Abstract
T helper 1 (Th1) immunity is typically viewed as a critical adaptation by vertebrates against intracellular pathogens. Identifying novel targets to enhance Th1 cell differentiation and function is increasingly important for anti-infection immunity. Here, through small-molecule screening focusing on epigenetic modifiers during the in vitro Th1 cell differentiation process, we identified that the selective histone deacetylase 6 (HDAC6) inhibitors ricolinostat and nexturastat A (Nex A) promoted Th1 cell differentiation. HDAC6-depleted mice exhibit elevation of Th1 cell differentiation, and decreased severity of Listeria monocytogenes infection. Mechanistically, HDAC6 directly deacetylated CBP-catalyzed acetylation of signal transducer and activator of transcription 4 (STAT4)-lysine (K) 667 via its enzymatic activity. Acetylation of STAT4-K667 is required for JAK2-mediated phosphorylation and activation of STAT4. Stat4K667R mutant mice lost the ability to normally differentiate into Th1 cells and developed severe Listeria infection. Our study identifies acetylation of STAT4-K667 as an essential signaling event for Th1 cell differentiation and defense against intracellular pathogen infections, and highlights the therapeutic potential of HDAC6 inhibitors for controlling intracellular pathogen infections.
Similar content being viewed by others
Introduction
IL-12 promotes Th1 cell differentiation and augments IFNγ production through STAT4 activation. In mice, deficiency or inactivation of STAT4 significantly impairs Th1 cell differentiation [1,2,3]. As research on STATs has progressed over the years, it has become clear that STAT proteins are subject to important posttranslational modifications (PTMs), such as acetylation which heavily affects their dimerization, nuclear translocation and DNA-binding ability [4]. However, STAT4 is the only STAT family member that has not been studied for acetylation.
Histone acetyltransferases (HATs) and HDACs/sirtuins control the acetylation of lysine residues.. HDAC6 plays multiple roles in immune pathway and exerts different effects on different T cell subsets [5,6,7,8]. In Hdac6–/– mice, the percentage of interleukin-17-secreting γδT cells increased [9]. Tregs from Hdac6–/– mice exhibit enhanced immunosuppressive activity, and administration of HDAC6 inhibitors substantially ameliorates colitis progression in mice [10]. To date, whether HDAC6 is involved in Th1 cell differentiation and bacterial disease remains elusive. Two studies reported that the ricolionstat significantly augments the percentage and the antitumor characteristics of IFNγ+ CD4+ T cells in myeloma [11, 12]. Although both pan- and selective-HDACis have the potential to modulate the functions of all types of cells, their effects on T cells and on IFNγ-secreting Th1 cells in particular are not fully illustrated.
Listeria monocytogenes is an intracellular pathogen of humans and animals and is widely used as a model organism in studying bacterial pathogenesis and host defense [13]. A previous study showed that IL-12 does not play a direct role in the early innate immune clearance of L. monocytogenes, but IL-12-dependent IFNγ production is important for generating Th1 cells and shaping the ensuing inflammatory response [3, 14]. Individuals with IFNγ deficiency are more susceptible to environmental pathogens. Therefore, identifying new mechanisms involved in Th1 cell differentiation is crucial for the development of new therapeutic strategies against Listeria infection.
Here, we screened a library of chemical compounds targeting epigenetic enzymes for their potential roles in the Th1 cell differentiation process and identified ricolinostat and Nex A obviously promote Th1 cell differentiation and decrease Listeria infection in mice. With a series of in vivo and in vitro assays, we proved that HDAC6 directly deacetylates CBP-catalyzed acetylation of STAT4-K667, established HDAC6’s function in regulating the Th1 cell differentiation process, and provided selective HDAC6 inhibitors as potential therapies for intracellular bacterial infectious diseases.
Results
Epigenetic compound library screening identifies HDAC6 as an inhibitor of Th1 cell differentiation
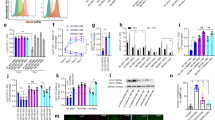
To explore the protein modifier landscape of Th1 cell differentiation, mouse primary CD4+ T cells were treated with a library of compounds targeting epigenetic enzymes, including HATs, HDACs, methyltransferases, demethylases and so on, while they were primed to the Th1 subtype. We used quantitative polymerase chain reaction (qPCR) indexing of Ifng to identify compounds that interfered with the Th1 cell differentiation process (Fig. S1A). We identified a set of compounds that enhanced Ifng expression over vehicle treatment by sixteen fold or more (Fig. 1A). Among these lead compounds were four inhibitors that target HDACs: quisinostat, oxamflatin, vorinostat and ricolinostat. The former three are paninhibitors of HDACs; however, the last one, ricolinostat, is a selective inhibitor of HDAC6. We then analyzed all the selective inhibitors of HDAC6 and three of them significantly promoted Th1 cell differentiation (Fig. 1B). The convergent effects of chemically dissimilar compounds suggested that the effects were exerted through HDAC6 inhibition. Through flow cytometry analysis, we confirmed that the HDAC6 inhibitors enhanced Th1 cell differentiation (Fig. 1C, D).
A Relative expression of Ifng in CD4+ T cells on day 3 of induction into Th1 cell subsets treated with epigenetic compounds (black dots). The top eight compounds that induced expression of Ifng by more than sixteen-fold over that induced by vehicle (mock) treatment are listed on the right. Blue dots, compounds that preferentially target HDAC. B qPCR analysis of the mRNA level of Ifng in CD4+ T cells on day 3 of induction into Th1 cell subsets in the absence or presence of HDACs inhibitors. TSA, trichostatin. Representative fluorescence-activated cell sorting (FACS) plots (C) and population frequencies (D) of Th1 cells polarized in the presence or absence of the indicated inhibitors are shown. E Listeria CFUs per spleen (left) and liver (right) of WT mice infected with 1 ×104 L. monocytogenes i.v. and treated with vehicle saline (mock), TSA (1 mg/kg), Nex A (3 mg/kg) or Ricolinostat (3 mg/kg) accordingly i.p. Mice were sacrificed for the following analysis on day 5 post infection. Colored dots represent individual mice (n = 6). Bars represent the mean log CFU/organ. F Survival rate of mice from Fig. 1E (n = 7). G, H Representative FACS plots (G) and cell number (H) of CD4+ IFNγ+ T cells in the spleen and liver of mice from Fig. 1E are shown. The data are shown as the mean ± SD of three independent experiments. *p < 0.05, **p < 0.01 and ***p < 0.001, n.s means not significant (Student’s t test).
With Listeria infection model, we found that Nex A and ricolinostat obviously reduced Listeria burdens and increased the survival rate of WT mice over time (Fig. 1E, F and Fig. S1B). Compared with saline treatment, the administration of HDAC6 inhibitors profoundly increased the number of Th1 cells in the spleen and liver of Listeria-infected mice (Fig. 1G, H); however, the HDAC6 inhibitors displayed a minor effect on the amount of cytotoxic CD8+ IFNγ+ T cells (Fig. S1C, D). These findings provide Nex A and ricolinostat as potential therapeutic drugs for preventing intracellular pathogen infection and suggest that HDAC6 is involved in the Th1 cell differentiation process.
