Abstract
Activation of oval cells (OCs) has been related to hepatocyte injury during chronic liver diseases including non-alcoholic fatty liver disease (NAFLD). However, OCs plasticity can be affected under pathological environments. We previously found protection against hepatocyte cell death by inhibiting protein tyrosine phosphatase 1B (PTP1B). Herein, we investigated the molecular and cellular processes involved in the lipotoxic susceptibility in OCs expressing or not PTP1B. Palmitic acid (PA) induced apoptotic cell death in wild-type (Ptpn1+/+) OCs in parallel to oxidative stress and impaired autophagy. This lipotoxic effect was attenuated in OCs lacking Ptpn1 that showed upregulated antioxidant defences, increased unfolded protein response (UPR) signaling, higher endoplasmic reticulum (ER) content and elevated stearoyl CoA desaturase (Scd1) expression and activity. These effects in Ptpn1−/− OCs concurred with an active autophagy, higher mitochondrial efficiency and a molecular signature of starvation, favoring lipid droplet (LD) formation and dynamics. Autophagy blockade in Ptpn1−/− OCs reduced Scd1 expression, mitochondrial fitness, LD formation and restored lipoapoptosis, an effect also recapitulated by Scd1 silencing. PTP1B immunostaining was detected in OCs from mouse liver and, importantly, LDs were found in OCs from Ptpn1−/− mice with NAFLD. In conclusion, we demonstrated that Ptpn1 deficiency restrains lipoapoptosis in OCs through a metabolic rewiring towards a “starvation-like” fate, favoring autophagy, mitochondrial fitness and LD formation. Dynamic LD-lysosomal interations likely ensure lipid recycling and, overall, these adaptations protect against lipotoxicity. The identification of LDs in OCs from Ptpn1−/− mice with NAFLD opens therapeutic perspectives to ensure OC viability and plasticity under lipotoxic liver damage.
Similar content being viewed by others
Introduction
Non-alcoholic fatty liver disease (NAFLD) is the most common chronic liver disease in adults [1]. NAFLD ranges from benign and reversible ectopic accumulation of fat in the liver, accompanied by elevation in circulating free fatty acids (FFAs), to non-alcoholic steatohepatitis (NASH), cirrhosis, and in some cases to hepatocellular carcinoma (HCC) [1]. Hepatocyte lipoapoptosis in NAFLD mainly arises from FFA-induced lipotoxic stress in organelles, particularly mitochondria and endoplasmic reticulum (ER) [2, 3]. Triglycerides (TG) buffer toxic lipid species, including saturated FFAs (SFAs) [4] and stearoyl CoA desaturase 1 (SCD1) is essential for liver TG formation easily packable into lipid droplets (LDs) [5]. Originated from the ER, during their life cycle LDs contact with cellular organelles, not only ER and mitochondria, but also autophagosomes, lysosomes and peroxisomes through membrane contact sites [6].
Oval cells (OCs) (hepatic progenitor cells in humans) constitute 1–3% of the adult liver cell population. Upon liver damage, they proliferate and differentiate into hepatocytes or cholangiocytes in a process known as ductular reaction [7]. It has been suggested that OC activation is an adaptive response to liver damage in adult and pediatric NASH [7,8,9]. However, whether lipotoxicity affects OC viability has not been addressed.
Protein tyrosine phosphatase 1B (PTP1B), encoded by the Ptpn1 gene, modulates signaling cascades triggered by hormones/growth factors such as insulin [10], insulin-like growth factor-1 (IGF-1), epidermal growth factor (EGF) [11] and hepatocyte growth factor (HGF) [12], all relevant for liver regeneration [13]. In this context, PTP1B has been proposed as a promising therapeutic target for hepatoprotection [14, 15]. In a recent study, we found enhanced HGF-mediated signaling in OCs lacking Ptpn1 [16].
Since there is not an approved treatment for NAFLD and OC expansion is a challenging strategy against chronic liver damage, we aimed to investigate the impact of lipotoxicity in these progenitor liver cells and the role of PTP1B in this process.
Results
Ptpn1 deficiency prevents oxidative stress and PA-induced lipoapoptosis in liver OCs
We first analyzed the sensitivity of wild-type (Ptpn1+/+) OCs to PA (400 µM) for 24 h. A marked increase in cell death concurrently with Caspase-3 and PARP-1 cleavage and increased Bax/Bcl-xL ratio was found (Fig. 1A, Fig. S1). However, Ptpn1−/− or Ptpn1-silenced (shPtpn1) OCs were protected against lipoapoptosis (Fig. 1A, Fig. S2A–C). Interestingly, these effects were less evident in PA-treated Ptpn1+/+ primary hepatocytes (Fig. S3).
A Representative bright field microscopy images of Ptpn1+/+ and Ptpn1−/− OCs after 24 h of PA treatment; quantification of apoptotic cells by flow cytometry upon 24 h of PA treatment (n = 4–7); representative Western blot images of cleaved Caspase-3 and PARP-1 at 8 h of PA treatment (n = 4); representative Western blot images of Bax/Bcl-xL ratio at 24 h of PA treatment (n = 3). B Principal component analysis (PCA), volcano plots comparing PA versus C in Ptpn1+/+ and Ptpn1−/− OCs, principal categories of gene families upregulated at 8 h of PA treatment in Ptpn1−/− versus Ptpn1+/+ OCs and dot plots with significant functional results (n = 4). C DHE staining (n = 4) analyzed by confocal microscopy and lipid peroxidation (n = 4–6) after 2 or 16 h of PA treatment, respectively, in Ptpn1+/+ and Ptpn1−/− OCs. D Representative confocal microscopy immunofluorescence images of NRF2 after 4 h of PA treatment (n = 3); Nqo1 and Hmox1 mRNA levels (n = 4) at 4 h of PA treatment; representative Western blot images showing nuclear and cytosolic NRF2 and its targets HO-1 and Catalase at different periods of treatment. E Representative confocal immunofluorescence images using MitoTracker Red CM-H2XRos (MTR) and MitoTracker Green FM (MTG) probes and quantification by flow cytometry after 8 h of PA treatment (n = 4); representative TEM images showing mitochondria morphology at 16 h of PA treatment. A–E Data are presented as mean ± SEM. ***p < 0.001 versus each C group; ###p < 0.001 versus PA group; ++p < 0.01 Ptpn1+/+ C versus Ptpn1−/− C group according to two-way ANOVA with post hoc Bonferroni test.
RNA-Seq analysis in OCs treated with PA for 8 h, a time period prior to cell death, revealed a homogeneous expression level, while samples were well separated in the groups of interest in hierarchical clustering and principal component analysis (PCA) (Fig. 1B, Fig. S4A, B). Volcano plots showed differential gene expression between PA versus controls in both genotypes of OCs (Fig. 1B, Tables S3 and S4). Of interest, 1446 genes were commonly upregulated in PA-treated OCs whereas 1825 genes were commonly down-regulated (Fig. S4C). These genes were clustered in 116 upregulated and 604 down-regulated gene families. The comparison between PA-treated OCs revealed 171 gene families upregulated in Ptpn1−/− OCs (Table S4). Enrichment and dotplot analysis (Fig. 1B, S4D, Tables S5–S9) show 38 functions related to lipid metabolism, 20 to autophagy and lysosomes, 17 to catabolic processes, 14 to reticulum and vesicle metabolism, 14 to bile acids and cholesterol metabolism, 4 to peroxisomes and 5 to gluthatione metabolism.
Oxidative stress is an early hallmark of lipotoxic cell death [17]. As shown in Fig. 1C, Fig. S5A, treatment with PA for 2 or 16 h increased DHE staining and lipid peroxidation (MDA), respectively, only in Ptpn1+/+ OCs. Of note, basal DHE staining was higher in Ptpn1−/− OCs, but did not increase upon PA treatment. RNA-Seq revealed upregulated antioxidant gene expression in Ptpn1−/− OCs (Fig. S5B). Likewise, nuclear and cytosolic NRF2 levels, expression of NRF2-target genes (Nqo1, Hmox1) and Heme oxygenase (HO-1) and Catalase protein levels, showed that Ptpn1 deficiency upregulated NRF2-mediated antioxidant defences and triggered a rapid antioxidant response against lipotoxic stress (Fig. 1D and Fig. S5C) which was confirmed in shPtpn1 OCs (Fig. S5D).
Confocal and flow cytometry analysis of OCs stained with MitoTracker Red CM-H2XRos (MTR) evidenced accumulation of damaged mitochondrial in PA-treated Ptpn1+/+ OCs (Fig. 1E) that was also assessed by quantification of mtDNA damage (Fig. S5E). Damaged mitochondria in PA-treated Ptpn1+/+ OCs were visualized by transmission electronic microscopy (TEM) (Fig. 1E). Importantly, in Ptpn1−/− OCs, increased mitochondrial mass, assessed by MitoTracker Green FM (MTG) staining, and higher Ppargc1a mRNA were found under basal conditions (Fig. 1E and Fig. S5F), pointing to enhanced mitochondrial biogenesis. Also, in Ptpn1−/− cells PA did not induce mitochondrial damage (Fig. 1E and Fig. S5E). As reported in hepatocytes [18], treatment of Ptpn1+/+ OCs with PA and sulforaphane increased nuclear NRF2, enhanced antioxidant gene expression and reduced DHE staining and lipid peroxidation (Fig. S6A–D). As a result, apoptosis was partly reverted (Fig. S6E).
Ptpn1 −/− OCs have a higher capacity to form LDs to prevent PA-induced lipoapoptosis
Our observations (Fig. 1A) evidenced LD accumulation in Ptpn1−/− OCs exposed to PA, an effect also analyzed by adipose differentiation-related protein (ADRP) or Bodipy and Nile Red staining (Fig. 2A). Accumulation of LDs was not found in PA-treated Ptpn1+/+ OCs and, in this regard, marked differences were found between genotypes in TG metabolism-related gene expression. In PA-treated Ptpn1−/− OCs, LD formation was not associated to more PA uptake since 14C 2-Bromopalmitate incorporation and FFA transporter/receptor-related gene/protein expression were reduced compared to Ptpn1+/+ cells (Fig. S7A). Notably, Plin2 (encoding ADRP) mRNA and protein levels of DGAT2 and ADRP, involved in TG biosynthesis and LD biogenesis, respectively, were upregulated in Ptpn1−/− OCs under basal conditions and further elevated by PA (Fig. 2B and Fig. S7B).
