Abstract
Although the Hedgehog (Hh) pathway plays an evolutionarily conserved role from Drosophila to mammals, some divergences also exist. Loss of Sufu, an important component of the Hh pathway, does not lead to an obvious developmental defect in Drosophila. However, in mammals, loss of SUFU results in serious disorder, even various cancers. This divergence suggests that SUFU plays additional roles in mammalian cells, besides regulating the Hh pathway. Here, we identify that the transcription factor ZNF281 is a novel binding partner of SUFU. Intriguingly, the Drosophila genome does not encode any homologs of ZNF281. SUFU is able to suppress ZNF281-induced tumor cell migration and DNA damage repair by inhibiting ZNF281 activity. Mechanistically, SUFU binds ZNF281 to mask the nuclear localization signal of ZNF281, culminating in ZNF281 cytoplasmic retention. In addition, SUFU also hampers the interactions between ZNF281 and promoters of target genes. Finally, we show that SUFU is able to inhibit ZNF281-induced tumor cell migration using an in vivo model. Taken together, these results uncover a Hh-independent mechanism of SUFU exerting the anti-tumor role, in which SUFU suppresses tumor cell migration through antagonizing ZNF281. Therefore, this study provides a possible explanation for the functional divergence of SUFU in mammals and Drosophila.
Similar content being viewed by others
Introduction
The Hedgehog (Hh) pathway plays a pivotal role in controlling embryogenesis and adult tissue homeostasis by regulating Ci/GLI family of transcription factors [1,2,3,4]. Suppressor of fused (Sufu/SUFU) is one of the key components of the Hh pathway and acts as a negative regulator both in vertebrate and invertebrate to ensure appropriate signal output [5, 6]. In Drosophila, Sufu forms a complex with Costal2 (Cos2) and Fused (Fu) to block Ci nuclear import [2, 7, 8]. However, the previous study also shows that Sufu enters the nucleus to suppress Ci transcriptional activity by inhibiting Ci-CBP interaction [9]. In addition, Sufu is able to stabilize Ci by inhibiting the interaction between Ci and its E3 ligase Rdx [10]. Although Sufu plays multiple roles in regulating the Hh pathway in Drosophila, loss of sufu or overexpression of sufu does not lead to obvious developmental defects [11, 12]. In mammals, SUFU is a major negative regulator of the Hh pathway. SUFU binds GLI to mask the nuclear localization signal (NLS) on GLI, culminating in GLI cytoplasmic retention [13,14,15]. Unlike Drosophila, mammalian SUFU is indispensable for embryonic development. Mutation of SUFU in mouse leads to embryonic lethality possibly due to defects in the neural tubes and cephalic vesicles [11,12,13]. Loss of SUFU tightly links with several human cancers, including medulloblastoma [16]. The functional divergence between Drosophila Sufu and mammal SUFU indicates that SUFU likely plays additional roles, besides regulating the Hh pathway. To date, the biological function of SUFU, other than the effect upon Hh signaling, remains unclear.
To identify SUFU-interacting proteins, we expressed Flag-tagged SUFU in 293T cells and immunoprecipitated the cell lysates with Flag (Fg) antibody. The immunoprecipitation (IP) was then subjected to mass spectrometry assay. In the candidates, we selected the oncogenic factor ZNF281 for subsequent study because it has no homologs in Drosophila. ZNF281 belongs to the family of Krüppel-type zinc-finger transcription factors. ZNF281 widely expresses in many tissues, such as the kidney, liver, and peripheral blood lymphocytes [17, 18]. It has been shown that deletion of ZNF281 results in mouse embryonic lethality due to its important role in the establishment and maintenance of the pluripotency of embryonic stem cells [19, 20]. Besides, the expression of ZNF281 increases in cells after treatment with DNA-damaging drugs. ZNF281 contributes to DNA damage repair by activating the expression of XRCC2 and XRCC4, two DNA damage response genes [21]. In addition, ZNF281 induced epithelial-mesenchymal transition (EMT) by triggering the expression of SNAIL in colorectal, breast, lung, and oral cancers [19, 22,23,24]. On the other hand, the abundance of ZNF281 is strictly controlled via multiple mechanisms. The transcription factor SNAIL is able to activate ZNF281 expression to form a positive feedback loop [25]. The miR-34 is reported to suppress ZNF281 expression via direct targeting its 3′-UTR [25]. The ZNF281 protein is phosphorylated by glycogen synthase kinase 3β, and subsequent ubiquitinated by β-transducin repeat-containing protein 2 for degradation [26]. However, the mechanism to govern the nuclear localization of ZNF281 is still unknown.
Here, we provided biochemical evidence to support that ZNF281 was a bona fide interacting partner of SUFU. ZNF281 promoted migration and invasion of hepatocellular carcinoma (HCC) cells, which was attenuated by SUFU. In addition, ZNF281-mediated DNA damage repair was also inhibited by SUFU. Mechanistically, we found that ZNF281 shuttled between the cytoplasm and the nucleus. SUFU bound ZNF281 to mask its NLS, resulting in ZNF281 cytoplasmic accumulation. Alternatively, SUFU was able to decrease the affinities between ZNF281 and promoters of target genes, including SNAIL and XRCC2. Overall, we provided a possible explanation for why Sufu/SUFU functions divergently in Drosophila and mammals.
Results
ZNF281 is a novel SUFU-binging protein in mammalian cells
Since the functional divergence of Sufu/SUFU in Drosophila and mammals, we sought to identify novel binding partners of SUFU using mass spectrometry assay and found that ZNF281 was a candidate. Phylogenetic analysis showed that ZNF281 appeared only in vertebrates and lacked homologs in invertebrates (Supplementary Fig. S1). To validate this interaction, we performed coimmunoprecipitation (coIP) assays. The coIP results revealed that Fg-tagged SUFU protein (Fg-SUFU) was able to pull down Myc-tagged ZNF281 (Myc-ZNF281) in 293T cells (Fig. 1A). Reciprocally, Myc-ZNF281 also pulled down Fg-SUFU (Fig. 1B). We further confirmed this interaction in HCC HepG2 cells (Fig. 1C). In addition, endogenous ZNF281 reciprocally interacted with endogenous SUFU in HepG2 and SK-Hep1 cells (Fig. 1D, E), together suggesting ZNF281 is a bona fide interacting partner of SUFU in mammalian cells.
A Immunoblots of immunoprecipitates (IP, top two panels) or whole cell lysates (WCL, bottom two panels) from 293T cells expressing indicated constructs. Of note, Fg-SUFU interacted with Myc-ZNF281 in 293T cells. B Myc-ZNF281 was able to pull down Fg-SUFU in 293T cells. C The coIP result showed that Fg-SUFU immunoprecipitated Myc-ZNF281 in HepG2 cells. D, E There was an interaction of endogenous ZNF281 and SUFU in HepG2 cells and SK-Hep1 cells. F ZNF281 did not affect SUFU-GLI2 interaction. G ZNF281 failed to regulate SUFU-GLI3 interaction. Above all, arrowheads indicate heavy IgG bands.
