Abstract
Blockade of programmed cell death 1 (PD-1)/programmed cell death 1 ligand (PD-L1) has evolved into one of the most promising immunotherapy strategies for cancer patients. Tumor cells frequently overexpress PD-L1 to evade T cell-mediated immune surveillance. However, the specific genetic alterations that drive aberrant overexpression of PD-L1 in cancer cells remain poorly understood. The gene encoding the E3 ubiquitin ligase substrate-binding adaptor SPOP is frequently mutated in endometrial cancer (EC). Here, we report that SPOP negatively regulates PD-L1 expression at the transcriptional level. Wild-type SPOP binds to IRF1, a primary transcription factor responsible for the inducible expression of PD-L1, and subsequently triggers its ubiquitin- proteasomal degradation to suppress IRF1-mediated transcriptional upregulation of PD-L1. In contrast, EC-associated SPOP mutants lose their capacity to degrade IRF1 but stabilize IRF1, and upregulate PD-L1 expression. EC-associated SPOP mutations accelerate xenograft tumor growth partially by increasing IRF1 and PD-L1 expression. Together, we identify SPOP as a negative regulator of the IRF1–PD-L1 axis and characterize the critical roles of IRF1 and PD-L1 in SPOP mutation-driven tumor immune evasion in EC.
Similar content being viewed by others
Introduction
Endometrial cancer (EC) is the most common gynecological malignancy of the female genital tract with an increasing incidence globally [1]. At the time of diagnosis, about 50–60% of women have a disease confined to the uterus and a 5-year survival rate of 95%. In contrast, 5–10% of patients with distant metastases at the time of diagnosis have a 5-year survival rate of 17% [2]. The complete response rates reported for radiotherapy and cytotoxic chemotherapy in late-stage EC patients are generally low [3]. Therefore, innovative approaches to derive more efficacious therapies for patients with chemotherapy-refractory advanced or recurrent EC are urgently required.
The co-inhibitory PD-1 pathway has received considerable attention because of its role in modulating the immune checkpoint response of cytotoxic T cells, resulting in tumor cells capable of evading immune surveillance. Therapeutic antibodies that block the PD-1 pathway by targeting PD1 or its ligand PD-L1 have resulted in immense breakthroughs in cancer therapeutics [4]. Although checkpoint blockade immunotherapy has been shown to be effective in some cancer types, including EC, patient response rates vary, and only a small subset of patients among a large cohort respond favorably to the treatment [5]. PD-L1 levels in tumors have emerged as valuable biomarkers to determine appropriate cancer patients that can benefit from PD-1/PD-L1 blockade immunotherapy [6]. PD-L1 expression is tightly regulated at the transcriptional, translational, and post-translational levels, but the underlying molecular mechanisms are still not fully understood [7, 8].
Cullin-RING E3 ligases (CRLs) are modular, multi-subunit complexes that constitute a major class of E3 ubiquitin ligases that mediate ubiquitination and degradation of various cellular regulators involved in a plethora of physiological and pathological processes. SPOP is a substrate-binding adaptor of the Cullin 3-RING E3 ubiquitin ligase complex (CRL3). SPOP uses its N-terminal MATH domain to specifically recognize a short motif named SPOP-binding consensus (SBC) in its substrates. The BTB and BACK domains of SPOP mediate dimerization, interaction with CUL3, and self-oligomerization [9]. Interestingly, heterozygous SPOP mutations have been recurrently found in two hormone-related cancers, endometrial cancer and prostate cancer (PCa), but such mutations are relatively rare in cancers of other tissue types [10, 11]. The majority of EC- or PCa-associated SPOP mutations lie in the substrate-binding MATH domain, suggesting that a change in binding affinity between SPOP and its substrate partners may occur. We and others have identified multiple oncoproteins, such as SRC-3 [12], AR [13], ERα [14], ERG [15, 16], DEK [17], BRD2/3/4 [18,19,20], PD-L1 [21], SENP7 [22], Caprin1 [23], c-MYC [24], TRIM24 [25], and Nanog [26, 27], that are ubiquitinated and degraded by CRL3–SPOP ubiquitin ligase complexes. Moreover, CRL3-SPOP ubiquitin ligase complexes exert tumor-suppressive function by regulating non-degradative ubiquitination of some substrates, such as INF2 [28] and MyD88 [29]. Cancer-associated SPOP mutants generally display impaired substrate-binding capacity and oligomerization with wild-type SPOP, which disrupts the functions of the entire CRL3-SPOP complex in a dominant-negative manner [17]. Accumulating evidence from cancer cell lines and animal models support the notion that SPOP mutations promote the initiation and progression of EC and PCa, potentially owing to the dysregulation of their ubiquitinated substrates [30]. However, the underlying molecular mechanisms are not fully understood.
Interferon regulatory factor-1 (IRF1) is a primary transcription factor responsible for PD-L1 induction via the IFN-γ-driven JAK/STAT–IRF1 signaling cascade. IFN-γ has been shown to play a central role in regulating the response of tumor cells to IFN-γ signaling [14, 31]. In this study, we identified IRF1 as a proteolytic substrate for the CRL3–SPOP complexes. SPOP mutations lead to aberrant IRF1 protein stabilization, enhanced PD-L1 induction, and tumor immune escape in EC.
Results
Identification of IRF1 as a novel SPOP interactor
Previously, we performed a Y2H screen in a human fetal liver tissue cDNA library using full-length SPOP as bait29. Among the positive clones, we identified two clones that corresponded to IRF1 fragments (Fig. 1A). Given that IRF1 serves as a signaling hub that coordinates PD-L1 induction, we explored whether IRF1 is an authentic SPOP substrate, and whether PD-L1 levels are dysregulated in SPOP-mutated ECs. First, we verified that the ectopically expressed SPOP interacted with IRF1 (Fig. 1B). Moreover, endogenous SPOP and IRF1 interacted with each other in KLE endometrial cancer cells (Fig. 1C, D). The SPOP–IRF1 interaction was specific because SPOP did not bind to other IRFs (IRF2/3/4/5/6/8/9) or STATs (STAT1-6) we tested (Fig. 1E, F). Only SPOP, but none of the other CUL3-based BTB domain-containing adaptors examined, showed any interaction with IRF1 (Fig. 1G). In accordance with a previous study showing that the MATH domain of SPOP is responsible for recruiting substrates [9], we demonstrated that deletion of the MATH domain, but not the CUL3-binding BTB domain, completely abolished the interaction between SPOP and IRF1 (Fig. 1H, I). Moreover, IFN-γ treatment had no effect on the binding affinity of the SPOP-IRF1 interaction (Fig. 1J).
A Diagram showing the portion of IRF1 protein identified by yeast two-hybrid Y2H screen using full-length SPOP as bait. The region between 2 vertical dashed red lines is the interacting region shown by positive clones, and the red rectangles represent the SBC motif. B Western blots of indicated proteins in WCL and co- IP samples of anti-FLAG antibody obtained from 293 T cells transfected with indicated plasmids. C Western blots of indicated proteins in WCL and co- IP samples of IgG or anti-SPOP antibody obtained from the cell extracts of KLE cells treated with 20 μM of MG132 for 8 h. D Western blots of indicated proteins in WCL and co- IP samples of IgG or anti- IRF1 antibody obtained from the cell extracts of KLE cells treated with 20 μM of MG132 for 8 h. E–G Western blots of indicated proteins in WCL and co-IP samples of anti-FLAG antibody obtained from 293 T cells transfected with indicated plasmids and treated with 20 µM MG132 for 8 h. H Schematic representation of SPOP deletion mutants indicating their binding capability with IRF1. I Western blots of indicated proteins in WCL and co- IP samples of anti-FLAG antibody obtained from 293 T cells transfected with indicated plasmids and treated with 20 µM MG132 for 8 h. J Western blots of indicated proteins in WCL and co-IP samples of anti-FLAG antibody obtained from KLE cells transfected with indicated plasmids and treated with DMSO or IFN-γ ((200 ng/ml) for 6 h. K Representative IF images of KLE cells transfected with indicated plasmids, stained with HA (SPOP), FLAG (IRF1) and DAPI. Scale bar, 20 μm.
Immunofluorescence (IF) analysis showed that IRF1 was dispersed throughout the nucleoplasm, but was recruited into SPOP speckles when co-transfected with SPOP (Fig. 1K). We were interested in investigating whether endogenous SPOP and IRF1 co-localized. However, no commercially available SPOP antibodies are suitable for IF detection of endogenous SPOP. Alternatively, we generated cell lines that stably overexpressing FLAG-SPOP. Endogenous IRF1 protein expression was relatively weak at basal levels but can be strongly induced by IFN-γ treatment. In this condition, we observed that endogenous IRF1 was recruited to SPOP speckles (Supplementary Fig. 1A). The in situ co-localization of endogenous IRF1 and FLAG-SPOP was verified using proximity ligation assays (Supplementary Fig. 1B).