A Representative confocal microscopy images of ADRP immunofluorescence and Bodipy staining in Ptpn1+/+ and Ptpn1−/− OCs at 24 h of PA treatment (n = 4); quantification of Nile Red staining analyzed by flow cytometry at 24 h of PA treatment (n = 3); RNA-Seq data heatmap of TG synthesis-related genes at 8 h post-PA treatment (B) mRNA levels of Plin2 (n = 3); representative Western blot images of DGAT2 at different time-periods of PA treatment (n = 4) and ADRP (n = 4) at 8 h of PA treatment. C Quantification of UFA/SFA ratio and FFA content in different lipid species (DG, TG, PL) (n = 3); mRNA levels of Scd1 (n = 3) in Ptpn1+/+ and Ptpn1−/− OCs at 4 h post-PA addition. D Representative western blot images of p-mTOR (n = 5) and LC3II/I (n = 4) in Ptpn1+/+ and Ptpn1−/− OCs at different time points of PA treatment; (upper) P62 levels analyzed at 8 h upon PA addition (n = 4); (lower) LC3II/I and P62 in Ptpn1+/+ OCs after 8 h of treatment with PA or PA plus chloroquine (CQ) (n = 4–7). Representative confocal microscopy images of P62 and LC3 at 8 h of PA treatment in Ptpn1+/+ and Ptpn1−/− OCs (n = 3). E Representative TEM images showing accumulation of autophagosomes in Ptpn1+/+ OCs at 16 h of PA treatment. F mRNA levels of Ddit3 (n = 3) at 24 h post-PA treatment; GRP78 protein (n = 4) and Atf6 mRNA levels (n = 3) at 8 h post-PA treatment. p-PERK, p-eIF2α and p-JNK (n = 5) Western blots in Ptpn1+/+ and Ptpn1−/− OCs treated with PA at the indicated time-periods and their respective densitometric analysis; A–F Data are presented as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001 versus each C group; #p < 0.05, ##p < 0.01, ###p < 0.001 versus PA group; +p < 0.05, ++p < 0.01 Ptpn1+/+ C versus Ptpn1−/− C group according to two-way ANOVA with post hoc Bonferroni test.
Lipidomic analysis showed increased unsaturated FA (UFA)/saturated (SFA) ratio in different lipid species (diacylglycerols (DG), TG and phospholipids (PL)) in Ptpn1−/− OCs exposed to PA for 16 h (Fig. 2C, Table S10) together with increased Scd1 mRNA. All these results were confirmed in PA-treated Ptpn1+/+-silenced OCs (Fig. S8A–D). Of relevance, Ptpn1+/+ OCs treated with the monounsaturated oleic acid (OA) were able to form LDs and preserve their viability (Fig. S9A, B), excluding defects in the LD formation machinery. LD formation, Plin2 and Scd1 mRNA levels were opposite modulated by PA in Ptpn1−/− primary hepatocytes (Fig. S10), pointing to the cell-specificity of our results in OCs.
PA blocks autophagy in Ptpn1 +/+ but not in Ptpn1 −/− OCs
Blockade of autophagy is a hallmark of lipotoxicity [3], but autophagy has not been studied in OCs exposed to a lipotoxic environment. Since activation of mTORC1 suppresses autophagy [19], we evaluated its phosphorylation in PA-treated OCs. Figure 2D, and Fig. S11A, B show higher mTORC1 phosphorylation in Ptpn1+/+ cells in parallel with elevations in P62 and LC3II/I ratio at 8 h upon PA treatment, the later response being enhanced in the presence of the lysosomal inhibitor chloroquine (CQ). Moreover, accumulation of autophagosomes was visualized by TEM in Ptpn1+/+ cells treated with PA for 16 h (Fig. 2E) indicating blockade of the autophagic flux in Ptpn1+/+ OCs.
Enrichment in ER content and higher UPR, but not ER stress-mediated apoptosis, concur with LD formation in PA-treated Ptpn1 −/− OCs
Since sustained ER stress has been related to defective autophagy in the liver [20], we analyzed this response in OCs. The proapoptotic ER stress-related transcription factor Ddit3, encoding CHOP, was increased in Ptpn1+/+ OCs exposed to PA for 24 h; this effect being attenuated in Ptpn1−/− cells (Fig. 2F). However, GRP78 protein levels, Atf6 mRNA levels and UPR-mediated signaling (PERK, eiF2α, JNK phosphorylation) increased in the basal state and in the early response (2–8 h) to PA in Ptpn1−/− OCs and Ptpn1-silenced OCs (Fig. 2F, Fig. S12A–D).
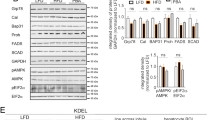
As LDs emerge from the ER surface [6] and UPR was enhanced in Ptpn1−/− OCs, we studied in detail the ER features. RNA-Seq revealed constitutive upregulation of ER-related genes in Ptpn1−/− OCs (Fig. 3A, Table S11), confirmed determining Canx (encoding Calnexin), Slc35b3 (encoding Solute carrier 35B3), and Lpin2 (encoding Lipin 2) mRNAs by RT-qPCR. More abundance of ER in Ptpn1−/− OCs under basal conditions or PA treatment was also analyzed by Calnexin immunostaining and ER-Tracker staining (Fig. 3A, B and Fig. S13A, B). Next we purified both small and large LDs as described [21] in Ptpn1−/− OCs treated with PA for 16 h and associated proteins were identified by Western blot. Figures 3C, S13C show Calnexin mainly associated with small (likely nascent) LDs. In this line, ER-Tracker/Bodipy stained small/nascent LDs surrounded by ER (Fig. 3D, Video S1). This effect was also observed by ADRP/Calnexin and ADRP/GRP78 immunostaining (Fig. 3D). The close contact between ER and LDs was visualized in detail by TEM in PA-treated Ptpn1−/− OCs (Fig. 3E). Upregulation of genes related to ER-LD interactions was also found in the RNA-Seq and validation of Bscl2, encoding Seipin, confirmed these results (Fig. 3F). Of note, Calnexin protein content was enriched in small/nascent LDs from OA-treated Ptpn1+/+ OCs (Fig. S13D).
A RNA-Seq data heatmap of ER-related genes in Ptpn1+/+ and Ptpn1−/− OCs under basal condition and validation of Canx, Slc35b3 and Lpin2 mRNAs by RT-qPCR (n = 4); representative confocal microscopy images of ER-Tracker staining and Calnexin immunofluorescence in Ptpn1+/+ and Ptpn1−/− OCs under basal conditions (n = 4). B Representative confocal microscopy images of ER-Tracker and Calnexin staining (n = 4) in Ptpn1+/+ and Ptpn1−/− OCs at 4 h of PA treatment. C Representative Western blot images of ADRP, Calnexin, LAMP1, LAMP2A, ATGL, MFN2, and GAPDH in large (LD1) and small (LD2) lipid droplets upon 16 h of PA treatment in Ptpn1−/− OCs. GAPDH was also analyzed in post-nuclear supernatant (PNS). D Live-cell confocal imaging of Ptpn1−/− OCs stained with Bodipy and ER Tracker upon 4 h of PA treatment; representative confocal microscopy images of ADRP/Calnexin (n = 3) and ADRP/GRP78 (n = 4) in Ptpn1−/− OCs at 4 h of PA treatment. E Representative TEM images in Ptpn1+/+ and Ptpn1−/− OCs at 16 h post-PA treatment. F Heatmap of LD–ER interaction-related genes and mRNA levels of Bscl2 (n = 3) at 8 h of PA treatment. A–F Data are presented as mean ± SEM. *p < 0.05, ***p < 0.001 versus each C group; ###p < 0.001 versus PA group; +++p < 0.001 Ptpn1+/+ C versus Ptpn1−/− C group according to two-way ANOVA with post hoc Bonferroni test and Student’s t test (A).
Characterization of LD catabolism by the lysosomes in PA-loaded Ptpn1 −/− OCs
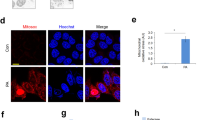
Considering that in hepatocytes LD-lysosome interactions are relevant for LD catabolism [21, 22], we characterized this process in OCs. The lysosomal proteins LAMP1/2A were found associated to small LDs in Ptpn1−/− OCs (Fig. 3C). Also, Lysotracker/Bodipy staining identified lysosomes surrounding LDs in Ptpn1−/− OCs either in basal conditions or upon PA treatment (Fig. 4A). Moreover, live-cell imaging showed lysosome-LD stable contacts (Fig. 4B, Video S2) and LDs engulfment into the lysosomes (Fig. 4C, Video S3). This close interaction (synapsis), as well as presence of LDs into the lysosomes, were also visualized by TEM (Fig. 4D). At the molecular level, upregulation of lysosome- (Fig. S14, Table S12) and lysosome-LD interaction-related genes (Fig. 4E), the latter validated by increased Lipa mRNA, was found in the RNA-Seq of PA-treated Ptpn1−/− OCs. Lysosome-LD synapsis was also visualized by long-term live-cell imaging in Ptpn1−/− OCs transiently expressing the dual fluorescence mRFP1-EGFP-ADRP/PLIN2 reporter that exhibits differential fluorescence dependent on the pH of its surrounding environment as described [22]. Figure 4F and Videos S4, S5 show instances of prolonged interactions between lysosomes and reporter-coated LDs indicating that a considerable quantity of the PLIN2/ADRP reporter is continually being sampled by the lysosomes.
A Representative confocal microscopy images of Ptpn1+/+ (left) and Ptpn1−/− (right) OCs stained with Bodipy and Lysotracker at 16 h of PA treatment. B Live-cell confocal imaging of Ptpn1−/− OCs stained with Bodipy and Lysotracker at 16 h of PA treatment showing the appearance of Bodipy-positive signal within a lysosome throughout a persistent contact lasting >30 min (see Video S2). C Live-cell confocal imaging of Ptpn1−/− OCs stained with Bodipy and Lysotracker at 16 h of PA treatment, showing an example of a lysosome directly engulfing and quickly degrading the internalized LD within less than 12 min (see Video S3). To the right of each panels B and C a kymograph shows lysosome(LYS)-LD contact. D Representative TEM images of LYS-LD interactions in Ptpn1+/+ and Ptpn1−/− OCs at 16 h of PA treatment. E Heatmap of LYS-LD interactions-related genes at 8 h of PA treatment; mRNA levels of Lipa (n = 3) in Ptpn1+/+ and Ptpn1−/− OCs at 16 h of PA treatment. F Live-cell confocal imaging of Ptpn1−/− OCs overexpressing mRFP1-EGFP-ADRP/PLIN2 fusion protein stained with Dextran-A647 at 16 h of PA treatment (see Videos S4, S5). Data are presented as mean ± SEM. ***p < 0.001 versus C group; ###p < 0.001 versus PA group according to two-way ANOVA with post hoc Bonferroni test.
Blockade of LD catabolism by cytosolic or lysosomal lipases did not induce apoptosis in Ptpn1 −/−OCs
Once studied LD formation and interaction with lysosomes, we evaluated a possible role of LD catabolism, either by cytosolic or lysosomal lipases, in OC lipotoxicity. Figure 3C shows enrichment in adipose TG lipase (ATGL) in protein extracts from large LDs in Ptpn1−/− OCs. Therefore, Ptpn1−/− OCs were treated with PA in presence of the lysosomal acid lipase inhibitor lalistat 1 (LALi) or/and the adipose triglyceride lipase inhibitor atglistatin (ATGLi). Although these inhibitors added individually did not affect total neutral lipid content (Fig. 5A), LDs with different average size were found with a substantial accumulation of small or large LDs in the presence of LALi or ATGLi, respectively, in comparison with LDs accumulated with PA alone. Of note, in Ptpn1−/− OCs, cellular ATP content decreased in presence of the inhibitors confirming the blockade of LD catabolism, but interestingly, cellular viability was retained (Fig. 5B).