The well-documented function of SUFU is to bind GLI2/3 transcription factors to impede GLI2/3 entry into the nucleus [15]. We next wanted to examine whether ZNF281 competes with GLI2/3 to bind SUFU. The coIP results showed that ZNF281 did not affect the interaction between SUFU and GLI2 (Fig. 1F). On the other hand, ZNF281 also failed to influence SUFU-GLI3 association (Fig. 1G). Taken together, the novel SUFU-binding protein ZNF281 does not affect SUFU-GLI2/3 interactions.
SUFU suppresses ZNF281-induced tumor cell invasion and migration
Increasing studies have demonstrated that ZNF281 contributes to EMT, an important process for tumor migration [25, 27]. Since our above results reveal an interaction between SUFU and ZNF281 in HCC cells, it is necessary to test the functional relevance. We first checked the expression levels of SUFU and ZNF281 in seven HCC cell lines and found that ZNF281 was highly expressed in SMMC-7721 and BEL-7402 cells, while low in HepG2 and SK-Hep1 cells (Fig. 2A). Consistent with the previous reports [25], ZNF281 was able to improve the invasive ability of SK-Hep1 cells, as measured by the transwell assay (Fig. 2B). The enhancement invasion of HepG2 by ZNF281 was substantially restored by SUFU (Fig. 2B). Although several studies have shown that SUFU inhibits EMT, such as gastric cancer [28,29,30] and cervical squamous cell carcinoma [31], our results revealed that overexpression of SUFU alone exerted a marginal effect on the invasive ability of SK-Hep1 cells (Fig. 2B). Furthermore, we confirmed the inhibitory effect of SUFU on ZNF281-induced cell invasion in HepG2 cells (Fig. 2D, E). It is well known that SUFU promotes GLI2/3 degradation through recruiting the E3 ligase β-TrCP [32], we tested whether SUFU modulates the stability of ZNF281. In SK-Hep1 cells, overexpression of SUFU did not affect exogenous Myc-ZNF281 protein level (Fig. 2C). In SK-Hep1 cells, dose-dependent expression of SUFU failed to regulate Myc-ZNF281 protein (Supplementary Fig. S2A). SUFU also could not modulate the endogenous ZNF281 protein level in SMMC-7721 cells (Supplementary Fig. S2B). In addition, we checked the stability of ZNF281 protein via chase experiments and found that SUFU was unable to regulate ZNF281 stability (Supplementary Fig. S2C, D). In sum, these results show that SUFU suppresses ZNF281-induced HCC cell invasion not due to controlling ZNF281 protein abundance.
A The protein levels of ZNF281 and SUFU in seven types of liver cancer cell lines. Actin is used as a loading control. B The transwell analysis showed that SUFU inhibited ZNF281-induced SK-Hep1 cell invasion. Quantification analysis was shown on the right. Of note, overexpression of SUFU alone cannot affect cell invasion. C The expression of Myc-ZNF281 and Fg-SUFU in SK-Hep1 cells was determined by western blot. Overexpression of ZNF281 elevated SNAIL and VIM protein levels, which were restored by SUFU co-expression. Actin acts as a loading control. D SUFU hampered ZNF281-triggered cell invasion in HepG2 cells. Quantification analysis was shown on the right. E The expression of Myc-ZNF281 and Fg-SUFU in HepG2 cells was examined by western blot. Overexpression of ZNF281 elevated SNAIL and VIM protein levels, which were restored by SUFU co-expression. Actin acts as a loading control. F, G Wound-healing assays revealed that ZNF281-induced cell migrations were attenuated by co-expression of SUFU in SK-Hep1 and HepG2 cells. Quantification analyses were shown on the right. H Western blot analysis to test the expression of SUFU and ZNF281 in SMMC-7721 cells stably expressing indicated constructs. Actin acts as a loading control. I Representative image showed bioluminescence signals of the resected lungs 4 weeks after the intravenous injection of SMMC-7721 cells stably expressing ZNF281 alone or ZNF281 plus SUFU. Quantification of bioluminescence signals was shown on the right (n = 5).
On the other hand, we examined cell migration ability, another tumor characteristic, using a wound-healing assay. In SK-Hep1 cells, ZNF281 promoted wound healing, which was attenuated by SUFU co-expression (Fig. 2F). We got a similar result in HepG2 cells (Fig. 2G). In addition, we tested the expression of two well-known cell migration markers, SNAIL and Vimentin (VIM), to reflect the tumorigenicity. ZNF281 elevated SNAIL and VIM protein levels, which were decreased by SUFU co-expression (Fig. 2C, E). In a mouse pulmonary metastatic model, 1 × 106 SMMC-7721 cells stably expressing ZNF281 alone or ZNF281 plus SUFU (Fig. 2H) were injected into the caudal veins of five BALB/C nude male mice. The lungs of the mice were analyzed for metastatic nodules after four weeks. The SMMC-7721 cells with ZNF281 overexpression showed increased lung metastatic tumors compared with control cells, which were rescued by SUFU co-expression (Fig. 2I).
To validate the role of SUFU inhibiting ZNF281-induced tumor cell invasion and migration, we chose SMMC-7721 and BEL-7402 cells for loss-of-function assay, due to their high levels of ZNF281 and SUFU (Fig. 2A). First, we synthesized two ZNF281 siRNAs and three SUFU siRNAs to silence endogenous genes. WB results showed that siRNAs could effectively silence ZNF281 and SUFU expression in SMMC-7721 cells (Supplementary Fig. S3A). Compared to control cells, knockdown of ZNF281 substantially suppressed cell invasion, while SUFU siRNAs showed a subtle effect (Supplementary Fig. S3B). ZNF281 siRNA-induced cell invasion inhibition was restored by exogenous ZNF281 expression (Supplementary Fig. S3C), removing the off-target effect. In SMMC-7721 cells, knockdown of ZNF281 apparently hampered cell invasion, which was restored by SUFU siRNA co-transfection (Fig. 3A). This result was repeated in BEL-7402 cells (Fig. 3C). The wound-healing assays showed that knockdown of SUFU was able to rescue ZNF281 siRNA-induced cell migration inhibition in SMMC-7721 cells (Fig. 3E) and BEL-7402 cells (Fig. 3F). In addition, ZNF281 siRNA decreased SNAIL and VIM levels, which was rescued by SUFU siRNA (Fig. 3B). The previous study has shown that ZNF281 exclusively promotes tumor cell migration, without affecting cell proliferation [25]. Consistent with this result, neither overexpression nor knockdown of ZNF281 failed to regulate cell proliferation in HepG2 cells (Supplementary Fig. S4A) and BEL-7402 cells (Supplementary Fig. S4B). The BrdU incorporation assays also revealed that ZNF281 overexpression or knockdown could not affect the proliferation of SMMC-7721 cells (Supplementary Fig. S3C, D). Furthermore, the flow cytometry analyses confirmed that both ZNF281 and SUFU were unable to modulate SMMC-7721 cell proliferation (Supplementary Fig. S5A, B), removing the possibility that SUFU hampers ZNF281-mediated cell migration through cell proliferation arrest. Taken together, these results support that SUFU is capable of suppressing ZNF281-induced cell invasion and migration.
