Abstract
Primary ovarian insufficiency (POI) causes female infertility by abolishing normal ovarian function. Although its genetic etiology has been extensively investigated, most POI cases remain unexplained. Using whole-exome sequencing, we identified a homozygous variant in RAD51B –(c.92delT) in two sisters with POI. In vitro studies revealed that this variant leads to translation reinitiation at methionine 64. Here, we show that this is a pathogenic hypomorphic variant in a mouse model. Rad51bc.92delT/c.92delT mice exhibited meiotic DNA repair defects due to RAD51 and HSF2BP/BMRE1 accumulation in the chromosome axes leading to a reduction in the number of crossovers. Interestingly, the interaction of RAD51B-c.92delT with RAD51C and with its newly identified interactors RAD51 and HELQ was abrogated or diminished. Repair of mitomycin-C-induced chromosomal aberrations was impaired in RAD51B/Rad51b-c.92delT human and mouse somatic cells in vitro and in explanted mouse bone marrow cells. Accordingly, Rad51b-c.92delT variant reduced replication fork progression of patient-derived lymphoblastoid cell lines and pluripotent reprogramming efficiency of primary mouse embryonic fibroblasts. Finally, Rad51bc.92delT/c.92delT mice displayed increased incidence of pituitary gland hyperplasia. These results provide new mechanistic insights into the role of RAD51B not only in meiosis but in the maintenance of somatic genome stability.
Similar content being viewed by others
Introduction
Meiosis is a sexual division that halves the diploid somatic chromosomal complement to a haploid state. During this process, each chromosome associates with its corresponding homologue (pairing) whose ultimate physical hallmark is a chiasma. In mammals, this process relies on the repair of DNA double-strand breaks (DSBs) by homologous recombination (HR) that generates crossover recombination events (COs) [1].
In molecular terms, the nuclease SPO11 generates DSBs throughout the genome. After DNA resection of 5´termini [2], the generated 3′ssDNA ends recruit the recombinases RAD51 and DMC1. The resulting recombinase-coated ssDNA filaments can invade the homologous chromosome dsDNA, searching for a complementary sequence (chromosome pairing), which will foster genetic material exchange. The nucleation of RAD51 is mediated by its direct interaction with BRCA2. This supramolecular complex promotes the initial binding of RPA to the resected DNA and, importantly, fosters the displacement of RPA from ssDNA by RAD51 nucleofilaments [3, 4]. However, the presence of many paralogues of RAD51 in most taxa including vertebrates (RAD51B, RAD51C, XRCC3, XRCC2, RAD51D, and the meiotic factor DMC1) and the lethality of the constitutional mutant mice has hampered the study of the physiological role of each RAD51 paralogue to somatic and meiotic HR [5,6,7,8,9,10].
In mammals, functional analysis of the individual RAD51 paralogues in cell lines has shown similar but non-redundant contributions in DNA repair processes such as HR efficiency, RAD51 nuclear focus formation, sensitization to mitomycin C (MMC) and protection of perturbed replications forks [11]. Accordingly, mutations in genes on this pathway can lead to genome instability, which can result in both cancer- and infertility-associated genetic syndromes [12, 13].
Infertility is defined as a failure of a couple to conceive and affects 10–15% of couples [14]. POI is a disorder associated with female infertility that affects about 1% of women before the age of 40 years. The genetic etiology of isolated POI is highly heterogeneous. In fact, over 75 genes associated with POI have been implicated in ovarian development and meiosis [15]. Particularly, pathogenic variants in genes involved in meiotic recombination, such as STAG3, SYCE1, HFM1, MSH4, MSH5, DMC1, MEIOB, BRCA2 and HSF2BP, have been associated with this disorder [15,16,17,18,19,20,21,22,23,24]. Nevertheless, the presence of genetic defects in the majority of POI patients remains to be established [15].
Here, we report the identification of a homozygous variant in RAD51B- (c.92delT) in two sisters with POI. This RAD51B variant leads to a premature termination codon (PTC) resulting in translation reinitiation at codon AUG in position 64. The “humanized” knock-in (KI; Rad51bc.92delT/c.92delT) mouse shows meiotic DNA repair defects due to RAD51 and HSF2BP/BRME1 accumulation in the meiotic chromosome axes leading to a significant reduction in the number of COs. Interestingly, RAD51, RAD51C and HELQ interaction with RAD51B was diminished with the N-terminally truncated RAD51B protein. Moreover, RAD51B-c.92delT human lymphoblastoid cells, humanized MEFs and explanted bone marrow cells from mutant mice displayed an increased sensitivity to MMC-induced chromosome instability (CIN). The RAD51b-c.92delT variant was also found to decrease replication fork progression of lymphoblastoid cell lines and the reprogramming efficiency of primary mouse embryonic fibroblasts (MEFs) to produce induced pluripotent stem cells (iPSCs). Finally, Rad51bc.92delT/c.92delT mice exhibited an increased incidence of hyperplasia of the pituitary gland. These results provide new mechanistic insights into RAD51B role in human ovarian insufficiency and in the maintenance of genome stability in the soma.
Results
Genetic analysis
Whole-exome sequencing was performed in two sisters presenting with POI (II-1 and II-4) and in one unaffected sister (II-2). The mean coverage of the capture regions was ≥150× in all samples with at least 99.38% of the targeted positions being sequenced at ≥10× (Supplementary Table 1). Consistent with an autosomal recessive inheritance, homozygous candidate variants were selected in both affected sisters and a minor allele frequency cut-off of 0.01% was applied. After an in-depth assessment of coding (missense, nonsense and frameshift) and splice-site variants, a homozygous frameshift pathogenic variant in the RAD51B gene was identified in the two affected sisters and retained as the best candidate (Supplementary Table 2). RAD51B is located in chromosome 14, position 14q23-q24.2, and the c.92delT variant is located in exon 3. As predicted by Mutalyzer, the RAD51B-c.92delT variant creates a PTC at amino acid #39 (predicted alteration: p.Leu31Tyrfs*9). The variant c.92delT (i.e., 14-68292183-CT-C) is reported in gnomAD with an extremely low allele frequency of 4.05 × 10−6, which represents one allele count of 1 in 247,158 alleles and has not been reported to exist in a homozygous state [25, 26]. The presence of this variant was screened by Sanger sequencing in other members of this family (Fig. 1A). The father (I-1) and unaffected siblings (II-2, II-3, II-6, and II-8) were heterozygous. DNA of mother (I-2) and one brother (II-5) were unavailable (Fig. S1A). Furthermore, the RAD51B-c.92delT variant was not found in 235 fertile Brazilian women controls screened by Sanger sequencing. In order to rule out any additional pathogenic variants, we evaluated 107 genes already associated with POI [27]. No other clearly pathogenic variant, which could underlie the phenotype, could be identified in this family (Supplementary Table 9).
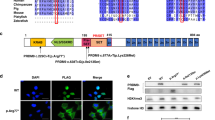
A Family Pedigree. A homozygous recessive variant in RAD51B was shown to be present in two sisters from Brazil affected with POI. The black arrow indicates proband (II-1). Pedigree numbers of individuals are indicated below the symbols. Samples sent for whole-exome sequencing (WES) are indicated by a hash and samples sent for Sanger sequencing are indicated by an asterisk. Sanger electropherograms confirmed the presence of the homozygous variant in both affected sisters II-1 and II-4. B HEK293T cells were transfected or not (Vehicle) with the different variants of RAD51B: the WT form (WT), the c.92delT form and both the WT and c.92delT forms in which the three secondary Methionines were mutated into Alanines (individual Met to Ala substituting the M39 to A39, the M55 to A55 and the M64 to A64; double Met to Ala substitutions following the above argument, both M39A39 and M55A55, both M39A39 and M64A64 and both M55A55 and M64A64. Of note, the bigger size of the RAD51B products is due to the presence of GFP tag (27 kDa). C COS7 cells were transfected to express human WT or mutant RAD51B fused to GFP tag. The WT construct showed a robust nuclear signal in addition to a faint cytoplasmic pattern. In contrast, the mutant variant displayed a partial delocalization of the nuclear signal to the cytoplasm. Quantification of the nucleus/cytoplasm signal rate is shown in the lower plot. Scale bars: 20 µm. D Fertility assessment in female mice of WT and mutant Rad51b. The plots show the number of pups per litter and the litters per month. Rad51bc.92delT/c.92delT variant is referred as Rad51bKI/KI for simplicity. Welch´s t-test analysis: ns, non-significant differences; ****p < 0.0001.
The c.92delT variant in RAD51B leads to translation re-initiation at AUG64
In silico analyses predict that the c.92delT variant would provoke the appearance of a PTC leading to a truncated protein of 39 residues lacking functional motifs (Fig. S1B) [28]. Given that the depletion of Rad51b in the mice is embryonic lethal [8] and that three close downstream in-frame AUG codons (AUG39, AUG55 and AUG64) exist, we hypothesized that the translation might restart at one of them. To test this, HEK293 cells were transfected with constructs encoding RAD51B: wild-type (WT), c.92delT and a mutant or WT RAD51B in which the three secondary Methionine codons were replaced by GCN encoding Alanines (individual and double Met to Ala substitutions) and protein expression was analyzed by Western blot (Fig. 1B). This experiment showed the production of a band in the c.92delT that was smaller than the main product resulting from the expression of the WT construct. This band disappeared only when replacing the codons underlying M64 to A64 but not when the M39 or M55 were replaced by Alanines (individually or double mutated; Fig. 1B). To demonstrate the translation of the RAD51B-c.92delT endogenous protein and given that we failed to detect by Western blot and immunofluorescence (IF) endogenous RAD51B protein with commercial or even home-made antibodies, we analyzed by IP-coupled with Mass spectrometry (MS) human lymphoblastoid cell extracts from RAD51BWT/c.92delT and RAD51Bc.92delT /c.92delT. Our results clearly show the presence of peptides corresponding to the C-Term of the protein, demonstrating that translation is indeed taking place (Supplementary Table 3). Altogether, these results reveal that the variant c.92delT leads translation re-initiation at M64. To assess the impact of the RAD51B-c.92delT variant on its subcellular localization, the human and murine RAD51B variant and the WT forms (fused to GFP and untagged) were expressed in Cos7 cells. RAD51B-c.92delT displayed lower ratio of nuclear to cytoplasmic labelling in comparison to the WT (Figs. 1C and S1C).