Taken together, these findings indicate that SPOP interacts with IRF1 in cells.
SPOP controls IRF1 stability via the ubiquitin–proteasomal pathway
We then investigated whether IRF1 stability is regulated by the ubiquitin–proteasomal pathway. Treatment of KLE cells with the proteasome inhibitor MG132 increased IRF1 protein levels, but not the corresponding mRNA levels. MLN4924, a small-molecule inhibitor of the NEDD8-activating enzymes required for the activation of CRL complexes, also caused the accumulation of IRF1 proteins, but not mRNA transcripts (Fig. 2A, B). We found that ectopically expressed SPOP resulted in a marked decrease in IRF1 protein levels, which was completely blocked by MG132 treatment (Fig. 2C). Only SPOP-WT, but not the ∆MATH or ∆BTB mutants, promoted IRF1 degradation (Fig. 2D). In contrast, SPOP had no effect on the protein stability of IRF3/4/5/8 (Fig. 2E). Furthermore, all other CRL3 adaptors that we tested failed to degrade IRF1 (Fig. 2F).
A Western blots of indicated proteins in WCL from KLE cells treated with DMSO, MG132 (20 μM) or with MLN4924 (100 nM) for 8 h. B RT-qPCR assessment of IRF1 mRNA expression in KLE cells treated with DMSO or MG132 (20 μM) or with MLN4924 (100 nM) for 8 h. The mRNA level of GAPDH was used for normalization. Data are shown as means ± SD (n = 3). C Western blots of indicated proteins in WCL from 293 T cells transfected with indicated plasmids with DMSO or MG132 (20 μM) for 8 h. D–F Western blots of indicated proteins in WCL from 293 T cells transfected with indicated plasmids. G Western blots of indicated proteins in WCL from FLAG-SPOP inducible KLE cells treated with tetracycline (10 ng/ml) with indicated times. H Western blots of indicated proteins in WCL from KLE or SPEC-2 cells with SPOP knockout through CRISPR/Cas9 methods. Parental cells were used as the control. I Western blots of indicated proteins in WCL from KLE cells infected with lentivirus expressing SPOP specific shRNA or negative control (NC). J RT-qPCR measurement of IRF1 mRNA expression in KLE or SPEC-2 cells with SPOP knockout through CRISPR/Cas9 methods. Parental cells were used as the control. The mRNA level of GAPDH was used for normalization. Data are shown as means ± SD (n = 3). K, L Western blots of indicated proteins in WCL of KLE cells with SPOP knockout through CRISPR/Cas9 methods or parental cells for 48 h and then treated with 50 μg/ml cycloheximide (CHX) and harvested at different time points (K). At each time point, the intensity of IRF1 was normalized to the intensity of actin and then to the value at 0 h (L). Data are shown as means ± SD (n = 3). M Western blots of the products of in vivo ubiquitination assays from 293 T cells transfected with the indicated plasmids and treated with 20 µM MG132 for 8 h. N 293 T cells were co-transfected with indicated plasmids. 24 h after transfection, cell lysates were prepared and Ni–NTA beads pulldown was performed as described in Materials and Methods. The polyubiquitinated forms of IRF1 were detected by western blots with anti-FLAG antibody. O Western blots of the products of in vitro ubiquitination assays performed by incubating the reconstituted SPOP–CUL3–RBX1 E3 ubiquitin ligase complex with E1 and E2 enzymes, ubiquitin and GST-IRF1 at 30 °C for 2 h. P Parental or SPOP KO KLE cells were transfected with indicated plasmids treated with 20 µM MG132 for 8 h, then cell lysates were prepared for co- IP assay with IgG or anti- IRF1 antibody. The polyubiquitinated forms of IRF1 were detected by western blots with anti-HA antibody. P values are calculated using One-way ANOVA test in (B, J) and Two-way ANOVA test in (L). *p < 0.05, **p < 0.01, ***p < 0.001, n.s. non-significant.
To further characterize the effect of SPOP on endogenous IRF1, we generated a tet-on-inducible endometrial cancer KLE cell line that conditionally expressed FLAG-SPOP. The induction of FLAG-SPOP by doxycycline led to a time-dependent decrease in the expression of endogenous IRF1 and TRIM24, a previously reported SPOP substrate [25] (Fig. 2G). Depletion of SPOP by shRNA-mediated knockdown or CRISPR/Cas9-mediated knockout (KO) in EC cell lines resulted in an increase in IRF1 protein levels (Fig. 2H, I), whereas IRF1 mRNA levels were not affected by SPOP KO (Fig. 2J). SPOP KO markedly prolonged the half-life of the IRF1 protein in KLE cells (Fig. 2K, L). SPOP-WT, but not the ∆MATH or ΔBTB mutants, promoted IRF1 polyubiquitination in vivo (Fig. 2M, N). We further demonstrated that the SPOP–CUL3–RBX1 E3 ubiquitin ligase complexes catalyzed IRF1 polyubiquitination in vitro (Fig. 2O). Accordingly, the endogenous ubiquitination levels of IRF1 was decreased in SPOP-deficient cells (Fig. 2P).
Together, these findings indicate that SPOP negatively regulates IRF1 protein stability via the ubiquitin–proteasomal pathway.
SPOP-binding consensus motif in IRF1 is required for SPOP-mediated IRF1 destruction
Previous studies have reported that one or more SBC motifs are present in SPOP substrates [9]. We sought to determine the protein sequence within IRF1 that is responsible for interacting with SPOP. First, we deduced the minimal interacting region from the two SPOP-bound IRF1 fragments obtained from the Y2H screen (Fig. 1A). We found that an IRF1 fragment (amino acids 208–212) corresponded to the minimal interacting region necessary for the SPOP interaction (Fig. 3A). Subsequently, we performed a protein motif search of this region and discovered a perfectly matched SBC motif. This motif is similar to that observed in previously reported SPOP substrates (Fig. 3B). To examine whether this potential motif is required for the SPOP–IRF1 interaction, we generated an IRF1 mutant in which the motif sequence was deleted. SPOP only bound IRF1-WT but not the ΔSBC mutant (Fig. 3C), indicating that the SBC motif in IRF1 is required for SPOP binding. Deletion of the SBC motif in IRF1 completely abolished SPOP-mediated IRF1 degradation (Fig. 3D), ubiquitination (Fig. 3E), and recruitment IRF1 into SPOP speckles (Fig. 3F), and substantially prolonged the half-life of IRF1 (Fig. 3G, H). Mutagenesis of the amino acids in the SBC motif considerably reduced the IRF1–SPOP interaction (Fig. 3I) and SPOP-mediated IRF1 degradation/ubiquitination (Fig. 3J, K). Taken together, our findings indicate that the conserved SBC motif present in IRF1 acts as a degron recognized by SPOP.
A Diagram showing wild-type and SBC motif-deleted IRF1 proteins. B Amino acid sequence alignment of the potential SBC motif of IRF1 in human, mouse and known SPOP substrates. C Western blots of WCL and co- IP samples of anti-FLAG antibody from 293 T cells transfected with the indicated plasmids and treated with 20 µM MG132 for 8 h. D Western blots of WCL from 293 T cells transfected with the indicated plasmids. E Western blots of the products of in vivo ubiquitination assays from 293 T cells transfected with the indicated plasmids and treated with 20 µM MG132 for 8 h. F Representative IF images of KLE cells transfected with indicated plasmids, stained with HA (SPOP), FLAG (IRF1-ΔSBC) and DAPI. Scale bar, 20 μm. G, H Western blots of WCL of 293 T cells transfected with indicated plasmids and treated with 50 μg/ml of CHX and harvested at different time points for western blots (E). Quantification of IRF1 protein from western blots normalized to actin and then to 0-h time point (F). Data are shown as means ± SD (n = 3). I Western blots of WCL and co-IP samples of anti-FLAG antibody from 293 T cells transfected with the indicated plasmids and treated with 20 µM MG132 for 8 h. J Western blots of WCL from 293 T cells transfected with the indicated plasmids. K Western blots of the products of in vivo ubiquitination assays from 293 T cells transfected with the indicated plasmids and treated with 20 µM MG132 for 8 h. P values are calculated using Two-way ANOVA test in (H). *p < 0.05, **p < 0.01, ***p < 0.001, n.s. non-significant.