A Representative bright field microscopy images and representative confocal microscopy images of Ptpn1−/− OCs after 24 h of PA treatment with or without CQ, LALi, ATGLi, and LALi+ATGLi; quantification of Nile Red staining analyzed by flow cytometry (n = 4); quantification of LD abundance distributed by LD size (µm2) in Ptpn1−/− OCs stained with Bodipy. B Analysis of ATP content (n = 4) and quantification of the apoptotic cells by flow cytometry (n = 3) after 24 h of PA treatment with or without LALi, ATGLi and LALi+ATGLi in Ptpn1−/− OCs. C Representative TEM images and quantification of MT-LD interaction in Ptpn1−/− OCs at 16 h of PA treatment (n = 12); representative confocal microscopy images of ADRP and MFN2 at 16 h of PA treatment; heatmap of MT-LD interaction-related genes comparing Ptpn1+/+ and Ptpn1−/− OCs and mRNA levels of Vps13d (n = 3). D Seahorse analysis (ATP production, Maximal respiration, Proton leak, and Coupling efficiency) in Ptpn1+/+ and Ptpn1−/− OCs under basal conditions (n = 10). E Representative western blot images of SIRT1 and p-AMPK (n = 6) in Ptpn1+/+ and Ptpn1−/− OCs under basal conditions and analysis of ATP content. A–E Data are presented as mean ± SEM. **p < 0.01, ***p < 0.001 versus C group; #p < 0.05, ##p < 0.01, ###p < 0.001 versus PA group; ++p < 0.01, versus PA + ATGLi group; †††p < 0.001 versus PA + CQ according to one (A, B) and two-way ANOVA (C) with post hoc Bonferroni test and Student’s t test (D, E).
Mitochondrial fitness is essential for protecting OCs against lipotoxicity
Preserved mitochodrial fitness protects cells against lipotoxicity [23]. Evidence of direct trafficking of FFAs from LDs to mitochondria through specific contact sites was recently reported [24,25,26]. Mitochondria-LD contacts in PA-treated Ptpn1−/− OCs were visualized by TEM, Mitofusin-2 (MFN2)-ADRP immunofluorescence and by detecting MFN2 in protein extracts from large LDs (Figs. 5C, 3C). Moreover, genes related with this interaction were upregulated in Ptpn1−/− OCs and validation of Vps13d, encoding a protein responsible for FFA trafficking from LDs to mitochondria [27], supported these results (Fig. 5C). Taking this into account, mitochondrial bioenergetics was examined and Seahorse analysis (Fig. 5D) showed similar basal ATP production between both genotypes, but less maximal respiration and proton leak in Ptpn1−/− OCs and, as a consequence, higher coupling efficiency as occurred in starvation-resistant cells [28]. These energy status in Ptpn1−/− OCs under basal conditions resembled a molecular starvation-like signature characterized by increased phospho-AMPK and Sirtuin 1 (SIRT1) levels, the latter in inverse relationship with PTP1B as we and others reported [29, 30] and a decrease in total ATP content (Fig. 5E, Fig. S15A). This rewiring was also coincident with a gene signature of autophagy, validated by measuring Vamp8, Atg13, and Lamp2a mRNAs by RT-qPCR (Fig. S15B, Table S13). In fact, treatment with CQ led to a higher increase in P62 and LC3II/I ratio in OCs lacking Ptpn1, confirming an exacerbated autophagy flux compared to Ptpn1+/+ OCs that was also assessed by P62 and LC3 immunostaining (Fig. 6A, B).
Representative Western blot and quantification (n = 6) (A) and immunofluorescence microscopy images (n = 5) (B) of P62 and LC3II/I at 8 h of CQ treatment in Ptpn1+/+ and Ptpn1−/− OCs. C Representative images of bright field microscopy; quantification of the number of apoptotic cells analyzed by flow cytometry (n = 4); representative Western blot images of cleaved Caspase-3 and PARP-1 (n = 4) in Ptpn1−/− OCs at 24 h of treatment with PA with or without CQ. D Seahorse analysis (Basal and Maximal respiration) in Ptpn1−/− OCs treated with PA in the absence or presence of CQ for 4 h (n = 10); fatty acid oxidation (n = 6) and ATP production (n = 9) in Ptpn1−/− OCs treated with PA with or without CQ for 4 h. E Representative confocal immunofluorescence images in Ptpn1−/− OCs using MTR CM-H2XRos and MTG probes and quantification by flow cytometry after 8 h of PA treatment with or without CQ (n = 4); mitochondrial DNA damage analyzed by RT-qPCR after 8 h of PA treatment with or without CQ (n = 4). A–E Data are presented as mean ± SEM. In A ***p < 0.001 versus each C; +++p < 0.001 versus each CQ group according to two-way ANOVA with post hoc Bonferroni test. In C–E *p < 0.05, **p < 0.01, ***p < 0.001 versus C group; #p < 0.05, ##p < 0.01, ###p < 0.001 versus PA group; +p < 0.05, ++p < 0.01, +++p < 0.001 versus PA + CQ group according to one-way ANOVA with post hoc Bonferroni test.
Elevated autophagic capacity of Ptpn1 −/− OCs is necessary for LD formation and survival upon PA treatment
In order to unravel if the exacerbated autophagic flux found in Ptpn1−/− OCs favors survival under a lipotoxic insult, these cells were cotreated with PA/CQ. Under these conditions, Ptpn1−/− (Fig. 6C) and Ptpn1-silenced (Fig. S16A) OCs were susceptible to lipoapoptosis.
We next evaluated the role of macroautophagy in mitochondrial fitness in Ptpn1−/− OCs. Seahorse analysis showed that PA/CQ cotreatment decreased both basal and maximal respiration, β-oxidation and ATP production compared to cells treated only with PA (Fig. 6D, E). Under these conditions, accumulation of damaged mitochondria was evidenced (Fig. 6E). In fact, addition of CQ to PA-loaded Ptpn1−/− OCs was able to induce mitochondrial oxidative stress and apoptosis, pointing to the essential role of macroautophagy to maintain mitochondrial fitness and ensure viability in LD-enriched OCs (Fig. S16B, C).
We analyzed whether autophagy was also necessary for LD formation in Ptpn1−/− OCs to preserve survival. Neither LDs nor increased Nile Red staining were observed in Ptpn1−/− OCs cotreated with PA/CQ (Fig. 7A), confirming the requirement of autophagy for LD formation in OCs. Interestingly, PA/CQ cotreatment abolished PA-induced Scd1 mRNA levels and reduced UFA/SFA ratio in lipid species (Fig. 7B, C, Table S14). Of note, the effect of CQ in preventing LD accumulation and inducing apoptosis was absent in OA-treated Ptpn1−/− cells (Fig. 7D). Finally, to further demonstrate the relevance of these findings we knocked down Scd1 in Ptpn1−/− OCs. As shown in Fig. 7E, Scd1 silencing in Ptpn1−/− OCs led to lipoapoptosis by PA which paralleled to their inability to accumulate LDs and a decrease in UFA/SFA ratio in lipid species (Fig. 7E, Table S15). Overall, these results extended to OCs lipotoxicity the relevance of the relationship between autophagy and Scd1 previously reported in liver tissue during fasting [31].
A Representative confocal microscopy images of ADRP and Bodipy in Ptpn1−/− OCs at 24 h of treatment with PA with or without CQ; quantification of Nile Red staining analyzed by flow cytometry (n = 4). B mRNA levels of Scd1 in Ptpn1−/− OCs at 4 h of PA with or without CQ treatment (n = 3). C Quantification of UFA/SFA ratio and FFA content in different lipid species (DG, TG, PL) in Ptpn1−/− OCs at 16 h of PA with or without CQ treatment (n = 4). D Representative bright field microscopy images and quantification of the apoptotic Ptpn1−/− OCs analyzed by flow cytometry upon 24 h of PA or oleic acid (OA) treatment with or without CQ (n = 4); quantification of Nile Red staining in Ptpn1−/− OCs analyzed by flow cytometry upon 24 h of PA or OA treatment with or without CQ (n = 3). E mRNA levels of Scd1 in Ptpn1−/− OCs transduced with scrambled (Scr) or Scd1 lentiviral particles (n = 3); representative Western blot images and quantification of cleaved Caspase-3 at 8 h of PA treatment (n = 4); quantification of the apoptotic cells by flow cytometry upon 24 h of PA treatment (n = 4). F Representative images of bright field microscopy of Ptpn1−/− OCs transduced with scrambled (Scr) or Scd1 lentiviral particles after 24 h of PA treatment; quantification of Nile Red staining analyzed by flow cytometry at 24 h of PA treatment (n = 3); quantification of UFA/SFA ratio in different lipid species (DG, TG, PL) (n = 4). A–F Data are presented in mean ± SEM. **p < 0.01, ***p < 0.001 versus C group; #p < 0.05, ##p < 0.01, ###p < 0.001 versus PA group; +p < 0.05, ++p < 0.01, +++p < 0.001, versus PA + CQ group according to one (A, B, D) and two-way (F) ANOVA with post hoc Bonferroni test and Student’s t test (C and Scd1 mRNA levels in E).
LDs were found in OCs in livers from Ptpn1 −/− mice with advanced NASH
To provide a pathophysiological context to this study, Ptpn1+/+ and Ptpn1−/− mice were submitted to the NAFLD protocol recently reported [32] and detailed in Marerials and Methods. As shown in Fig. 8A, NASH was induced in both genotypes of mice without differences in body weight or liver-to-body weight ratio (Fig. S17A). Importantly, less fibrosis and ALT levels were found in Ptpn1−/− mice with NAFLD.
A Images of Hematoxylin and Eosin (HE) and Sirius Red staining of liver sections from Ptpn1+/+ and Ptpn1−/− mice with or without NAFLD; NAS score (n = 6–11 mice/group) and plasma ALT (n = 10–11 mice/group) from Ptpn1+/+ and Ptpn1−/− mice; fibrosis score (n = 6–11 mice/group); hydroxyproline content (n = 6 mice/group) and mRNA levels of Col1a1 from Ptpn1+/+ and Ptpn1−/− mice (n = 6–10 mice/group). B Representative confocal microscopy images of PTP1B and A6 in liver sections from Ptpn1+/+ mice (n = 3–4 mice/group and 4 fields per mice). C Representative confocal microscopy images of A6 and SOX9 of liver sections from Ptpn1+/+ and Ptpn1−/− mice (n = 4–5 mice/group and 4 fields per mice). D Quantification of images from panel C. mRNA levels of Epcam (n = 6–11 mice/group) and Krt19 (n = 6–10 mice/group) from Ptpn1+/+ and Ptpn1−/− mice. E Representative confocal microscopy images of A6 and ADRP in liver sections from Ptpn1+/+ and Ptpn1−/− mice; quantification of LDs/total cell number per field in liver sections from Ptpn1+/+ and Ptpn1−/− mice (n = 4 mice/group and 4 fields per mice); A–E Data are presented in mean ± SEM. *p < 0.05; **p < 0.01; ***p < 0.001 versus C group; #p < 0.05, ##p < 0.01, ###p < 0.001 versus WD + CCl4 group according to two-way ANOVA with post hoc Bonferroni test and Student’s t test in E. F Expression levels of hepatic progenitor cells-related genes from the National Cancer for Biotechnology Information Gene Expression Omnibus database based on the gene chips of fibrotic liver tissues associated to NAFLD with mild (0–1) or advanced (≥2) fibrosis; levels of genes commonly upregulated in advanced fibrosis NAFLD patients from these studies (GSE130970 n = 53 mild and n = 25 advanced; GSE49541 n = 40 mild and n = 32 advanced; GSE16294 n = 103 mild and n = 40 advanced; GSE48452 n = 23 mild and n = 8 advanced). Data are presented in mean ± SEM. *p < 0.05; **p < 0.01; ***p < 0.001 versus mild fibrosis according to Student’s t test.