Rad51b c.92delT/c.92delT mice have no obvious ovarian and testis morphological abnormalities
In order to evaluate the physiological relevance of the RAD51B-c.92delT variant, we generated a humanized KI mouse Rad51bc.92delT/c.92delT by CRISPR/Cas9 editing (Fig. S2A). Although RAD51B is essential for mouse development [8], humanized KI mice showed no somatic phenotype and were born with the expected Mendelian ratios (Supplementary Table 4), which strongly suggests that the reinitiation of the translation is also operating in the mouse model. RT-PCR analysis using testis cDNA derived from Rad51bc.92delT/c.92delT mice confirmed transcription of the mutated and WT alleles (Fig. S2B).
Rad51bc.92delT/c.92delT male and female mice were able to reproduce. Accordingly, the histological analysis of adult Rad51bc.92delT/c.92delT testes revealed no apparent abnormalities in the seminiferous tubules (Fig. S2C). Ovaries from Rad51bc.92delT/c.92delT exhibited no differences in the number of follicles in comparison to WT mice (Fig. S2D). Haematoxylin-eosin staining patterns of mouse ovaries were similar at two time-points explored (5 dpp, in which primordial oocytes are expected to be found, and 3-months). The quantification of the primordial oocyte pool was also similar between genotypes (Fig. S2D). Fertility assessment in females also showed no statistical differences in litters per month and in pups per litter between mutant and WT mice, though a trend towards a reduction in the number of pups per litter is observed in the former (Fig. 1D).
Rad51b c.92delT/c.92delT mice show several defects in the DNA repair process
RAD51B is ubiquitously expressed (see www.uniprotorg) in somatic and reproductive tissues including meiocytes, though its meiotic function has not been established. To further characterize the involvement of RAD51B in meiosis, we analyzed Rad51bc.92delT/c.92delT meiocytes for the assembly of the synaptonemal complex by monitoring the distribution of SYCP1 and SYCP3. Even though, no defects in synapsis and desynapsis from leptotene to diakinesis were observed (Fig. S3A, B), a reduced number of pachynemas accompanied by an elevated number of diplonemas were found in both Rad51bc.92delT/c.92delT males and females. These results display subtle alteration of normal prophase I progression (Fig. S3C, D).
We assessed DSBs generation and repair by detecting phosphorylated histone γH2AX in Rad51bc.92delT/c.92delT meiocytes (Quantifications at Supplementary Table 5) [29, 30]. We observed similar labelling of γH2AX from leptotene to zygotene spermatocytes (Fig. S4A), but increased levels of labelling were detected in meiocytes at pachytene. This accumulation was also observed in oocytes at diplotene (Fig. 2A).
A Double immunolabelling of γH2AX (green) and SYCP3 (red) of spermatocyte and oocyte spreads from WT and KI mice showing the accumulation of γH2AX patches in the mutant pachynemas. Plots below the panels show the quantification of γH2AX intensity. B Double immunolabelling of RAD51 (green) and SYCP3 (red) of spermatocyte and oocyte spreads from Rad51WT/WT and Rad51bKI/KI. RAD51 foci accumulate at pachytene in KI spermatocytes and at diplotene in KI oocytes (small green dots). Plots below the panels represents the quantification of RAD51 foci on each genotype. Rad51bc.92delT/c.92delT variant is referred to as Rad51bKI/KI for simplicity. Welch´s t-test analysis: ns, non-significant differences; **p < 0.01; ***p < 0.001; ****p < 0.0001. Bar in panels, 10 μm. See Supplementary Table 5 for raw data quantification.
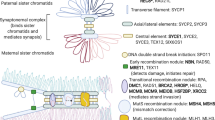
Meiotic DSB repair is orchestrated by a BRCA2-containing supramolecular complex that dictates the sequential recruitment of proteins to the DSBs, such as the binding of RPA for end resection, exchange of RPA by RAD51/DMC1 and loading of the complex MEIOB-SPATA22 to the RPA complexes with the interplay of HSF2BP and its interactor/stabilizer BRME1 (meiotic double-stranded break BRCA2/HSF2BP complex associated protein) [3, 4, 24, 31]. We first analyzed the initial loading of RPA. Our results showed that the recruitment of RPA and its associated protein SPATA22 was unaltered in mutant RAD51B meiocytes (Fig. S4B, C), suggesting that DSBs resection is unaffected. Then, we analyzed the recombinases RAD51 and DMC1 to assess the strand invasion process in our Rad51b mutant model. RAD51 foci formation was similar at leptotene and zygotene (Fig. S5A) but foci persisted in pachytene nuclei in spermatocytes and in diplotene nuclei in oocytes, indicating a slight defect in DSB repair (Fig. 2B). By contrast, labelling of DMC1 foci was not affected (Fig. S5B). This fact could reflect the activation of a somatic-like HR DNA repair pathway at late-pachytene involving RAD51 but not DMC1 [32], as described for other mouse mutants (i.e., HSF2BP and BRME1 [33]). We next analyzed by IF these essential meiotic recombination proteins in Rad51bc.92delT/c.92delT mice (Fig. S5C). BRME1 and HSF2BP labelling accumulated at late pachytene in Rad51bc.92delT/c.92delT mice meiocytes and persisted at diplotene (Fig. 3A). DNA repair can eventually culminate with the formation of COs; therefore, we analyzed the distribution of MLH1 foci as a direct measure of CO formation. A statistically significant difference in the number of CO events was found in Rad51bc.92delT/c.92delT mice compared to WTs in both spermatocytes (KI: 20.70 ± 1.79 vs WT: 22.98 ± 1.61) and oocytes (KI:22.39 ± 3.01 vs WT: 23.82 ± 2.02) (Fig. 3B). We also evaluated the functional relevance of this reduction of COs by measuring the number of bivalents without MLH1 foci that would lead to univalents. Our results revealed an increased number of bivalents that did not show MLH1 foci in both Rad51bc.92delT/c.92delT oocytes and spermatocytes (both sexual and autosomal bivalents) (Fig. 3B). Taken altogether, these results suggest that RAD51B variant provokes an increase of DSBs that are not effectively repaired as COs in both oocytes and spermatocytes.
A Double labelling of BRME1 (green) and SYCP3 (red) of spermatocyte and oocyte spreads from WT and KI mice showing the accumulation of BRME1 in mutant late pachynemas and diplonemas. Double labelling of HSF2BP (green) and SYCP3 (red) of meiocyte spreads from Rad51bWT/WT and Rad51bKI/KI mice showing the accumulation of HSF2BP in mutant late pachynemas and diplonemas. Plots on the right side of the panels represent the quantification of BRME1 and HSF2BP foci and intensity. B Double immunolabelling of MLH1 (green) and SYCP3 (red) of spermatocyte and oocyte spreads from Rad51bWT/WT and Rad51bKI/KI. MLH1 foci are significant reduced in mutant Rad51b meiocytes. The plots on the right of the panels represent the quantification of MLH1 foci at pachytene in both male and female meiocytes. Quantification of the % of spermatocytes with any autosome or the sexual bivalent without MLH1 foci. n = 108–114 (autosomes), n = 66–96 (X-Y bivalent). Quantification of the % of oocytes with any autosome without MLH1 foci. n = 51–54 (autosomes). Rad51bc.92delT/c.92delT variant is referred as Rad51bKI/KI for simplicity. Welch´s t-test analysis: ns, non-significant differences; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. Bar in panels, 10 μm. See Supplementary Table 5 for raw data quantification.
RAD51B interacts with the meiotic recombination machinery
We next immuno-precipitate RAD51B from mouse testis extracts coupled with MS. We identified in addition to RAD51B itself, RAD51C, a well-known interactor of RAD51B (Supplementary Table 6) [34], which is essential for meiotic recombination [35]. Next, we adopted a candidate approach to identify additional putative interactors of RAD51B. Thus, we co-transfected RAD51B with cDNAs encoding BRCA2, PALB2, DMC1, RAD51, MEIOB, BRME1, HSF2BP, HELQ, and RPA (Fig. S6) and also the MS-identified protein RAD51C. We detected positive co-immunoprecipitations between RAD51B and the paralogues, RAD51C, RAD51, HELQ and DMC1 (Fig. 4A–D).
HEK293T cells were co-transfected with either RAD51B-WT or RAD51B-c.92delT and A RAD51C-HA, B Flag-RAD51, C HA-HELQ or D Flag-DMC1. Protein complexes were immunoprecipitated with either an anti-RAD51B, anti-Flag, anti-HA or IgGs (negative control), and analyzed by immunoblotting with the indicated antibody. The red > indicates the band corresponding to RAD51B.
Given that the N-term part of RAD51B (residues 1–75) interacts with RAD51C [34], we analyzed the ability of mutant RAD51B to maintain its interaction with the identified partners. RAD51C immunoprecipitated WT RAD51B but not mutant RAD51B indicating that the c.92delT variant drastically diminishes or abrogates their interaction (Fig. 4A). This loss of interaction was also verified by Co-IP coupled with MS of RAD51Bc.92delT/c.92delT lymphoblastoid cells (Supplementary Table 3). Similarly, RAD51B-c.92delT interaction was weaker with RAD51 and with the helicase HELQ, but not with DMC1 (Fig. 4B–D). These results suggest that RAD51B acts in concert with DMC1/RAD51/RAD51C recombinases to promote/mediate meiotic recombination.
