RNA-binding complexes were eluted.
Full-length amplification and coding ability verification of IFFD
A 5′/3′-RACE Kit (Roche, USA) was used to obtain the full-length sequences of IFFD. Total RNA from GCs was used as the template for nested-PCR, and the corresponding products of RACE PCR were sequenced.
To verify the coding ability of IFFD, the Open Reading Frame (ORF) of EGFP (GFP-WT), the deleted EGFP start codon ATG (GFP-MUT), and ORF of IFFD with GFP-MUT were amplified and inserted into pcDNA3.1 by using the NheI and HindIII restriction sites. All primers used for vector construction were listed in Table 4 and oligonucleotide sequences were listed in Table 5.
Statistical analysis
All data are shown as mean ± standard deviation from at least three independent replicate experiments. Differences were analyzed by Student’s t-test, and P < 0.05 or P < 0.01 represents statistically significant.
Data availability
The RNA-seq data supporting the results of this study have been deposited in the NCBI BioProject database under accession number PRJNA906921.
References
McGee EA, Hsueh AJ. Initial and cyclic recruitment of ovarian follicles. Endocr Rev. 2000;21:200–14.
Matsuda F, Inoue N, Manabe N, Ohkura S. Follicular growth and atresia in mammalian ovaries: regulation by survival and death of granulosa cells. J Reprod Dev. 2012;58:44–50.
Kumariya S, Ubba V, Jha RK, Gayen JR. Autophagy in ovary and polycystic ovary syndrome: role, dispute and future perspective. Autophagy. 2021;17:2706–33.
Richards JS, Ren YA, Candelaria N, Adams JE, Rajkovic A. Ovarian follicular theca cell recruitment, differentiation, and impact on fertility: 2017 update. Endocr Rev. 2018;39:1–20.
Jin M, Yu Y, Huang H. An update on primary ovarian insufficiency. Sci China Life Sci. 2012;55:677–86.
Richards JS. From follicular development and ovulation to ovarian cancers: an unexpected journey. Vitam Horm. 2018;107:453–72.
Azziz R. PCOS in 2015: new insights into the genetics of polycystic ovary syndrome. Nat Rev Endocrinol. 2016;12:183.
De Vos M, Devroey P, Fauser BCJM. Primary ovarian insufficiency. Lancet. 2010;376:911–21.
Visser JA, Schipper I, Laven JSE, Themmen APN. Anti-Mullerian hormone: an ovarian reserve marker in primary ovarian insufficiency. Nat Rev Endocrinol. 2012;8:331–41.
Zhou X, He Y, Li N, Bai G, Pan X, Zhang Z, et al. DNA methylation mediated RSPO2 to promote follicular development in mammals. Cell Death Dis. 2021;12:653.
Yeung CK, Wang G, Yao Y, Liang J, Tenny Chung CY, Chuai M, et al. BRE modulates granulosa cell death to affect ovarian follicle development and atresia in the mouse. Cell Death Dis. 2017;8:e2697.
Khristi V, Chakravarthi VP, Singh P, Ghosh S, Pramanik A, Ratri A, et al. ESR2 regulates granulosa cell genes essential for follicle maturation and ovulation. Mol Cell Endocrinol. 2018;474:214–26.
Zhang H, Luo Q, Lu X, Yin N, Zhou D, Zhang L, et al. Effects of hPMSCs on granulosa cell apoptosis and AMH expression and their role in the restoration of ovary function in premature ovarian failure mice. Stem Cell Res Ther. 2018;9:20.
Pan Z, Zhang J, Li Q, Li Y, Shi F, Xie Z, et al. Current advances in epigenetic modification and alteration during mammalian ovarian folliculogenesis. J Genet Genomics. 2012;39:111–23.
Liu YN, Qin Y, Wu B, Peng H, Li M, Luo H, et al. DNA methylation in polycystic ovary syndrome: emerging evidence and challenges. Reprod Toxicol. 2022;111:11–19.
He C, Wang K, Gao Y, Wang C, Li L, Liao Y, et al. Roles of noncoding RNA in reproduction. Front Genet. 2021;12:777510.