A Knockdown of ZNF281 inhibited SMMC-7721 cell invasion, which was rescued by SUFU siRNA. Quantification analysis was shown on the right. Notably, knockdown of SUFU alone is failed to affect cell invasion. B Immunoblots of whole cell lysates from SMMC-7721 cells transfected with indicated siRNAs. Knockdown of ZNF281 decreased SNAIL and VIM proteins, which were rescued by SUFU siRNA. Actin acts as a loading control. C SUFU siRNA was able to suppress ZNF281-siRNA-induced cell invasion inhibition in BEL-7402 cells. Quantification analysis was shown on the right. D Immunoblots of whole cell lysates from BEL-7402 cells transfected with indicated siRNAs. Actin acts as a loading control. E, F Wound-healing assays revealed that ZNF281-siRNA-induced cell migration inhibitions were attenuated by SUFU siRNA in SMMC-7721 and BEL-7402 cells. Quantification analyses were shown on the right.
SUFU inhibits ZNF281-mediated DNA damage repair
During cell division, DNA damage is difficult to avoid due to physiological and environmental stimuli [33]. Eukaryotic cells have evolved a rigorous mechanism to respond DNA lesions [34]. However, if DNA lesions are too severe, apoptosis is triggered to clear damaged cells [35]. Defect in DNA damage repair is a key cause of cancers. The previous study has demonstrated that ZNF281 is involved in DNA damage repair through activating XRCC2 and XRCC4, two genes that respectively regulate homologous recombination and non-homologous end joining [21]. We next sought to examine whether SUFU affects ZNF281-mediated DNA damage repair through three methods (Supplementary Fig. S6A). After treatment with DNA-damaging drug etoposide (Eto), remarkable DNA damage was induced in SMMC-7721 and HepG2 cells, judged by neutral comet assay (Fig. 4A, C). Overexpression of ZNF281 substantially inhibited DNA damage, which was rescued by SUFU co-expression (Fig. 4A, C). The results also showed that overexpression of SUFU alone did not modulate DNA damage repair (Fig. 4A). Next, we examined the level of γH2AX, a biomarker of DNA damage, and found that ZNF281 decreased γH2AX, which was restored by SUFU co-expression (Fig. 4B). Conversely, knockdown of ZNF281 was able to upregulate DNA lesions under Eto treatment, which was relieved by SUFU siRNA in both SMMC-7721 cells and HepG2 cells (Fig. 4D, F). Consistently, the upregulation of γH2AX caused by ZNF281 knockdown was attenuated by SUFU siRNA (Fig. 4E). Furthermore, we also examined the γH2AX using immunostaining. Without Eto treatment, SMMC-7721 cells and HepG2 cells did not express γH2AX, suggesting that Eto indeed induces DNA damage (Supplementary Fig. S6B, D). In line with the western blot results, SUFU was able to suppress ZNF281-mediated DNA damage repair (Supplementary Fig. S6C, E).
A The comet assay showed that ZNF281 triggered DNA damage repair in SMMC-7721 cells, which was inhibited by co-expression of SUFU. Cells were treated with 50 μM etoposide for 1 h, and after 3 h recovery, the comet images were captured using the fluorescence microscopy. Quantification analyses were shown on the right. Overexpression of SUFU alone did not affect DNA damage repair. B Immunoblots of whole cell lysates from SMMC-7721 cells expressing indicated constructs. Of note, ZNF281 decreased γH2AX, which was restored by SUFU co-expression. Actin acts as a loading control. C The comet assay showed that ZNF281 triggered DNA damage repair in HepG2 cells, which was inhibited by co-expression of SUFU. D, F The comet assay showed that knockdown of ZNF281 impaired DNA damage repair in SMMC-7721 (D) and HepG2 (F) cells, which was rescued by SUFU siRNA. Quantification analyses were shown on the right. E Knockdown of ZNF281 elevated γH2AX, which could be decreased by SUFU siRNA. Actin acts as a loading control.
SUFU impedes ZNF281 nuclear accumulation
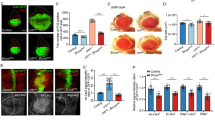
It is known that Sufu/SUFU negatively regulates the Hh pathway through tethering Ci/GLI proteins in the cytoplasm [15]. Our above studies have revealed that SUFU is able to bind the transcription factor ZNF281 and inhibits its biological functions. Thus, it is necessary to test whether SUFU controls ZNF281 localizations. Immunostaining showed that ZNF281 is predominantly located in the nucleus, with a small amount in the cytoplasm (Fig. 5A). Upon treatment with Leptomycin B (LMB), which is a specific nuclear export inhibitor, we found that ZNF281 is exclusively located in the nucleus (Fig. 5B), suggesting that ZNF281 is a shuttle protein. When ZNF281 and SUFU were co-transfected in cells, ZNF281 accumulated in the cytoplasm (Fig. 5C). After LMB treatment, ZNF281 entered the nucleus in the presence of SUFU (Fig. 5D). In addition, we separated the cytoplasmic and nuclear protein to test whether SUFU suppresses ZNF281 nuclear accumulation. In 293T cells, co-expression of SUFU indeed decreased nuclear ZNF281 (Fig. 5E). SUFU also diminished the nuclear ZNF281 in HepG2 cells (Fig. 5F). In contrast, knockdown of SUFU elevated nuclear ZNF281 in HepG2 cells (Fig. 5G). Together, these observations suggest that SUFU possibly reduces the ability of ZNF281 to enter the nucleus.
A–D HEK-293T cells transfected with indicated constructs were carried out with immunofluorescence analyses with or without LMB treatment. Of note, Myc-ZNF281 protein mainly locates in the nucleus (A) in the absence of SUFU, while locates in the cytoplasm in the presence of SUFU (C). Scale bars: 10 μm for all images. E Immunoblots of nuclear protein and cytoplasmic protein of 293T cells transfected with indicated constructs. Lamin B acts as a loading control for nuclear protein, whereas β-Tubulin (β-Tub) is a loading control for cytoplasmic protein. Notably, SUFU decreases ZNF281 nuclear localization. F SUFU decreased nuclear ZNF281 protein level in HepG2 cells. G Knockdown of SUFU promoted ZNF281 nuclear accumulation in HepG2 cells.