RAD51B-c.92delT human and mouse cells are sensitive to MMC-induced DNA damage
Typically, RAD51-deficient cells are hyper-sensitive to DNA interstrand crosslinking (ICL) agents such as MMC. Accordingly, Rad51bc.92delT/c.92delT MEFs showed a relatively lower growth rate in the presence of MMC (Fig. 5A). Clonogenic survival assays of MMC-treated MEFsc.92delT/c.92delT also showed a reduced number of colonies (Fig. 5B). We next monitored the efficiency of DNA repair. MEFs were treated with MMC and analyzed at different time points. The results of this experiment showed a delay in the disappearance of γH2AX foci even at 72 h after MMC treatment, indicating the presence of unrepaired DNA (Fig. 5C). In addition, MMC-treated MEFs from Rad51bc.92delT/c.92delT showed more chromosome break events (passage 2 and passage 4) (Fig. 6A). The assessment of the impact of the variant affected on genome stability in vivo showed that bone marrow-derived metaphase plates from Rad51bc.92delT/c.92delT MMC-treated mice had a significantly increased number of chromosomal breaks events per cell compared to WT mice (Fig. 6B).
A Cell proliferation assay of WT and mutant Rad51b primary MEFs at passage 2 (p + 2) incubated in presence of a continuous treatment with MMC. The results are expressed as a percentage relative to the control (not treated with MMC). Each point on the graph represents the mean ± SD. B Percentage of colonies obtained by clonogenic cell survival assays after treatment with MMC. The results are expressed as a percentage relative to the control (untreated) of Rad51bWT/WT and Rad51bKI/KI immortalized cells. C Representative γH2AX immunolabelling of WT and mutant Rad51b at 72 h. Quantification of γH2AX foci in Rad51b WT and mutant MEFs. Rad51bWT/WT and Rad51bKI/KI MEFs at p + 2 were incubated in presence of MMC at 1 µg/ml for 1 h and then supplemented with fresh medium without MMC. The quantification was performed at different time points: 0 h: 0 h, 6 h: 6 h, 24 h: 24 h, 48 h; 48 h and 72 h: 72 h. Cells were classified in 5 groups: 0 foci, 1 to 10 foci, 10 to 30 foci, 30 to 150 foci and >150 foci. n = 3. Rad51bc.92delT/c.92delT variant is referred as Rad51bKI/KI for simplicity. Welch´s t-test analysis: *p < 0.05; **p < 0.01. Bar in panel, 10 µm.
A Evaluation of metaphase chromosome breaks/gaps from Rad51WT/WT and Rad51bKI/KI MEFs after MMC treatment (150 nM). Lower panel (graphs) shows the quantification of breaks/gaps at passage 2 (p + 2) and passage 4 (p + 4). B Evaluation of metaphase chromosome aberrations from bone marrow of Rad51bWT/WT and Rad51bKI/KI after intraperitoneal injection of MMC (4 mg/kg). In addition, to breaks/gaps, triradial chromosomes were observed only in the mutant mice (shown by red asterisks). C Homozygous RAD51B-c.92delT human-derived lymphoblastoid cells showed more chromosome alterations with and without MMC treatment (200 nM) in comparison with the corresponding heterozygous sister. n = 3. Rad51bc.92delT/c.92delT variant is referred to as Rad51bKI/KI for simplicity. Welch´s t-test analysis: ns, non-significant differences; *p < 0.5; **p < 0.01; ***p < 0.001. Bar in panels, 10 μm.
RAD51Bc.92delT/c.92delT immortalized lymphoblastoid cells from the affected patient (II-4) also displayed more chromosome breaks events in comparison with the heterozygous RAD51BWT/c.92delT cells from the unaffected sister (II-2) in the presence of MMC and even in their absence (Fig. 6C). Taken together, these results confirm that the homozygous RAD51B-c.92delT variant leads to an impaired repair of MMC-induced DNA damage and as a consequence increase the levels of CIN in vitro and in vivo.
MMC stress leads to ICL lesions that inhibits the unwinding of DNA strands and blocks the progression of replicative DNA helicases. To further assess the involvement of Rad51b-c.92delT in this phenotype, we also analyzed its susceptibility to hydroxyurea (HU) and the specific DNA polymerase inhibitor Aphidicolin, two inducers of replication fork DNA damage [36]. Our results show that both drugs produce a lower percentage of surveillance in the c.92delT MEFs (Fig. 7A).
A Cell proliferation assay of WT and mutant Rad51b primary MEFs at passage 2 (p + 2) and 3 (p + 3) incubated in presence of a continuous treatment with hydroxyurea (HU) and in presence of a continuous treatment with Aphidicolin. The results are expressed as a percentage relative to the control (not treated). Each point on the graph represents the mean ± SD. B Top: Schematic of the stretched DNA fiber assay. Bottom: Examples of DNA fiber images from the indicated cells. Bar in panels, 10 µM. Histogram shows the fork progression rate (median and distribution) in each experimental condition. n = 3 experimental replicates (data pooled together). >400 structures scored per condition and replica. ***p < 0.001; in Mann-Whitney test. C Sister chromatid exchange (SCE) per chromosome after treatment with MMC. RAD51BWT/c.92delT and RAD51Bc.92delT/c.92delT variants are referred to as RAD51BWT/KI and RAD51BKI/KI for simplicity. Welch´s t-test analysis: ns, non-significant differences, *p < 0.5; **p < 0.01.
Given these observations and that RAD51 paralogues participate in the response to replication stress [37, 38], we used stretched DNA fibers to monitor fork speed in RAD51Bc.92delT/c.92delT and RAD51BWT/c.92delT in immortalized lymphoblastoid cells treated with MMC. In these conditions, cells derived from the affected patient displayed shorter tracks and reduced fork progression rate (Fig. 7B), which likely reflect a high frequency of fork stalling events and/or impaired fork restart [38, 39]. This observation suggests that Rad51b-c.92delT mutant protein is defective in the DNA damage tolerance pathways that restart DNA synthesis at stalled forks, impairing their subsequent repair and potentially increasing the frequency of DNA breaks.
Given the involvement of the Rad51 family of paralogues in the canonical pathway of HR between sister chromatids [40], we analyzed this parameter in RAD51B-c.92delT lymphoblastoid cells by the classical 5BrdU incorporation procedure. We observed no differences between the WT and mutant RAD51B even in presence of MMC indicating that the canonical HR pathway remains unaffected (Fig. 7C).
Rad51b-c.92delT decreases reprogramming efficiency in mouse embryonic fibroblasts
The expression of the Yamanaka factors [41] induce genome instability providing a plausible explanation for the requirement of an intact HR pathway and low replication stress to achieve efficient reprogramming [42, 43]. In the light of this, we evaluated the role of RAD51B during reprogramming using homozygous Rad51bc92delT-c.92delT MEFs. The numbers of alkaline phosphatase positive colonies were significantly reduced (up to ~2 fold) in Rad51b-c.92delT homozygous mutant MEFs in comparison with the WT (Fig. 8A). By picking colonies with iPS-like morphology (3D shape, defined borders and refringence), we were able to establish Rad51b-c.92delT mutant iPS cell lines with comparable efficiency (~30%) to WT controls, suggesting they are similarly functional bona fide iPS clones.
A MEFs from the indicated genotypes were infected with the 3 reprogramming factors and the numbers of alkaline phosphatase positive colonies were counted showing a significantly reduction (up to ~2 fold) in Rad51b-c.92delT homozygous mutant MEFs in comparison with the wild-type control. n = 9. Welch´s t-test analysis: ***p < 0.001. B Macroscopic image of adenohypophysis from Rad51bWT/WT and adenohypophysis rom Rad51bKI/KI mice. Reticulin staining (C–H) and Prolactin IHQ (I, J) of pituitary adenohypophysis from Rad51bWT/WT and Rad51bKI/KI. C, D Normal adenohypophysis from Rad51bWT/WT show a reticulin staining pattern that is partially lost in zones of hyperplasia where the cell size is increased (magnified in D, indicated by an arrow). E, F Microadenoma from a Rad51bKI/KI showing a complete loss of reticulin staining pattern that is still persistent in the neighbor misplaced adenohypophysis (magnified in F). G, H Macroadenoma from a Rad51bKI/KI showing total loss of reticulin staining pattern and complete absence of normal adenohypophysis tissue. The neoplasm shows different adenoid and pseudopapillary growing patterns (magnified in H). I, J IHQ of prolactin in normal pituitary glands from Rad51bWT/WT showing labelled cells unevenly distributed whereas adenomas from Rad51bKI/KI revealed a high density of prolactin expressing cells. Bar in panels, 250 μm (C, E, G) and 50 μm (D, F, H, I, J). Rad51bc.92delT/c.92delT variant is referred as Rad51bKI/KI for simplicity.
Rad51b c.92delT/c.92delT mice show increased incidence of pituitary hyperplasia
The somatic phenotypes observed are all hallmarks of genome instability. Thus, we evaluated the incidence of spontaneous neoplasia in aged mice (from 18 to 22 months). We observed a pituitary hyperplasia and frequent adenomas of the pituitary gland in mutant homozygous mice (9 out of 14 Rad51bc.92delT/c.92delT mice vs 1 out of 12 in wild type controls) (Fig. 8B). These benign tumors ranged from micro- to macroadenomas (Fig. 8C–H) and showed immuno-histochemical overexpression of prolactin (Fig. 8I, J) in the cytoplasm of tumor cells (but not of ACTH, GH, FSH and LH shown in Fig. S7). Overall, the mice developed pituitary hyperplasia and frequent adenomas, with typical features of prolactinomas. Interestingly, a genetic variant in human RAD51B has been recently associated with pituitary tumors [44].