Hdac6-deficient mice are resistant to Listeria infection
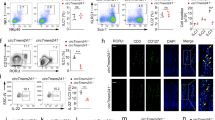
We further confirmed that HDAC6 deficiency leads to enhanced Th1 cell polarization (Fig. 2A, B and Fig. S2A). Hdac6 deficiency in mice was confirmed with qPCR (Fig. S2B). Knockdown of HDAC6 in WT CD4+ T cells consistently enhanced Th1 cell differentiation (Fig. S2C–E). HDAC6 depletion also increased Th17 cell differentiation but exhibited less of an effect on Treg cell differentiation (Fig. S2F, G). Since Th17 cells mainly mediate adaptive immunity against extracellular bacterial infection, we focused on how HDAC6 regulates Th1 cell-mediated immunity in the following study.
Representative FACS plots (A) and population frequencies (B) of Th1 cells polarized from wild-type (WT) or Hdac6–/– naïve CD4+ T cells are shown. C Listeria CFUs per spleen (left) and liver (right) of WT and Hdac6−/− mice infected with 1 ×104 L. monocytogenes i.v (n = 6). D Survival rate of mice from Fig. 2C (n = 7). Representative FACS plots (E) and cell number (F) of CD4+ IFNγ+ T cells in the spleen and liver of mice from Fig. 2C are shown. G Listeria CFUs per spleen (left) and liver (right) of infected Rag2−/− mice reconstituted with either PBS or WT or Hdac6−/− naïve CD4+ T cells (n = 4). Reconstituted Rag2−/− mice were infected with 1 × 104 Listeria and sacrificed on day 5 for further analysis. Recon: reconstitution. Representative FACS plots (H) and cell number (I) of CD4+ IFNγ+ T cells in the spleen and liver of mice from Fig. 2G are shown. The data are shown as the mean ± SD of three independent experiments. *p < 0.05, **p < 0.01 and ***p < 0.001, n.s means not significant (Student’s t test).
Olga Moreno-Gonzalo et al. reported in 2017 that during the first hours to 24 h of Listeria infection, Hdac6–/– bone marrow-derived dendritic cells (BMDCs) were less efficient in clearing Listeria than WT BMDCs. However, they observed that HDAC6 depletion exhibited a protective effect against Listeria infection from 3 to 10 days postinoculation [15], strongly indicating that HDAC6 plays different roles in regulating innate and adaptive immunity. In our experimental setting, we found that bone marrow-derived macrophages (BMDMs) from WT and Hdac6–/– mice under IFNγ−treatment expressed comparable amounts of the IFN-stimulated genes Cxcl9, Cxcl10, Ccl5, Icam and iNOS2 and the proinflammatory cytokines Il6, Tnfα and Il1β (Fig. S2H). However, Hdac6−/−- mice were more resistant to Listeria infection, had less bacterial load in the spleen and liver on day 5 postinfection and exhibited an increased survival rate compared with WT mice (Fig. 2C, D). Meanwhile, the accumulation of Th1 cells increased obviously in the spleen and liver of Listeria-infected Hdac6–/– mice compared with WT mice (Fig. 2E, F).
We further isolated naïve CD4+ T cells from WT or Hdac6−/− mice and adoptively transferred them into Rag2−/− mice devoid of T and B cells. After successful reconstitution, Hdac6−/− CD4+ T cell-reconstituted Rag2−/− mice accumulated more Th1 cells than WT CD4+ T cell-reconstituted mice (Fig. S2I). None of the reconstituted Rag2−/− mice died during the 5 days post Listeria infection. However, we found that compared with WT CD4+ T cell-transferred mice, adoptive transfer of Hdac6−/− CD4+ T cells into Rag2−/− mice significantly decreased bacterial burden (Fig. 2G). As expected, Th1 cells were more abundant in the spleen and liver of Hdac6−/− CD4+ T cells reconstituted Rag2−/− mice (Fig. 2H, I), suggesting that Hdac6−/− CD4+ T cells have a higher potential to differentiate into Th1 cells.
These results demonstrate a critical role of HDAC6 in inhibiting the differentiation and antibacterial immunity of Th1 cells; however, the underlying mechanism needs to be investigated further.
HDAC6 and CBP reversibly modify the acetylation pattern of STAT4
With mass spectrometric analysis. HDAC6 was identified as a STAT4-binding protein (Fig. S3A). The interaction between STAT4 and HDAC6 was confirmed through a co-IP assay, both ectopically and endogenously (Fig. 3A, B). In CD4+ T cells, IL-12 induced acetylation eof STAT4, which was recognized by a pan acetyl-lysine antibody (Fig. 3C). Meanwhile, STAT4 acetylation intensity was preferentially elevated in differentiated Th1 cells compared with Th0 cells (Fig. 3D). Interestingly, HATs inhibitors from the epigenetic compound library evenly decreased Th1 cell polarization (Fig. 3E, F and Fig. S2B). We further found that CBP is the major acetyltransferase of STAT4 (Fig. 3G). STAT4 and CBP formed complexes in CD4+ T cells (Fig. 3H) and in HEK293T cells overexpressed with STAT4 and CBP (Fig. S3C). STAT4 acetylation level was obviously decreased in SGC-CBP30-treated CD4 + T cells compared with that in control cells (Fig. S3D). CBP greatly enhanced the transcriptional activity of STAT4 in luciferase assay (Fig. S3E). Knockdown of CBP in CD4+ T cells obviously decreased Th1 cell polarization (Fig. 3I and Fig. S3F). The results indicated that CBP enhanced STAT4 transcriptional activity by acetylating its lysine residues.
Coimmunoprecipitation (CoIP) and immunoblot analysis of HEK293T cells (A) transfected with the indicated constructs or primary CD4+ T cells (B). Immunoblot analysis of IL-12-treated CD4+ T cells (C) or polarized CD4+ T cells (D) after immunoprecipitation with an anti-STAT4 antibody. E qPCR analysis of the mRNA level of Ifng in CD4+ T cells on day 3 of induction into Th1 cell subsets in the absence or presence of HATs inhibitors. F Representative FACS plots of Th1 cells polarized in the presence or absence of HATs inhibitors are shown. G Immunoblot analysis of HEK293T cells transfected with the indicated constructs followed by immunoprecipitation with an anti-myc antibody. H CoIP and immunoblot analysis of primary CD4+ T cells treated with or without IL-12. I Representative FACS plots of Th1 cells polarized from CBP-knockdown and control naïve CD4+ T cells are shown. J Immunoblot analysis of STAT4 acetylation in HEK293T cells transfected with the indicated constructs. K Immunoblot analysis of STAT4 acetylation incubated in an in vitro system. The data are representative of three independent experiments with similar results or shown as the mean ± SD of three independent experiments. *p < 0.05, **p < 0.01 and ***p < 0.001 (Student’s t test).
As expected, CBP-mediated STAT4 acetylation was abolished by HDAC6 both in vivo and in vitro (Fig. 3J, K). We constructed inactive HDAC6 by mutating the critical histidine residue H216 and H611 into alanine (A) separately or simultaneously. HDAC6-H216A did not jeopardize HDAC6 enzymatic activity; however, HDAC6-H611A and HDAC6-H216, 611A completely lost deacetylase activity, indicating that the HDAC6 H611 enzymatic site was responsible for STAT4 deacetylation (Fig. 3J). Therefore, these observations show that HDAC6 and CBP are involved in the Th1 cell differentiation process by reversibly manipulating STAT4 acetylation.