The analysis of Ptpn1 mRNA in OCs and mouse primary hepatocytes showed comparable levels (1 ± 0.03 in hepatocytes versus 0.91 ± 0.01 in OCs, n = 3). Moreover, PTP1B immunostaining was visualized in A6 (OC marker) positive cells in livers from Ptpn1+/+ mice under normal or NAFLD conditions (Fig. 8B). A more detailed analysis of liver samples revealed a marked increase in A6/SOX9 positive cells in all mice with NASH (Fig. 8C, D). Interestingly, less A6/SOX9 immunostaining together with lower mRNA levels of the progenitor markers Epcam and Krt19 was found in livers from Ptpn1−/− mice with NASH, pointing to a major presence of OCs in livers with more advanced NASH as reported [33].
Since Ptpn1−/− OCs accumulated LDs in response to a lipotoxic insult, we tested whether this effect also occurred in vivo. As Fig. 8E shows, LDs were found into the OCs from Ptpn1−/− mice with NASH, an effect absent in Ptpn1+/+ mice. This capacity of Ptpn1−/− OCs to accumulate LDs both in vivo and in vitro concurred with a gene signature of a more differentiated state upon PA treatment compared to Ptpn1+/+ OCs (Fig. S17B).
Finally, gene expression levels related to OCs were obtained from the NCBI Gene Expression Omnibus database based on the gene chips of fibrotic livers associated to NAFLD (GSE130970, GSE49541, GSE162694 and GSE48452, references in Materials and Methods). In line with our data in mice, the expression of SOX9, and EPCAM as well as KRT7, is significantly increased in NAFLD patients with advanced fibrosis compared to those with mild fibrosis (Fig. 8F).
Discussion
The main findings of this study relate to the impact of lipoapoptosis in liver OCs, an issue not investigated in NAFLD, and the protection conferred by Ptpn1 deficiency through the convergence of relevant molecular events leading to LD biogenesis and catabolism.
Lipotoxicity is a common hallmark of NAFLD [34, 35]. Our data herein revealed that, compared to hepatocytes, OCs are highly sensitive to PA-induced lipoapoptosis, an effect mediated by the classical mitochondrial intrinsic pathway involving oxidative stress [7, 36] that led to accumulation of damaged mitochondria due, at least in part, to autophagy blockade. These data uncovered OCs as a new hepatic cell type sensitive to a lipotoxic environment and, therefore, preventing OCs lipoapoptosis might enhance their proliferative/differentiation capacity in advanced NAFLD. This strategy was successfully achieved herein by deleting Ptpn1 based in previous studies from we [15, 37, 38] and others [39] on hepatoprotection. Although basal ROS levels were higher in Ptpn1−/− compared to Ptpn1+/+ OCs likely due to higher mitochondrial mass and activity (as discussed below), the enhancement of NRF2-mediated antioxidant gene expression found in Ptpn1−/− OCs under basal or PA treatment counteracted ROS effects, thereby protecting against PA-mediated lipoapoptosis. These results are in line with the protection against acetaminophen toxicity found in our previous study in PTP1B-deficient hepatocytes [15]. Notably, PTP1B inhibition protects against apoptosis in adipose tissue-derived stem cells [40].
An unexpected result was the capacity of Ptpn1−/− OCs to accumulate LDs upon PA treatment, an effect not found in Ptpn1−/− primary hepatocytes. In fact, hepatocyte-specific PTP1B-deficient mice are protected against steatosis [41]. One hallmark of the LDs is their capacity to buffer excess of FFAs or other toxic lipids as reviewed [42]. TGs are the main lipid specie in neutral LDs and RNA-Seq revealed upregulation of TG metabolism-related genes in PA-treated Ptpn1−/− OCs; a result confirmed by measuring total TGs. Moreover, endogenously generated UFA by desaturases are critical for TG synthesis [5]. As such, overexpression of SCD1 protects proximal tubular cells against lipoapoptosis by enhancing LD formation [43]. Herein we found elevated Scd1 expression and activity in Ptpn1−/− OCs upon PA exposure. This increase could be mediated by an upregulation of SIRT1 as reported [44]. In fact, AMPK phosphorylation and SIRT1 levels were higher in Ptpn1−/− OCs highlighting the reciprocal regulation of PTP1B and SIRT1 previously described [30]. Of relevance, increased AMPK/SIRT1 axis resembles a state of cellular calorie restriction which could also enhance LD formation as found in cells exposed to nutrient restriction [24].
LDs emerge from the ER membrane [6] and their accumulation has been observed as a general downstream effect of ER stress, including that induced by lipid overload [45]. Our study uncovered a new role of Ptpn1 in restraining LD biogenesis in OCs by regulating both ER content and UPR. In this regard, an upregulation of ER-related gene expression and ER content was found in Ptpn1−/− OCs under basal conditions. Importantly, these cells showed increased basal and PA-induced UPR-mediated signaling. This is particularly relevant since PERK is a direct PTP1B substrate [46]. However, in PA-treated Ptpn1−/− OCs, UPR was not associated with higher levels of ER stress-related apoptosis, again reinforcing lipoprotection. Moreover, close ER-nascent LD contacts were visualized in Ptpn1−/− OCs at an early time period upon PA addition, suggesting for the first time a key role of Ptpn1 in ER-derived LD biogenesis.
Using different methodologies we also demonstrated dynamic LD-lysosome interactions in PA-loaded Ptpn1−/− OCs, pointing to autophagy of the LDs (lipophagy) as recently described by McNiven’s group in hepatocytes [21, 22]. However, this direct LD processing by the lysosome is dispensable for OCs survival, as discussed below. As expected, LAL inhibition reduced the cellular ATP content, but this response might not reach a threshold enough to trigger cell death. Nevertheless, we cannot rule out lysosomes in OCs as regulators of lipid recycling and redistribution to various cellular compartments as reviewed [47].
As stated above, Ptpn1−/− OCs resembled a “starvation-like” metabolic phenotype associated with a very active autophagic flux, another feature of nutrient deprivation. LD biogenesis is a protective cellular response during periods of elevated autophagy [24]. Under these conditions, LDs sequester FFAs released during the autophagic breakdown of organelles to prevent lipotoxicity. In agreement, our data demonstrated that autophagy blockade suppressed the protection against lipoapoptosis conferred by Ptpn1 deletion, at least in part through accumulating damaged mitochondria and reducing β-oxidation and respiration. In addition, LD formation protects cells committed to die [48]. In fact, in Ptpn1−/− OCs autophagy blockade totally abolished LD formation, an effect that led to mitochondrial damage and apoptosis. Importantly, under these conditions, downregulation of Scd1 was very rapid (4 h post-PA/CQ addition) and preceded cell death. These results strongly suggest that reduced Scd1 limits TG synthesis and LD formation in OCs. The regulation of Scd1 expression by autophagy has been previously described in the liver [31], highlighting the relevance of our results in OCs. This was confirmed by (i) Scd1 silencing in Ptpn1−/− OCs that induced a phenotype resembling the Ptpn1+/+ counterparts, (ii) treating Ptpn1+/+ OCs with OA that resulted in LD formation and preservation of cellular viability, and (iii) co-treating Ptpn1−/− OCs with OA/CQ that also preserved LD biogenesis and survival. In addition, the fact that PA-loaded Ptpn1−/− OCs die by the subsequent treatment with CQ suggest another key role of autophagy for LD-mediated energy supply to ensure cell viability.
Expansion of OCs has been identified in livers from mice and humans with NAFLD or alcoholic fatty liver disease in positive correlation with fibrosis [33]. Interestingly, the expression of the hepatic progenitor cell markers KRT7, SOX9, and EPCAM was upregulated in NAFLD patients with advanced fibrosis. However, whether these progenitor cells are damaged by the lipotoxic environment and/or present limited cell plasticity is unknown. Our preclinical in vivo data showed attenuation of fibrosis in Ptpn1−/− mice with NASH in parallel with less presence of A6/SOX9 positive OCs and reduced Epcam and Krt19 expression. Nevertheless, the presence of LDs in OCs from Ptpn1−/− livers might be a feature of protection against lipotoxicity and/or other components of chronic liver damage. These results together with higher gene expression related to hepatocyte/cholangiocyte fate under lipotoxic injury in Ptpn1−/− OCs strongly suggest that LDs-containing OCs might have better capacity to repopulate the damaged liver. This relevant issue deserves future research.
In conclusion, this study has unraveled lipotoxicity as a trigger of apoptosis in OCs which is restrained by Ptpn1 deletion, an effect concurring with a metabolic rewiring towards a “starvation-like” cell fate favoring LD formation and autophagy. This regulation protects against FFA-mediated toxicity and ensures mitochondrial fitness. In this scenario, dynamic interactions of LDs with the lysosomes likely allow a lipid recycling probably by other cellular compartments and, overall, these adaptations avoid lipotoxic death. The fact that LDs were found in OCs from Ptpn1−/− mice with NASH opens new therapeutic perspective to preserve OCs proliferation under chronic liver damage, but favoring their plasticity.
Materials and methods
Human studies
Human subjects
The expression levels of hepatic progenitor cell-related genes were obtained from the National Cancer for Biotechnology Information Gene Expression Omnibus database based on the gene chips of fibrotic liver tissues associated to NAFLD after a search and selection of transcriptomic studies (GSE130970 [49], GSE49541 [50], GSE162694 [51] and GSE48452 [52]). We then selected those samples belonging to the experimental groups NAFLD patients with mild fibrosis (0–1 fibrosis stage) or advanced fibrosis (≥2 fibrosis stage). The differential expression of commonly upregulated genes (KRT7, SOX9, and EPCAM) was analyzed with GEO2R for GSE49541 and GSE48452. For GSE130970 and GSE162694 we obtained the normalized data directly from GEO.
Animal studies
Animal housing
Animal experiments were approved by the Animal Ethics Committees of the Spanish National Research Council and Comunidad de Madrid (Reference PROEX 007/19) in accordance with Spanish (RD 53/2013) and European Union (63/2010/EU) legislation. Wild-type (Ptpn1+/+) and PTP1B-deficient (Ptpn1−/−) mice were maintained on the same mixed genetic background (C57Bl/6J × 129Sv/J), in light/dark (12 h light/12 h dark), temperature (22 °C) and humidity-controlled rooms. Mice were fed a chow diet (A04, U8220G10R, SAFE, Panlab, France) ad libitum, and with free access to drinking water at the animal facilities at IIBm Alberto Sols (CSIC-UAM, Madrid) as previously described [16]. To induce NAFLD, 8-week-old male mice were fed a chow diet and normal tap water or a Western Diet (WD) containing 21.1% fat, 41% sucrose, and 1.25% cholesterol by weight (Teklad diets, TD 120528, Envigo, Spain) and a high sugar solution (23.1 g/L d-fructose (F0127, Sigma Aldrich, St. Lois, MO, USA) and 18.9 g/L d-glucose (G8270, Sigma Aldrich,) as decribed [32]. Additionally, CCl4 (289116, Sigma Aldrich) at the dose of 0.32 μg/g of body weight, was injected intra-peritoneally (i.p.) once per week, starting one week after the WD. Mice were euthanized at 12 weeks of intervention and tissues were removed, weighed, and frozen at −80 °C or fixed with 4 % paraformaldehyde (PFA) (16005, Sigma Aldrich) for further analysis. Body weight-matched animals were randomly allocated into the experimental groups. The number of mice included is indicated in the corresponding figure legend. Experimenters were not blind to the group allocation during data collection and/or data analysis.