Herman JG, Baylin SB. Mechanisms of disease: gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–54.
Lomniczi A, Loche A, Castellano JM, Ronnekleiv OK, Bosch M, Kaidar G, et al. Epigenetic control of female puberty. Nat Neurosci. 2013;16:281–9.
Kawai T, Richards JS, Shimada M. The cell type-specific expression of lhcgr in mouse ovarian cells: evidence for a DNA-demethylation-dependent mechanism. Endocrinology. 2018;159:2062–74.
Kawai T, Richards JS, Shimada M. Large-scale DNA demethylation occurs in proliferating ovarian granulosa cells during mouse follicular development. Commun Biol. 2021;4:1334.
Jandura A, Krause HM. The new RNA world: growing evidence for long noncoding RNA functionality. Trends Genet. 2017;33:665–76.
Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–52.
Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–8.
Zhang D, Tang HY, Tan L, Zhao DM. MALAT1 is involved in the pathophysiological process of PCOS by modulating TGFbeta signaling in granulosa cells. Mol Cell Endocrinol. 2020;499:110589.
Zhao J, Xu J, Wang W, Zhao H, Liu H, Liu X, et al. Long non-coding RNA LINC-01572:28 inhibits granulosa cell growth via a decrease in p27 (Kip1) degradation in patients with polycystic ovary syndrome. Ebiomedicine. 2018;36:526–38.
Geng X, Zhao J, Huang J, Li S, Chu W, Wang WS, et al. lnc-MAP3K13-7:1 inhibits ovarian GC proliferation in PCOS via DNMT1 downregulation-mediated CDKN1A promoter hypomethylation. Mol Ther. 2021;29:1279–93.
Adriaens C, Rambow F, Bervoets G, Silla T, Mito M, Chiba T, et al. The long noncoding RNA NEAT1_1 is seemingly dispensable for normal tissue homeostasis and cancer cell growth. RNA. 2019;25:1681–95.
Nakagawa S, Shimada M, Yanaka K, Mito M, Arai T, Takahashi E, et al. The lncRNA Neat1 is required for corpus luteum formation and the establishment of pregnancy in a subpopulation of mice. Development. 2014;141:4618–27.
Wijgerde M, Ooms M, Hoogerbrugge JW, Grootegoed JA. Hedgehog signaling in mouse ovary: Indian hedgehog and desert hedgehog from granulosa cells induce target gene expression in developing theca cells. Endocrinology. 2005;146:3558–66.
Dewailly D, Robin G, Peigne M, Decanter C, Pigny P, Catteau-Jonard S. Interactions between androgens, FSH, anti-Mullerian hormone and estradiol during folliculogenesis in the human normal and polycystic ovary. Hum Reprod Update. 2016;22:709–24.
Chen J, Wu L, Xu H, Cheng S. 5-Aza-CdR regulates RASSF1A By inhibiting DNMT1 to affect colon cancer cell proliferation, migration and apoptosis. Cancer Manag Res. 2019;11:9517–28.
Lu C, Wei Y, Wang X, Zhang Z, Yin J, Li W, et al. DNA-methylation-mediated activating of lncRNA SNHG12 promotes temozolomide resistance in glioblastoma. Mol Cancer. 2020;19:28.
Ma Y, Yang Y, Wang F, Moyer MP, Wei Q, Zhang P, et al. Long non-coding RNA CCAL regulates colorectal cancer progression by activating Wnt/beta-catenin signalling pathway via suppression of activator protein 2alpha. Gut. 2016;65:1494–504.
Sang B, Zhang YY, Guo ST, Kong LF, Cheng Q, Liu GZ, et al. Dual functions for OVAAL in initiation of RAF/MEK/ERK prosurvival signals and evasion of p27-mediated cellular senescence. Proc Natl Acad Sci USA. 2018;115:E11661–E11670.
Yu T, Zhao Y, Hu Z, Li J, Chu D, Zhang J, et al. MetaLnc9 facilitates lung cancer metastasis via a PGK1-activated AKT/mTOR pathway. Cancer Res. 2017;77:5782–94.