Acetylation of STAT4-K667 is required for Th1 cell differentiation
We constructed vectors expressing STAT4 with K-to-arginine (R) mutations that mimic the loss of acetylation status or a tyrosine (Y)-to-phenylalanine (F) mutation that abrogates tyrosine phosphorylation. Among all the K-to-R mutants tested, only STAT4-K667R lost the transcriptional activity, which was quite similar to STAT4-Y693F (Fig. 4A). K667 and K691 of STAT4 were demonstrated to be acetylated lysine residues via mass spectrometric analysis (Fig. 4B). However, STAT4-K691R exhibited normal transcriptional activity (Fig. 4A). Only STAT4-K667R reduced the CBP-catalyzed acetylation intensity, and neither the K691R, K673R mutant nor C-terminal domain-deleted STAT4 affected the acetylation level of STAT4 (Fig. S4A, B). STAT4-K667 is located in its SH2 domain and is highly conserved among STAT family proteins and among different species (Fig. 4C and Fig. S4C). Then, we generated a rabbit polyclonal antibody specifically recognizing acetylation on STAT4-K667 (Fig. S4D). We validated that CBP indeed acetylated STAT4-K667 (Fig. 4D). Moreover, acetylation of STAT4-K667 was observed in Th1 cells (Fig. 4E). SGC-CBP30 significantly decreased STAT4-K667 acetylation (Fig. 4F); however, TSA and Nex A obviously enhanced STAT4-K667 acetylation intensity in Th1 cells (Fig. 4G). Furthermore, HDAC6 directly deacetylated the STAT4-K667 acetyl peptide (Fig. 4H). These findings prove that CBP and HDAC6 reversibly modify the acetylation of STAT4-K667 in Th1 cells.
A The IRF-luciferase reporter activity assay was performed in HEK293T cells transfected with empty vector (EV), WT or mutant STAT4 constructs. B Mass spectrometry analysis of purified STAT4 protein revealed the acetylation of STAT4 at two lysine residues. C Sequence alignment showing that the acetylated lysine residue 667 of STAT4 is conserved in both humans and mice species, as well as in the STAT protein family. D Immunoblot analysis of STAT4-K667 acetylation in HEK293T cells transfected with the indicated constructs. E Immunoblot analysis of polarized CD4+ T cells after immunoprecipitation with an anti-STAT4 antibody. F Immunoblot analysis of STAT4-K667 acetylation in CD4+ T cells pretreated with PBS or SGC-CBP30 (1 μM) for 12 h followed by IL-12 treatment or not. G Immunoblot analysis of STAT4-K667 acetylation in CD4+ T cells primed into Th1 cells in the absence or presence of the indicated inhibitors. H Mass spectrometry analysis of acetylated STAT4-K667 peptide which were incubated with HDAC6 or not under in vitro system. I, J Representative FACS plots (I) and population frequencies (J) of Th1 cells polarized from naïve Stat4−/− CD4+ T cells reconstituted with WT or mutant STAT4 are shown. K qPCR analysis of the mRNA levels of Stat4 (upper) and Ifng (lower) in naïve Stat4−/− CD4+ T cells reconstituted with WT or mutant STAT4, which were induced to differentiate into Th1 cells. The data are representative of three independent experiments with similar results or shown as the mean ± SD of three independent experiments. **p < 0.01 and ***p < 0.001, n.s means not significant (Student’s t test).
STAT4 deficiency significantly impaired Th1 cell differentiation (Fig. S4E, F). By reconstituting WT or mutant STAT4 in Stat4−/− CD4+ T cells, we found that neither STAT4-K667R nor Y693F could rescue Th1 cell differentiation; however, the gain-of-function mutant STAT4-K667A partially rescued Th1 cell differentiation, as did WT STAT4 (Fig. 4I, J). STAT4-K673R and K691R possessed the ability to maintain Th1 differentiation (Fig. 4A, K). Taken together, our results demonstrate that STAT4-K667 is a bona fide acetylation site, which is important for STAT4 functionality.
STAT4-K667 acetylation is necessary for Y693 phosphorylation and STAT4 nuclear translocation
We further delineated the relationship between K667 acetylation and Y693 phosphorylation to investigate the mechanism by which acetylation of STAT4-K667 regulates its function. STAT4-K667R abolished JAK2-catalyzed phosphorylation of Y693 (Fig. S5A). However, STAT4-Y693F displayed the same acetylation level as WT STAT4 (Fig. S5B). In the WT or mutant STAT4 stable-transfected A1.1 cell line, which is a mouse-derived helper T cell hybridoma, STAT4-K667R markedly decreased phosphorylation of STAT4-Y693 (Fig. 5A). The acetylation mimic mutants K667Q (glutamine) and K667A partially rescued STAT4 phosphorylation intensity (Fig. 5B). Consistently, WT HDAC6 substantially decreased STAT4 phosphorylation intensity, while HDAC6 -H611A had no such effect (Fig. 5C). In primary CD4+ T cells, knockdown of CBP resulted in decreased phosphorylation levels of STAT4-Y693 (Fig. 5D). Both HDAC6 knockdown and depletion in CD4+ T cells increased STAT4-K667 acetylation and STAT4-Y693 phosphorylation (Fig. 5E–G). In Listeria-infected mice, HDAC6 inhibitors obviously enhanced the STAT4-K667 acetylation intensity (Fig. 5H). These data suggest that there is crosstalk between STAT4-K667 acetylation and STAT4-Y693 phosphorylation.
A Immunoblot analysis of STAT4-Y693 phosphorylation levels in A1.1 cells stably transfected with STAT4-constructs treated with or without IL-12. B, C Immunoblot analysis of STAT4-Y693 phosphorylation levels in HEK 293 T cells transfected with the indicated constructs. Immunoblot analysis of STAT4-Y693 phosphorylation levels in control or CBP-knockdown (D) or HDAC6-knockdown (E) CD4+ T cells treated with IL-12 or not. F Left, immunoblot analysis of STAT4 acetylation and phosphorylation levels in WT or HDAC6-deficient CD4+ T cells treated with or without IL-12. Right, quantification of the relative modified STAT4 level from three independent experiments. G Mean flour intensity of phosphorylated STAT4-Y693 in WT and Hdac6−/− naïve CD4+ T cells on day 4 of induction into Th1 cell subsets. H Mean flour intensity of acetylated STAT4-K667 in CD4+ T cells from the spleen and liver of mice from Fig. 1E. I CoIP and immunoblot analysis of HEK293T cells transfected with the indicated constructs. J Immunofluorescence analysis of CD4+ T cells from WT or Stat4K667R mice treated with or without IL-12 for 30 min. Scale bar, 50 mm. K, L The IRF-luciferase reporter activity assay was performed in HEK 293 T cells transfected with the indicated constructs. The data are representative of three independent experiments with similar results or shown as the mean ± SD of three independent experiments. *p < 0.05, **p < 0.01 and ***p < 0.001 (Student’s t test).