Histopathological assessment and Sirius Red staining
Paraffin-embedded liver biopsy sections (5 μm) were stained with Hematoxylin & Eosin and Sirius Red and evaluated by a single-blinded hepatopathologist. All liver sections measured >1.5 cm in length and showed more than 10 complete portal tracts. The percentage of hepatocytes containing lipid droplets was determined for each 10x field. An average percentage of steatosis was then determined for the entire specimen. Steatosis was assessed as outlined by Kleiner et al. [53] grading percentage involvement by steatotic hepatocytes as follows: grade 0, <5%; grade 1, 5–33%; grade 2, >33–66%; and grade 3, >66%. In addition, Brunt’s histological scoring system was used to evaluate the degree of hepatocellular ballooning and lobular inflammation (grade of activity) [54]. Also, the fibrosis stage was assessed using the system for mice models validated by Liang et al. [55], which defined fibrosis staging as 0, none; 1, perisinusoidal and/or pericentral; 2, incomplete central/central bridging fibrosis; 3, complete central/central bridging fibrosis.
Determination of serum transaminase levels
Serum ALT activity was determined using Reflotron strips (Cat. 10745138203, Roche Diagnostics, Barcelona, Spain), accordingly with the manufacturer’s instructions.
In vitro studies
Culture of OCs and treatments
The isolation of Ptpn1+/+ and Ptpn1−/− OCs from mice fed a 3,5-diethoxycarbonil-1,4-dihydrocollidine (DDC) diet for 14 days was previously described [16]. Once established, OCs were routinely maintained in DMEM supplemented with 100 U/mL penicillin, 100 µg/mL streptomycin, 10 mM HEPES and 10% fetal bovine serum (FBS) in a humidified incubator at 37 °C and a 5% CO2 atmosphere. Medium was replaced every 3 days, and cells were harvested at 80%-90% confluence using trypsin-EDTA and replated at 1:10 dilution. OCs were routinely tested for mycoplasma contamination and used until 12–14 passages. For experiments, OCs were treated with PA (P0500, Sigma Aldrich) or OA (O1008-1G, Sigma Aldrich) at 400 µM concentration in DMEM-2.5% FBS and 1% free fatty acid (FFA) bovine serum albumin (BSA) at different time points. In another set of experiments, OCs were treated with either sulforaphane (S8044, LKT Laboratories, St. Paul, MN, USA) (15 μM), CQ (C6628, Sigma Aldrich) (50 μM), lalistat 1 (23891, Cayman Chemical, Ann Arbor, MI, USA) (75 μM) or atglistatin (SML1075, Sigma Aldrich) (20 μM), for several time-periods in the absence or presence of PA.
Lentiviral silencing of Ptpn1 and Scd1 in OCs
Stable oval cell lines silenced for Ptpn1 gene, encoding PTP1B protein, or Scd1 gene, encoding SCD1 were generated by transduction of Ptpn1+/+ or Ptpn1−/− OCs, respectively, with a battery of lentiviral particles (TRCN0000218522, TRCN0000226109, TRCN0000226108, TRCN0000226107, and TRCN0000226106; Mission Ptpn1 shRNA Transduction Particles, Sigma Aldrich) for Ptpn1 silencing or lentiviral particles generated from Mission Scd1 shRNA Bacterial Glycerol Stock (TRCN0000337878 and TRCN0000114331, Sigma Aldrich) following the manufacturer’s protocol. OCs transduced with scrambled (Scr) shRNA lentiviral particles (SHC001V, Sigma Aldrich) were used as a control. Transduced cells were selected with puromycin (10 μg/mL) for 2 weeks. Transduction was confirmed by measuring PTP1B protein levels and Scd1 expression, respectively.
Culture of primary mouse hepatocytes
Primary mouse hepatocytes were isolated from non-fasting male Ptpn1+/+ and Ptpn1−/− mice by a two-step collagenase perfusion as described [56]. Briefly, cells were seeded on collagen-coated 12-well plates and cultured at a density of 35 ×104 cells/well in DMEM and Ham’s F-12 medium (1:1) supplemented with 10% FBS, 2 mM glutamine, 100 U/mL penicillin, 100 μg/mL streptomycin, 20 mM HEPES and 1 mM sodium pyruvate (attachment medium) and maintained under humidity conditions in 95% air and 5% CO2 at 37 °C for 24 h before treatment with PA at 400 µM concentration in 5.5 mM glucose DMEM-2.5% FBS and 1% BSA (FFA).
Methods details
Analysis of protein expression by western blotting
To obtain total cellular extracts, the cells were lysed in lysis buffer (10 mM Tris, pH 7.5, 5 mM EDTA, 50 mM Tris-HCl, 30 μM sodium pyrophosphate, 50 mM NaF, 100 mM o-vanadate sodium, 1% Triton X100, 1 mM PMSF, and 10 μg/mL protease inhibitors (P8340, Sigma Aldrich) at pH 7.4–7.6. The cells were then centrifuged twice at 11,200 × g for 10 min at 4 °C and the supernatants were collected. Nuclear and cytosolic protein extracts were prepared as previously described [15].
Protein levels were quantified using the Bio-Rad Protein Assay Dye Reagent (500-0006, Bio-Rad Laboratories, Hercules, CA, USA). Protein extracts (30–70 μg) were boiled at 95 °C for 5 min in loading buffer (100 mM Tris, pH 6.8, 10% glycerol, 4% sodium dodecyl sulfate (SDS), 0.2% bromophenol blue, and 2 mM β-mercaptoethanol), subjected to 8–15% SDS polyacrylamide gel electrophoresis (SDS-PAGE), and then transferred to PVDF membranes (Immobilon-P IPVH00010, Merck, Darmstadt, Germany). Once transferred, Ponceau solution (0.1% Ponceau S (P3504, Sigma Aldrich) and 5% acetic acid in water) was added to the membranes to visualize the protein loading. After washing with Tris-buffered saline-0.05% Tween-20 (T1379, Sigma Aldrich) (TBS-T), the membranes were blocked with 5% skim milk powder in TBS-T for 2 h at room temperature (RT) and incubated with primary antibodies at the indicated dilutions (Table S1) at 4 °C with waving overnight (o/n). After 4–5 washes with TBS-T, the membranes were incubated with the corresponding secondary antibody (Table S1) for 1 h at RT. Membranes were developed with chemiluminescent substrate (Clarity Western ECL Substrate, 170-5060 Bio-Rad Laboratories) and different exposure times were performed for each primary antibody with radiographic films in a radiology cassette (Medical X-ray film blue, EWPJH AGFA, Mortsel, Belgium) or developed in a ChemiDoc imager (733BR-3548, Bio-Rad Laboratories). Blots were normalized using antibodies against housekeeping proteins. Values of densitometry were determined using Image J or Image Lab (Bio-Rad Laboratories).
RNA library construction and sequencing
RNA was extracted using the RNeasy Mini kit (QIAGEN 217004, Hilden, Germany) following the manufacturer’s instructions and assessed for quality. The highest-quality RNA samples were further anonymized and sent to the Centro Nacional de Análisis Genómico (CNAG-CRG, Barcelona, Spain) for analysis by RNA sequencing (RNA-Seq). The starting input material for the construction of the RNA library was DNA-free total RNA from FFPE samples, obtained using the KAPA RNA HyperPrep Kit with RiboErase (HMR; Kapa Biosystems, Wilmington, MA, USA), according to the manufacturer’s protocol. Briefly, the ribosomal RNA (rRNA) was depleted from 0.8 to 1 μg of total RNA using RiboErase. rRNA-depleted RNA samples were fragmented in the presence of divalent ions at 65 °C for 1 min due to the low quality of the initial total RNA. Following RNA fragmentation (first- and second-strand synthesis), the Illumina-compatible bar-coded adapters were ligated at a concentration of 1.5 μM. Libraries were enriched with 15 cycles of PCR using KAPA HiFi HotStart ReadyMix (Kapa Biosystems). The size and quality of the libraries were assessed in a High Sensitivity DNA Bioanalyzer Assay (Agilent Technologies, Santa Clara, CA, USA). The libraries were sequenced on HiSeq2000 (Illumina, San Diego, CA, USA) in paired-end mode with a read length of 2 × 76 bp using TruSeq SBS Kit v4. Each sample was sequenced in a fraction of a sequencing v4 flow cell lane, following the manufacturer’s protocol. Sequencing was performed with a minimum of 65 mol/L paired-end reads (2 × 75 bp) per RNA-Seq sample. Image analysis, base calling, and quality scoring of the run were processed using real-time analysis (RTA 1.18.66.3) software and followed by the generation of FASTQ sequence files by CASAVA (RRID:SCR_001802).
RNA-Seq processing and data analysis
RNA-Seq paired-end reads were mapped against the mouse reference genome using STAR (RRID:SCR_015899, version 2.5.2a) with ENCODE parameters for long RNA. Genes were quantified using RSEM (RRID:SCR_013027, version 1.2.2.8) and GENCODE (RRID:SCR_014966, version 24). Mapping quality metrics were calculated with GEMtools (http://gemtools.github.io/) and RSeQC (RRID:SCR_005275). Normalization and transformation of the expression data for plotting and analysis were done with the variance stabilizing transformation function of DESeq2 (RRID:SCR_015687). Only the protein-coding genes from the normalized count matrix were selected for further analysis. After low-expressed genes were filtered out (at least 1 count in 70% of samples) and genes with duplicated gene symbol were averaged by the median of their expression. Finally, counts were log2 transformed for Limma analysis. Exploratory analysis. Boxplots, unsupervised clustering, and principal component analysis (PCA) of the processed data were used to detect patterns of expression between samples. Differential expression analysis was performed using the Limma package [57]. The p values were corrected with the Benjamini & Hochberg method [58], considering significant those genes with an adjusted p value lower than 0.05. Functional enrichment analysis was carried out with the gene set analysis (GSA) method implemented in the mdgsa package [59]. As functional annotations, the propagated GO terms of the three ontologies were used [60], as well as, the KEGG [61] and Reactome pathways [62]. Those gene sets with less than 10 genes of more than 500 were filtered out. Functions and pathways with an adjusted p value lower than 0.05 were considered significant. Also, the Benjamini & Hochberg method were used in this analysis. RNA-Seq data have been deposited on Gene Expression Omnibus (GEO) database (Accession ID: GSE199644).
Real-time quantitative PCR (RT-qPCR)
Total RNA was extracted with Trizol® reagent (15596026, Thermo Fisher Scientific, Walthmam, MA, USA) and reverse transcription performed using a High-Capacity cDNA Reverse Transcription Kit (4368813, Thermo Fisher Scientific) using random primers and Superscript III enzyme (18080093, Thermo Fisher Scientific) according to the manufacturer’s instructions. RT-qPCR was performed in a 7900 HT-Fast real-time PCR (Thermo Fisher Scientific) with TaqMan Universal PCR Master Mix. RT-qPCR was carried out at the at IIBm Alberto Sols Genomics Unit using 5 ng of cDNA. Probes and primers used are listed in Table S2. The relative changes in gene expression were calculated using the 2−ΔΔCt (cycle threshold) quantification method normalized to the expression levels of the Actb and Rplp0 genes.