SUFU masks the N-terminal NLS in ZNF281
To investigate the mechanism of SUFU inhibiting ZNF281 nuclear accumulation, we analyzed ZNF281 protein sequence and found three potential NLSs, named S1, S2, and S3 (Fig. 6A). We divided ZNF281 into two parts, ZNF281-N and ZNF281-C (Fig. 6A). As shown in Fig. 6A, ZNF281-N contained S1 and S2, whereas ZNF281-C harbored S3. The coIP results showed that SUFU could bind both ZNF281-N and ZNF281-C (Fig. 6B). Although ZNF281-N and ZNF281-C are mainly located in the nucleus, SUFU selectively suppressed ZNF281-N nuclear accumulation (Fig. 6C–F). To further map SUFU-binding site in ZNF281-N, we divided ZNF281-N into two parts, ZNF281-N1 and ZNF281-N2. As shown in Fig. 6A, S1 and S2 are located in ZNF281-N2. The coIP experiment revealed that SUFU interacted with ZNF281-N2, not ZNF281-N1 (Fig. 6G). Possibly due to lack of NLS, ZNF281-N1 is exclusively located in the cytoplasm, irrespective of with or without SUFU (Fig. 6H, I). In contrast, ZNF281-N2 is mainly located in the nucleus (Fig. 6J). Co-expression of SUFU resulted in ZNF281-N2 cytoplasmic retention (Fig. 6K), suggesting that SUFU likely binds ZNF281-N2 to mask the NLS.
A Schematic diagram showing the full-length and truncated ZNF281 used in subsequent assays. The predicted NLS sequences were highlighted in red and listed on the right. B Immunoblots of immunoprecipitates (IP, top two panels) or whole cell lysates (WCL, bottom two panels) from 293T cells expressing indicated constructs. Of note, both ZNF281-N and ZNF281-C bind SUFU. The arrowhead indicates the heavy IgG band. C–F HEK-293T cells transfected with indicated constructs were carried out immunofluorescence analyses. SUFU is able to sequester ZNF281-N in the cytoplasm, without affecting ZNF281-C localization. The DAPI staining marks the nucleus. G The coIP analysis showing the interaction between Myc-ZNF281-N1 or Myc-ZNF281-N2 and Fg-SUFU in 293T cells. Of note, ZNF281-N2, not ZNF281-N1 pulls down SUFU. The arrowhead indicates the heavy IgG band. H–K Immunofluorescence analyses of 293T cells transfected with indicated constructs. SUFU is able to tether ZNF281-N2 in the cytoplasm, without affecting ZNF281-N1 localization. The DAPI staining marks the nucleus. Scale bars: 10 μm for all immunostaining images.
SUFU inhibits the transcriptional activity of ZNF281
The transcription factor ZNF281 exerts its biological functions by activating the expression of various target genes. Therefore, the binding between ZNF281 and promoter regions is important for its activity. We next sought to test whether SUFU affects ZNF281’s binding with promoters. We selected three well-documented ZNF281 target genes for subsequent studies. ChIP-PCR assay showed that ZNF281 was able to pull down the promoters of SNAIL, XRCC2, and AXIN2 in SMMC-7721 cells (Fig. 7A). The affinities between ZNF281 and SNAIL promoter or XRCC2 promoter were robustly decreased by co-expression of SUFU (Fig. 7A). However, the interaction between ZNF281 and AXIN2 promoter was not influenced by SUFU (Fig. 7A), indicating that the inhibition of ZNF281-promoter interaction by SUFU cannot simply be attributed to cytoplasmic retention of ZNF281. In addition, we carried out ChIP-PCR assays using HepG2 cells and found that SUFU reduced the interaction between ZNF281 and all three promoters (Fig. 7B), suggesting that the regulation of ZNF281-promoter affinity by SUFU is context dependent.
A ChIP-PCR assays showed the interaction between ZNF281 and promoters. SMMC-7721 cells transfected with indicated constructs were carried out ChIP-PCR to test the interaction of ZNF281 and promoters. The expressions of constructs were confirmed via western blot assays below. B The affinities of ZNF281 and promoters were decreased by co-expression of SUFU. The expressions of constructs were shown below. C ZNF281 increased SNAIL-Luc activity, which was decreased by co-expression of SUFU in SMMC-7721 cells. The expressions of constructs were shown on the right. D SUFU attenuated ZNF281-induced SNAIL-Luc enhancement. The expressions of constructs were shown on the right. E The relative mRNA levels of SNAIL and XRCC2 were detected by RT-PCR.
To further validate the inhibition of ZNF281-promoter interaction by SUFU, we performed luciferase analyses, in which ZNF281 was used to drive luciferase expression from a DNA fragment containing SNAIL promoter (refers to SNAIL-Luc hereafter). In SMMC-7721 cells, ZNF281 enhanced SNAIL-Luc activity, which was attenuated by SUFU co-expression (Fig. 7C). In addition, we confirmed this result in HepG2 cells (Fig. 7D). Finally, RT-PCR results showed that overexpression of ZNF281 activated the expression of endogenous SNAIL and XRCC2, which was restored by SUFU co-expression (Fig. 7E). Taken together, SUFU suppresses the transcription activity of ZNF281, at least in part through decreasing ZNF281’s affinity to promoters.
SUFU inhibits ZNF281-induced tumor cell migration in vivo
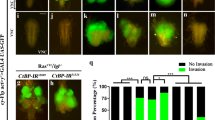
Our above studies have demonstrated that SUFU inhibits ZNF281-induced HCC cell migration. We next wanted to investigate its anti-tumor role using an in vivo model. In Drosophila wing discs, knockdown of the neoplastic tumor suppressor gene scribble (scrib) along the anterior/posterior (A/P) boundary using the ptc-gal4 driver induces an invasive cell migration phenotype, which has been widely used as an ideal in vivo model to study tumor cell migration [36]. Compared with the control disc (Fig. 8A), knockdown of scrib robustly triggered cell migration (Fig. 8B), judged by numbers of dissociative GFP-positive cells. Consistent with the result in HCC cell lines, overexpression of ZNF281 indeed promoted scrib-RNAi-induced cell migration (Fig. 8C, F). However, the enhancement of tumor cell migration by ZNF281 was inhibited by SUFU co-expression in wing discs (Fig. 8E, F). Overexpression of SUFU alone was unable to affect scrib-RNAi-induced cell migration (Fig. 8D, F). Furthermore, we examined the inhibitory effect of SUFU upon ZNF281 under physiological background. Compared to the control disc (Supplementary Fig. S7A), overexpression of ZNF281 slightly triggered cell migration (Supplementary Fig. S7C), which was compromised by SUFU co-expression (Supplementary Fig. S7D). In sum, SUFU suppresses ZNF281-induced tumor cell migration in vivo.
A A control wing disc was stained to show GFP (green) and DAPI (blue). The ptc-gal4 drives gene expression along the A/P boundary. B Knockdown of scrib triggered cell migration. C Overexpression of ZNF281 promoted scrib-RNAi-induced cell migration. D Overexpression of SUFU alone did not affect scrib-RNAi-induced cell migration. E The enhancement cell migration by ZNF281 was attenuated via expressing SUFU. F Quantification analysis of migrating cell numbers in A–E (N = 15). G Proposed schematic model to summarize the divergence of Sufu/SUFU in Drosophila and mammals. In Drosophila, Sufu exerts its function by inhibiting Ci nuclear accumulation. In addition to regulating the Hh pathway, the mammalian SUFU suppresses the function of ZNF281 via two mechanisms. Scale bars: 50 μm for all images.