Interestingly, we found that compared with the binding between JAK2 and WT STAT4, the interaction between JAK2 and STAT4-K667R was dramatically reduced (Fig. 5I), indicating that K667 acetylation is required for JAK2-STAT4 complex formation. With immunofluorescence assay, we found that the intensity of phosphorylated STAT4 decreased obviously, and STAT4-K667R protein exhibited less sharp accumulation in the nucleus of IL-12-treated Stat4K667R CD4+ T cells compared with that in IL-12-treated WT CD4+ T cells (Fig. 5J). It has been reported that leukemia inhibitory factor (LIF) and IFNα can induce STAT4-Y693 phosphorylation and activation [16,17,18]. Here, we found that the STAT4-K667R also exhibited reduced phosphorylation intensity and nuclear translocation in LIF- or IFNα-treated HeLa cells (Fig. S5C, D). Meanwhild, STAT4-K667R displayed a low level of transcriptional activity; however, the K667Q and K667A mutants partially rescued STAT4 transcriptional activity (Fig. 5K). HDAC6 substantially decreased the transcriptional activity of STAT4, while HDAC6-H611A exhibited a much weaker effect on the transcriptional activity of STAT4 (Fig. 5L). Collectively, these results demonstrated that acetylation of STAT4-K667 is required for its interaction with JAK2, which safeguards the subsequent phosphorylation of Y693, nuclear translocation and transcription activity.
Stat4 K667R mice display impaired Th1 cell differentiation and low expression of Th1 cell polarization-related genes
We further generated Stat4K667R mice with the crisp/cas9 system (Fig. S6A). The frequencies of CD4+ and CD8+ T cells were similar under homeostatic conditions in the peripheral lymph organs of WT and Stat4K667R mice (Fig. S6B, C). Stat4K667R and Stat4−/− CD4+ T cells comparably lost the ability to differentiate into Th1 cells compared with WT CD4+ T cells (Fig. 6A, B). However, STAT4-K667R did not affect Th17 cell and Treg cell differentiation (Fig. S6D).
Representative FACS plots (A) and population frequencies (B) of Th1 cells polarized from WT, Stat4K667R or Stat4–/– naïve CD4+ T cells are shown. C Top 100 differentially expressed genes from RNA-seq among WT, KO (Stat4–/–) and KR (Stat4K667R) Th1 cells. D Volcano plot of significantly altered genes between WT and KR Th1 cells. E Hierarchical clustering of Pearson correlations of DESeq2 normalized counts for the RNA-seq libraries among WT, KO and KR Th1 cells. Colors represent the Pearson correlation coefficient. F GSEA plots of KR Th1 cells show downregulation of Th1 cell differentiation. G GO analysis of WT and KR Th1 cells for the top differentially regulated gene sets. H Listeria CFUs per spleen (left) and liver (right) of WT, Stat4−/− and Stat4K667R mice infected with 1 ×104 L. monocytogenes (n = 6). Representative FACS plots (I) and cell number (J) of CD4+ IFNγ+ T cells in the spleen and liver of mice from Fig. 6H are shown. The data are representative of three independent experiments with similar results or shown as the mean ± SD of three independent experiments. ***p < 0.001 (Student’s t test).
By interrogating the gene expression signatures of WT, Stat4–/– and Stat4K667R Th0 and Th1 cells, we determined that in Stat4K667R Th0 cells, the expression of Th1 cell marker genes Ifng, Tbx21, Il12rb2 and genes associated with T cell cytotoxicity (Prf1 and Nkg7) significantly decreased, while the transcriptome profiles were quite similar between WT and Stat4–/– Th0 cells (Fig. S6E–G). Gene set enrichment analysis (GSEA) revealed that the IL-12-mediated signaling pathway was substantially downregulated in Stat4K667R Th0 cells (Fig. S6H). Both hierarchical correlation clustering analysis and principal component analysis (PCA) showed different gene expression signatures in wild-type, Stat4–/– and Stat4K667R Th0 and Th1 cells (Fig. S6I, J). The expression levels of Ifng and Tbx21 decreased more significantly in both Stat4K667R and Stat4–/– Th1 cells than in WT Th1 cells (Fig. 6C, D). Consistently, hierarchical correlation clustering analysis showed an obvious correlation between Stat4K667R and Stat4–/– Th1 cells, which separately clustered from WT Th1 cells (Fig. 6E). GSEA and GO analysis revealed that the genes downregulated in Stat4K667R cells were enriched in Th1 cell differentiation, cytokine signaling and the JAK-STAT signaling pathway (Fig. 6F, G). Genes related to T cell activation and proliferation were also downregulated in Stat4K667R Th1 cells (Fig. 6G). Within the CD4+ T cell compartment from both the spleen and lymph nodes, we observed a significant decrease in the proportion of effector and memory cells (CD62Llow CD44high) and proliferative Ki67+ cells in Stat4K667R CD4+ T cells (Fig. S6I, J). Between the WT and Stat4K667R CD8+ T cell compartments, however, we noticed less obvious differences in the proportions of effector and memory cells and proliferating cells (Fig. S6K, L). However, T cell proliferation inhibitory genes and myeloid differentiation signaling effectors, such as proteoglycan (Prg2), leucine rich repeat containing 32 (Lrrc32), and platelet factor 4 (Pf4), were upregulated in Stat4K667R Th1 cells (Fig. 6D, G).
Together, these data indicate that acetylation of STAT4-K667 is required for the expression of Th1 cell differentiation-related genes.
ChIP-Seq analysis revealed STAT4 binding in the promoter of Th1 cell polarization-related genes
We noticed that the expression of Th1 cell polarization-related genes consistently decreased in Stat4–/– and Stat4K667R Th1 cells (Fig. S7A), suggesting that the expression of those genes was dependent on STAT4-K667 acetylation. qPCR analysis confirmed a reduction in the STAT4 acetylation-dependent genes (Fig. S7B). Analysis of publicly available chromatin immunoprecipitation (ChIP)-seq data from polarized WT and Stat4–/– Th1 cells [19], revealed a strong enrichment of STAT4 occupancy in the promoter/enhancer regions of Ifng, Ccl4, Csf1 and so on(Fig. S7C),and STAT4 deficiency resulted in a significant reduction in H3K4me3 and H3K4me1 marks (Fig. S7D) [20, 21]. On the other hand, we analyzed the publicly available data of the assay for transposase-accessible chromatin using sequencing (ATAC-seq) from WT and Stat4–/– T follicular helper (Tfh) cells, which are a functionally and phenotypically distinct subset of CD4+ Th cells that produce the cytokines IL-21 and IFNγ [22]. The results showed that the chromatin accessibility in the promoters and proximal enhancers of the genes Ccl4, Tbx21, Lag3 and Ifng revealed no significant differences between WT and Stat4–/– Tfh cells (Fig. S7E). We proposed that neither STAT4 deficiency nor STAT4-K667R mutation would affect the accessible chromatin landscape of Th1 cell differentiation-related genes. With ChIP-qPCR, we found that STAT4-K667R displayed less occupancy on the promoters of Ifng, Ccl4 and Csf1 (Fig. S7F). These findings suggested that acetylation of K667 promoted STAT4 targeting of Th1 cell differentiation-related genes.