Discussion
We describe for the first time a homozygous variant in RAD51B (c.92delT) found in two sisters affected with isolated POI. RAD51B seems to be involved in the assembly of RAD51 nucleoprotein filaments during HR and in replication fork remodeling and restart [10, 38, 45]. Thus, RAD51B has been associated with CIN and cancer predisposition [44, 46, 47]. However, its role on female fertility and meiotic DSB repair has not been reported yet. Herein, a perfect segregation of RAD51B-c.92delT and female infertility was found in a family. Specifically, all affected subjects were homozygous for the variant, while heterozygosity or absence of this variant was observed in unaffected members.
The variant was predicted to lead to a PTC leading to the expression of a short N-term protein of only 39 residues lacking the two walker motifs essential for its ATPase activity [28], which would result in null allele known to be lethal in the mouse [8]. Such a PTC might also generate a null allele by activating the nonsense mediated decay (NMD), a surveillance pathway that reduces the production of truncated proteins translated from mRNAs bearing PTCs. NMD is more likely to be triggered when a PTC is located at least 50 nucleotides upstream of the last exon-junction [48]. This rule fits well for the location of the RAD51B-c.92delT variant. However, the observed expression of the RAD51B mRNA in Rad51bc.92delT/c.92delT mice and RAD51B-c.92delT human-derived lymphoblastoid cells indicates that NMD is not operating, probably abrogated by the restart of translation according to a recent model based on truncating mutations in tumors [49]. This model would explain the absence of NMD in the RAD51B-c.92delT variant because translation reinitiation at AUG64 would abrogate NMD and consequently allow the expression of a truncated RAD51B protein lacking its N-terminal domain. This domain is involved in protein-protein interactions with its paralogues, such as RAD51C and in nuclear localization through a NLS [34]. This is consistent with the observed altered nuclear localization of the RAD51B-c.92delT in comparison to the WT in transfected cells and would suggest a similar dysfunction of the endogenous protein in vivo.
The pathogenicity of the human RAD51B variant is supported by the observed meiotic DNA repair defects during mouse prophase I, the increased incidence of pituitary adenomas, and the defects of MMC-induced damage repair in c.92delT human and mouse cells. In addition, RAD51B-c.92delT variant reduced the reprogramming efficiency of primary MEFs into iPSCs.
Similar to its Rad51 paralogues [6, 35], the disruption of Rad51b in mice has shown early embryonic lethality [8]. The Rad51b hypomorphic allele analyzed in this study causes a loss of RAD51B-c.92delT interaction with RAD51C and a partial loss of interaction with RAD51 and HELQ. In vivo, Rad51b-c.92delT mice did not show any effect on DMC1 loading, but revealed accumulation of foci containing γH2AX, RAD51 and the BRCA2-associated proteins HSF2BP/BRME1 on the chromosome axes from pachytene onward [24, 31, 33, 35]. The accumulation of these non-meiotic specific repair proteins could be explained by the reactivation of a somatic-like HR DNA-repair pathway at pachytene as a consequence of the persistence of unrepaired meiotic DSBs [32]. Regarding HELQ, this helicase promotes efficient HR at damaged replication forks by interacting directly with the RAD51-containing BCDX2 complex. This complex and HELQ act together in the resolution of DNA ICLs upon exposure to MMC [50]. The diminished interaction between the mutant RAD51B-c.92delT variant and HELQ sheds light into the presence of unrepaired DNA in MEFs, bone marrow and human lymphoblastoid cells but also into the persistence of unrepaired meiotic DSBs and reduced number of COs. Interestingly, mice lacking HELQ exhibit MMC sensitivity, pituitary adenomas as well as germ cell attrition with subfertility [50] and worms lacking both HELQ and the RAD51 paralogue RFS1 results in a block to meiotic DSB repair after strand invasion [51]. These inefficient interactions of RAD51B-c.92delT together with the altered nuclear localization of the RAD51B variant synergistically lead to a reduction of COs in both spermatocytes and oocytes.
A recent study carried by our laboratory evaluating a POI-inducing variant in the gene HSF2BP showed a comparable reduction in the number of COs in comparison with the Rad51b-c.92delT mice, which led to a very slight reduced female fertility [24]. This trend of the reduction of litter size in the RAD51B-c.92delT, albeit not statistically significant, could be due to non-identical genetic backgrounds between both mouse models and would support the idea of a slight impact on fertility.
RAD51 and its paralogues have been also associated with cancer predisposition and Fanconi anemia [52]. RAD51B has been involved in male and female breast cancer, ovary cancer, prostate cancer, and pituitary adenoma [44, 46, 47, 53,54,55,56,57]. Consistently, RAD51B-c.92delT somatic human-derived lymphoblastoid cells, as well as MEFs and explanted bone marrow cells from humanized mice are sensitive to MMC-induced CIN. Moreover, Rad51bc.92delT/c.92delT and HELQ-deficient mice display an increased incidence of pituitary hyperplasia suggesting a common pathological mechanism of action. Of note, patient II-4 was diagnosed with a duodenal tubulovillous adenoma with high-grade dysplasia (details in case report section). Altogether, these data provide evidence for a critical role of RAD51B in somatic genome instability and tumorigenesis.
Previous studies have shown that efficient reprogramming of MEFs to a pluripotent stage requires a proficient HR pathway. Accordingly, efficient reprogramming requires key HR genes [42]. Interestingly, the expression of the 3 F reprogramming factors in MEFs induces replication stress, and genetic reduction of stress (overexpressing Chk1) increases the efficiency of iPSC generation [43]. Besides, it is known that replication forks may be stalled at DNA lesions generated by cellular metabolism (aldehydes) or DNA damaging agents including MMC [58]. One pathway to resume DNA synthesis involves fork remodeling events mediated in part by RAD51 paralogues [38]. Our in vivo and in vitro observations of the effects caused by Rad51b-c.92delT in reprogramming, fork progression, chromosomal breakage and clonogenic survival suggest also a somatic function of this paralogue in assisting fork progression through certain DNA lesions, as part of the DNA damage tolerance response.
In summary, we describe for the first time two sisters with an isolated POI phenotype caused by a novel homozygous variant in RAD51B (c.92delT). Humanized mice reveal that the RAD51B pathogenic variant evades NMD probably owing to the reinitiation of the translation at a secondary downstream AUG. The resulting truncated protein lacks most of the N-term domain involved in protein-protein interactions and proper nuclear localization. As a result, homozygous Rad51b-c.92delT mice shows meiotic DNA repair defects with RAD51 and HSF2BP/BRME1 accumulation in the chromosome axes which leads to a significant reduction in COs. In addition, RAD51B-c.92delT human-derived lymphoblastoid cells and mutant MEFs are sensitive to MMC-induced CIN and revealed a reduced reprogramming efficiency to iPSCs as well as a decrease in replication fork progression. Of interest, humanized mice exhibit an increased incidence of hyperplasia of the pituitary gland likely as a consequence of the RAD51B-c.92delT-induced genome instability.
Methods
Case report
Written informed consent was obtained from all subjects before sample collection for DNA analysis. This study was approved by the Ethics Committee of Hospital das Clínicas, Sao Paulo University School of Medicine, Brazil (protocol number 2015/12837/1.015.223). The proposita (II-1) and her younger sister (II-4) were born from first-degree cousins from Northeastern of Brazil (Fig. 1A). On examination at 23 and 21 years of age respectively, they were diagnosed with POI presenting with primary amenorrhea. These siblings had no menarche and physical examination revealed Tanner stage 1 for breast development (II-1 and II-4) and Tanner stage IV pubic hair. Consistent with the diagnosis of hypergonadotropic hypogonadism, basal gonadotropin levels were elevated in the proband and her affected sister (FSH = 44 and 31 U/L, LH = 21 and 17 U/L, respectively) while estradiol levels were undetectable. At first appointments, physical examination showed normal height (149 and 156 cm respectively), and body weight (60 and 47 Kg, respectively). Pelvic ultrasound scans showed an infantile uterus, and the ovaries could not be visualized. Thyroid, adrenal or ovarian autoimmune disorders had been excluded in these patients. On karyotyping analysis performed in 30 metaphases, both sisters were 46,XX. Treatment with conjugated estrogens followed by progesterone replacement resulted in complete breast development and menstrual bleeding. However, the proband and her affected sister developed hypertension, insulin resistance, primary hypothyroidism with absence of thyroid antibodies, and obesity over the years, being current BMI = 41 and 34 kg/m2, respectively. During the course of this investigation, the affected sister was also diagnosed with a duodenal tubulovillous adenoma with high-grade dysplasia and underwent surgery. Thereafter, she was found to have an indeterminate hepatic nodule and hepatic steatosis. She has been followed and no additional radiotherapy or chemotherapy has been done. The proband has not been diagnosed with any neoplasia. Regarding their family’s history, one first-degree cousin and three-second degree cousins were found to have 46,XY DSD due to 5-alpha-reductase type 2 deficiency.
Genetic analysis
DNA extraction: Genomic DNA was extracted from peripheral blood leukocytes from all patients using standard procedures. Whole-exome sequencing: Libraries were prepared on both sisters (II-1 and II-4) and her unaffected sister (II-2, as shown in Fig. 1A). Briefly, genomic DNA was sheared to 200–300 bp using the Covaris acoustic adaptor. Exons were captured using SureSelect Human All exons kit (Agilent Technologies) and sequenced by Illumina platform (HiSeq2500, Illumina). Alignment of raw data and variant calling were performed following the steps described by Franca and collaborators [59]). Briefly, the reads were aligned to the human reference genome GRCh37/hg19 using Burrows-Wheeler aligner [60]. Variant calling was performed with Freebeys and annotated ANNOVAR. SNVs were run through independent protein pathogenicity predictors: Polyphen-2, SIFT, Mutation Taster, Mutation Assessor, FATHMM, Radial SVN, and LRT. Sanger sequencing: Sanger sequencing was done to validate only the primary variant of interest in all subjects. Primers flanking the RAD51B variant (ENSG00000182185/NM_133509: exon3:c.92delT:p.Leu31Tyrfs*9) were used for PCR amplification. Sanger sequencing was performed to validate only the primary variant of interest in all family subjects for whom DNA was available. Primers flanking the RAD51B variant (ENSG00000182185/NM_133509: exon3:c.92delT:p.Leu31Tyrfs*9) were used for PCR amplification. Moreover, Sanger sequencing was used to screen 235 fertile Brazilian control women for the presence of the putative damaging variant. All PCR products were sequencing using BigDye terminator v1.1 followed by automated sequencing at the ABI PRISM 310 (Applied Biosystems, Foster City, CA).