Hematoxylin and eosin (HE) staining of liver sections
Liver tissue was fixed in 4% PFA for 24 h, washed twice with PBS and dehydrated with ascending ethanol solutions, incubated with xylene, and then embedded in paraffin. Blocks were cut in 5 µm sections. Prior to HE staining, sections were deparaffinized in xylene and hydrated in descending ethanol solutions and distilled water. The slides were stained with Mayer’s hematoxylin (MHS32–1L, Sigma Aldrich) for 15–20 min and eosin (1.15935.0025, Merck) for 1 min. After dried and mounted, images were captured with an Axiophot light microscope (Zeiss, Oberkochen, Germany) using a 20× objective.
Immunofluorescence analysis in livers and OCs
For immunofluorescence, fresh frozen liver sections (6 μm, mounted in Tissue-Tek OCT Compound, Sakura Finetek) or OCs were fixed using 4% PFA, washed, and permeabilized with 0.5% Triton X100 before blocking (1% BSA, 5% horse serum, 0.2% Triton X100). Samples were incubated o/n with primary antibodies (Table S1) at 4 °C. After washing, Alexa-conjugated secondary antibodies (Table S1) were applied for 1 h at RT with DAPI (5 μg/mL) prior to mounting. Confocal images were obtained using a Leica TCS-SP6 (Leica, Wetzlar, Germany) or Zeiss LSM710 confocal microscopes and analyzed using ZEN Blue and ImageJ softwares.
Quantification of hydroxyproline in liver samples
To quantify collagen, hepatic hydroxyproline content was assayed using a hydroxyproline kit according to the manufacturer’s instructions (MAK008, Hydroxyproline Assay Kit, Sigma Aldrich).
In vivo staining of OCs and analysis by confocal microscopy
Cells were cultured on µ-slide 8-well IbiTreat plates (80826, Ibidi, Gräfelfing, Germany). After treatments, OCs were washed with PBS and marked with the following probes: 10 μM dyhidroethidium (DHE, 104821–25–2, Sigma Aldrich), 1 μM Lysotracker (L7528, Thermo Fisher Scientific), 5 μM Bodipy (D3835 and D3922, Thermo Fisher Scientific), 100 nM MitoTracker Green (MTG, M7514, Thermo Fisher Scientific), 100 nM MitoTracker Red CM-H2XRos (M7512, Thermo Fisher Scientific), 1 μM ER-Tracker (34250, Thermo Fisher Scientific), for 30 min at 37 °C in darkness. Then, cells were analyzed using a Zeiss LSM780 confocal microscope and ZEN Blue software.
MitoTracker Green FM and MitoTracker Red CM-H2XRos flow cytometry
Cells were incubated with 1 mM MitoTracker Green FM or 100 nM MitoTracker Red CM-H2XROS for 30 min at 37 °C in darkness. After washing with PBS, the cells were trypsinized, neutralized, and centrifuged at 238 x g. The pellet was resuspended in PBS and the relative fluorescence was determined by flow cytometry (FACSCanto, Becton Dickinson) and analyzed with the FACS Diva software (Becton Dickinson).
Mitochondrial oxidative damage estimation
After 8 h of PA (400 µM) treatment, total RNA was extracted with Trizol® reagent and reverse transcription was performed as we described above. The amplification reaction of the long fragment of mtDNA was carried out using the primers HVII-FOR260 and HVII-C339 and for the short fragment the primers used were HVII-FOR260 and HVII-L421 (Table S2). The absolute value of the oxidative damage of the mtDNA was calculated as the ratio between the copies of the long fragment and those of the short fragment.
Analysis of neutral lipid content with Nile Red
Cells were collected using trypsin-EDTA and washed with ice-cold PBS. Afterward, the cells were centrifuged at 13400 × g for 5 min at 4°C. PBS was discarded, and the pellets were resuspended in 1 mL 4% PFA and incubated on ice for 1 h. Then, cells were centrifuged at 13,400 × g for 5 min, PFA was removed, and the cells were resuspended into 300 μL of Nile Red (0.4 µg/µL) (3013, Sigma Aldrich). The samples were analyzed by flow cytometry.
Isolation of lipid droplets (LDs)
LD isolation was conducted following the procedure previously described by Schott et al. [21]. Briefly, OCs were treated with PA (400 µM) for 16 h. Cells were washed, scraped, and incubated in a hypotonic lysis medium (HLM) before Dounce homogenization. Then the post-nuclear supernatant (PNS) was placed at the bottom of a 25% to 0% OptiPrep density gradient (D1556; Sigma Aldrich) in HLM. Following centrifugation for 30 min spin at 36,000 × g, the floating fat layer (LD1) was collected and washed. After that, 400 µL of HLM were carefully added to the top of the gradient and samples were centrifuged for an additional 60 min at 182,000 × g. The LD2 fraction was then collected from the top (200–400 µL). LD1 and LD2 samples were loaded onto SDS-PAGE gels.
Live-cell imaging
Cells were cultured on µ-slide 8-well IbiTreat plates (80826, Ibidi). To analyze ER-LD interactions, Ptpn1−/− OCs were treated 4 h with PA and then stained with 1 μM ER-Tracker and 5 μM Bodipy for 30 min at 37 °C. To measure lysosome-LD interactions, Ptpn1−/− OCs were treated with PA for 16 h and then stained with 1 μM Lysotracker and 5 μM Bodipy for 30 min at 37 °C. After the incubations, cells were washed twice with PBS and placed into regular culture media during measurements. Intensity of the Lysotracker channel within the thresholder lysosomal area was then calculated. In another experiments, Ptpn1−/− OCs were transfected to transiently express mRFP1-EGFP-ADRP/PLIN2, a dual-fluorescent variant of ADRP/PLIN2 that exhibits differential fluorescence dependent upon the pH of its surrounding environment, as described [22]. Briefly, cells were transfected with 5 µg of DNA with Lipofectamine 2000 (52887, Thermo Fisher Scientific). Sixteen h after transfection, lysosomes were loaded with Dextran-A647 at 250 µg/ ml (D22914, Thermo Fisher Scientific) following a loading period of 4 h and a chase period of 16 h in the absence or presence of PA. Imaging was performed on a Zeiss LSM710 confocal microscope using a 63× oil 1.4 numerical aperture (NA) objective lens with frames acquired approximately every 3 sec for a minimum of 5 min. Raw.czi files were imported into Fiji v.1.52 for downstream analysis. A Gaussian blur was applied to remove background noise. To visualize the LD contact with the lysosome a kymograph analysis was performed using a kymograph plugin.
Transmission electron microscopy (TEM)
Cells were fixed in glutaraldehyde (2%) plus 4% PFA for 60 min. Samples were then post-fixed in 1% osmium tetroxide for 60 min at 25 °C, stained with uranyl acetate (5 mg/mL) for 1 h at 25 °C, dehydrated in acetone and embedded in Epon 812 (EMbed 812; Electron Microscopy Science, Hatfield, PA, USA). Ultrathin sections, unstained or post-stained with uranyl acetate and lead hydroxide, were examined under a Morgagni 268D TE microscope (FEI, Hillsboro, OR, USA) equipped with a Mega View II charge-coupled device camera (SIS, Soft Imaging System GmbH, Munster, Germany) and analyzed with AnalySIS software (SIS).
Fatty acid uptake assay
Confluent Ptpn1+/+ and Ptpn1−/− OCs were washed with PBS and incubated for 50 min in Krebs-Ringer buffer without glucose, containing 125 mM NaCl, 5 mM KCl, 2 mM CaCl2, 1.25 mM KH2PO4, 1.25 mM MgSO4.7H2O, 25 mM NaHCO3 pH 7.4, 3% BSA (FFA), and saturated oxygen. At the end of the incubation, 1 mM (final concentration) l-carnitine (Sigma Aldrich), 80 μM PA (Sigma-Aldrich), and 20 μM 14C 2-Bromopalmitate (58 μCi/mmol, PerkinElmer, Waltham, MA) were added for a further 10 min. Then, cells were washed with PBS and scraped in cold buffer (0.25 M sucrose, 10 mM Tris-HCl, 1 mM EDTA, and 1 mM dithiothreitol, pH 7.4). After 2 cycles of freezing/thawing in liquid nitrogen, radioactivity was counted in a Wallac 1409 counter (EG&G Company, Bedford, MA, USA). Results were normalized to total protein content of cell extracts and expressed as nmol/mg protein.
Fatty acid oxidation assay
Palmitate oxidation to acid-soluble products (ASPs), essentially acyl-carnitine, Krebs cycle intermediates, acetyl-CoA and ketone bodies, was measured in OCs grown in 12-well plates in presence or absence of 400 µM of PA for 4 h. The day of the assay, cells were washed in KRB-HEPES buffer (KRBH buffer: 135 mM NaCl, 3.6 mM KCl, 0.5 mM NaH2PO4, 0.5 mM MgSO4, 1.5 mM CaCl2, 2 mM NaHCO3, and 10 mM HEPES, pH 7.4) supplemented with 0.1% BSA, preincubated at 37 °C for 30 min in KRBH-1% BSA and washed again in KRBH-0.1% BSA. Cells were then incubated for 3 h at 37 °C with fresh KRBH containing 25 mM glucose and 8 mM carnitine (C0283, Sigma Aldrich), 2.5 mM palmitate and 1 μCi/mL [1–14C] palmitate (CFA23, GE Healthcare, Chicago, IL, USA) bound to BSA. Oxidation measurements were performed as previously described [63] with minor modifications by trapping the radioactive CO2 in a 1 cm2 Whatman paper soaked with 40 µL 0.1 N KOH using parafilm-sealed system. The reaction was stopped by the addition of 40% perchloric acid through a syringe that pierced the parafilm. The following day, the system was opened, and the papers were left to dry out, before transferring them into tubes with 5 mL scintillation liquid (6013159, PerkinElmer). Then, an amount of 300 µl of the reaction mix with perchloric acid was removed and centrifuged at maximum speed for 10 min. 150 µl of the supernatant, which contained the ASPs, were transferred into a tube containing 5 mL scintillation liquid. After 2 h of stabilization, samples were analyzed and data for CO2 or ASPs are expressed as: nmol palmitate mg−1 prot* h−1 as previously described.
Measurement of the cellular bioenergetic profile in OCs
For analysis of oxygen consumption rate (OCR), OCs were seeded in customized Seahorse 24-well plates and then treated with 400 µM PA and/or 50 µM CQ for 4 h. Before any measurement, cells were incubated for 1 h with XF Assay Medium (Seahorse Bioscience, North Billerica, MA, USA) plus 5 mM glucose. We followed the protocol and injection strategies of the Mito Stress Assay (Seahorse Bioscience). The concentration of the inhibitors was: 2 µM oligomycin (75351, Sigma Aldrich), 1 µM uncoupler FCCP (C2920, Sigma Aldrich) and a mix of 1 µM rotenone (R8875) and 1 µM antimycin-A (A8674, Sigma Aldrich). OCR was calculated by plotting the O2 tension of media as a function of time (pmol/min) and data were normalized by the protein concentration measured in each individual well by the BCA method. Calculations were performed as previously described [64] using the Agilent Seahorse Wave desktop software.