Stat4 K667R mice are more susceptible to Listeria infection
In Listeria infection model, Stat4–/– mice showed dramatically increased bacterial burdens in the spleen and liver and a significantly decreased survival rate compared with WT mice; however, the disease severity of Stat4K667R mice was slightly milder than that of Stat4–/– mice but was more severe than WT mice (Fig. 6H and S8A). The percentage of Th1 cells from the spleen and liver of both Stat4K667R and Stat4–/– mice was significantly less than the amount of Th1 cells from the WT group (Fig. 6I, J). The expression of chemokines and proinflammatory cytokines was comparably increased between IFNγ-treated WT and Stat4K667R BMDMs compared with untreated control cells (Fig. S8B). These results suggest that acetylation of STAT4-K667 plays a critical role in maintaining Th1 cell-mediated immunity against Listeria infection.
HDAC6 inhibitors reduce Listeria infection in a STAT4-K667-dependent manner
We further found that the inhibitors TSA, Nex A and ricolinostat significantly promoted Th1 cell differentiation in WT CD4+ T cells but not in Stat4K667R CD4+ T cells (Fig. 7A–C). Meanwhile, the infection-preventing effect of HDAC6 inhibitors only existed in WT mice but not in Stat4–/– or Stat4K667R mice (Fig. 7D), indicating that the protective effect of HDAC6 inhibitors in Listeria infection was dependent on STAT4-K667.
Representative FACS plots (A) and population frequencies (B) of Th1 cells polarized from WT or Stat4K667R naïve CD4+ T cells are shown. WT or Stat4K667R naïve CD4+ T cells were cultured in the absence or presence of HDACs inhibitors. C qPCR analysis of the mRNA level of Ifng in the CD4+ T cells from Fig. 7A. D Listeria CFUs per spleen (left) and liver (right) of WT, Stat4−/− and Stat4K667R mice infected with L. monocytogenes i.v. and treated with vehicle saline, TSA, Nex A or Ricolinostat accordingly i.p. (n = 6). E Schematic diagram of Th1 polarization induced by STAT4-K667 acetylation during Listeria infection. The data are representative of three independent experiments with similar results or shown as the mean ± SD of three independent experiments. *p < 0.05, **p < 0.01 and ***p < 0.001 (Student’s t test).
Taken together, the study proved that by promoting STAT4-K667 acetylation with HDAC6 inhibitors, Th1 cells accumulated significantly in the mouse spleen and liver, which effectively ameliorated L. monocytogenes infection in mice (Fig. 7E).
Discussion
In this study, we aimed to identify new pharmacological molecules involved in the Th1 cell differentiation process. We found that ricolionstat and Nex A dramatically elevated Th1 cell differentiation and significantly ameliorated Listeria-burden in mice by inhibiting HDAC6 enzymatic activity and decreasing the acetylation level of STAT4. Moreover, Hdac6–/– mice exhibited more potential for Th1 cell development and were more resistant to Listeria infection than WT mice. Mice with defects in STAT4 acetylation failed to upregulate Th1 cell polarization and developed more severe Listeria infection. Our study highlights an important yet previously unappreciated role of HDAC6 in orchestrating Th1 cell differentiation and the related antibacterial response.
Mechanistically, CBP catalyzed the acetylation of STAT4-K667 in Th1 cells, which is located in the SH2 domain. The SH2 domain of STAT is composed of “αβββα” motifs discrete with positive lysine residues. The acetylation of these lysine residues may therefore remodel the confirmation of the SH2 domain to enable it to form a complex with phosphorylated tyrosine. We identified that the binding between STAT4-K667R and JAK2 decreased significantly, which explains why STAT4-K667R blocked phosphorylation of STAT4-Y693. The transcriptomes of WT, Stat4–/– and Stat4K667R Th0 and Th1 cells revealed that most Th1 cell differentiation-related genes were largely dependent on STAT4-K667 acetylation for their expression. This study identifies that STAT4 acetylation positively regulates STAT4 activity and primes Th1 cell development.
In the classical dichotomy hematopoiesis model, T cells and B cells arise from common lymphoid progenitors, while macrophages, neutrophils and other leukocytes arise from common myeloid progenitors. However, several studies have reported that T cell progenitors retain substantial B cell and myeloid potential in the adult thymus [23, 24]. Intriguingly, we noticed that the genes involved in the positive regulation of B cell activation, macrophage activation and monocyte chemotaxis and neutrophil chemotaxis were significantly upregulated in Stat4K667R CD4+ T cells (Fig. S6E, F). We speculate that STAT4-K667R might play a role in sustaining the myeloid potential of T cells in peripheral lymphoid organs.
CD8+ T cells are supposed to be the main T cells involved in the clearance of intracellular pathogens with cytotoxicity; however, we found that ricolionstat and Nex A exerted less significant effects on IFNγ-secreting CD8+ T cells. HDAC6 protein was expressed at the same level in both CD4+ and CD8+ T cells (data not shown). We excluded the possibility that the differential effects of HDAC6 on CD4+ and CD8+ T cells were caused by differential expression levels of HDAC6. Although IL-12 can augment both CD4+ and CD8+ T cells to secrete IFNγ during intracellular infection, the mechanisms involved in the two processes are quite different. In CD4+ T cells, IL-12-activated STAT4 is the key for Th1 cell differentiation and IFNγ secretion. However, it has been reported that in CD8+ T cells, the IL-12-mediated increase in IFNγ secretion is due to increased MAPKs and AKT activation and T-bet expression but not due to STAT4 activation [25,26,27]. Thus, we suggest that the reason HDAC6 shows less regulation of CD8+ T cells during Listeria infection is that HDAC6’s downstream substrate STAT4 displays a minor regulatory effect on CD8+ T cell function.
L. monocytogenes is a food-borne bacterium that causes listeriosis. Since listeriosis occurs more frequently in immunocompromised individuals and pregnant women, exploring new strategies to enhance Th1 cell-mediated immunity is of obvious significance. Our study shows that the HDAC6 inhibitors ricolinostat and Nex A could directly promote Th1 cell differentiation by enhancing acetylation of STAT4-K667 and ameliorate the L. monocytogenes infection in mice. This mechanism may be targeted in human intracellular bacterial infection treatment, and ricolinostat and Nex A could serve as potential therapies for treating intracellular pathogen infection.
Materials and methods
Mice
Stat4–/– and Rag2–/– mice were purchased from Nanjing Biomedical Research Institute of Nanjing University, and back crossed to C57BL6 background. Stat4K667R mutant mice were generated with the Crisp-cas9 system. Hdac6–/– mice were provided by the Yu Zhang laboratory from the National Institute of Biological Sciences, Beijing. Stat4–/–, Stat4K667R, Hdac6–/–, Rag2–/– and their representative wild-type control mice were maintained in a specific-pathogen-free (SPF) facility. 6–12 weeks old mice were used in all animal-related experiments. Age and sex matched mice were used in all animal-related experiments.
Culture and maintenance of cell lines
HEK293T, NIH 3T3 and Plat E cell lines were cultured in DMEM (HyClone) supplemented with 10% fetal bovine serum (Genmini), 100 units/ml penicillin, and 100 μg/ml streptomycin (Shenggong). A1.1 cell line was cultured in RPMI (HyClone) supplemented with 10% fetal bovine serum (Genmini), 100 units/ml penicillin, and 100 μg/ml streptomycin (Shenggong). All cell lines were purchased from the Cell Bank of the Chinese Academy of Sciences and routinely tested for mycoplasma contamination with PCR.