RNA extraction from patient blood samples
Blood samples were centrifuged at 400 × g for 10 min at room temperature with the brake off for leukocyte isolation. The buffy coats were removed, and RNA extractions were performed by adding TRI-reagent according to the manufacturer’s recommendation (Sigma). Total RNA was reverse-transcribed using MMLV RT enzyme (Takara) and standard procedures. The expression and integrity of patients RAD51B transcript were studied by RT-PCR using the high-fidelity Herculase II fusion DNA polymerase (Agilent) and the couple of primers RAD51B-Fext: 5′-GCATGGGTAGCAAGAAACTAAAA-3′ and RAD51B-Rext: 5′-GATCCACAAGCCACACCAC-3′ encompassing the c.92delT variant. The oligo RAD51B-Fint: 5′-GAGCTGTGTGACCGTCTGAG-3′ was used to verify the PCR product by Sanger sequencing (Eurofins genomics).
Generation of CRISPR/Cas9-edited mice
For developing the Rad51bKI/KI model, Rad51b-sgRNA1 5′-CTAGAACTTATGAAAGTGAC-3′ and sgRNA2 5′-GAGACTTAAAAAGTGCTAAA-3′ targeting the exon 3 were predicted at https://eu.idtdna.com/site/order/designtool/index/CRISPR_SEQUENCE. The designed ssODN contains the muted base (c.92delT) and mutations that humanize the locus (TTTTAAGTCTCTCCCCACTAGAACTTATGAAA > TTTATGTCTCTCCCCACTGGAGCTTATGAAG). The crRNAs, the tracrRNAs and the ssODNs (Supplementary Table 7) were produced by chemical synthesis by IDT. The crRNA and tracrRNA were annealed to obtain the mature sgRNA. A mixture containing the sgRNAs (20 ng/µl of each annealed sgRNA), 30 ng/µl of recombinant Cas9 protein (IDT) and 10 ng/µl of the ssODN were microinjected into B6/CBA F2 zygotes (hybrids between strains C57BL/6 J and CBA/J) [61] at the Transgenic Facility of the University of Salamanca. Edited founders were identified by PCR amplification (Taq polymerase, NZYtech) with primers flanking the edited region (Supplementary Table 7). PCR products were directly sequenced or subcloned into pBlueScript (Stratagene) followed by Sanger sequencing. Selected founders, carrying the desired alleles, were crossed with wild-type C57BL/6 J to eliminate possible unwanted off-targets. Heterozygous mice were re-sequenced and crossed to generate the edited homozygotes. Genotyping was performed by agarose gels analysis or Sanger sequencing (in Rad51b humanized mutant) of PCR products from genomic DNA extracted from tail biopsies.
Histology
For histological analysis, after the necropsy of the mice their testes were fixed in Bouin’s fixative, ovaries and pituitary glands were fixed in 10% formol. Tissues were processed into serial paraffin sections and stained with haematoxylin-eosin (ovaries) or Periodic acid–Schiff (PAS) and haematoxylin (testes). Jones’ reticulin staining was performed automatically on the ArtisanTM Link Pro Special Staining System (Dako). Prolactin and ACTH detection by IHC was carried out in an Artisan Link Pro (DAKO) at pH6 using a polyclonal rabbit against prolactin (A0569) and a mouse monoclonal against ACTH (clone 02A3; 1:100), respectively. GH, FSH, and LH detection was carried out in an ultra-view BenchMark (Roche) using a rabbit polyclonal antibody against ACTH (206A-74), a rabbit polyclonal against GH (208A-74), a rabbit polyclonal against SH (EP257), and a rabbit polyclonal against LH (209A-14). All the immunohistochemical stainings for pituitary hormones ACTH, TSH, GH, LH, FSH were negative, using normal pituitary as a control. The samples were analyzed using a microscope OLYMPUS BX51 and images were taken with a digital camera OLYMPUS DP70. Primordial cell evaluation was carried out by serially slicing into 5 µ thick sections the inner third of each ovary.
Fertility assessment
Rad51bWT/WT and Rad51bKI/KI females (8 weeks old) were mated with wild type males, respectively, over the course of 4–12 months. 8 mice per the wild type and 10 mice in the case of Rad51bKI/KI were crossed. The presence of copulatory plug was examined daily and the number of pups per litter was recorded.
RAD51B localization in Cos7 cells
Localization of RAD51B variants was studied in COS7 cells transfected with Jetpei (PolyPlus) according to the manufacturer’s protocol. 5 × 104 cells were plated on 0.5% fish gelatin-treated (Sigma-Aldrich) 35 mm culture dishes and were subsequently transfected the day after with 0.2 µg of GFP-fused expression vectors. The culture media was replaced after 24 h. COS7 cells were fixed 48 h post-transfection with 4% formaldehyde solution (ThermoFisher) for 7 min at 4 °C. They were washed three times in 1X PBS before a permeabilization with 0.2% Triton X-100 in KB buffer (0.1 M NaCl, 20 mM Tris-HCl pH 7.5, 0.1% BSA). Cells were then washed again and blocked in 7% FBS-supplemented 1X PBS for 30 min at RT. After that, they were incubated with a rabbit anti-GFP antibody (Invitrogen) 1:300-diluted in 7% FBS in PBS for 1 h in a wet chamber at RT. After three washes in 1X PBS, the cells were incubated with a secondary FITC anti-rabbit antibody (Jackson Immunoresearch) diluted 1:100 in 7% FBS in PBS for 1 h. The slides were rinsed 3 times in PBS and finally mounted with Vectashield® and DAPI. Images were taken at 63X magnification with the Laser Scan Confocal Microscopy Leica SP8 (Leica).
Immunocytology and antibodies
Testes were detunicated and processed for spreading using a conventional “dry-down” technique. Oocytes from fetal ovaries (E18 and E18.5 embryos) were digested with collagenase, incubated in a hypotonic buffer, disaggregated, and fixed in paraformaldehyde. Rabbit polyclonal antibodies (R1 and R2 generated from two different host rabbits) against HSF2BP and BRME1 were developed by ProteintechTM against a fusion protein of poly-His with full length HSF2BP or BRME1 (pUC57 vector) of mouse origin. Rabbit polyclonal antibody against DMC1 was developed by ProteintechTM against a DMC1 peptide (EESGFQDDEESLFQDIDLLQKHGINMADIKKLKSVGICTIKG). Both meiocytes were incubated with the following primary antibodies for IF: rabbit anti-γH2AX (ser139) IgG #07–164 (1:500, Millipore), mouse anti-γH2AX (ser139) IgG #05–636 (1:400, Millipore), rabbit αRAD51 PC130 (1:100, Calbiochem), rabbit αBRME1 R2 (1:200, ProteintechTM), rabbit αHSF2BP R2 (1:30, ProteintechTM), mouse αMLH1 51-1327GR (1:30, BD Biosciences), rabbit αRPA serum ¨Molly¨ (1:30, provided by Dr. Edyta Marcon, Medical Research University of Toronto, Canada), rat RPA2 2208 S (1:100, Cell Signaling), rabbit αSPATA22 16989-1-AP (1:60, Proteintech), rabbit αDMC1 (1:500, ProteintechTM), mouse αSYCP3 IgG sc-74569 (1:1000, Santa Cruz), rabbit serum αSYCP3 K921 (1:500), rabbit αSYCP1 IgG ab15090 (1:200, Abcam). We could not find any reliable antibody that detect endogenous mouse HELQ in chromosome spreads, squashes or cytospin preps. The secondary antibodies used were goat Alexa 555 α-mouse A-32727, goat Alexa 488 α-mouse A-11001, donkey Alexa 555 α-rabbit A-31572, goat Alexa 488 α-rat A-11006 (1:200, ThermoFisher), goat Alexa 488- Fab α-rabbit 111-547-003 (1:100, Jackson Immunoresearch). Slides were mounted with DAPI and visualized at room temperature using a microscope (Axioplan2; Carl Zeiss, Inc.) with 63x objectives with an aperture of 1.4 (Carl Zeiss, Inc.). Images were taken with a digital camera (ORCA-ER C4742-80; Hamamatsu) and processed with Leica LAS X Life Science Software and Adobe Photoshop CS6 (Adobe). Quantification of fluorescence signals was performed using Fiji (ImageJ) software.
Generation of plasmids
The cDNAs encoding RAD51B (full length and truncated constructs) were RT-PCR amplified from lymphoblastoid cells (derived from patients) RNA. Full-length cDNAs encoding RAD51B (WT and mutant constructs), RPA1, BRCA2 (N, M and C constructs), PALB2, RAD51, BRME1, RAD21, DMC1, HELQ, RAD51C, MEIOB and HSF2BP were RT-PCR amplified from murine testis RNA using the primers listed in Supplementary Table 7. The cDNAs were cloned into the EcoRV pcDNA3, EcoRV pcDNA3-2XFlag, SmaI pcDNA3-2XHA, SmaI pEGFP-C1 or SmaI pEGFP-N1 expression vectors under the CMV promoter. In frame cloning was verified by Sanger sequencing.