EC-associated SPOP mutants are defective in promoting IRF1 destruction
To date, the vast majority of SPOP mutaions identified in ECs primarily occur in the MATH domain, which is responsible for substrate binding (Fig. 4A). We postulated that EC-associated SPOP mutants might be defective in mediating IRF1 destruction. We examined the interactions between EC-associated SPOP mutants and IRF1. As shown in Fig. 4B, the IRF1 binding ability of all SPOP mutants was abolished or severely impaired compared to that of SPOP-WT. SPOP-mediated degradation and ubiquitination of the IRF1 protein were also markedly attenuated in all mutants (Fig. 4C, D). Interestingly, SPOP mutants that occur near the C-terminal region of the MATH domain (M117V, R121Q, and D140G) retained a partial capacity to bind, ubiquitinate, and degrade IRF1 compared to other mutants (Fig. 4B-D). In contrast to PCa-associated SPOP mutations that are frequently present in the C-terminal part of the MATH domain, EC-associated SPOP mutations are enriched in the N-terminal part of the MATH domain. The mutation sites of SPOP showed largely no overlap between the two tumor types, even though they were confined to the same substrate-binding MATH domain, implying that the degree of impact of EC- or Pca-associated SPOP mutations on the same substrate may be different [19]. Indeed, Pca-associated hot spot mutants of SPOP (F125V, F133V/L, W131G, and K134N) retained a partial capacity to degrade IRF1 (Supplementary Fig. 2A).
A Distribution of the point mutations on the SPOP gene found in EC specimens. B Western blots of indicated proteins in WCL and samples from co- IP with anti-FLAG antibody in 293 T cells transfected with the indicated plasmids and treated with 20 µM MG132 for 8 h. C Western blots of indicated proteins in WCL from 293 T cells transfected with the indicated plasmids. D Western blots of the products of in vivo ubiquitination assays from 293 T cells transfected with the indicated plasmids and treated with 20 µM MG132 for 8 h. E Representative IF images of KLE cells transfected with indicated plasmids, stained with HA (SPOP-E47K or E50K), FLAG (IRF1) and DAPI. Scale bar, 20 μm. F Western blots of the indicated proteins in WCL from KLE cells infected with EV or lentivirus expressing wild-type or EC-associated SPOP mutants. G Western blots of indicated proteins in WCL and samples from co- IP with anti-FLAG antibody in 293 T cells transfected with the indicated plasmids and treated with 20 µM MG132 for 8 h. H Western blots of indicated proteins in WCL from 293T cells transfected with the indicated plasmids. I Western blots of the products of in vivo ubiquitination assays from 293 T cells transfected with the indicated plasmids and treated with 20 µM MG132 for 8 h.
IF analysis showed that two EC-associated hotspot mutants of SPOP (E47K and E50K) failed to recruit IRF1 into nuclear speckles (Fig. 4E). Stable overexpression of SPOP-E47K or E50K mutant failed to degrade IRF1 but led to elevated endogenous IRF1 levels, demonstrating a dominant-negative effect similar to that of known SPOP substrates such as TRIM24 and BRD4 (Fig. 4F). In accordance with previous studies showing that mutant SPOP acts as a dominant negative regulator of its substrates [17, 30], we also found that co-expression of SPOP-E47K or E50K mutant markedly reduced the interaction between SPOP-WT and IRF1 (Fig. 4G), resulting in the suppression of SPOP-mediated IRF1 degradation and ubiquitination (Fig. 4H, I). To further test the impact of EC-associated SPOP mutants on endogenous IRF1 protein levels, we generated SPOP-E78K, S80R knockin (KI) KLE cells using CRISPR/Cas9-mediated gene editing (Supplementary Fig. 2B, C). We found that endogenous IRF1 levels were elevated in SPOP-MUT KI KLE cells, compared to parental cells (Supplementary Fig. 2D), and the endogenous SPOP-IRF1 interaction was abrogated in SPOP KI KLE cells (Supplementary Fig. 2E).
Taken together, our findings indicate that EC-associated SPOP mutations cause aberrant accumulation of IRF1 due to impaired protein degradation.
EC-associated SPOP mutants elevate PD-L1 expression through IRF1
Having established the regulation of IRF1 protein stability by SPOP, we next sought to investigate whether SPOP has any impact on IRF1-regulated gene transcription when JAK–STAT–IRF1 signaling is activated. We tested six endometrial cancer cell lines and found that only KLE and SPEC-2 cells showed inducible IRF1 expression upon IFN-γ treatment; such effects were not observed in Ishikawa, RL95-2, HEC1-A, and HEC-1B cells (Supplementary Fig. 3A). These results were not surprising since JAK1 truncating mutations are extraordinarily frequent in ECs [32]. The sequencing data retrieved from the Cancer Cell Line Encyclopedia (TCGA) showed that JAK1 truncating mutations are present in four IFN-γ-unresponsive EC cell lines (Supplementary Fig. 3A). Ectopically overexpressed SPOP promoted IRF1 degradation in both JAK-WT and JAK1-MUT EC cell lines, indicating that JAK1 is not required for SPOP-mediated IRF1 degradation (Supplementary Fig. 3B).
We measured multiple reported IRF1 transcriptional targets [33] and found that the mRNA expression of PD-L1, TRAIL, and PSMA6 could be induced in KLE cells upon IFN-γ treatment; however, this effect was attenuated by the stable overexpression of SPOP-WT, but potentiated by the SPOP-E50K mutant. In contrast, neither SPOP-WT nor SPOP-E50K overexpression influenced IFN-γ-induced upregulation of IRF1 mRNA, whose induction was primarily mediated by STAT1 (Fig. 5A). The fact that SPOP negatively regulates PD-L1 mRNA expression at the basal level and IFN-γ-treated condition is of particular interest because PD-L1 elevation usually leads to tumor immune escape [4]. Western blot (WB) analyses also showed that overexpression of SPOP-WT reduced the protein levels of IRF1 and PD-L1 in both IFN-γ-treated and untreated cells. In contrast, overexpression of the SPOP-E50K mutant increased the protein levels of IRF1/PD-L1 in both IFN-γ-treated and untreated cells (Fig. 5B). Flow cytometric measurements showed that SPOP KO led to increased PD-L1 expression on the cell surface in both IFN-γ-treated and untreated cells (Fig. 5C, D). Moreover, IFN-γ-induced IRF1 and PD-L1 proteins gradually decreased when IFN-γ was removed from the cell media, but this process was prolonged in SPOP-KO cells compared to parental cells (Fig. 5E). SPOP KO upregulated the mRNA and protein levels of PD-L1, but this effect was abrogated by co-depletion of IRF1, suggesting that SPOP modulates PD-L1 expression in an IRF1-dependent manner (Fig. 5F, G). Lastly, PD-L1 promoter activity was activated by IRF1, but this effect was attenuated by co-expression of SPOP-WT, but not the E50K and E87K mutants (Fig. 5H, I).
A KLE cells infected with EV, SPOP-WT or SPOP-E50K were treated with DMSO or IFN-γ (200 ng/ml, 12 h). The mRNA levels of IRF1 and its transcriptional targets were measured by RT-qPCR. The mRNA level of GAPDH was used for normalization. Data are shown as means ± SD (n = 3). B Western blots of the indicated proteins in WCL from KLE or SPEC-2 cells that infected with lentivirus expressing EV, SPOP-WT or SPOP-E50K and treated with DMSO or IFN-γ (200 ng/ml) for 12 h. C, D PD-L1mean fluorescence intensity (MFI) of parental or SPOP-KO KLE cells treated with DMSO or IFN-γ (200 ng/ml) for 12 h. Representative profiles (C) and MFI PD-L1 (D) are shown. Data are shown as means ± SD (n = 3). E Parental and SPOP-KO KLE cells were treated with IFN-γ (200 ng/ml) for 12 h, then the treatment media is removed and replaced with fresh media for indicated times. The WCL were prepared for western blots. T: IFN-γ treated, R: IFN-γ removed. F RT-qPCR measurement of IRF1, PD-L1 and SPOP mRNA levels in parental or SPOP-KO KLE cells infected with lentivirus expressing IRF1-specific shRNA or NC. Data are shown as means ± SD (n = 3). G Western blots of the indicated proteins in WCL from parental or SPOP-KO KLE cells infected with lentivirus expressing SPOP-specific shRNA or NC. Measurement of PD-L1 promoter activity in KLE cells transfected with indicated plasmids and treated (H) or not treated (I) with DMSO or IFN-γ (200 ng/ml) for 6 h. The results are expressed as the fold change compared with empty KLE cells. Data are shown as means ± SD (n = 3). P values are calculated using One-way ANOVA test in (A, D, F, H and I). *p < 0.05, **p < 0.01, ***p < 0.001, n.s. non-significant.
Together, our findings indicate that SPOP acts as a negative regulator of the IFNγ-IRF1-PD-L1 signaling, and that the nuclear localization of SPOP is indispensable for its regulation of the IRF1–PD-L1 axis in EC cells.