Discussion
The hh gene is first identified in Drosophila through EMS-induced screening [37]. Mutation of hh gene leads to disorder of Drosophila segments, resembling an Hh [37]. Subsequent studies have revealed that Hh protein acts as a morphogen, which is able to turn on a pathway, named the Hh pathway [38]. Although the framework of the Hh pathway is evolutionarily conserved from Drosophila to humans, fundamental differences also exist. In Drosophila, the negative regulator Sufu exerts a weak influence on Ci activity. Embryos with sufu deletion can develop into viable and fertile adults, without any obvious defects [11]. In contrast, ablation of mouse SUFU leads to embryonic lethality at about E9.5 with cephalic and neural tube defects [12], indicating its indispensable role for development. In adults, loss of SUFU has been implicated in various human cancers, such as breast cancer, medulloblastoma, lung cancer, and prostate cancer [39,40,41,42]. The functional divergence of Sufu/SUFU suggests that mammalian SUFU possibly plays other roles, in addition to regulating the Hh pathway. Here, we find that the transcription factor ZNF281 is a novel binding partner of SUFU in mammalian cells. Interestingly, the Drosophila genome does not encode any orthologs of ZNF281. ZNF281 is a multifunctional protein, which promotes EMT and DNA damage repair through activating a series of target genes. Our results show that the functions of ZNF281 are substantially suppressed by SUFU both in vitro and in vivo, at least through two mechanisms. First, SUFU binds ZNF281 to hamper ZNF281 nuclear accumulation. Alternatively, SUFU decreases the affinities of ZNF281 with promoters (Fig. 8G). Therefore, this study provides a possible explanation that SUFU exerts anti-tumor roles through antagonizing ZNF281.
Although a large number of studies have established that SUFU is closely related to tumorigenesis, it is difficult to develop drugs targeting SUFU due to two possible reasons. First, SUFU is a tumor suppressor, which is not suitable for tumor therapy. Second, in the past, it was thought that SUFU exerts the anti-tumor role mainly by inhibiting the Hh pathway, which is also indispensable for adult homeostasis. Drug inhibition of GLI proteins, the downstream factors of SUFU, will bring serious side effects. In this study, we demonstrate that SUFU suppresses HCC cell migration and invasion by antagonizing ZNF281, providing ZNF281 as an alternative target for SUFU-related HCC patients. Our results and other studies [25] together show that ZNF281 fails to regulate cell proliferation, removing the possibility that ZNF281 maintains adult homeostasis. Thus, ZNF281 is possible a considerable drug targets. Here, we choose HCC for study since the Hh pathway overactivation is not the key driver for HCC tumorigenesis, suggesting that SUFU harbors Hh-independent anti-tumor functions. In further, it will be fruitful to examine whether the SUFU-ZNF281 regulatory module is also applied to other types of cancers.
In the process of cell division, DNA lesion is inevitable due to endogenous and environmental stimuli [43]. A moderate amount of DNA lesion does not bring fateful consequences because the organism has evolved an effective repair system. Thus, DNA damage repair is important to keep genome stability. Its deregulation leads to various human diseases, including cancers [44]. ZNF281 contributes to DNA damage repair through activating XRCC2 and XRCC4 expression [21]. Although our results show that SUFU is able to suppress ZNF281-induced DNA repair in HCC cells, its role under physiological status is still unclear. The previous study has revealed that the Hh pathway promotes proteasome-mediated SUFU protein degradation in prostate and breast cancer cell lines, forming a positive feedback loop to aggravate tumorigenesis [45]. It is interesting to test whether the Hh pathway is involved in regulating DNA damage repair through controlling SUFU abundance.
Although our results show that SUFU suppresses ZNF281-mediated cell invasion and DNA damage repair, overexpression or knockdown of SUFU alone does not lead to noticeable phenotypes, suggesting that SUFU exerts anti-tumor roles in a context-dependent manner. On the other hand, the inhibition of SUFU on ZNF281 cannot explain the whole ZNF281’s effects on tumor invasion. Therefore, the oncogenic activity of ZNF281 can be controlled by other proteins other than SUFU.
ZNF281 consists of three NLSs, which contribute differently to ZNF281 nuclear accumulation. Although SUFU binds both N-terminus and C-terminus of ZNF281, SUFU only inhibits ZNF281-N nuclear accumulation. A possible explanation is that the C-terminal NLS locates at the end and cannot be masked by SUFU. Besides, SUFU suppresses ZNF281 transcription activity via decreasing ZNF281’s binding with promoters. This regulation is possible context-dependent since SUFU inhibits the interaction between ZNF281 and AXIN2 promoter in SMMC-7721 cells, not HepG2 cells. Therefore, the selection of SUFU or ZNF281 as drug targets for tumor treatment requires careful consideration of tumor backgrounds.
Materials and methods
Cell lines and transfection
HEK-293T, PLC/PRF/5, HepG2, SMMC-7721, BEL-7402, Hep3B, Huh-7, and SK-Hep1 cell lines were purchased from the ATCC. HEK-293T, PLC/PRF/5, HepG2, Hep3B, Huh-7, and SK-Hep1 cells were cultured in Dulbecco’s Modified Eagle’s medium, while SMMC-7721 and BEL-7402 cells were cultured in RPMI-1640 medium supplemented with 10% FBS at 37 °C and 5% CO2. These all cell lines have been examined to exclude the mycoplasma contamination.
For transfection, DNA RNA Advanced Transfection Reagent (Zeta Life) was used according to the manufacturer’s instructions. For siRNA-mediated gene silence, cells were transfected via lipo2000 (Invitrogen) according to the manufacturer’s instructions. The siRNA sequences were shown as follows: ZNF281-siRNA-1, 5′-CCA GAA UCU CAG GGA AUC AdTdT-3′; ZNF281-siRNA-2, 5′-GGU CAU CAA ACC AUA CCA AdTdT-3′; SUFU siRNA-1, 5′-GCA GCU UGA GAG CGU ACA UdTdT-3′; SUFU siRNA-2, 5′-CGG CCU GAG UGA UCU CUA UdTdT-3′; SUFU siRNA-3, 5′-GAU CCA CAC CUG CAA GAG AdTdT-3′; MOCK-siRNA, 5′-UUC UCC GAA CGU GUC ACG UUU dTdT-3′.
Constructs and transgenic flies
The ZNF281, SUFU, GLI2, and GLI3 were amplified via PCR using HepG2 cDNA as template, and then were inserted into the pcDNA3.1 vectors. Truncated ZNF281 plasmids were generated as previously described [46]. For pGL3-SNAIL-Luc reporter, the human SNAIL promoter (−663/−163) containing the ZNF281 binding site was amplified from HepG2 cDNA and cloned into pGL3-Basic vector (Promega). The Drosophila expression plasmids, including pUAST-Myc-ZNF281 and pUAST-Fg-SUFU, were constructed by inserting the sequences into pUAST vector. Fg-SUFU and Myc-ZNF281 transgenic flies were generated by injecting the constructs into w1118 eggs according to our previous described [46]. The information on fly stocks used in this study was described in our previous study [47].