Site-directed mutagenesis analysis
Point-mutations were introduced to hRAD51B cloned in pEGFP-N1 using primers listed in Supplementary Table 8 (with the desired mutation) in which the three secondary methionine codons were PCR replaced by CGN encoding alanine codons (individual and double Met to Ala substitutions). The parent template was removed using a methylation-dependent endonuclease DpnI. Plasmids are isolated from the resulting colonies and screened for the desired modification. Positive clones are verified by Sanger sequencing.
Immunoprecipitation
HEK293T cells were transiently transfected with WT or KI constructs of RAD51B in combination with full-length cDNAs encoding RPA1, BRCA2 (N, M and C constructs), PALB2, RAD51, BRME1, RAD21, DMC1, HELQ, RAD51C, MEIOB and HSF2BP. Whole cell extracts were prepared in a 50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100 buffer supplemented with protease inhibitors. Those extracts were cleared with protein G Sepharose beads (GE Healthcare) for 1 h. Immunoprecipitations were performed using rabbit αFlag IgG (3.2 µg; F7425, Sigma-Aldrich), rabbit αRad51b R2 IgG (3 µg; ProteintechTM), ChromPure rabbit IgG (3.4 µg/1 mg prot; 011-000-003). These were incubated with the extracts for 2 h and immunocomplexes were isolated by adsorption to protein G-Sepharose beads overnight. After washing, the proteins were eluted from the beads with 2xSDS gel-loading buffer 100 mM Tris-HCl (pH 7.0), 4% SDS, 0.2% bromophenol blue, 200 mM β-mercaptoethanol and 20% glycerol, and loaded onto reducing polyacrylamide SDS gels.
Western blot
HEK293T cells were transfected with WT and mutated constructs encoding human RAD51B generated with the site-directed mutagenesis. They were cultured for 48 h before lysis in RIPA buffer (50 mM Tris HCl pH 7.4. 150 mM NaCl, 1 mM EDTA, 1% Triton X-100) supplemented with protease inhibitors. Cell lysates were sonicated to disrupt cell membranes and shear DNA. After total protein quantification using Bradford assay, 5 µg of protein extracts were loaded onto reducing polyacrylamide SDS gels. Primary antibodies used for western blotting were mouse αFlag IgG (1:3000; F1804, Sigma-Aldrich), mouse αHA IgG (1:3000; 11 101 R, Covance), rabbit αRAD51B R2 (1:2000, ProteintechTM), rabbit αGFP IgG (1:3000; A11122, Invitrogen), rabbit αHSF2BP R2 (1:2000, ProteintechTM). Secondary fluorochrome-conjugated α-mouse DyLightTM 680 (35518, ThermoFisher), α-rabbit DyLightTM 800 (35571, ThermoFisher) antibodies were used at 1:10000 dilution and incubated in dark. The fluorescent signal of the antibodies was obtained through Odyssey Infrared Imaging system. Full length original western blots for these results are provided in Supplementary information.
Immunoprecipitation coupled to Mass spectrometry analysis
Testis extracts and human lymphoblastoid cells were prepared in 50 mM Tris-HCL (pH8), 500 mM NaCl, 1 mM EDTA, 1% Triton-X100 and 10 mg of extracts were incubated 2 h with 30 µg of antibody against mouse RAD51B (residues 122 to 350, ProteintechTM) or IgG from rabbit. The corresponding immunocomplexes were incubated with 60 µl of sepharose beads overnight. After washing, beads were eluted in 100 mM glycine pH 2.5–3 and analyzed by Lc-MS/MS shotgun in LTQ Velos Orbitrap at the Proteomics facility of Centro de Investigación del Cáncer (CSIC/University of Salamanca). Raw data were analyzed using MaxQuant v 1.6.2.6 (Cox and Mann, 2008) against SwissProt Mouse database (UP000000589, Oct, 2019) and MaxQuant contaminants. All FDRs were of 1%. Variable modifications considered were oxidation of M and acetylation of the N-term, while fixed modifications included only carbamidomethylation of C. The maximum number of modifications allowed per peptide was 5. The proteins related with DNA repair were quantified using iBAQ [62].
Cell culture
Primary MEFs were derived from embryonic day 13.5 (E13.5) embryos following standard procedures. MEFs, HEK 293 T and Cos7 cell lines were cultured at atmospheric oxygen pressure in Dulbecco’s modified Eagle’s medium (GIBCO) supplemented with 10% fetal bovine serum (GIBCO), and 2 mM Glutamine. MEFs were immortalized at passage 2 using SV40 and hTERT for clonogenical survival assay. Lymphoblast cell lines were derived from donor B-lymphocytes by Epstein Bar virus transformation according to standard procedures and cultured in RPMI medium (GIBCO) supplemented with 10% inactivated fetal bovine serum (GIBCO), and 2 mM Glutamine. Cell lines were tested for mycoplasma contamination using the Mycoplasma PCR ELISA (Sigma).
Cell proliferation assay and DNA damage recuperation
For cell proliferation assays, 2.5 × 104 cells/well were seeded in 12-well plates in duplicates in complete media. Continuous treatment was started in 18 h at the following doses: 0, 5, 10, 20, and 30 ng/ml for MMC; 0, 5, 10 and 20 µM for HU; 0, 50, 100, 500 nM for Aphidicolin. Cells from one plate were trypsinized and counted as a “before treatment” day1 reference. Three days later (4th day after seeding) the remaining cells were counted the same way. Day1 reference numbers were subtracted from day 3 cell numbers to evaluate growth of each cell line. The resulting cell counts were expressed as percentages from the untreated wells. For monitoring the efficiency of DNA repair, 1.4 × 104 MEFs cells/well were treated with 1 µg/ml for one hour and the recuperation of the DNA was measured by quantification of γH2AX foci at 0, 6, 12, 48 and 72 h.
Clonogenic survival assay
Clonogenic survival following an exposure to MMC was assessed in immortalized wild-type and mutant Rad51b MEFs. 700 cells were plated per well in 60 mm plates. The day after plating, cells were treated with 0, 50, 100, 150 and 300 nM of MMC and allowed to incubate for 2 h. After drug treatment, media was removed, cells were washed several times with phosphate buffered saline (PBS), and fresh media was added. Once colonies were detectable by the naked eye (7–12 days), media was removed, and cells were washed with PBS and incubated for 30 min at room temperature in a fixation solution (8% paraformaldehyde in cell media). The fixation solution was removed, and colonies were stained with Giemsa (0.02% Giemsa solution in PBS) for 30 min at room temperature. Plates were then rinsed with water and allowed to dry. Colonies were counted by hand.
Karyotyping of mice bone marrow, lymphoblastoid cells and MEFs
Mice between 6–12 weeks old were injected intraperitoneally with a single dose of 4 mg of MMC per kilogram of body weight. After 24 h were injected with 0.1 ml of 0.5% colchicine solution intraperitoneally and the karyotyping derived from bone marrow was realized following standard procedures protocol [63]. Metaphases were obtained after MMC treatment, 150 nM for MEFs at passage 2–5 and 200 nM for lymphoblastoid cells by standard procedures. Active growing cultures were arrested using colcemide (1 µg/mL) for 4 h for MEFs and 3 h for lymphoblastoid cells, trypsinized, treated with hypotonic solution (0.75 mM KCl), and fixed with Methanol/Acetic. Metaphase spreads were Giemsa-stained and analyzed for microscopically visible chromosomal aberrations. Chromatid gap, chromatid break and triradial/quatriradial chromosomes were classified as previously described [64]. At least 100 metaphases were counted from three independent mice and embryos of each genotype.
Single-molecule analysis of DNA replication
Lymphoblastoid cells were treated with 1 µg/ml MMC for 2 h. In the last 40 min, cells were pulse-labelled sequentially with 50 μM CldU (20 min) and 250 μM IdU (20 min). Labelled cells were resuspended in PBS and lysed in 0.2 M Tris pH 7.4, 50 mM EDTA, 0.5% SDS (6 min/ RT). DNA fibers were prepared and stained as described [65] with the following modification: slides were incubated in stringency buffer (10 mM Tris-HCl pH 7.4; 0.1 M NaCl; 0.2% Triton X-100) for 6 min at RT, washed and incubated in blocking solution for 15 min at RT prior to secondary antibodies incubation. Images were obtained in a DM6000 B Leica microscope with an HCX PL APO 40x, 0.75 NA objective. Conversion factor 1 μm = 2.59 kb was used [66]. Fork rate values were obtained by dividing the length of green tracks in ongoing forks by the time of the IdU pulse. Primary antibodies and dilutions used: CldU (rat monoclonal anti-BrdU, 1:100, ab6326, Abcam), IdU (mouse monoclonal anti-BrdU, 1:100, BD 347580), ssDNA (1:100, MAB3034, Millipore). Secondary antibodies and dilutions used: goat anti-rat IgG AF-594 (1:300, A11007, Molecular Probes); goat anti-mouse IgG AF-488 (1:300, A121121, Molecular Probes), goat anti-mouse IgG2a AF-647 (1:300, A21241, Molecular Probes).
Sister chromatid exchange analysis
Exponentially growing lymphoblastoid cells were inoculated at a density of 3 × 105 and were treated with 10 µM of BrdU (5-bromodeoxyuridine, Sigma) for 48 h (approximately two cell divisions). 5 and 10 ng/ml of MMC was added 24 h after BrdU treatment followed by a 3 h incubation with 1 µg/ml of colcemide. Metaphase spreads were stained with 0.1 mg/ml of acridine orange (ThermoFisher) in dH2O for 5 min at RT. Slides were washed for 2 min under running dH2O tap water, incubated 1 min in Sorenson Buffer (0.1 M Na2HPO4, 0.1 M NaH2PO4, pH 6.8) and mounted in Sorenson Buffer. Slides were immediately visualized under FITC filter and at least 30 reciprocal exchange events were counted of each genotype.
iPSC generation from MEFs
For iPSC generation, 2.5 × 105 MEFs were infected with retroviral particles produced by HEK293T transfected with constitutive retroviral expression vectors pMXs KLF4, OCT4 and SOX2. The iPSC media (DMEM, GIBCO; 15% KSR, Invitrogen; 1% Non-essential aminoacids, MEM NEE 100X GIBCO; 1% PSG; 0,002% β-mercaptoethanol 50 mM, GIBCO; 1000 units/mL LIF, Merck) was changed every 24 h until iPS cell colonies appeared (after ∼14 days of treatment). Three weeks after plating the MEFs, reprogramming plates were stained for alkaline phosphatase activity (AP detection kit, MERCK).