SPOP exerts an antitumor immunity effect via the IRF1–PD-L1 axis
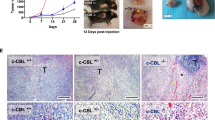
Binding of PD-L1 to its receptor PD-1 suppresses the production of effector cytokines and induces T-cell apoptosis [4]. We explored the influence of SPOP mutations on tumor growth in the presence of a functional immune system and in response to an immune checkpoint blockade. CT26 and MC38 cells (murine colon carcinoma) and ID8 cells (murine ovarian carcinoma) are commonly used to study tumor immune escape in immunocompetent mouse models. We showed that the SPOP–IRF1–PD-L1 axis worked in these cell lines (Supplementary Fig. 4A, B). We then generated CT26 cells stably overexpressing empty vector (EV), SPOP-WT, or E50K mutant. SPOP-WT largely limited CT26 tumor growth in vivo, whereas the SPOP-E50K mutant had an opposing effect to accelerate tumor growth. Strikingly, the difference in tumor weights between the EV and SPOP-E50K groups was largely alleviated following treatment with an anti-PD-L1 mAb, indicating that SPOP mutations promote tumor immune evasion (Fig. 6A–C). As PD-L1 has a strong influence on tumor-infiltrating lymphocytes (TILs), which are crucial for the efficacy of cancer immunotherapy [4], we examined the number of apoptotic cells, PD-L1 expression, and the absolute number of TILs in xenograft tumors. Antibodies against CD3 and CD8 were used to identify T cells and cytotoxic T cells, respectively. IF analysis of tumor tissues showed that tumors expressing the SPOP-E50K mutant displayed elevated PD-L1 levels and significantly reduced numbers of CD3+ and CD8+ TILs (Fig. 6D, E, Supplementary Fig. 4C–F). Given that antitumor immunity is accompanied by apoptosis caused by cytotoxic T lymphocytes (CTLs) in tumor tissues, we compared the levels of cleaved caspase 3 (CCA3) using IF analysis. Although a small number of cells underwent apoptosis in EV tumor tissues, strong clustered apoptotic signals were observed in SPOP-WT tumors (Supplementary Fig. 4D, G). We also found that the addition of an anti-PD-L1 mAb led to a moderate increase in CD3+ and CD8+ TILs in the tumor tissues (Supplementary Fig. 4C–F).
A–C CT26 cells that infected with lentivirus expressing EV, SPOP-WT or E50K mutant were injected subcutaneously (s.c.) into the right flank of BALB/c mice and treated with anti-PD-L1 antibody or IgG2b isotype control at different day points. Tumor growth was measured every other day for 16 days. 8 mice per experimental group. Tumors in each group at day 16 were harvested and photographed (A), tumor volume (B) and tumor weight (C) at each time point was documented. Data are shown as means ± SD (n = 8). D, E Immunostaining of PD-L1 in the CT26 tumor mass (A–C). DAPI: nuclear counterstaining (D). Scale bar, 200μm. The intensity of PD-L1 were quantified using Image J and shown in (E). Data are shown as means ± SD (n = 8). 1 tissue slide per tumor, 6 mice per group. Unit = 182, 047 μm2. F Specific CD8+ T cell-mediated cancer cell killing assay in vitro. OVA-peptide coated parental, SPOP-KO, IRF1-KO, PD-L1-KO, SPOP/IRF1-KO or SPOP/PD-L1DKO MC38 cells co-cultured with activated CD8 + T cells isolated from OT-1 C57BL/6 mice for 12 h were subjected to crystal violet staining. MC38 cells to T cells ratio, 1:5 (left) or 1:10 (right). Data are shown as means ± SD (n = 4). Cells co-culture supernatants in (F) were collected and the cytokines were assessed with ELISA kit for testing TNF-α (G) or IFN-γ (H). Data are shown as means ± SD (n = 4). I–K Parental, SPOP-KO, IRF1-KO, PD-L1-KO, SPOP/IRF1-KO or SPOP/PD-L1-KO CT26 cells were injected s.c. into the right flank of BALB/c mice. Tumor growth was measured every other day for 16 days. 6 mice per experimental group. Tumors in each group at day 16 were harvested and photographed (I), tumor volume (J) and tumor weight (K) at each time point was documented. Data are shown as means ± SD (n = 6). P values are calculated using One-way ANOVA test in (C, E, F, G, H and K) and Two-way ANOVA test in (B, J). *p < 0.05, **p < 0.01, ***p < 0.001, n.s. non-significant.
To further test the hypothesis that SPOP and its mutant exert a role in the regulation of systemic resident immune effector cells, we assessed the relative sizes of T cell populations represented in splenocytes by flow cytometry. Splenocytes from mice bearing SPOP-E50K tumors showed a decreased percentage of CD3+ or CD8+ T cells compared to splenocytes from mice bearing SPOP-WT or EV tumors. Furthermore, the percentage of CD3+ or CD8+ T cells in splenocytes was mildly increased by anti-PD-L1 mAb treatment (Supplementary Fig. 4H, I). Similar results were obtained when ID8 cells were used to generate stable cell lines overexpressing EV, SPOP-WT, or E50K mutant (Supplementary Fig. 4J, K).
To further investigate whether SPOP exerts an antitumor immune effect via the IRF1-PD-L1 axis, we prepared in vitro-activated chicken ovalbumin (OVA)-specific OT1 CTLs that were co-cultured with parental, SPOP-KO, IRF1-KO, PD-L1-KO, SPOP/IRF1 double KO (DKO), or SPOP/PD-L1 DKO MC38 cells coated with OVA peptide (Supplementary Fig. 5). To assess the serial killing efficacy of OT1 CD8+ CTLs, we analyzed the survival rate of MC38 cells by setting different effector-target (ET) ratios. SPOP KO markedly reduced OVA-specific-induced T cell-mediated cancer cell death at different ET ratios in MC38 cells; however, these effects could be reversed by co-ablation of IRF1 or PD-L1 (Fig. 6F). ELISA analysis of co-culture media showed that SPOP KO markedly reduced the levels of secreted cytokines (TNF-α and IFN-γ), which are indicators of T cell activation, in mouse CD8+ T cells; however, these effects could be reversed by co-ablation of IRF1 or PD-L1 (Fig. 6G, H). Consistently, the enhanced MC38 tumor growth caused by SPOP KO could be partially reversed by co-ablation of IRF1 or PD-L1 (Fig. 6I–K), suggesting the existence of a coordinated SPOP–IRF1–PD-L1 pathway in regulating antitumor immunity.
Taken together, our findings indicate that SPOP affects the priming or activation of T cells and the infiltration of T cells into EC tumors by regulating the surface levels of PD-L1 expression on tumor cells.
SPOP mutations are correlated with IRF1/PD-L1 expression in ECs
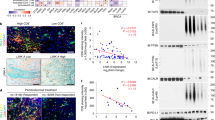
To examine the effect of SPOP mutations on the protein levels of IRF1 and PD-L1 in EC patient specimens, we performed the immunohistochemistry (IHC) of IRF1 and PD-L1 proteins in a patient cohort that included a total of 99 primary EC specimens (Supplementary Table 1). We identified 18 SPOP-mutated tumors using Sanger sequencing. The frequency of SPOP mutations was 18. 1% in our samples and higher than previous findings of different EC cohorts, including those of TCGA dataset. IHC analysis showed that approximately 60% of SPOP-mutated tumors exhibited strong or intermediate staining for IRF1 and PD-L1 (Fig. 7A–D). In contrast, less than 30% of tumors with WT SPOP exhibited strong or intermediate staining for IRF1 and PD-L1 (Fig. 7A–D). There was a trend that PD-L1 expression correlated with IRF-1 expression in ECs with a Pearson’s correlation coefficient of 0.8163 (Supplementary. Fig. 6A, p < 0.001). Moreover, the number of CD8+ TILs was reduced in SPOP-mutated tumors compared to that in SPOP-WT tumors (Supplementary Fig. 6B).
Representative IHC images of IRF1 staining in 99 EC patient specimens including 81 SPOP-WT and 18 SPOP mutant (MUT) cases (A) and the quantitative data of IRF1 staining (B). Scale bar, 50 μm. Representative IHC images of PD-L1 staining in 99 EC patient specimens including 81 SPOP-WT and 18 SPOP-MUT cases (C) and the quantitative data of PD-L1 staining (D). Scale bar, 50 μm. E Schematic of the proposed mechanism through which SPOP mutants enhance PD-L1 transcription in SPOP-mutated ECs. P values are calculated using Two-way ANOVA test in (B and D). *p < 0.05, **p < 0.01, ***p < 0.001, n.s. non-significant.