Western blotting and immunofluorescence
After transfection, cells were lysed for immunoblotting (IB) or IP analysis according to standard protocols [48]. The antibodies used were as following: rabbit anti-ZNF281 (1:3000 for IB, 1:300 for IP; Abcam), rabbit anti-SUFU (1:1000 for IB, 1:100 for IP; ABclonal), rabbit anti-SNAIL (1:1000 for IB; ABclonal), rabbit anti-Vimentin (1:2000 for IB; ProteinTech), rabbit anti-N-cadherin (1:1000 for IB; ABclonal), rabbit anti-Lamin B (1:2000 for IB; ProteinTech), rabbit anti-β-Tubulin (1:1000 for IB; ABclonal), mouse anti-Actin (1:5000 for IB; Genscript), mouse anti-Myc (1:2000 for IB, 1:200 for IP; Santa Cruz), mouse anti-Fg (1:5000 for IB, 1:500 for IP; Sigma), mouse anti-HA (1:2000 for IB, 1:200 for IP; Santa Cruz) and rabbit anti-γH2AX (1:1000 for IB; ABclonal).
For cell-based immunofluorescence, HEK-293T cells were washed with PBS for 10 min and then fixed with 4% fresh-made formaldehyde for 20 min at room temperature. After three washes in PBS, cells were permeabilized in 0.1% Triton X-100 for 10 min. Then cells were incubated with rabbit anti-Myc antibody (1:100, ABclonal), mouse anti-Flag (1:500, Sigma) antibody, or rabbit anti-γH2AX (1:100, ABclonal) antibody at 4 °C overnight. After three washes in PBS, cells were incubated with secondary antibody (1:500, Jackson ImmunoResearch) for 2 h and DAPI (1:1000, Sigma) for 15 min at room temperature. After three washes in PBS, cells were mounted with 40% glycerol, and images of cells were captured using the Zeiss confocal microscope. For BrdU incorporation assays, SMMC-7721 cells with indicated transfection were incubated with 30 μM BrdU (Sigma) for 1 h before cell harvesting, and the subsequent immunofluorescence was carried out as 293T cells.
RNA extraction and qRT-PCR
Total RNAs were extracted from cultured cells with TRIzol (Invitrogen) following standard protocols and reverse transcribed using HiScript® Q RT SuperMix with gDNA wiper (Vazyme) according to the manufacturer’s instructions. Quantitative real-time PCR was carried out on Bio-Rad CFX96TM using the ChamQ SYBR® Color qPCR Master Mix (Vazyme). Relative expression of SNAIL and XRCC2 were detected using the 2-∆∆Ct method [49]. The primer pairs used were as follows: SNAIL, 5′-GCA CAT CCG AAG CCA CAC-3′ (forward) and 5′-GGA GAA GGT CCG AGC ACA-3′ (reverse); XRCC2, 5′-CAG AAA GCC TCG AGC TCA TCA-3′ (forward) and 5′-GCT GCC ATG CCT TAC AGA GAT-3′ (reverse); ACTIN, 5′-TGA CAT TAA GGA GAA GCT GTG CTA C-3′ (forward) and 5′-GAG TTG AAG GTA GTT TCG TGG ATG-3′ (reverse). Data are presented as means ± SD of values from at least three experiments.
Wound-healing assay and transwell assay
For the wound-healing assay, HCC cells were seeded into 6-well plates and grown to confluence for 48 h after transfection. Two perpendicular straight-line scratches were generated by sterile pipette tip scratching on the bottom of the plate. The fraction of cell coverage was measured at 24 or 48 h.
For transwell assays, 1 × 105 cells were mixed with 0.3 mL of serum-free medium and seeded into upper transwell chambers (BD Biosciences) at 48 h after transfection. 0.5 mL of medium with 10% FBS was added to the lower chambers of the 24-well plate (Corning). After 48 h, the migrating cells adhered to the bottom of the chambers were fixed with 20% methanol for 20 min and stained with 0.1% crystal violet (Sangon Biotech). Thoroughly clean the inside of the inserts with a damp cotton swab. Numbers of migrating cells per well were counted under a microscope in eight fields, and data are presented as means ± SD of values from these fields.
Mouse pulmonary metastasis assay
For mouse pulmonary metastasis experiments, we first constructed SMMC-7721 cells stably expressing ZNF281 or ZNF281 plus SUFU using lentiviral infection. Briefly, cDNAs of ZNF281 and SUFU were inserted into lentiviral vectors pLVX-IRES-ZsGFP (a gift from Dr Yong Li, Nanchang University, China) and pLVX-IRES-puro, respectively. The above plasmids were co-transfected with the packaging plasmids psPAX2 and pMD2G into HEK-293T cells using Lipofectamine 3000 (Thermo Fisher Scientific). Infectious lentiviruses were harvested 48 h after transfection. These lentiviruses were designated Lenti-GFP-ZNF281 and Lenti-SUFU-puro. We used the empty vectors as a negative control and named the harvested lentiviruses Lenti-GFP. SMMC-7721 cells were infected with Lenti-GFP and Lenti-GFP-ZNF281, respectively, in the presence of 10 µg/mL polybrene (Hanbio Biotechnology). We selected several single clones expressing green fluorescence for expansion and used western blotting to identify the constructed 7721-Control and 7721-ZNF281 cell lines. 7721-ZNF281/SUFU cells were 7721-ZNF281 cells infected with Lenti-SUFU-puro. After being selected with 2 μg/mL puromycin for 2 weeks, the infected cells were identified using western blot. The pulmonary metastasis study was performed using BALB/C nude male mice (6 weeks old), which were obtained from Nanjing University and were randomly divided into indicated groups. The mice in the groups were injected with the indicated SMMC-7721 cells (1 × 106) expressing GFP through a caudal vein. After 4 weeks, all animals were sacrificed and the lung tissues were isolated for IVIS Lumina Series III imaging analysis. The protocols for animal experiments were approved by the Institutional Animal Care Committee of China Pharmaceutical University.
Flow cytometry analysis
SMMC-7721 cells with indicated transfection were incubated with 30 μM BrdU (Sigma) for 1 h before cell harvesting. Proliferating cells were stained using the FITC-BrdU cell proliferation Detection Kit (KeyGEN; BioTECH) according to the manufacturer’s instructions. After suspending in 500 μL, PBS cells were subjected to flow cytometry analysis.
Comet assay
Comet assay was performed as previously described [50]. In brief, HCC cells were transfected with indicated plasmids or siRNAs for 48 h, then treated with etoposide (MedChemExpress) for an additional 1 h and collected for DNA damage analysis. Data are presented as means ± SD of values from at least three experiments.