In vitro T cell culture
Spleens and lymph nodes were collected from mice of different genotypes, and single-cell suspensions were prepared by mechanical disruption in PBS. Naïve CD4+ T cells were isolated by magnetic sorting with a Miltenyi Biotec CD4+ CD62L+ T Cell Isolation kit, according to the manufacturer’s directions (Miltenyi Biotec, 130-106-643). CD4+ T cells were cultured in IMDM (HyClone) supplemented with 10% FBS (Genmini), 100 units/ml penicillin, 100 μg/ml streptomycin (Shenggong), 4 mM L-glutamine, and 50 μM β-mercaptoethanol, and activated with 5 μg/ml precoated anti-CD3 (eBioscience) and 2 μg/ml anti-CD28 (eBioscience). Th1 cells were differentiated by the addition of recombinant IL-12 (1 ng/ml, R&D Systems) and anti-IL-4 (10 μg/ml, BD Pharmingen). Th17 cells were differentiated by the addition of recombinant IL-6 (20 ng/ml, R&D Systems), recombinant TGF-β (1 ng/ml, R&D Systems), anti-IFNγ (10 μg/ml, BD Pharmingen) and anti-IL-4 (10 μg/ml, BD Pharmingen), followed by recombinant IL-23 (30 ng/ml, R&D Systems) on day 3. Treg cells were differentiated by the addition of TGF-β (5 ng/ml,R&D Systems) and recombinant IL-2 (40 ng/ml, PeproTech). CD4+ T cell cultures were split at a ratio of 1:2 on day 3 after activation.
Flow cytometry
Single cell suspensions were pelleted, resuspended and stained with Live/Dead-violet (Life Technologies) in PBS for 30 min on ice. Cells were washed and resuspended with surface-staining antibodies in PBS supplemented with 1% FBS. For transcription factor staining, cells were fixed and permeabilized with the FoxP3 staining buffer set from eBioscience according to the manufacturer’s protocol. Intracellular stains were prepared in 1X eBioscience permwash buffer containing conjugated antibodies. Staining was performed for 30–60 min on ice. For cytokine analysis, CD4+ or CD8+ T cells were activated for 4–5 h with PMA (50 ng/ml, Sigma) and ionomycin (500 ng/ml, Sigma), and Golgi Stop (BD Pharmingen) was added. After live/dead and surface staining, cells were treated with the Cytofix/Cytoperm solution set (BD Pharmingen) according to the manufacturer’s protocol. Intracellular stains were prepared in BD permwah. Absolute numbers of isolated cells from peripheral mouse tissues in all studies were determined by comparing the ratio of cell events to bead events of CountBright absolute counting beads. Samples were acquired on a Gallios (Beckman Coulter), and the data were analyzed with FlowJo software.
Epigenetic compound library screening
96-round-bottom well plates were coated with anti-CD3 antibody overnight at 4 °C. Naïve CD4+ T cells were isolated and primed into the Th1 cell subtype as described in the in vitro T cell culture section. A compound library of 188 chemicals targeting different epigenetic modifiers. Each compound in the epigenetic compound library (MCE) was diluted to 0.02 to 10 μM accordingly in IMDM medium. Naïve CD4+ T cells were treated with each compound or untreated from the scratch while they were cultured in Th1 cell conditions. The cells were collected 72 h later by centrifugation at 300 x g for 5 min, washed with PBS, and then treated with the Turbo Capture 96 mRNA Kit (Qiagen, 72251) following the manufacturer’s instructions. The cell lysates from each well were collected for qRT-PCR using the Hifair® II 1st Strand cDNA Synthesis Kit (Yeasen, 11121ES60), following the manufacturer’s instructions. Each compound was tested in triplicate.
RNA isolation and cDNA preparation
Total RNA from in vitro polarized T cells was extracted using TRIzol (Invitrogen) followed by DNase I (Qiagen) treatment and cleanup with the RNeasy MiniElute kit (Qiagen) following the manufacturer’s protocols. cDNA was generated using the Hifair® II 1st Strand cDNA Synthesis Kit (Yeasen, 11121ES60).
qRT-PCR
Quantitative RT-PCR was performed using the Hieff® qPCR SYBR Green Master Mix (Low Rox Plus) (Yeasen, 11202ES08) on the Real-Time PCR System (ABI QuantStudio™ 7 Flex). Gene specific primers spanning exons were used. Values were normalized to β-actin for each sample. ‘Fold differences’ were calculated with the ΔΔCt method.
Whole cell extraction, immunoprecipitation and western blot analysis
Cells were collected and lysed in ice-cold RIPA buffer (Millipore, 20–188) containing inhibitors (1 mM phenylmethylsulfonyl fluoride, cocktails of protease inhibitors, 1 mM Na3VO4, 1 mM NaF). Cell debris was removed by centrifugation at 4 °C, 13,000 r.p.m. for 15 min. The cell supernatants were subjected to immunoprecipitation using the appropriate antibodies and protein A or protein G-Sepharose beads. Protein extracts were analyzed by SDS-PAGE and immunoblotting with the indicated antibodies. The following primary antibodies were commercially obtained: acetylated-lysine (9441; Cell Signaling Transduction), Stat4 pY693 (554002; BD Biosciences), Stat4 (sc-398228; Santa Cruz), Myc, HA, Flag (F7425; Sigma-Aldrich), CBP, HDAC6 (sc-5258; Santa Cruz), actin and Gapdh. A polyclonal antibody against acetyl-lysine 667 STAT4 was generated by immunizing rabbits at Ab land company. Primary antibodies were visualized using corresponding secondary a-mouse or a-rabbit Dylight 800/680 antibodies (Li-COR) and signal intensities were quantified using the Li-COR Odyssey Infrared Imaging system. All of the full length original western blots for these results are provide in Supplementary File 1.
Luciferase reporter assay
HEK293T or NIH 3T3 cells were plated in 48-well plates and transfected with a DNA mixture containing 0.2 μg of 2x-IRF-luciferase reporter plasmid (Promega, pGL3-basic), 2 ng of a plasmid containing Renilla luciferase (Promega, pRL-CMV), and 0.2 μg of STAT4 constructs. Transfection was performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Chemical inhibitors or vehicle control was added 36 h after transfection and luciferase activity was measured 8 h later using a dual-luciferase reporter kit (Promega).
In vitro deacetylation assay
An in vitro deacetylation assay was performed as described previously [28]. Briefly, the Flag-SIRT1, Flag-HDAC6 and Flag-HDAC3 constructs were expressed in HEK293T cells individually. The proteins were purified with M2 beads and eluted with Flag peptide. STAT4-Flag and CBP constructs were coexpressed in HEK293T cells at a ratio of 1:5. Acetylated STAT4 was purified with M2 beads and eluted with Flag peptide. A total of 1 μg of the purified SIRT1, HDAC6 or HDAC3 was incubated with 1 μg of purified ac-STAT4 in reaction buffer (10 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM MgCl2, 10% glycerol and protease inhibitor cocktail) with or without NAD at 30 °C for 1 h. Samples were analyzed by western blot.