Together, these findings indicate that the protein levels of IRF1 and PD-L1 were higher in SPOP-mutated EC specimens than that in SPOP WT specimens.
Discussion
PD-L1 expression levels in tumor cells and the tumor microenvironment affect the clinical response to PD-1/PD-L1 blockade immunotherapy, highlighting the need for a better understanding of the biological processes that regulate PD-L1 expression at multiple levels [8]. In the current study, we demonstrated that SPOP negatively regulates PD-L1 mRNA transcription by targeting IRF1 for ubiquitination-dependent destruction, whereas EC-associated SPOP mutations led to enhanced expression of IRF1 and its major transcriptional target, PD-L1 (Fig. 7E). Moreover, SPOP mutation-driven EC xenograft tumor growth can be mitigated by anti-PD-L1 mAb. These results suggest that PD-L1-mediated immune evasion could be a possible mechanism driving tumor growth in SPOP-mutated ECs. Although there was no significant correlation between SPOP mutation status and EC prognosis in TCGA EC cohort (data not shown), our study suggests that detection of SPOP mutation status may facilitate the identification of a subset of ECs responsive to PD-1/PD-L1 blockade immunotherapy.
The IFN-γ-JAK-STAT1-IRF1 axis is the predominant pathway that regulates PD-L1 mRNA transcription, with IRF1 directly binding to the PD-L1 promoter [30]. IRF1 was first discovered as the main transcription factor mediating responses to IFN-α/β/γ, and subsequently, it has been shown that IRF1 is capable of preventing oncogene-mediated malignant transformations [34]. Accumulating evidence has demonstrated that multiple oncogenes such as BRD4 [18,19,20], EZH2 [35], EGFR [36], and CDK5 [37] modulate PD-L1 mRNA expression in an IRF1-dependent manner. Moreover, IRF1 is required for PD-L1 induction in cells upon genotoxic insults [38]. A recent study conducted in melanoma models demonstrated that IRF1 KO in tumor cells results in the loss of in vivo tumor growth without affecting in vitro cell growth. Exogenous expression of PD-L1 in IRF1 deficient cells restored tumor progression [39]. These studies suggested that IRF1 acts as a double-edged sword with the capacity not only to promote antitumor effects, but also to facilitate cancer immune evasion by the upregulation of PD-L1. Since the identification of transcriptional dependence of PD-L1 on IRF1, nuclear IRF1 expression has been suggested as a surrogate for PD-L1 expression to assess the response to anti-PD-1 therapy in melanoma [40]. Analysis of RNA-sequencing data from TCGA cohorts revealed a positive and statistically significant correlations between the mRNA expression levels of IRF1 and PD-L1 in several cancers, including ovarian cancer and breast cancer. However, no such correlation was observed in the EC cohort [40]. Abnormal protein turnover of IRF1 might partially explain this discrepancy. Interestingly, IRF1 can be ubiquitinated and degraded by FBXW7, a CRL1 E3 ubiquitin ligase adaptor that is frequently mutated in ECs [41, 42]. Thus, enhanced IRF1 protein stability due to specific genetic alterations may lead to PD-L1 upregulation in ECs.
Zhang et al. have reported that PD-L1 is a direct proteolytic substrate in PCa cells [21]. In the present study, we provide evidence for a model whereby SPOP downregulates PD-L1 expression at the mRNA level by targeting IRF1 for ubiquitin-dependent degradation. Therefore, SPOP may exert multiple layers of regulation towards PD-L1 expression at the transcriptional, translational, and post-translational levels in a cancer content-dependent manner. SPOP controls the stability of multiple substrates, which may directly or indirectly affect PD-L1 mRNA expression. The influence of SPOP on PD-L1 may not be solely attributed to its activity towards IRF1. One potential candidate is BRD4, a key factor in the transcriptional activation of distinct sets of cancer-related genes. Two recent studies have demonstrated that BET inhibitors repress constitutively expressed and IFN-γ-induced PD-L1 expression across different human tumor cell lines, mouse tumor cell lines, and primary patient samples. Mechanistically, BRD4 and IRF1 co-regulate IFN-γ-induced PD-L1 transcription, and treatment with BETi remarkably decreased BRD4 recruitment to PD-L1 loci [41, 43]. Therefore, the accumulated BRD4 may act synergistically with IRF1 to promote PD-L1 expression in SPOP-mutated ECs.
The differential effects of EC and PCa-associated SPOP mutants on their ubiquitylated substrates have been reported in many other studies. For example, PCa-associated SPOP mutants were defective in prompting DEK degradation, whereas EC-associated SPOP mutants retained similar capacity to degrade DEK as SPOP-WT [17]. PCa-associated SPOP mutants impaired the degradation of BET proteins, whereas EC-associated SPOP mutants enhanced BET protein degradation through a gain-of-function mechanism [19]. We noted that EC-associated SPOP mutations are prone to localize to the N-terminal part of the MATH domain, whereas PCa-associated SPOP mutations are prone to localize to the C-terminal part of the MATH domain. Our results indicated that three EC-associated SPOP mutants (M117V, R121Q, and D140G) that retained partial IRF1-binding capacity are all localized to the C-terminal part of the MATH domain. In contrast, other EC-associated SPOP mutants incapable of binding to IRF1 are localized to the N-terminal part of the MATH domain (Fig. 4A, B). A possible explanation for this phenomenon was proposed by Theurillat et al. that PCa-associated mutations are found to be confined to amino acid residues of the substrate-binding cleft of the MATH domain, whereas EC-associated SPOP mutations occur in another territory of the MATH domain [9]. However, a structural biology study is required to draw a clear conclusion in the future.
In summary, we demonstrate that EC-associated SPOP mutations elevate PD-L1 expression by directly upregulating IRF1 expression. Elucidation of the genetic alterations that lead to abnormal PD-L1 expression will guide PD-1/PD-L1 blockade immunotherapy in EC.
Materials and methods
Cell culture and reagents
293 T, KLE, SPEC-2, Ishikawa, RL95-2, HEC-1A, HEC-1B, MC38, ID8, and CT26 cells were obtained from American Type Culture Collection. 293 T, KLE, MC38, CT26, and ID8 cells were maintained in DMEM supplemented with 10%(v/v) FBS. Ishikawa, SPEC-2, ECC- 1, HEC- 1 A, and HEC-1B cells were maintained in DMEM/F12 supplemented with 10% (v/v) fetal bovine serum (FBS). The cells were maintained in at 37 °C humidified incubator supplied with 5% CO2. Transient transfections were performed using Lipofectamine 3000 (Thermo Fisher Scientific). For lentivirus transfection, pLKO-base gene shRNA knockdown plasmid or pCDH-SPOP-WT/mutant and virus packing constructs were transfected into 293 T cells. The viral supernatant was collected 48 h after transfection. EC cells were infected with the viral supernatant in the presence of polybrene (8 µg/ml) and were then selected in growth media containing 1.5 µg/ml puromycin. All the cell lines were tested and authenticated by karyotyping. Plasmocin (InvivoGen) was added to the cell culture medium to prevent mycoplasma contamination. Mycoplasma contamination was tested regularly using a Lookout Mycoplasma PCR Detection Kit (Sigma-Aldrich). Antibodies and other resources are listed in Supplementary Table 2.
CRISPR/Cas9-mediated gene KO
The PX459 plasmid was used to clone guide oligos targeting SPOP, CD274, or IRF1 gene. The cells were plated and transfected overnight with pX459 constructs. 24 h after transfection, 1 µg/ml puromycin was used to screen the cells for 3 days. Living cells were seeded in 96 well plate at a limited dilution to isolate the monoclonal cell line. KO cell clones were screened by WB and validated by Sanger sequencing. The sequences of gene-specific sgRNAs are listed in Supplementary Table 3.
CRISPR/Cas9-mediated gene KI
The SPOP KI cell lines were generated following the protocol described previously. The sgRNA targeting the genomic sequence close to the mutation site was designed using the CRISPR design tool (http://crispr.mit.edu) and cloned into pX459 constructs. A donor sequence was used as the template with SPOP-E78K or S80R mutation was cloned into pDor04 constructs. The two constructs were co-transfected into KLE cells at a 1:1 molar ratio. 24 h after transfection, 1 µg/ml puromycin was used to screen the cells for 3 days. Then the GFP-positive cells were enriched by FACS sorting and seeded into a 96-well plate with one cell per well. After about two weeks, the genomic DNA of individual clone was extracted and used as the template to amplify the DNA fragment containing the mutation site. Finally, Knockin positive cells were verified by Sanger sequencing.