Philipsen S, Suske G. A tale of three fingers: the family of mammalian Sp/XKLF transcription factors. Nucleic Acids Res. 1999;27:2991–3000.
Sekiguchi T, Mizutani T, Yamada K, Yazawa T, Kawata H, Yoshino M, et al. Transcriptional regulation of the epiregulin gene in the rat ovary. Endocrinology. 2002;143:4718–29.
Yu C, Li M, Wang Y, Liu Y, Yan C, Pan J, et al. MiR-375 mediates CRH signaling pathway in inhibiting E2 synthesis in porcine ovary. Reproduction. 2016;153:63–73.
Carlevaro-Fita J, Johnson R. Global positioning system: understanding long noncoding RNAs through subcellular localization. Mol Cell. 2019;73:869–83.
Cirillo F, Catellani C, Lazzeroni P, Sartori C, Nicoli A, Amarri S, et al. MiRNAs regulating insulin sensitivity are dysregulated in polycystic ovary syndrome (PCOS) ovaries and are associated with markers of inflammation and insulin sensitivity. Front Endocrinol. 2019;10:879.
Casarini L, Crepieux P. Molecular mechanisms of action of FSH. Front Endocrinol. 2019;10:305.
Jing D, Li C, Yao K, Xie X, Wang P, Zhao H, et al. The vital role of Gli1(+) mesenchymal stem cells in tissue development and homeostasis. J Cell Physiol. 2021;236:6077–89.
Russell MC, Cowan RG, Harman RM, Walker AL, Quirk SM. The hedgehog signaling pathway in the mouse ovary. Biol Reprod. 2007;77:226–36.
Galvin KE, Ye H, Erstad DJ, Feddersen R, Wetmore C. Gli1 induces G2/M arrest and apoptosis in hippocampal but not tumor-derived neural stem cells. Stem Cells. 2008;26:1027–36.
Liu C, Peng J, Matzuk MM, Yao HH. Lineage specification of ovarian theca cells requires multicellular interactions via oocyte and granulosa cells. Nat Commun. 2015;6:6934.
Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–60.
Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33:290–5.
Kang YJ, Yang DC, Kong L, Hou M, Meng YQ, Wei L, et al. CPC2: a fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res. 2017;45:W12–W16.
Sun L, Luo H, Bu D, Zhao G, Yu K, Zhang C, et al. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013;41:e166.
Wucher V, Legeai F, Hedan B, Rizk G, Lagoutte L, Leeb T, et al. FEELnc: a tool for long non-coding RNA annotation and its application to the dog transcriptome. Nucleic Acids Res. 2017;45:e57.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550.
Acknowledgements
This project was supported by the earmarked fund for China Agriculture Research System (CARS-35), the National Natural Science Foundation of China (32072694), the Guangdong Basic and Applied Basic Research Foundation (2021A1515012396), and the Science and Technology Project of Guangzhou (202002030071).
Author information
Authors and Affiliations
Contributions
XZ, ZZ, HZ, JL, and XY conceived the study. XZ, YH, GB, HQ, and YL conducted in vivo experiments. XZ, YH, BH, and NL conducted in vitro experiments. XZ and XP conducted the RNA-seq data analysis. XZ summarized results and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
All experiments were conducted with strict reference to the Regulations for Administration of Affairs Concerning Experimental Animals, and approved by the Animal Care and Use Committee of South China Agricultural University with approval number: SYXK 2019-0136.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by M. Hardwick
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhou, X., He, Y., Pan, X. et al. DNMT1-mediated lncRNA IFFD controls the follicular development via targeting GLI1 by sponging miR-370. Cell Death Differ 30, 576–588 (2023). https://doi.org/10.1038/s41418-022-01103-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-022-01103-y
This article is cited by
-
miR-423 sponged by lncRNA NORHA inhibits granulosa cell apoptosis
Journal of Animal Science and Biotechnology (2023)
-
A novel long non-coding RNA XLOC_004787, is associated with migration and promotes cancer cell proliferation by downregulating mir-203a-3p in gastric cancer
BMC Gastroenterology (2023)