A total of 100 ng of acetylated-STAT4 K667 peptide (PDIPK(ac)DKAFGKHYS) was incubated with or without 1 μg of purified HDAC6 in the above reaction buffer at 30 °C for 1 h. Samples were desalted using C18 Zip Tips and were then subjected to LC-mass spectrometry for analysis.
Mass spectrometric analysis of lysine acetylation
MS analysis was performed as described previously ([16]). In brief, Myc-tagged STAT4 constructs of mouse origin and CBP constructs were transfected into HEK293T cells that were cultured in 10-cm Petri dishes using Lipofectamine 2000. Twenty-four hours later, the cells from two dishes were collected to immunoprecipitate STAT4 with 2 μg of anti-myc antibody and protein G agarose beads. The immunoprecipitated STAT4 from the above HEK293T cells was separated via SDS-PAGE. The Coomassie blue-stained STAT4 band was excised from the gel for protease digestion followed by liquid chromatography-tandem mass spectrometric analysis with a Thermo LC-MS/MS system. The spectra from MS were analyzed with Protein Discovery software (version 2.2).
Retrovirus package and reconstitution of T cells
Plat E cells were seeded into 10-cm petri dishes and transfected with a DNA mixture containing 21 μg of wild-type or mutant STAT4 plasmids (pMSCV-PIG-puro) and 3 μg of pcl-eco plasmid to generate retrovirus. 24 h later, the cell media were collected and filtered. Naïve CD4+ T cells were seeded into anti-CD3 precoated 48-well plates and cultured under Th1 cell differentiation conditions. 16 h after anti-CD3/28 activation, CD4+ T cell media were removed, and media containing retrovirus was gently added to CD4+ T cells. The 48-well plate was centrifuged at 900 g and 37 °C for 90 min. 4 h later, media containing retrovirus was discarded, and Th1 cell conditioned media were added back to CD4+ T cells. The above procedure was repeated once 24 h later. Puromycin was used to eliminate uninfected cells. The CD4+ T cell phenotype was analyzed on day4 with flow cytometry and qRT-PCR.
RNA-sequencing and data analysis of T cells
Th0 and Th1 cells from wild-type, Stat4K667R mutant or Stat4–/– mice were cultured as described above. Total RNA from Th0 or Th1 cells was extracted, cleaned up as described above and used for RNA-seq analysis. cDNA library construction and sequencing were performed by Beijing Novogene Corporation. Briefly, a total amount of 3 μg RNA per sample was used as input material for the RNA sample preparations. Sequence libraries were generated using the NEBNext UltraTM RNA Library Prep Kit for Illumina (NEB) following the manufacturer’s instructions, and index codes were added to attribute sequences to each sample. The clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS (Illumina) according to the manufacturer’s instructions. After cluster generation, the library preparations were sequenced on an Illumina HiSeq platform and 125 bp/150 bp paired-end reads were generated. We used featureCounts v1.5.0-p3 to count the reads numbers mapped to each gene. The FPKM of each gene was calculated based on the length of the gene and reads count mapped to this gene. Differential expression analysis of the two groups was performed using the DESeq2 R package (1.16.1). Heatmaps of gene expression were generated using the R language (version 3.2.1) and were generated on the basis of log2 (FPKM). Gene ontology (GO) enrichment analysis of differentially expressed genes was implemented using Gene Set Enrichment Analysis (GSEA; http://www.broadinstitute.org/gsea/index.jsp).
CUT&RUN
CUT&RUN experiments were carried out as described previously [29]. Briefly, 5 × 105 fresh cells were harvested, captured with concanavalin A-coated magnetic beads and incubated with anti-H3K27ac antibody or control IgG antibody overnight at 4 °C. After washing away the unbound antibody with Dig-wash buffer (20 mM HEPES-NaOH pH 7.5, 150 mM NaCl, 0.5 mM spermidine, 0.025% digitonin and 1x protease inhibitor cocktails), protein A-MNase was added and incubated for 1 h at 4 °C. After washing, CaCl2 was added to each sample at a final concentration of 2 mM to activate protein A-MNase. The reaction was carried out in ice water bath for 30 min and stopped by addition of 2xSTOP buffer (340 mM NaCl, 20 mM EDTA, 4 mM EGTA, 0.05% digitonin, 100 μg/mL RNase A and 50 μg/mL glycogen). DNA was extracted with MinElute PCR Purification Kit (Qiagen, 28004).
L. monocytogenes infection and enumeration of bacterial load
The L.monocytogenes strain 10403 s (provided by Y. Qian, Institute of Health Sciences, Shanghai Institutes for Biological Sciences) was used in this study. L. monocytogenes were stored as glycerol stocks at −80 °C and cultured in brain heart infusion (BHI) broth containing 50 μg/ml streptomycin overnight at 37 °C with shaking. For infections, L. monocytogenes were grown to mid-log phase and then diluted into pyrogen-free PBS. Age- and sex-matched mice (8–10 weeks of age) were injected i.v. with 1 × 104 CFU of L. monocytogenes and killed on day 5. To determine organ Listeria burden, spleens and livers were homogenized separately in PBS. Serial dilutions of homogenate were plated on BHI agar plates, and L. monocytogenes CFUs were assessed after overnight growth at 37 °C.
CD4+ T cell reconstitution of Rag2 –/– mice
CD4+ T cell reconstitution of Rag2–/– mice was carried out as described previously [29, 30]. Negatively isolated naïve CD4+ T cells were purified from the spleen and lymph nodes of WT or Hdac6–/– mice as described in the in vitro T cell culture section. 2 × 105 CD4+ T cells were then injected into Rag2–/– mice via tail vein injection. Mice were left for 8 weeks and then tested for the success of reconstitution by FACS staining of spleen and liver cell suspensions for the presence of CD4.
Quantification and statistical analysis
The statistical significance of comparisons between two groups was performed using a two-tailed, unpaired t test, *p < 0.05, **p < 0.01 and ***p < 0.001, n.s means not significant (Student’s t test). GraphPad Prism 7 software (GraphPad Software) was used to plot all the graphs and calculate statistical significance.
Data availability
RNA-seq datasets are deposited in the Gene Expression Omnibus (GEO) with accession number GEO: GSE146253. We collected ChIP-seq and ATAC-seq data from the GEO public resource (http://www.ncbi.nlm.nih.gov/geo) and the accession numbers are GSM550303, GSM550304, GSM2828376, GSM2828377, GSM2828378, GSM2828379, GSM836118, GSM836120, GSM994512, and GSM994513. All of the full length original western blots for these results are provide in Supplementary File 1.
References
Kaplan MH, Sun YL, Hoey T, Grusby MJ. Impaired IL-12 responses and enhanced development of Th2 cells in Stat4-deficient mice. Nature 1996;382:174–7.
Thierfelder WE, van Deursen JM, Yamamoto K, Tripp RA, Sarawar SR, Carson RT, et al. Requirement for Stat4 in interleukin-12-mediated responses of natural killer and T cells. Nature 1996;382:171–4.
Wagner RD, Steinberg H, Brown JF, Czuprynski CJ. Recombinant interleukin-12 enhances resistance of mice to Listeria monocytogenes infection. Micro Pathogenesis. 1994;17:175–86.