Lipidomic analysis
Lipidomic analysis was carried out as previously described with slight modifications [65, 66]. Briefly, cells are washed twice with PBS, and a cell extract corresponding to 107 cells was scraped in ice-cold water and sonicated in a tip homogenizer twice for 15 s and, before extraction and separation of lipid classes, internal standards are added as follows: total phospholipids, 10 nmol of 1,2-diheptadecanoyl-sn-glycero-3-phosphocholine; TAG, 10 nmol of 1,2,3-triheptadecanoylglycerol; DAG, 10 nmol of 1,2-diheptadecanoylglycerol; FFA, 15 nmol of heptadecanoic acid; CE, 10 nmol of cholesteryl erucate; phospholipid classes: 10 nmol of 1,2-dipentadecanoyl-sn-glycero-3-phosphocholine (PC), 10 nmol of 1,2-diheptadecanoyl-sn-glycero-3-phosphoethanolamine (PE), 10 nmol of 1,2-ditetradecanoyl-sn-glycero-3-phosphoinositol (PI) and 10 nmol of 1,2-ditetradecanoyl-sn-glycero-3-phosphoserine (PS). Total lipids are extracted according to the method of Bligh and Dyer [67] and the lipids are separated by thin-layer chromatography, with n-hexane/diethyl ether/acetic acid (70:30:1 [v/v/v]) as the mobile phase for separation of neutral lipids. For analysis of phospholipid subclasses, the plates are pre-developed with chloroform/methanol (1:1, v/v) and, after being air-dried, they are impregnated with 1.5% boric acid in ethanol/water (1:1, v/v). Phospholipids are separated twice with chloroform/methanol/28% ammonium hydroxide (60:37.5:4, v/v/v) as the mobile phase. Spots corresponding to the various lipid classes are extracted from the silica with 1 mL of chloroform/methanol (1:1 [v/v]) followed by 1 mL of chloroform/methanol (2:1 [v/v]). Glycerolipids and CE are transmethylated with 500 mL of 0.5 M KOH in methanol for 30 min at 37 °C. 500 mL of 0.5 M HCl will be added to neutralize. Afterward, samples are neutralized with 450 mL of KOH 0.14 M and 500 mL of water. Extraction of FAMEs is carried out with 1 mL of n-hexane twice. Hexane extracts are subjected to GC/MS analysis using an Agilent 7890 A gas chromatograph coupled to an Agilent 5975 C mass-selective dPBSetector operated in electron impact mode (EI, 70 eV), equipped with an Agilent 7693 autosampler and an Agilent DB23 column (60 m length × 0.25 mm internal diameter × 0.15 mm film thickness). Data analysis was carried out with the Agilent G1701EA MSD Productivity Chemstation software, revision E.02.00.
Cell cycle analysis by propidium iodide staining
Cells were collected using trypsin-EDTA and washed with ice-cold PBS. Afterward, cells were centrifuged at 500 × g for 5 min at 4 °C. PBS was removed, and the pellet was resuspended in 100 μL of ice-cold PBS. Then, 1 mL of 70% ethanol was added dropwise and the cells were put on ice for at least 30 min. Then, the cells were centrifuged at 500 × g for 5 min at 4 °C and washed with 500 mL ice-cold PBS to remove ethanol and centrifuged at 500 × g for 5 min at 4 °C. Finally, cells were resuspended in 500 μL of 0.1 mg/mL propidium iodide solution (4087, Cell Signaling Technology, Danvers, MA, USA) and incubated during 30 min at 37 °C protected from light. Flow cytometry data were acquired with a FACSCanto II (BD Biosciences, Madrid, Spain) and data analysis was performed using Cytomics FC500 with the CXP program.
Measurement of ATP levels
Cells were lysed in cold Milli-Q water, heated to 80 °C for 5 min, and centrifuged at 134 × g for 5 min. ATP levels were detected using a bioluminescence kit (Cat. LBR-PO10, ATP determination kit, PRO) following manufacturer’s instructions (Biaffin GMbH & Co. KG, Kassel, Germany). The chemiluminescence generated was detected at a 560 nm wavelength in a luminometer (GloMax 96 Microplate Luminometer, 9101-002, Promega, Madison, WI, USA). A curve with increasing concentrations of ATP was used to calculate the amount of ATP in each sample.
Measurement of Malondialdehyde (MDA)
MDA levels in OCs were determined using a colorimetric commercial kit (108383, Sigma Aldrich) according to manufacturer´s instructions with slight modifications. Briefly, after treatments, cells were washed with PBS and scraped in 0.15 M KCl solution. The homogenate was centrifuged at 1000 × g at RT for 5 min and the supernatant was kept. After, the samples were vortexed and heated at 95 °C for 1 h and cooled during 30 min. Samples were then centrifuged at 1000 × g for 5 min at RT and MDA content was determined in the supernatants.
Statistical analysis
Data are presented as mean ± SEM. Sample size is indicated in each figure legend. Normal distribution was assessed by two different tests, Shapiro–Wilk and Kolmogorov–Smirnov tests. Groups with similar variance were statistically compared. Data were compared by using one-way ANOVA, two-way ANOVA followed by a Bonferroni post hoc test, or Student’s t test. The p values presented in figures and tables corresponded to post hoc test. All statistical analyses were performed using the GraphPad Prism 5.0 software (GraphPad Software Inc., San Diego, CA, USA) and SPSS 15.0 software (SPSS, Inc., Chicago, IL, USA). Differences were considered statistically significant at p value < 0.05.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding authors under reasonable request.
References
Younossi ZM, Koenig AB, Abdelatif D, Fazel Y, Henry L, Wymer M. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology. 2016;64:73–84.
Feldstein AE, Canbay A, Angulo P, Taniai M, Burgart LJ, Lindor KD, et al. Hepatocyte apoptosis and fas expression are prominent features of human nonalcoholic steatohepatitis. Gastroenterology. 2003;125:437–43.
Gonzalez-Rodriguez A, Mayoral R, Agra N, Valdecantos MP, Pardo V, Miquilena-Colina ME, et al. Impaired autophagic flux is associated with increased endoplasmic reticulum stress during the development of NAFLD. Cell Death Dis. 2014;5:e1179.
Listenberger LL, Han X, Lewis SE, Cases S, Farese RV Jr., Ory DS, et al. Triglyceride accumulation protects against fatty acid-induced lipotoxicity. Proc Natl Acad Sci USA. 2003;100:3077–82.
Miyazaki M, Kim YC, Gray-Keller MP, Attie AD, Ntambi JM. The biosynthesis of hepatic cholesterol esters and triglycerides is impaired in mice with a disruption of the gene for stearoyl-CoA desaturase 1. J Biol Chem. 2000;275:30132–8.
Olzmann JA, Carvalho P. Dynamics and functions of lipid droplets. Nat Rev Mol Cell Biol. 2019;20:137–55.
Akazawa Y, Cazanave S, Mott JL, Elmi N, Bronk SF, Kohno S, et al. Palmitoleate attenuates palmitate-induced Bim and PUMA up-regulation and hepatocyte lipoapoptosis. J Hepatol. 2010;52:586–93.
Richardson MM, Jonsson JR, Powell EE, Brunt EM, Neuschwander-Tetri BA, Bhathal PS, et al. Progressive fibrosis in nonalcoholic steatohepatitis: association with altered regeneration and a ductular reaction. Gastroenterology. 2007;133:80–90.
Nobili V, Carpino G, Alisi A, Franchitto A, Alpini G, De Vito R, et al. Hepatic progenitor cells activation, fibrosis, and adipokines production in pediatric nonalcoholic fatty liver disease. Hepatology. 2012;56:2142–53.
Seely BL, Staubs PA, Reichart DR, Berhanu P, Milarski KL, Saltiel AR, et al. Protein tyrosine phosphatase 1B interacts with the activated insulin receptor. Diabetes. 1996;45:1379–85.
Haj FG, Markova B, Klaman LD, Bohmer FD, Neel BG. Regulation of receptor tyrosine kinase signaling by protein tyrosine phosphatase-1B. J Biol Chem. 2003;278:739–44.
Sangwan V, Paliouras GN, Abella JV, Dube N, Monast A, Tremblay ML, et al. Regulation of the Met receptor-tyrosine kinase by the protein-tyrosine phosphatase 1B and T-cell phosphatase. J Biol Chem. 2008;283:34374–83.
Fausto N, Campbell JS, Riehle KJ. Liver regeneration. Hepatology. 2006;43:S45–53.
Revuelta-Cervantes J, Mayoral R, Miranda S, Gonzalez-Rodriguez A, Fernandez M, Martin-Sanz P, et al. Protein Tyrosine Phosphatase 1B (PTP1B) deficiency accelerates hepatic regeneration in mice. Am J Pathol. 2011;178:1591–604.
Mobasher MA, Gonzalez-Rodriguez A, Santamaria B, Ramos S, Martin MA, Goya L, et al. Protein tyrosine phosphatase 1B modulates GSK3beta/Nrf2 and IGFIR signaling pathways in acetaminophen-induced hepatotoxicity. Cell Death Dis. 2013;4:e626.
Gonzalez-Rodriguez A, Valdecantos MP, Rada P, Addante A, Barahona I, Rey E, et al. Dual role of protein tyrosine phosphatase 1B in the progression and reversion of non-alcoholic steatohepatitis. Mol Metab. 2018;7:132–46.
Spahis S, Delvin E, Borys JM, Levy E. Oxidative stress as a critical factor in nonalcoholic fatty liver disease pathogenesis. Antioxid Redox Signal. 2017;26:519–41.
Liu P, Wang W, Tang J, Bowater RP, Bao Y. Antioxidant effects of sulforaphane in human HepG2 cells and immortalised hepatocytes. Food Chem Toxicol. 2019;128:129–36.
Neufeld TP. TOR-dependent control of autophagy: biting the hand that feeds. Curr Opin Cell Biol. 2010;22:157–68.
Yang L, Li P, Fu S, Calay ES, Hotamisligil GS. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab. 2010;11:467–78.
Schott MB, Weller SG, Schulze RJ, Krueger EW, Drizyte-Miller K, Casey CA, et al. Lipid droplet size directs lipolysis and lipophagy catabolism in hepatocytes. J Cell Biol. 2019;218:3320–35.
Schulze RJ, Krueger EW, Weller SG, Johnson KM, Casey CA, Schott MB, et al. Direct lysosome-based autophagy of lipid droplets in hepatocytes. Proc Natl Acad Sci USA. 2020;117:32443–52.
Schrauwen P, Hesselink MK. Oxidative capacity, lipotoxicity, and mitochondrial damage in type 2 diabetes. Diabetes. 2004;53:1412–7.
Nguyen TB, Louie SM, Daniele JR, Tran Q, Dillin A, Zoncu R, et al. DGAT1-dependent lipid droplet biogenesis protects mitochondrial function during starvation-induced autophagy. Dev Cell. 2017;42:9–21 e25.
Rambold AS, Cohen S, Lippincott-Schwartz J. Fatty acid trafficking in starved cells: regulation by lipid droplet lipolysis, autophagy, and mitochondrial fusion dynamics. Dev Cell. 2015;32:678–92.
Hernandez-Alvarez MI, Sebastian D, Vives S, Ivanova S, Bartoccioni P, Kakimoto P, et al. Deficient endoplasmic reticulum-mitochondrial phosphatidylserine transfer causes liver disease. Cell. 2019;177:881–95 e817.