ChIP-PCR assay
SMMC-7721 and HepG2 cells were transfected with indicated plasmids for 48 h. Then, after washing with PBS, these cells were fixed in a culture dish with 1% fresh-made formaldehyde for 10 min at room temperature. After washing with 0.125 M glycine and PBS, cells were extracted for 20 min on ice with lysis buffer containing protease inhibitor cocktail (#GK10014, Glpbio) and PMSF (#93482, Sigma). Then, the chromatin was sheared by sonication to obtain fragments of an average size of 200–500 bp. The chromatin fragments were incubated with mouse IgG (ABclonal) or mouse anti-Myc antibody for 3 h. Complexes were incubated with Protein-A/G agarose beads (Santa Cruz) for 3 h. After multiple washes, the DNA was reverse cross-linked at 65 °C for 6 h. The following primers were used for ChIP-PCR analysis: SNAIL promoter, 5′-GGA GTA CTT AAG GGA GTT GGC GG-3′ (forward), and 5′-GAA CCA CTC GCT AGG CCG T-3′ (reverse); XRCC2 promoter, 5′-CGA ACC AAA GTT TAC AAC AGA CAA G-3′ (forward) and 5′-CAT GCG CAA TAG GGT GTG C-3′ (reverse); AXIN2 promoter, 5′-CCA ACT CAC TCA GGG GAG AC-3′ (forward) and 5′-GAT TCT TGG CAC AGG CAG TAG -3′ (reverse).
Luciferase reporter assay
Cells were co-transfected with indicated plasmids and pGL3-SNAIL-Luc reporter for 48 h. The luciferase activity was assayed by the Dual-Luciferase Reporter assay (Vazyme). All luciferase activity data are presented as means ± SD of values from at least five experiments.
Statistical analysis
All statistical analyses were performed using GraphPad Prism software. Two-tailed unpaired Student’s t-test was performed to evaluate the differences between the two groups. The results are expressed as the mean ± SD from at least three to five independent experiments, and differences with P < 0.05 was considered statistically significant (*P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001).
Data availability
All relevant data are available from the corresponding author upon reasonable request.
References
Jiang J, Hui CC. Hedgehog signaling in development and cancer. Dev Cell. 2008;15:801–12.
Varjosalo M, Li SP, Taipale J. Divergence of hedgehog signal transduction mechanism between Drosophila and mammals. Dev Cell. 2006;10:177–86.
Ingham PW, Nakano Y, Seger C. Mechanisms and functions of Hedgehog signalling across the metazoa. Nat Rev Genet. 2011;12:393–406.
Briscoe J, Therond PP. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat Rev Mol Cell Biol. 2013;14:416–29.
Kogerman P, Grimm T, Kogerman L, Krause D, Undén AB, Sandstedt§ B, et al. Mammalian suppressor-of-fused modulates nuclear-cytoplasmic shuttling of Gli-1. Nat Cell Biol. 1999;1:312–9.
Pearse VR,II, Collier LS, Scott MP, Tabin CJ. Vertebrate homologs of Drosophila suppressor of fused interact with the gli family of transcriptional regulators. Dev Biol. 1999;212:323–36.
Monnier V, Dussillol F, Alves G, Lamour-Isnard C, Plessis A. Suppressor of fused links fused and Cubitus interruptus on the hedgehog signalling pathway. Curr Biol. 1998;8:583–6.
Me´thot N KB. Hedgehog controls limb development by regulating the activities of distinct transcriptional activator and repressor forms of Cubitus interruptus. Cell 1999;96:819–31.
Han YH, Shi Q, Jiang J. Multisite interaction with Sufu regulates Ci/Gli activity through distinct mechanisms in Hh signal transduction. Proc Natl Acad Sci USA. 2015;112:6383–8.
Zhang Q, Zhang L, Wang B, Ou C-Y, Chien C-T, Jiang J. A hedgehog-induced BTB protein modulates hedgehog signaling by degrading Ci/Gli transcription factor. Dev Cell. 2006;10:719–29.
Préat T. Characterization of Suppressor of fused, a complete suppressor of the fused segment polarity gene of Drosophila melanogaster. Genetics 1992;132:725–36.
Svard J, Heby-Henricson K, Persson-Lek M, Rozell B, Lauth M, Bergstrom A, et al. Genetic elimination of Suppressor of fused reveals an essential repressor function in the mammalian Hedgehog signaling pathway. Dev Cell. 2006;10:187–97.
Ding Q, Fukami S-I, Meng X, Nishizaki§ Y, Zhang X, Sasaki§ H, et al. Mouse suppressor of fused is a negative regulator of sonic hedgehog signaling and alters the subcellular distribution of Gli1. Curr Biol. 1999;9:1119–22.
Humke EW, Dorn KV, Milenkovic L, Scott MP, Rohatgi R. The output of Hedgehog signaling is controlled by the dynamic association between Suppressor of Fused and the Gli proteins. Genes Dev. 2010;24:670–82.
Shi Q, Han Y, Jiang J. Suppressor of fused impedes Ci/Gli nuclear import by opposing Trn/Kapbeta2 in Hedgehog signaling. J Cell Sci. 2014;127:1092–103.
Taylor MD, Liu L, Raffel C, Hui CC, Mainprize TG, Zhang X, et al. Mutations in SUFU predispose to medulloblastoma. Nat Genet. 2002;31:306–10.
Law DJ, Du M, Law GL, Merchant JL. ZBP-99 defines a conserved family of transcription factors and regulates ornithine decarboxylase gene expression. Biochem Biophys Res Commun. 1999;262:113–20.
Lisowskya T, Polosab PL, Saglianob A, Robertib M, Gadaletab MN, Cantatoreb P. Identification of human GC-box-binding zinc finger protein, a new Krüppel-like zinc finger protein, by the yeast one-hybrid screening with a GC-rich target sequence. FEBS Lett. 1999;45:369–74.
Fidalgo M, Huang X, Guallar D, Sanchez-Priego C, Valdes VJ, Saunders A, et al. Zfp281 coordinates opposing functions of Tet1 and Tet2 in pluripotent states. Cell Stem Cell. 2016;19:355–69.
Ishiuchi T, Ohishi H, Sato T, Kamimura S, Yorino M, Abe S, et al. Zfp281 shapes the transcriptome of trophoblast stem cells and is essential for placental development. Cell Rep. 2019;27:1742–54.e6
Pieraccioli M, Nicolai S, Antonov A, Somers J, Malewicz M, Melino G, et al. ZNF281 contributes to the DNA damage response by controlling the expression of XRCC2 and XRCC4. Oncogene 2016;35:2592–601.
Xue Y, Ding M, Xue L, Luo J. CircAGFG1 sponges miR-203 to promote EMT and metastasis of non-small-cell lung cancer by upregulating ZNF281 expression. Thorac Cancer. 2019;10:1692–701.
Ji W, Mu Q, Liu XY, Cao XC, Yu Y. ZNF281-miR-543 feedback loop regulates transforming growth factor-beta-induced breast cancer metastasis. Mol Ther Nucleic Acids. 2020;21:98–107.
Starzynska A, Sobocki BK, Sejda A, Sakowicz-Burkiewicz M, Szot O, Jereczek-Fossa BA. ZNF-281 as the potential diagnostic marker of oral squamous cell carcinoma. Cancers (Basel). 2021;13:2661.
Hahn S, Jackstadt R, Siemens H, Hunten S, Hermeking H. SNAIL and miR-34a feed-forward regulation of ZNF281/ZBP99 promotes epithelial-mesenchymal transition. EMBO J. 2013;32:3079–95.