Statistics
In order to compare counts between genotypes, we used the Welch´s t-test (unequal variances t-test), which was appropriate as the count data were not highly skewed (i.e., were reasonably approximated by a normal distribution) and in most cases showed unequal variance. We applied a two-sided test in all the cases. Asterisks denote statistical significance: *p-value < 0.05, **p-value < 0.01, ***p-value < 0.001 and ****p-value < 0.0001.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Genomic DNA sequences of H. sapiens (human, 317761), M. musculus (mouse, 75801) are available on GenBank (https://www.ncbi.nlm.nih.gov/genbank/). Amino acid sequences of H. sapiens (Q8N1H7), M. musculus (NP_083381) were obtained from the UniProt database (http://www.uniprot.org/). All remaining data generated during this study are included in this Article and its Supplementary Information files or available from the authors upon request from the authors.
References
Handel MA, Schimenti JC. Genetics of mammalian meiosis: regulation, dynamics and impact on fertility. Nat Rev Genet. 2010;11:124–36.
Paiano J, Wu W, Yamada S, Sciascia N, Callen E, Paola Cotrim A, et al. ATM and PRDM9 regulate SPO11-bound recombination intermediates during meiosis. Nat Commun. 2020;11:857.
Zhao W, Vaithiyalingam S, San Filippo J, Maranon DG, Jimenez-Sainz J, Fontenay GV, et al. Promotion of BRCA2-Dependent Homologous Recombination by DSS1 via RPA Targeting and DNA Mimicry. Mol Cell. 2015;59:176–87.
Martinez JS, von Nicolai C, Kim T, Ehlen A, Mazin AV, Kowalczykowski SC, et al. BRCA2 regulates DMC1-mediated recombination through the BRC repeats. Proc Natl Acad Sci USA. 2016;113:3515–20.
Chun J, Buechelmaier ES, Powell SN. Rad51 paralog complexes BCDX2 and CX3 act at different stages in the BRCA1-BRCA2-dependent homologous recombination pathway. Mol Cell Biol. 2013;33:387–95.
Dai J, Voloshin O, Potapova S, Camerini-Otero RD. Meiotic Knockdown and Complementation Reveals Essential Role of RAD51 in Mouse Spermatogenesis. Cell Rep. 2017;18:1383–94.
Pittman DL, Schimenti JC. Midgestation lethality in mice deficient for the RecA-related gene, Rad51d/Rad51l3. Genesis 2000;26:167–73.
Shu Z, Smith S, Wang L, Rice MC, Kmiec EB. Disruption of muREC2/RAD51L1 in mice results in early embryonic lethality which can Be partially rescued in a p53(-/-) background. Mol Cell Biol. 1999;19:8686–93.
Sullivan MR, Bernstein KA. RAD-ical New Insights into RAD51 Regulation. Genes (Basel). 2018;9:629. https://doi.org/10.3390/genes9120629.
Takata M, Sasaki MS, Sonoda E, Fukushima T, Morrison C, Albala JS, et al. The Rad51 paralog Rad51B promotes homologous recombinational repair. Mol Cell Biol. 2000;20:6476–82.
Garcin EB, Gon S, Sullivan MR, Brunette GJ, Cian A, Concordet JP, et al. Differential Requirements for the RAD51 Paralogs in Genome Repair and Maintenance in Human Cells. PLoS Genet. 2019;15:e1008355.
Huhtaniemi I, Hovatta O, La Marca A, Livera G, Monniaux D, Persani L, et al. Advances in the Molecular Pathophysiology, Genetics, and Treatment of Primary Ovarian Insufficiency. Trends Endocrinol Metab. 2018;29:400–19.
Tsui V, Crismani W. The Fanconi Anemia Pathway and Fertility. Trends Genet. 2019;35:199–214.
Thonneau P, Marchand S, Tallec A, Ferial ML, Ducot B, Lansac J, et al. Incidence and main causes of infertility in a resident population (1,850,000) of three French regions (1988-1989). Hum Reprod. 1991;6:811–6.
Franca MM, Mendonca BB. Genetics of Primary Ovarian Insufficiency in the Next-Generation Sequencing Era. J Endocr Soc. 2020;4:bvz037.
Caburet S, Todeschini AL, Petrillo C, Martini E, Farran ND, Legois B, et al. A truncating MEIOB mutation responsible for familial primary ovarian insufficiency abolishes its interaction with its partner SPATA22 and their recruitment to DNA double-strand breaks. EBioMedicine 2019;42:524–31.
de Vries L, Behar DM, Smirin-Yosef P, Lagovsky I, Tzur S, Basel-Vanagaite L. Exome sequencing reveals SYCE1 mutation associated with autosomal recessive primary ovarian insufficiency. J Clin Endocrinol Metab. 2014;99:E2129–32.
Wang J, Zhang W, Jiang H, Wu BL, Primary Ovarian Insufficiency C. Mutations in HFM1 in recessive primary ovarian insufficiency. N. Engl J Med. 2014;370:972–4.
Guo T, Zhao S, Zhao S, Chen M, Li G, Jiao X, et al. Mutations in MSH5 in primary ovarian insufficiency. Hum Mol Genet. 2017;26:1452–7.
Mandon-Pepin B, Touraine P, Kuttenn F, Derbois C, Rouxel A, Matsuda F, et al. Genetic investigation of four meiotic genes in women with premature ovarian failure. Eur J Endocrinol. 2008;158:107–15.
Caburet S, Arboleda VA, Llano E, Overbeek PA, Barbero JL, Oka K, et al. Mutant cohesin in premature ovarian failure. N. Engl J Med. 2014;370:943–9.
Carlosama C, Elzaiat M, Patino LC, Mateus HE, Veitia RA, Laissue P. A homozygous donor splice-site mutation in the meiotic gene MSH4 causes primary ovarian insufficiency. Hum Mol Genet. 2017;26:3161–6.
Franca MM, Funari MFA, Lerario AM, Santos MG, Nishi MY, Domenice S, et al. Screening of targeted panel genes in Brazilian patients with primary ovarian insufficiency. PLoS One. 2020;15:e0240795.
Felipe-Medina N, Caburet S, Sanchez-Saez F, Condezo YB, de Rooij DG, Gomez HL, et al. A missense in HSF2BP causing primary ovarian insufficiency affects meiotic recombination by its novel interactor C19ORF57/BRME1. Elife. 2020;9:e56996.
Naslavsky MS, Yamamoto GL, de Almeida TF, Ezquina SAM, Sunaga DY, Pho N, et al. Exomic variants of an elderly cohort of Brazilians in the ABraOM database. Hum Mutat. 2017;38:751–63.
Lerario AM, Mohan DR, Montenegro LR, Funari MFA, Nishi MY, Narcizo AM, et al. SELAdb: A database of exonic variants in a Brazilian population referred to a quaternary medical center in Sao Paulo. Clinics. 2020;75:e1913.
Franca MM, Mendonca BB. Genetics of ovarian insufficiency and defects of folliculogenesis. Best Pr Res Clin Endocrinol Metab. 2022;36:101594.
Wiese C, Hinz JM, Tebbs RS, Nham PB, Urbin SS, Collins DW, et al. Disparate requirements for the Walker A and B ATPase motifs of human RAD51D in homologous recombination. Nucleic Acids Res. 2006;34:2833–43.
Niu W, Spradling AC. Two distinct pathways of pregranulosa cell differentiation support follicle formation in the mouse ovary. Proc Natl Acad Sci USA. 2020;117:20015–26.
da Cruz I, Rodriguez-Casuriaga R, Santinaque FF, Farias J, Curti G, Capoano CA, et al. Transcriptome analysis of highly purified mouse spermatogenic cell populations: gene expression signatures switch from meiotic-to postmeiotic-related processes at pachytene stage. BMC Genomics. 2016;17:294.
Brandsma I, Sato K, van Rossum-Fikkert SE, van Vliet N, Sleddens E, Reuter M, et al. HSF2BP Interacts with a Conserved Domain of BRCA2 and Is Required for Mouse Spermatogenesis. Cell Rep. 2019;27:3790–8. e7
Enguita-Marruedo A, Martin-Ruiz M, Garcia E, Gil-Fernandez A, Parra MT, Viera A, et al. Transition from a meiotic to a somatic-like DNA damage response during the pachytene stage in mouse meiosis. PLoS Genet. 2019;15:e1007439.
Zhang J, Fujiwara Y, Yamamoto S, Shibuya H. A meiosis-specific BRCA2 binding protein recruits recombinases to DNA double-strand breaks to ensure homologous recombination. Nat Commun. 2019;10:722.
Miller KA, Sawicka D, Barsky D, Albala JS. Domain mapping of the Rad51 paralog protein complexes. Nucleic Acids Res. 2004;32:169–78.
Kuznetsov S, Pellegrini M, Shuda K, Fernandez-Capetillo O, Liu Y, Martin BK, et al. RAD51C deficiency in mice results in early prophase I arrest in males and sister chromatid separation at metaphase II in females. J Cell Biol. 2007;176:581–92.
Howlett NG, Taniguchi T, Durkin SG, D’Andrea AD, Glover TW. The Fanconi anemia pathway is required for the DNA replication stress response and for the regulation of common fragile site stability. Hum Mol Genet. 2005;14:693–701.