Western blot
Cell lysates or immunoprecipitates were subjected to SDS-PAGE and proteins were transferred to nitrocellulose membranes (GE Healthcare). The membranes were blocked in Tris-buffered saline (pH 7.4) containing 5% non-fat milk and 0.1% Tween-20, washed twice in TBS containing 0.1% Tween-20, and incubated with primary antibody for 2 h and followed by secondary antibody for 1 h at room temperature. The proteins of interest were visualized using ECL chemiluminescence system (Santa Cruz). Original Western blot images for all relevant figures are shown in “Supplementary Material—Original Blots”.
Real-time reverse transcription PCR (RT-qPCR)
Total RNAs was isolated from the indicated cells using TRIzol reagent (Tiangen) and cDNA was reverse-transcribed using the PrimeScript RT Master Mix Kit (Takara) according to the manufacturer’s instructions. PCR amplification was performed using an AceQ Universal SYBR qPCR Master Mix Kit (Vazyme). All quantifications were normalized to the level of the endogenous control GAPDH. The primer sequences are listed in Supplementary Table 4.
In vivo ubiquitination assays
293 T cells were transfected with HA-Ubiquitin and other indicated constructs. 36 h after transfection, cells were treated with 20 µM MG132 for 8 h and then lysed in 1% SDS buffer (Tris [pH 7.5], 0.5 mM EDTA, 1 mM DTT) and boiled for 10 min. For co-immunoprecipitation (Co-IP), the cell lysates were diluted 10-fold in Tris-HCl buffer and incubated with anti-FLAG M2 agarose beads (Sigma) for 4 h at 4 °C. The bound beads were then washed four times with BC100 buffer (20 mM Tris-Cl, pH 7.9, 100 mM NaCl, 0.2 mM EDTA, 20% glycerol) containing 0.2% Triton X-100. Proteins were eluted with 3X FLAG peptide for 2 h at 4 °C. The ubiquitinated form of IRF1 was detected by WB with anti-HA antibody.
In vitro ubiquitination assays
In vitro ubiquitination assays were performed using a protocol reported previously [17]. Briefly, 2 µg of APP-BP1/Uba3, 2 µg of His-UBE2M, and 5 µg of NEDD8 were incubated at 30 °C for 2 h in the presence of ATP. The thioester-loaded His-UBE2M–NEDD8 was further incubated with 3 µg of His-DCNL2 and 6 µg of CUL3–RBX1 at 4 °C for 2 h to obtain neddylated CUL3–RBX1. The neddylated CUL3–RBX1, 5 µg of GST-SPOP, 5 µg of ubiquitin, 500 ng of E1 enzyme, 750 ng of E2 enzyme (UbcH5a and UbcH5b), and 5 µg of GST-IRF1 were incubated with 0.6 µl of 100 mM ATP, 1.5 µl of 20 µM ubiquitin aldehyde, 3 µl of 10× ubiquitin reaction buffer (500 mM Tris-HCl (pH 7.5), 50 mM KCl, 50 mM NaF, 50 mM MgCl2, and 5 mM DTT), 3 µl of 10× energy regeneration mix (200 mM creatine phosphate and 2 µg/µl creatine phosphokinase) and 3 µl of 10× protease inhibitor cocktail at 30 °C for 2 h, followed by WB analysis.
Proximity ligation assays
The in situ interaction between IRF1 and SPOP in KLE cells stably overexpressing FLAG-SPOP was detected using Duolink® in situ detection Kit (Sigma). Briefly, KLE cells were attached to 24-well slides and treated with IFN-γ (200 ng/ml) for 4 h, and then the cells were fixed with 4% paraformaldehyde. and permeabilized by 0.5% Triton x-100 for 30 min. KLE cells were incubated with two primary antibodies (IRF1 and FLAG) and then incubated with a pair of PLA probes composed of species-specific secondary antibodies conjugated to complementary oligonucleotides. After adding hybridization solution and ligase, oligonucleotides form a circle when proteins were very close. Finally, the polymerase and nucleotides participate in the formation of rolling circle amplification, which is visualized by red fluorescence. The cells were imaged using a confocal laser-scanning microscope (LSM880, Zeiss).
Protein half-life assays
For the half-life study, 20 μg/ml cycloheximide (Sigma-Aldrich) was added to the media. At the indicated time points, the cells were collected and protein abundance was measured by WB.
Transcriptional reporter assays
Luciferase assays (Promega) were conducted in cells transiently transfected with the promoter reporter (PD-L1-Luc), pTK-galactosidase, and other indicated plasmids. Luciferase activity in the cell lysates was measured using a luciferase assay system on a Berthold Lumat LB 9507 luminometer (Promega). The luciferase activity was normalized to that of galactosidase as an internal control. Each assay was performed in triplicate and the results were confirmed by at least three individually repeated experiments.
IF and confocal microscopy
For cell staining, cells were plated on chamber slides and fixed with 4% paraformaldehyde at room temperature for 30 min. After washing with PBS, cells were permeabilized with 0.1% Triton-X100 in PBS for 15 min at room temperature. Cells were then washed with PBST, blocked with 5% donkey serum in PBS for 1 h, and incubated with primary antibodies in PBS at 4 °C for overnight in the dark. After washing with PBST, fluorescence-labelled secondary antibodies were applied and DAPI was counterstained for 1 h at room temperature in the dark. The slides were mounted in ProlongGold (Invitrogen). The cells were visualized and imaged using a confocal microscope (LSM880, Zeiss) with a 63*/1.4NA Oil PSF Objective.
For graft tumor tissue staining, the tumor tissues were isolated from mice after perfusion with 0.1 M PBS (pH7 .4) and fixed for 3 days with 4% paraformaldehyde at 4 °C. The tumor tissues were then placed in 30% sucrose solution for 2 days for dehydration. The tumor was embedded into the OCT block and frozen for cryostat sectioning. Cryostat sections (45-μm thick) were washed with PBS, and then incubated in blocking solution (PBS containing 10% goat serum, 0.3% Triton X-100, pH7.4) for 2 h at RT. In antibody reaction buffer (PBS plus 10% goat serum, 0.3% Triton X-100, pH7.4), samples were stained with primary antibodies against active caspase 3 (1:200), CD8 (1:100), Granzyme B (1:200), and PD-L1 (1:50) overnight at 4 °C, followed by Alexa 488, 594, and 647 secondary antibodies (1;2000) at RT for 3 h. DAPI was used for nuclear staining. The sections were then sealed with an anti-fluorescence quencher. The samples were visualized and imaged using a confocal microscope (Olympus FV3000 inverted confocal microscope) along the z-axis with a 40* Objective. The intensity of PD-L1/Granzyme B was quantified using ImageJ by computing the total positive staining area with at least eight units. The analysis was performed in triplicate in three different fields. The number of CD8/CD3 positive signals per unit area was calculated.
Mouse tumor implantation assays
Animals were housed under pathogen-free conditions with ad libitum access to food and water. All experimental protocols were approved by the Ethics Review Committee for Animal Experimentation at Tongji University. Female BALB/c mice were obtained from the SLAC Laboratory Animal Co., Ltd. At 6-8 weeks of age, the mice were injected subcutaneously into the flank with 1 × 105 parental, SPOP-KO, IRF1-KO, PD-L1-KO, SPOP/IRF1-KO, SPOP/PD-L1-KO MC38 cells, or CT26 cells stably expressing empty control (EV), SPOP-WT, or E50K mutant in 100 µl PBS. After 6 days and after the tumors reached an average volume of ~100 mm3, the EV, SPOP-WT, or E50K mutant mice received an intraperitoneal injection of IgG and anti-PD-L1 antibody (clone 10 F. 9G2, 200 μg per mouse in 200 μl HBSS saline buffer) every three days for a total of five injections. Tumor growth was monitored every two days in two dimensions using a digital caliper. Tumor volumes were calculated using the following ellipsoid volume formula: V = (L× W2)/2, where L is the length and W is the width. Mice were sacrificed when signs of ulceration in the tumor were evident or when tumors reached the maximum permitted area, and tumor weight was monitored as indicated. Tumor tissues were divided, and a portion was subjected to formalin fixation and embedded in paraffin for IF analysis. Mouse splenocytes were collected and analyzed using FACS.
1 × 106 ID8 cells stably expressing EV, SPOP-WT, or E50K mutant were intraperitoneally injected into the peritoneum of 6-week-old C57BL/6 female mice. Fourteen days after tumor cells were injected, the mice were sacrificed, and splenocytes were collected and analyzed by FACS.