Wang J, Fang N, Xiong J, Du Y, Cao Y, Ji WK. An ESCRT-dependent step in fatty acid transfer from lipid droplets to mitochondria through VPS13D-TSG101 interactions. Nat Commun. 2021;12:1252.
Isono T, Chano T, Yonese J, Yuasa T. Therapeutic inhibition of mitochondrial function induces cell death in starvation-resistant renal cell carcinomas. Sci Rep. 2016;6:25669.
Gonzalez-Rodriguez A, Santamaria B, Mas-Gutierrez JA, Rada P, Fernandez-Millan E, Pardo V, et al. Resveratrol treatment restores peripheral insulin sensitivity in diabetic mice in a sirt1-independent manner. Mol Nutr Food Res. 2015;59:1431–42.
Sun C, Zhang F, Ge X, Yan T, Chen X, Shi X, et al. SIRT1 improves insulin sensitivity under insulin-resistant conditions by repressing PTP1B. Cell Metab. 2007;6:307–19.
Saito T, Kuma A, Sugiura Y, Ichimura Y, Obata M, Kitamura H, et al. Autophagy regulates lipid metabolism through selective turnover of NCoR1. Nat Commun. 2019;10:1567.
Tsuchida T, Lee YA, Fujiwara N, Ybanez M, Allen B, Martins S, et al. A simple diet- and chemical-induced murine NASH model with rapid progression of steatohepatitis, fibrosis and liver cancer. J Hepatol. 2018;69:385–95.
Roskams T, Yang SQ, Koteish A, Durnez A, DeVos R, Huang X, et al. Oxidative stress and oval cell accumulation in mice and humans with alcoholic and nonalcoholic fatty liver disease. Am J Pathol. 2003;163:1301–11.
Gambino R, Bugianesi E, Rosso C, Mezzabotta L, Pinach S, Alemanno N, et al. Different serum free fatty acid profiles in NAFLD subjects and healthy controls after oral fat load. Int J Mol Sci. 2016;17:479.
de Almeida IT, Cortez-Pinto H, Fidalgo G, Rodrigues D, Camilo ME. Plasma total and free fatty acids composition in human non-alcoholic steatohepatitis. Clin Nutr. 2002;21:219–23.
Malhi H, Bronk SF, Werneburg NW, Gores GJ. Free fatty acids induce JNK-dependent hepatocyte lipoapoptosis. J Biol Chem. 2006;281:12093–101.
Gonzalez-Rodriguez A, Escribano O, Alba J, Rondinone CM, Benito M, Valverde AM. Levels of protein tyrosine phosphatase 1B determine susceptibility to apoptosis in serum-deprived hepatocytes. J Cell Physiol. 2007;212:76–88.
Garcia-Ruiz I, Blanes Ruiz N, Rada P, Pardo V, Ruiz L, Blas-Garcia A, et al. Protein tyrosine phosphatase 1b deficiency protects against hepatic fibrosis by modulating nadph oxidases. Redox Biol. 2019;26:101263.
Hsu MF, Koike S, Mello A, Nagy LE, Haj FG. Hepatic protein-tyrosine phosphatase 1B disruption and pharmacological inhibition attenuate ethanol-induced oxidative stress and ameliorate alcoholic liver disease in mice. Redox Biol. 2020;36:101658.
Bourebaba L, Kornicka-Garbowska K, Al Naem M, Rocken M, Lyczko J, Marycz K. MSI-1436 improves EMS adipose derived progenitor stem cells in the course of adipogenic differentiation through modulation of ER stress, apoptosis, and oxidative stress. Stem Cell Res Ther. 2021;12:97.
Delibegovic M, Zimmer D, Kauffman C, Rak K, Hong EG, Cho YR, et al. Liver-specific deletion of protein-tyrosine phosphatase 1B (PTP1B) improves metabolic syndrome and attenuates diet-induced endoplasmic reticulum stress. Diabetes. 2009;58:590–9.
Jarc E, Petan T. Lipid droplets and the management of cellular stress. Yale J Biol Med. 2019;92:435–52.
Iwai T, Kume S, Chin-Kanasaki M, Kuwagata S, Araki H, Takeda N, et al. Stearoyl-CoA Desaturase-1 protects cells against lipotoxicity-mediated apoptosis in proximal tubular cells. Int J Mol Sci. 2016;17:1868.
Qiang L, Kon N, Zhao W, Jiang L, Knight CM, Welch C, et al. Hepatic SirT1-dependent gain of function of stearoyl-CoA desaturase-1 conveys dysmetabolic and tumor progression functions. Cell Rep. 2015;11:1797–808.
Lee JS, Zheng Z, Mendez R, Ha SW, Xie Y, Zhang K. Pharmacologic ER stress induces non-alcoholic steatohepatitis in an animal model. Toxicol Lett. 2012;211:29–38.
Bettaieb A, Matsuo K, Matsuo I, Wang S, Melhem R, Koromilas AE, et al. Protein tyrosine phosphatase 1B deficiency potentiates PERK/eIF2alpha signaling in brown adipocytes. PLoS ONE. 2012;7:e34412.
Jaishy B, Abel ED. Lipids, lysosomes, and autophagy. J Lipid Res. 2016;57:1619–35.
Cabodevilla AG, Sanchez-Caballero L, Nintou E, Boiadjieva VG, Picatoste F, Gubern A, et al. Cell survival during complete nutrient deprivation depends on lipid droplet-fueled beta-oxidation of fatty acids. J Biol Chem. 2013;288:27777–88.
Hoang SA, Oseini A, Feaver RE, Cole BK, Asgharpour A, Vincent R, et al. Gene expression predicts histological severity and reveals distinct molecular profiles of nonalcoholic fatty liver disease. Sci Rep. 2019;9:12541.
Murphy SK, Yang H, Moylan CA, Pang H, Dellinger A, Abdelmalek MF, et al. Relationship between methylome and transcriptome in patients with nonalcoholic fatty liver disease. Gastroenterology. 2013;145:1076–87.
Salloum S, Jeyarajan AJ, Kruger AJ, Holmes JA, Shao T, Sojoodi M, et al. Fatty acids activate the transcriptional coactivator YAP1 to promote liver fibrosis via p38 mitogen-activated protein kinase. Cell Mol Gastroenterol Hepatol. 2021;12:1297–310.
Ahrens M, Ammerpohl O, von Schonfels W, Kolarova J, Bens S, Itzel T, et al. DNA methylation analysis in nonalcoholic fatty liver disease suggests distinct disease-specific and remodeling signatures after bariatric surgery. Cell Metab. 2013;18:296–302.
Kleiner DE, Brunt EM, Van Natta M, Behling C, Contos MJ, Cummings OW, et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41:1313–21.
Brunt EM, Janney CG, Di Bisceglie AM, Neuschwander-Tetri BA, Bacon BR. Nonalcoholic steatohepatitis: a proposal for grading and staging the histological lesions. Am J Gastroenterol. 1999;94:2467–74.
Liang W, Menke AL, Driessen A, Koek GH, Lindeman JH, Stoop R, et al. Establishment of a general NAFLD scoring system for rodent models and comparison to human liver pathology. PLoS ONE. 2014;9:e115922.
Benveniste R, Danoff TM, Ilekis J, Craig HR. Epidermal growth factor receptor numbers in male and female mouse primary hepatocyte cultures. Cell Biochem Funct. 1988;6:231–5.
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47.
Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125:279–84.
Montaner D, Dopazo J. Multidimensional gene set analysis of genomic data. PLoS ONE. 2010;5:e10348.
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29.
Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 1999;27:29–34.
Jassal B, Matthews L, Viteri G, Gong C, Lorente P, Fabregat A, et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2020;48:D498–D503.
Herrero L, Rubi B, Sebastian D, Serra D, Asins G, Maechler P, et al. Alteration of the malonyl-CoA/carnitine palmitoyltransferase I interaction in the beta-cell impairs glucose-induced insulin secretion. Diabetes. 2005;54:462–71.
Calderon-Dominguez M, Alcala M, Sebastian D, Zorzano A, Viana M, Serra D, et al. Brown Adipose Tissue Bioenergetics: A New Methodological Approach. Adv Sci. 2017;4:1600274.
Astudillo AM, Meana C, Guijas C, Pereira L, Lebrero P, Balboa MA, et al. Occurrence and biological activity of palmitoleic acid isomers in phagocytic cells. J Lipid Res. 2018;59:237–49.
Guijas C, Meana C, Astudillo AM, Balboa MA, Balsinde J. Foamy monocytes are enriched in cis-7-hexadecenoic fatty acid (16:1n-9), a possible biomarker for early detection of cardiovascular disease. Cell Chem Biol. 2016;23:689–99.
Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–7.
Acknowledgements
We acknowledge R. Schulze (Mayo Clinic, Rochester, MN) for providing the mRFP1-EGFP-ADRP/PLIN2 reporter plasmid and advice with LD isolation. We also acknowledge Monica Belinchón, Lucía Guerrero (IIBm, CSIC), and Laura Molero (UAM) for assistance with microscopy and flow cytometry, respectively, and all members of AMV’s laboratory for helpful discussions.
Funding
This work was funded by grants RTI2018-094052-B-100 (MICINN/AEI/FEDER, EU), S2017/BMD-3684 (Comunidad de Madrid, Spain) and Fundación Ramón Areces (Spain) to AMV, grant PID2019-105989RB-I00 (MICINN/AEI/FEDER, EU) to JB, grants SAF2017-83813-C3-1-R (MICINN/AEI/FEDER, EU, cofunded by the ERDF) and CB06/03/0001 (CIBEROBN, ISCIII, Spain) to LH and DS, grants PID2019-106982RB-I00 (MICINN/AEI/FEDER, EU) and CB12/03/30002 (CIBERobn, ISCIII, Spain) to MJM-A, grant PI20/00837 (ISCIII/FEDER, Spain) to CG-M, grant PI19/00123 (ISCIII/FEDER, Spain) to AG-R. We acknowledge support from CIBERDEM (ISCII) to JB, AMV, and MPV (grant DEM21 PI01/2021). MCS-V is a recipient of the Ajut de Personal Investigador Predoctoral en Formació (APIF) fellowship from the University of Barcelona.
Author information
Authors and Affiliations
Contributions
AMV, MPV and IB designed the project; IB, PR, SC, LP, MCS-V, LML, FG-G, and MPV carried out the experiments; CG-M and AG-R provided help with the histophatological analysis of mouse livers; JB provided help with the lipidomic analysis; RG-R and FG-G provided help with RNA-Seq analysis; LH and DS provided help with the seahorse and fatty acid oxidation analysis; MJM-A provided help with the fatty acid uptake analysis; AMV, MPV, and IB wrote the paper with the input of all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
Procedures involving mice were performed under licence number PROEX 007/19 (Comunidad de Madrid, Spain). Experiments were conducted in accordance with Spanish (RD 53/2013) and European Union (63/2010/EU) legislation. No human samples were used in this study.
Consent to participate
The authors have agreed to the publication in Cell Death & Differentiation.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by M. Piacentini
Rights and permissions
About this article
Cite this article
Barahona, I., Rada, P., Calero-Pérez, S. et al. Ptpn1 deletion protects oval cells against lipoapoptosis by favoring lipid droplet formation and dynamics. Cell Death Differ 29, 2362–2380 (2022). https://doi.org/10.1038/s41418-022-01023-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-022-01023-x