Zhu Y, Zhou Q, Zhu G, Xing Y, Li S, Ren N, et al. GSK-3β phosphorylation-dependent degradation of ZNF281 by β-TrCP2 suppresses colorectal cancer progression. Oncotarget 2017;8:88599–612.
Hahn S, Hermeking H. ZNF281/ZBP-99: a new player in epithelial-mesenchymal transition, stemness, and cancer. J Mol Med. 2014;92:571–81.
Peng Y, Qin Y, Zhang X, Deng S, Yuan Y, Feng X, et al. MiRNA-20b/SUFU/Wnt axis accelerates gastric cancer cell proliferation, migration and EMT. Heliyon 2021;7:e06695.
Peng Y, Zhang X, Lin H, Deng S, Qin Y, He J, et al. Dual activation of Hedgehog and Wnt/beta-catenin signaling pathway caused by downregulation of SUFU targeted by miRNA-150 in human gastric cancer. Aging (Albany NY). 2021;13:10749–69.
Peng Y, Zhang X, Lin H, Deng S, Qin Y, Yuan Y, et al. SUFU mediates EMT and Wnt/beta-catenin signaling pathway activation promoted by miRNA-324-5p in human gastric cancer. Cell Cycle. 2020;19:2720–33.
Zhang Z, Zou Y, Liang M, Chen Y, Luo Y, Yang B, et al. Suppressor of fused (Sufu) promotes epithelial-mesenchymal transition (EMT) in cervical squamous cell carcinoma. Oncotarget 2017;8:114226–38.
Liu A. Proteostasis in the Hedgehog signaling pathway. Semin Cell Dev Biol. 2019;93:153–63.
Nano M, Gemble S, Simon A, Pennetier C, Fraisier V, Marthiens V, et al. Cell-cycle asynchrony generates DNA damage at mitotic entry in polyploid cells. Curr Biol. 2019;29:3937–45.
Mladenov E, Magin S, Soni A, G I. DNA double-strand-break repair in higher eukaryotes and its role in genomic instability and cancer: cell cycle and proliferation-dependent regulation. Semin Cancer Biol. 2016;37-38:51–64.
Roos PW, Kaina B. DNA damage-induced cell death by apoptosis. Trends Mol Med. 2006;12:440–50.
Miles WO, Dyson NJ, Walker JA. Modeling tumor invasion and metastasis in Drosophila. Dis Model Mech. 2011;4:753–61.
Niisslein-Volhard C EW. Mutations affecting segment number and polarity in Drosophila. Nature 1980;287:795–801.
Heemskerk J SD. Drosophila hedgehog acts as a morphogen in cellular patterning. Cell. 1994;76:449–60.
Chi S, Huang S, Li C, Zhang X, He N, Bhutani MS, et al. Activation of the hedgehog pathway in a subset of lung cancers. Cancer Lett. 2019;244:53–60.
Sheng T, Li C, Zhang X, Chi S, He N, Chen K, et al. Activation of the hedgehog pathway in advanced prostate cancer. Mol Cancer. 2004;3:29.
Guerrini-Rousseau L, Dufour C, Varlet P, Masliah-Planchon J, Bourdeaut F, Guillaud-Bataille M, et al. Germline SUFU mutation carriers and medulloblastoma: clinical characteristics, cancer risk, and prognosis. Neuro Oncol. 2018;20:1122–32.
Xu F, Li H, C H. LIFR-AS1 modulates Sufu to inhibit cell proliferation and migration by miR-197-3p in breast cancer. Biosci Rep. 2019;39:BSR20180551.
Chatterjee N, Walker GC. Mechanisms of DNA damage, repair, and mutagenesis. Environ Mol mutagenesis. 2017;58:235–63.
O’Connor MJ. Targeting the DNA damage response in cancer. Mol cell. 2015;60:547–60.
Zhou F, Huang D, Li Y, Hu G, Rao H, Q L, et al. Nek2A/SuFu feedback loop regulates Gli-mediated Hedgehog signaling pathway. Int J Oncol. 2017;50:373–80.
Sun X, Ding Y, Zhan M, Li Y, Gao D, Wang G, et al. Usp7 regulates Hippo pathway through deubiquitinating the transcriptional coactivator Yorkie. Nat Commun. 2019;10:411.
Ding Y, Wang G, Zhan M, Sun X, Deng Y, Zhao Y, et al. Hippo signaling suppresses tumor cell metastasis via a Yki-Src42A positive feedback loop. Cell Death Dis. 2021;12:1126.
Zhou Z, Xu C, Chen P, Liu C, Pang S, Yao X, et al. Stability of HIB-Cul3 E3 ligase adaptor HIB is regulated by self-degradation and availability of its substrates. Sci Rep. 2015;5:12709.
Zhou Z, Yao X, Li S, Xiong Y, Dong X, Zhao Y, et al. Deubiquitination of Ci/Gli by Usp7/HAUSP regulates hedgehog signaling. Dev Cell. 2015;34:58–72.
Zhan M, Sun X, Liu J, Li Y, Li Y, He X, et al. Usp7 promotes medulloblastoma cell survival and metastasis by activating Shh pathway. Biochem Biophys Res Commun. 2017;484:429–34.
Acknowledgements
We sincerely thank Prof. Lei Xue (Tongji University, China) for providing scrib-RNAi fly and Prof. Yong Li (Nanchang University, China) for providing lentiviral vectors.
Funding
This work was supported by grants from the National Natural Science Foundation of China (31922011, 31802012, and ZR2021MH137), the National Key Research and Development Program of China (2017YFA0205200), Program for Scientific Research Innovation Team of Young Scholar in Colleges and Universities of Shandong Province (2019KJE009), and the Construction Engineering Special Fund of “Taishan Scholars” (no. Ts201712022).
Author information
Authors and Affiliations
Contributions
MZ, ZZ, and JD designed this study and provide financial support. YD, DP, JX, YZ, WD, SY, and LS carried out the experiments and analyzed the data. SY and LS provided HCC cell lines. YD and DP performed mouse experiments. YZ analyzed cell migration in Drosophila. YD and ZZ wrote the article with the assistance of all authors. MZ, ZZ, and JD supervised this study and modified the manuscript. All authors read and approved the final paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
All animal experiments were approved by the Institutional Animal Care Committee of China Pharmaceutical University.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by G. Del Sal
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Deng, Y., Peng, D., Xiao, J. et al. Inhibition of the transcription factor ZNF281 by SUFU to suppress tumor cell migration. Cell Death Differ 30, 702–715 (2023). https://doi.org/10.1038/s41418-022-01073-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-022-01073-1
This article is cited by
-
E3 ligase TRIM65 alleviates intestinal ischemia/reperfusion injury through inhibition of TOX4-mediated apoptosis
Cell Death & Disease (2024)
-
tRF3-IleAAT reduced extracellular matrix synthesis in diabetic kidney disease mice by targeting ZNF281 and inhibiting ferroptosis
Acta Pharmacologica Sinica (2024)
-
Deep dissection of stemness-related hierarchies in hepatocellular carcinoma
Journal of Translational Medicine (2023)