Somyajit K, Saxena S, Babu S, Mishra A, Nagaraju G. Mammalian RAD51 paralogs protect nascent DNA at stalled forks and mediate replication restart. Nucleic Acids Res. 2015;43:9835–55.
Berti M, Teloni F, Mijic S, Ursich S, Fuchs J, Palumbieri MD, et al. Sequential role of RAD51 paralog complexes in replication fork remodeling and restart. Nat Commun. 2020;11:3531.
Mouron S, Rodriguez-Acebes S, Martinez-Jimenez MI, Garcia-Gomez S, Chocron S, Blanco L, et al. Repriming of DNA synthesis at stalled replication forks by human PrimPol. Nat Struct Mol Biol. 2013;20:1383–9.
Takata M, Sasaki MS, Tachiiri S, Fukushima T, Sonoda E, Schild D, et al. Chromosome instability and defective recombinational repair in knockout mutants of the five Rad51 paralogs. Mol Cell Biol. 2001;21:2858–66.
Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–76.
Gonzalez F, Georgieva D, Vanoli F, Shi ZD, Stadtfeld M, Ludwig T, et al. Homologous recombination DNA repair genes play a critical role in reprogramming to a pluripotent state. Cell Rep. 2013;3:651–60.
Ruiz S, Lopez-Contreras AJ, Gabut M, Marion RM, Gutierrez-Martinez P, Bua S, et al. Limiting replication stress during somatic cell reprogramming reduces genomic instability in induced pluripotent stem cells. Nat Commun. 2015;6:8036.
Juknyt EG, Laurinaityt EI, VilkeviCiUt EA, Gedvilait EG, Glebauskien EB, KriauCiUnien EL, et al. TBX15 rs98422, DNM3 rs1011731, RAD51B rs8017304, and rs2588809 Gene Polymorphisms and Associations With Pituitary Adenoma. Vivo. 2021;35:815–26.
Havre PA, Rice M, Ramos R, Kmiec EB. HsRec2/Rad51L1, a protein influencing cell cycle progression, has protein kinase activity. Exp Cell Res. 2000;254:33–44.
Orr N, Lemnrau A, Cooke R, Fletcher O, Tomczyk K, Jones M, et al. Genome-wide association study identifies a common variant in RAD51B associated with male breast cancer risk. Nat Genet. 2012;44:1182–4.
Song H, Dicks E, Ramus SJ, Tyrer JP, Intermaggio MP, Hayward J, et al. Contribution of Germline Mutations in the RAD51B, RAD51C, and RAD51D Genes to Ovarian Cancer in the Population. J Clin Oncol. 2015;33:2901–7.
Thermann R, Neu-Yilik G, Deters A, Frede U, Wehr K, Hagemeier C, et al. Binary specification of nonsense codons by splicing and cytoplasmic translation. EMBO J. 1998;17:3484–94.
Lindeboom RG, Supek F, Lehner B. The rules and impact of nonsense-mediated mRNA decay in human cancers. Nat Genet. 2016;48:1112–8.
Adelman CA, Lolo RL, Birkbak NJ, Murina O, Matsuzaki K, Horejsi Z, et al. HELQ promotes RAD51 paralogue-dependent repair to avert germ cell loss and tumorigenesis. Nature. 2013;502:381–4.
Ward JD, Muzzini DM, Petalcorin MI, Martinez-Perez E, Martin JS, Plevani P, et al. Overlapping mechanisms promote postsynaptic RAD-51 filament disassembly during meiotic double-strand break repair. Mol Cell. 2010;37:259–72.
Vaz F, Hanenberg H, Schuster B, Barker K, Wiek C, Erven V, et al. Mutation of the RAD51C gene in a Fanconi anemia-like disorder. Nat Genet. 2010;42:406–9.
Thomas G, Jacobs KB, Kraft P, Yeager M, Wacholder S, Cox DG, et al. A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1). Nat Genet. 2009;41:579–84.
Date O, Katsura M, Ishida M, Yoshihara T, Kinomura A, Sueda T, et al. Haploinsufficiency of RAD51B causes centrosome fragmentation and aneuploidy in human cells. Cancer Res. 2006;66:6018–24.
Qin HD, Shugart YY, Bei JX, Pan QH, Chen L, Feng QS, et al. Comprehensive pathway-based association study of DNA repair gene variants and the risk of nasopharyngeal carcinoma. Cancer Res. 2011;71:3000–8.
Schoenmakers EF, Huysmans C, Van de Ven WJ. Allelic knockout of novel splice variants of human recombination repair gene RAD51B in t(12;14) uterine leiomyomas. Cancer Res. 1999;59:19–23.
Nowacka-Zawisza M, Wisnik E, Wasilewski A, Skowronska M, Forma E, Brys M, et al. Polymorphisms of homologous recombination RAD51, RAD51B, XRCC2, and XRCC3 genes and the risk of prostate cancer. Anal Cell Pathol. 2015;2015:828646.
Cortez D. Replication-Coupled DNA Repair. Mol Cell. 2019;74:866–76.
Franca MM, Lerario AM, Funari MFA, Nishi MY, Narcizo AM, de Mello MP, et al. A Novel Homozygous Missense FSHR Variant Associated with Hypergonadotropic Hypogonadism in Two Siblings from a Brazilian Family. Sex Dev. 2017;11:137–42.
Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–95.
Singh P, Schimenti JC, Bolcun-Filas E. A mouse geneticist’s practical guide to CRISPR applications. Genetics. 2015;199:1–15.
Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–42.
Akeson EC, Davisson MT. Mitotic chromosome preparations from mouse cells for karyotyping. Curr Protoc Hum Genet. 2001;Chapter 4:Unit4 10.
Li N, Ding L, Li B, Wang J, D’Andrea AD, Chen J. Functional analysis of Fanconi anemia mutations in China. Exp Hematol. 2018;66:32–41.
Rodriguez-Acebes S, Mouron S, Mendez J. Uncoupling fork speed and origin activity to identify the primary cause of replicative stress phenotypes. J Biol Chem. 2018;293:12855–61.
Jackson DA, Pombo A. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J Cell Biol. 1998;140:1285–95.
Acknowledgements
The authors thank the patients and their family for participating in this study. The authors are grateful to LIM42 and SELA teams for providing technical assistance. We thank also to Dr. Alex N. Zelensky for his useful comments and to Isabel Ramos and Marina Jiménez-Ruiz (Molecular Mechanisms Program, Centro de Investigación del Cáncer) for their help in genotyping and intraperitoneal injections of the mice used in this study. We also are indebted to Maria Daniela Corte Torres (Biobanco of the Principado de Asturias/Instituto de Investigación Sanitaria del Principado de Asturias) for her technical assistance. This work was supported by the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) Grant 2014/14231-0 (to MMF); FAPESP Grant 2013/02162-8, Nucleo de Estudos e Terapia Celular e Molecular (NETCEM), Conselho Nacional de Desenvolvimento Científico e Tecnológico Grant 303002/2016-6 (to BBM); and FAPESP Grant 2014/50137-5 (to SELA). This work was supported by MINECO (PID2020-120326RB-I00) and by Junta de Castilla y León (CSI239P18 and CSI148P20). NFM, FSS, and MRMH are supported by European Social Fund/JCyLe grants (EDU/310/2015, EDU/556/2019 and EDU/1992/2020). YBC and RSU are funded by a grant from MINECO (BS-2015–073993 and BFU2017-89408-R). Experiments performed at CNIO were supported by grant PID2019-106707-RB to JM, co-sponsored by EU ERDF funds. SM was supported by an international postdoctoral contract “CNIO Friends”. The proteomic analysis was performed in the Proteomics Facility of Centro de Investigación del Cáncer, Salamanca, Grant PRB3(IPT17/0019 -ISCIII-SGEFI/ERDF). CIC-IBMCC is supported by the Programa de Apoyo a Planes Estratégicos de Investigación de Estructuras de Investigación de Excelencia cofunded by the Castilla–León autonomous government and the European Regional Development Fund (CLC–2017–01). Veitia’s Lab is supported by the University of Paris and the Centre National de la Recherche Scientifique.
Author information
Authors and Affiliations
Contributions
MMF and ME contributed with the identification of the human variant of RAD51B. YBC, performed the validation of the truncating variant in vitro and characterization of the mutant mice including the cytological and biochemical analysis with the help of NFM, FSS, RSU, and MRMH, which contributed also with the formal analysis. SM and JM performed the stretched DNA fiber experiments. RGV contributed with the MS data analysis. MSM carried out the Cas9 injections. AA performed pituitary immunohistochemistry. EL, RV, BBM and AMP designed the experiments and wrote the paper with the input of the remaining authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics statement
Mice were housed in a temperature-controlled facility (specific pathogen free, spf) using individually ventilated cages, standard diet and a 12 h light/dark cycle, according to EU laws at the “Servicio de Experimentación Animal, SEA”. Mouse protocols have been approved by the Ethics Committee for Animal Experimentation of the University of Salamanca (USAL). We made every effort to minimize suffering and to improve animal welfare. Blinded experiments were applied when possible. No randomization methods were applied since the animals were not divided in groups/treatments. The minimum size used for each analysis was two animals/genotype. The mice analyzed were between 2 and 4 months of age, except in those experiments where it is indicated.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by D. Aberdam
Supplementary information
Rights and permissions
About this article
Cite this article
Franca, M.M., Condezo, Y.B., Elzaiat, M. et al. A truncating variant of RAD51B associated with primary ovarian insufficiency provides insights into its meiotic and somatic functions. Cell Death Differ 29, 2347–2361 (2022). https://doi.org/10.1038/s41418-022-01021-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-022-01021-z
This article is cited by
-
RAD51 and Infertility: A Review and Case-Control Study
Biochemical Genetics (2023)