Splenocytes profile analysis by FACS
Splenocytes were isolated from CT26 or ID8 injected mice. A single-cell suspension was prepared in PBS by grinding these tissues between the two rough ends of the microscopic slides. Red blood cell lysis buffer was used to ensure optimal lysis of RBCs in single-cell suspensions, with minimal effects on leukocytes. The cells were washed twice. After blocking with CD16/CD32 antibodies, single-cell suspensions were stained with CD45-FITC, CD3-APC and CD8-PE antibodies. Stained spleen T cells were analyzed using a Beckman Coulter Gallios flow cytometer (Fullerton). Compensation was performed according to single staining. Data were processed using FlowJo software.
T cell-mediated tumor cell killing assays
In biref, one ovalbumin-transgenic (OT-1) mice (kindly provided by Dr. Xiaofei Yu at Fudan University) were sacrificed, and spleens and peripheral (cervical, auxiliary, brachial, and inguinal) and mesenteric lymph nodes were harvested and pooled. A single-cell suspension was prepared in PBS by grinding these tissues between the two rough ends of the microscopic slides. The cells were washed twice and re-suspended in the RPMI medium. CD8+ T cells isolated using a mouse CD8+ T cell isolation kit (BioLegend) were used as effector cells. CD8+ T cells were activated with 20 ng/mL OVA peptide (SIINFEKL) and IL-2 (10 ng/mL) in RPMI medium for three days at 37 °C. CD8 + T cells were stained with CFSE (5 μM; BioLegend). target mouse cancer cell line, MC38, which was placed in a 96-well plate in advance, was coated with 10 μg/ml OVA peptide at 37 °C for 1 h. Excess OVA peptide was washed away with RPMI medium, and CD8+ T cells were added and co-cultured with target cancer cells at E:T ratios of 10:1 or 5:1 for 12 or 36 h. T cells were collected for T cell proliferation assays, and the medium was collected for ELISA analysis. Living cancer cells were quantified using a spectrometer at OD (570 nm) followed by crystal violet staining.
ELISA assays
ELISA kits for TNF-α and IFN-γ detection were purchased from BioLegend. Co-cultured cell supernatants were collected, and cytokines were assessed according to the manufacturer’s instructions. Capture antibody solution was added to microplate wells and incubated overnight (16-18 h) at 4 °C. The assay diluent buffer was used to block non-specific binding. Serially diluted protein standards and test samples were incubated for 2 h at room temperature with shaking. The trapped antibody was then added and detected using a substrate solution. The optical density of each well was determined within 20 min using a microplate reader at 450 nm.
Flow cytometry assays
Cells were detached using Versene (Thermo Fisher Scientific), resuspended in PBS containing Fc Block (BD Biosciences), centrifuged, incubated for 30 min with human PD-L1 antibody, and washed with PBS. Stained cells were analyzed using an LSRII flow cytometer (BD Biosciences). Data were analyzed using the FlowJo software. Data were collected, debris (low FSC and SCC) were excluded, and single cells that were negative for the live/dead discriminant were gated.
Detection of SPOP mutations in specimens from EC patients
Paraffin-embedded tissues from 105 EC patients in the study were obtained from Shanghai First Maternity and Infant Hospital between December 2010 and December 2018. Clinical information of the specimens is presented in Supplementary Table 1. The protection of the human subject board of the hospital approved the experimental protocols and informed consent was obtained from each patient. Genomic DNA was isolated using the GeneRead DNA FFPE kit (Qiagen) from tumor-rich regions (at least 50% of tumor cells) of formalin-fixed, paraffin-embedded (FFPE) samples assessed by two independent pathologists. In the samples with mutations, the non-tumor mucosa was also extracted from the same slide and subjected to the same approach as that used to validate the somatic mutation. SPOP mutations have been reported in the MATH domain (exons 5-7). Supplementary Table 3 lists the six pairs of primer sets covering these regions. PCR amplifications were performed using 2×Taq Master Mix (Vazyme), and the PCR products were purified using the QIAquick Gel Extraction Kit (Qiagen) following the manufacturer’s instructions and used for Sanger sequencing.
IHC for human EC specimens
For IHC, 4 μm sections were cut and mounted on platinum PRO Adhesive Glass slides (Matsunami), dried at 60 °C, dewaxed in xylene, and rehydrated in graded alcohols (100%, 95%, 85%, and 75%) to water. Antigen retrieval was performed at 95 °C for 20 min. The sections were then incubated with 3% H2O2 for 10 min at room temperature to block endogenous peroxidase activity. Incubation with the primary antibodies was done overnight at 4 °C. Immunostaining was automated using anti-rabbit or anti-mouse IgG secondary antibodies conjugated with HRP for 30 min. Specific samples were treated with SignalStain diaminobenzidine (DAB) substrate (Cell Signaling Technology), followed by counterstaining with hematoxylin and mounting. Images were captured using MoticEasyScan (Motic) and matching software. The expression levels of PD-L1 and IRF1 in the EC tumor samples were scored as 0 (negative), 1 (weak expression), 2 (intermediate expression), or 3 (strong expression). The antibodies used for the IHC analysis are listed in Supplementary Table 1.
Statistical analysis
All data are shown as mean values ± SD for experiments performed with at least three replicates. The differences between the two groups were analyzed using one-way analysis of variance (ANOVA) or two-way ANONA, unless otherwise specified. * represents p < 0. 05; **p < 0. 01; ***p < 0. 001.
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424.
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30.
Gebbia V, Testa A, Borsellino N, Ferrera P, Tirrito M, Palmeri S. Cisplatin and vinorelbine in advanced and/or metastatic adenocarcinoma of the endometrium: a new highly active chemotherapeutic regimen. Ann Oncol. 2001;12:767–72.
Zou W, Wolchok JD, Chen L. PD-L1 (B7-H1) and PD-1 pathway blockade for cancer therapy: Mechanisms, response biomarkers, and combinations. Sci Transl Med. 2016;8:328rv4.
Ott PA, Bang YJ, Berton-Rigaud D, Elez E, Pishvaian MJ, Rugo HS, et al. Safety and Antitumor Activity of Pembrolizumab in Advanced Programmed Death Ligand 1-Positive Endometrial Cancer: Results From the KEYNOTE-028 Study. J Clin Oncol. 2017;35:2535–41.
Davis AA, Patel VG. The role of PD-L1 expression as a predictive biomarker: an analysis of all US Food and Drug Administration (FDA) approvals of immune checkpoint inhibitors. J Immunother Cancer. 2019;7:278.
Zhang J, Dang F, Ren J, Wei W. Biochemical Aspects of PD-L1 Regulation in Cancer Immunotherapy: (Trends in Biochemical Sciences 43, 1014-1032; 2018). Trends Biochem Sci. 2019;44:557.
Sun C, Mezzadra R, Schumacher TN. Regulation and Function of the PD-L1 Checkpoint. Immunity 2018;48:434–52.
Zhuang M, Calabrese MF, Liu J, Waddell MB, Nourse A, Hammel M, et al. Structures of SPOP-substrate complexes: insights into molecular architectures of BTB-Cul3 ubiquitin ligases. Mol Cell. 2009;36:39–50.
Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat JP, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44:685–9.
Le Gallo M, O’Hara AJ, Rudd ML, Urick ME, Hansen NF, O’Neil NJ, et al. Exome sequencing of serous endometrial tumors identifies recurrent somatic mutations in chromatin-remodeling and ubiquitin ligase complex genes. Nat Genet. 2012;44:1310–5.
Li C, Ao J, Fu J, Lee DF, Xu J, Lonard D, et al. Tumor-suppressor role for the SPOP ubiquitin ligase in signal-dependent proteolysis of the oncogenic co-activator SRC-3/AIB1. Oncogene. 2011;30:4350–64.
An J, Wang C, Deng Y, Yu L, Huang H. Destruction of full-length androgen receptor by wild-type SPOP, but not prostate-cancer-associated mutants. Cell Rep. 2014;6:657–69.
Zhang P, Gao K, Jin X, Ma J, Peng J, Wumaier R, et al. Endometrial cancer-associated mutants of SPOP are defective in regulating estrogen receptor-α protein turnover. Cell Death Dis. 2015;6:e1687.
An J, Ren S, Murphy SJ, Dalangood S, Chang C, Pang X, et al. Truncated ERG Oncoproteins from TMPRSS2-ERG Fusions Are Resistant to SPOP-Mediated Proteasome Degradation. Mol Cell. 2015;59:904–16.
Gan W, Dai X, Lunardi A, Li Z, Inuzuka H, Liu P, et al. SPOP Promotes Ubiquitination and Degradation of the ERG Oncoprotein to Suppress Prostate Cancer Progression. Mol Cell. 2015;59:917–30.
Theurillat JP, Udeshi ND, Errington WJ, Svinkina T, Baca SC, Pop M, et al. Prostate cancer. Ubiquitylome analysis identifies dysregulation of effector substrates in SPOP-mutant prostate cancer. Science. 2014;346:85–9.
Dai X, Gan W, Li X, Wang S, Zhang W, Huang L, et al. Prostate cancer-associated SPOP mutations confer resistance to BET inhibitors through stabilization of BRD4. Nat Med. 2017;23:1063–71.
Janouskova H, El Tekle G, Bellini E, Udeshi ND, Rinaldi A, Ulbricht A, et al. Opposing effects of cancer-type-specific SPOP mutants on BET protein degradation and sensitivity to BET inhibitors. Nat Med. 2017;23:1046–54.
Zhang P, Wang D, Zhao Y, Ren S, Gao K, Ye Z, et al. Intrinsic BET inhibitor resistance in SPOP-mutated prostate cancer is mediated by BET protein stabilization and AKT-mTORC1 activation. Nat Med. 2017;23:1055–62.
Zhang J, Bu X, Wang H, Zhu Y, Geng Y, Nihira NT, et al. Cyclin D-CDK4 kinase destabilizes PD-L1 via cullin 3-SPOP to control cancer immune surveillance. Nature. 2018;553:91–5.
Zhu H, Ren S, Bitler BG, Aird KM, Tu Z, Skordalakes E, et al. SPOP E3 Ubiquitin Ligase Adaptor Promotes Cellular Senescence by Degrading the SENP7 deSUMOylase. Cell Rep. 2015;13:1183–93.
Shi Q, Zhu Y, Ma J, Chang K, Ding D, Bai Y, et al. Prostate Cancer-associated SPOP mutations enhance cancer cell survival and docetaxel resistance by upregulating Caprin1-dependent stress granule assembly. Mol Cancer. 2019;18:170.
Geng C, Kaochar S, Li M, Rajapakshe K, Fiskus W, Dong J, et al. SPOP regulates prostate epithelial cell proliferation and promotes ubiquitination and turnover of c-MYC oncoprotein. Oncogene. 2017;36:4767–77.
Ju LG, Zhu Y, Long QY, Li XJ, Lin X, Tang SB, et al. SPOP suppresses prostate cancer through regulation of CYCLIN E1 stability. Cell Death Differ. 2019;26:1156–68.
Wang X, Jin J, Wan F, Zhao L, Chu H, Chen C, et al. AMPK Promotes SPOP-Mediated NANOG Degradation to Regulate Prostate Cancer Cell Stemness. Dev Cell. 2019;48:345–60.e7.
Zhang J, Chen M, Zhu Y, Dai X, Dang F, Ren J, et al. SPOP Promotes Nanog Destruction to Suppress Stem Cell Traits and Prostate Cancer Progression. Dev Cell. 2019;48:329–44.e5.
Jin X, Wang J, Gao K, Zhang P, Yao L, Tang Y, et al. Dysregulation of INF2-mediated mitochondrial fission in SPOP-mutated prostate cancer. PLoS Genet. 2017;13:e1006748.
Jin X, Shi Q, Li Q, Zhou L, Wang J, Jiang L, et al. CRL3-SPOP ubiquitin ligase complex suppresses the growth of diffuse large B-cell lymphoma by negatively regulating the MyD88/NF-κB signaling. Leukemia. 2020;34:1305–14.
Blattner M, Liu D, Robinson BD, Huang D, Poliakov A, Gao D, et al. SPOP Mutation Drives Prostate Tumorigenesis In Vivo through Coordinate Regulation of PI3K/mTOR and AR Signaling. Cancer Cell. 2017;31:436–51.
Garcia-Diaz A, Shin DS, Moreno BH, Saco J, Escuin-Ordinas H, Rodriguez GA, et al. Interferon Receptor Signaling Pathways Regulating PD-L1 and PD-L2 Expression. Cell Rep. 2017;19:1189–201.
Ren Y, Zhang Y, Liu RZ, Fenstermacher DA, Wright KL, Teer JK, et al. JAK1 truncating mutations in gynecologic cancer define new role of cancer-associated protein tyrosine kinase aberrations. Sci Rep. 2013;3:3042.
Frontini M, Vijayakumar M, Garvin A, Clarke N. A ChIP-chip approach reveals a novel role for transcription factor IRF1 in the DNA damage response. Nucleic Acids Res. 2009;37:1073–85.
Kröger A, Köster M, Schroeder K, Hauser H, Mueller PP. Activities of IRF-1. J Interferon Cytokine Res. 2002;22:5–14.
Xiao G, Jin LL, Liu CQ, Wang YC, Meng YM, Zhou ZG, et al. EZH2 negatively regulates PD-L1 expression in hepatocellular carcinoma. J Immunother Cancer. 2019;7:300.
Chen N, Fang W, Zhan J, Hong S, Tang Y, Kang S, et al. Upregulation of PD-L1 by EGFR Activation Mediates the Immune Escape in EGFR-Driven NSCLC: Implication for Optional Immune Targeted Therapy for NSCLC Patients with EGFR Mutation. J Thorac Oncol. 2015;10:910–23.
Dorand RD, Nthale J, Myers JT, Barkauskas DS, Avril S, Chirieleison SM, et al. Cdk5 disruption attenuates tumor PD-L1 expression and promotes antitumor immunity. Science. 2016;353:399–403.
Sato H, Niimi A, Yasuhara T, Permata TBM, Hagiwara Y, Isono M, et al. DNA double-strand break repair pathway regulates PD-L1 expression in cancer cells. Nat Commun. 2017;8:1751.
Shao L, Hou W, Scharping NE, Vendetti FP, Srivastava R, Roy CN, et al. IRF1 Inhibits Antitumor Immunity through the Upregulation of PD-L1 in the Tumor Cell. Cancer Immunol Res. 2019;7:1258–66.
Smithy JW, Moore LM, Pelekanou V, Rehman J, Gaule P, Wong PF, et al. Nuclear IRF-1 expression as a mechanism to assess “Capability” to express PD-L1 and response to PD-1 therapy in metastatic melanoma. J Immunother Cancer. 2017;5:25.
Hogg SJ, Vervoort SJ, Deswal S, Ott CJ, Li J, Cluse LA, et al. BET-Bromodomain Inhibitors Engage the Host Immune System and Regulate Expression of the Immune Checkpoint Ligand PD-L1. Cell Rep. 2017;18:2162–74.
Garvin AJ, Khalaf AHA, Rettino A, Xicluna J, Butler L, Morris JR, et al. GSK3β-SCFFBXW7α mediated phosphorylation and ubiquitination of IRF1 are required for its transcription-dependent turnover. Nucleic Acids Res. 2019;47:4476–94.
Zhu H, Bengsch F, Svoronos N, Rutkowski MR, Bitler BG, Allegrezza MJ, et al. BET Bromodomain Inhibition Promotes Anti-tumor Immunity by Suppressing PD-L1 Expression. Cell Rep. 2016;16:2829–37.
Funding
This work was supported in part by the National Natural Science Foundation of China (No. 81872109, 82272992, 91954106 to KG; No. 91957125, 81972396 to CW; No. 81972438, 82172975 to XW), Shanghai Municipal Medical and Health Discipline Construction Projects (No. 2017ZZ02015 to XW), Natural Science Foundation of Shanghai (22ZR1449200 to KG; No. 22ZR1406600 to CW), the Clinical Science and Technology Innovation Project of Shanghai Shenkang Hospital Development Center (SHDC12020107 to XW), and Open Research Fund of the State Key Laboratory of Genetic Engineering, Fudan University (No. SKLGE-2111 to KG).
Author information
Authors and Affiliations
Contributions
KG, CW, and XW conceived of the project. KG, CW, and XW supervised the experiments, analyzed the data, and wrote the manuscript. QS, YG, WY, YH, ZL, YD, and WC performed molecular biological experiments. QS carried out the animal models and animal analyses. All the authors discussed the results and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics statement
This study involved human subjects and animal experiments and was approved by the Ethics Review Committee of Shanghai First Maternity and Infant Hospital.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by P Salomoni
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Gao, K., Shi, Q., Gu, Y. et al. SPOP mutations promote tumor immune escape in endometrial cancer via the IRF1–PD-L1 axis. Cell Death Differ 30, 475–487 (2023). https://doi.org/10.1038/s41418-022-01097-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-022-01097-7
This article is cited by
-
SPOP point mutations regulate substrate preference and affect its function
Cell Death & Disease (2024)
-
Comprehensive assessment of TECENTRIQ® and OPDIVO®: analyzing immunotherapy indications withdrawn in triple-negative breast cancer and hepatocellular carcinoma
Cancer and Metastasis Reviews (2024)
-
BCLAF1 binds SPOP to stabilize PD-L1 and promotes the development and immune escape of hepatocellular carcinoma
Cellular and Molecular Life Sciences (2024)