Wieczorek M, Ginter T, Brand P, Heinzel T, Krämer OH. Acetylation modulates the STAT signaling code. Cytokine Growth F R 2012;23:293–305.
Nijhuis L, Peeters JGC, Vastert SJ, van Loosdregt J. Restoring T Cell Tolerance, Exploring the Potential of Histone Deacetylase Inhibitors for the Treatment of Juvenile Idiopathic Arthritis. Front Immunol. 2019;10:151.
Nusinzon I, Horvath CM. Positive and negative regulation of the innate antiviral response and beta interferon gene expression by deacetylation. Mol Cell Biol. 2006;26:3106–13.
Into T, Inomata M, Niida S, Murakami Y, Shibata K-I. Regulation of MyD88 aggregation and the MyD88-dependent signaling pathway by sequestosome 1 and histone deacetylase 6. The J Biol Chem. 2010;285:35759–69.
Kovacs JJ, Murphy PJM, Gaillard S, Zhao X, Wu J-T, Nicchitta CV, et al. HDAC6 regulates Hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor. Mol cell. 2005;18:601–7.
Yan B, Liu Y, Bai H, Chen M, Xie S, Li D, et al. HDAC6 regulates IL-17 expression in T lymphocytes: implications for HDAC6-targeted therapies. Theranostics 2017;7:1002–9.
de Zoeten EF, Wang L, Butler K, Beier UH, Akimova T, Sai H, et al. Histone deacetylase 6 and heat shock protein 90 control the functions of Foxp3(+) T-regulatory cells. Mol Cell Biol. 2011;31:2066–78.
Laino AS, Betts BC, Veerapathran A, Dolgalev I, Sarnaik A, Quayle SN, et al. HDAC6 selective inhibition of melanoma patient T-cells augments anti-tumor characteristics. J Immunother Cancer. 2019;7:33.
Fukumoto T, Fatkhutdinov N, Zundell JA, Tcyganov EN, Nacarelli T, Karakashev S, et al. HDAC6 Inhibition Synergizes with Anti-PD-L1 Therapy in ARID1A-Inactivated Ovarian Cancer. Cancer Res. 2019;79:5482–9.
Pamer EG. Immune responses to Listeria monocytogenes. Nat Rev Immunol. 2004;4:812–23.
Zhan Y, Cheers C. Control of IL-12 and IFN-gamma production in response to live or dead bacteria by TNF and other factors. J Immunol (Baltim, Md: 1950). 1998;161:1447–53.
Moreno-Gonzalo O, Ramírez-Huesca M, Blas-Rus N HDAC6 controls innate immune and autophagy responses to TLR-mediated signalling by the intracellular bacteria Listeria monocytogenes. 2017; 13: e1006799.
Zhang YS, Xin DE, Wang Z, Song X, Sun Y, Zou QC, et al. STAT4 activation by leukemia inhibitory factor confers a therapeutic effect on intestinal inflammation. The EMBO journal. 2019;38.
Nguyen KB, Watford WT, Salomon R, Hofmann SR, Pien GC, Morinobu A, et al. Critical role for STAT4 activation by type 1 interferons in the interferon-gamma response to viral infection. Science 2002;297:2063–6.
Nguyen HN, Noss EH, Mizoguchi F, Huppertz C, Wei KS, Watts GF, et al. Autocrine Loop Involving IL-6 Family Member LIF, LIF Receptor, and STAT4 Drives Sustained Fibroblast Production of Inflammatory Mediators. Immunity 2017;46:220–32.
Wei L, Vahedi G, Sun H-W, Watford WT, Takatori H, Ramos HL, et al. Discrete roles of STAT4 and STAT6 transcription factors in tuning epigenetic modifications and transcription during T helper cell differentiation. Immunity 2010;32:840–51.
Vahedi G, Takahashi H, Nakayamada S, Sun H-W, Sartorelli V, Kanno Y, et al. STATs shape the active enhancer landscape of T cell populations. Cell 2012;151:981–93.
Nakayamada S, Kanno Y, Takahashi H, Jankovic D, Lu KT, Johnson TA, et al. Early Th1 cell differentiation is marked by a Tfh cell-like transition. Immunity 2011;35:919–31.
Weinstein JS, Laidlaw BJ, Lu Y, Wang JK, Schulz VP, Li N, et al. STAT4 and T-bet control follicular helper T cell development in viral infections. J Exp Med. 2018;215:337–55.
Wada H, Masuda K, Satoh R, Kakugawa K, Ikawa T, Katsura Y, et al. Adult T-cell progenitors retain myeloid potential. Nature 2008;452:768–72.
Luc S, Luis TC, Boukarabila H, Macaulay IC, Buza-Vidas N, Bouriez-Jones T, et al. The earliest thymic T cell progenitors sustain B cell and myeloid lineage potential. Nat Immunol. 2012;13:412–9.
Henry CJ, Ornelles DA, Mitchell LM, Brzoza-Lewis KL, Hiltbold EM. IL-12 produced by dendritic cells augments CD8 + T cell activation through the production of the chemokines CCL1 and CCL17. J Immunol. 2008;181:8576–84.
Vacaflores A, Freedman SN, Chapman NM, Houtman JC. Pretreatment of activated human CD8 T cells with IL-12 leads to enhanced TCR-induced signaling and cytokine production. Mol Immunol. 2017;81:1–15.
Wilson DC, Matthews S, Yap GS. IL-12 signaling drives CD8 + T cell IFN-gamma production and differentiation of KLRG1 + effector subpopulations during Toxoplasma gondii Infection. J Immunol. 2008;180:5935–45.
Ma L, Huang C, Wang XJ, Xin DE, Wang LS, Zou QC, et al. Lysyl Oxidase 3 Is a Dual-Specificity Enzyme Involved in STAT3 Deacetylation and Deacetylimination Modulation. Mol cell. 2017;65:296–309.
Skene PJ, Henikoff JG, Henikoff S. Targeted in situ genome-wide profiling with high efficiency for low cell numbers. Nat Protoc. 2018;13:1006–19.
Bergstrom KS, Morampudi V, Chan JM, Bhinder G, Lau J, Yang H, et al. Goblet Cell Derived RELM-beta Recruits CD4 + T Cells during Infectious Colitis to Promote Protective Intestinal Epithelial Cell Proliferation. PLoS Pathog. 2015;11:e1005108.
Acknowledgements
We are grateful to Yu Zhang for providing us with the Hdac6–/– mice. Jingjing Wang and Qi Li for technical help in L. monocytogenes infection experiments. We are grateful to Bing Su for manuscript review and discussion.
Funding
YZ was supported by the Young Scientists Fund of the National Natural Science Foundation Grant of China (32000620). This work was supported by National Natural Science Foundation Grant of China (81820108023, U1603284 and 81530083), Young Scientists Fund of the National Natural Science Foundation Grant of China (31700789), National Key Research and Development Program of China (2016YFC1302402), and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Author information
Authors and Affiliations
Contributions
YZ and DX contributed to the experimental design, conductions of experiments, and manuscript preparation. ZW and WP performed the experiments, data analysis and manuscript preparation. YZ, JL, MX, NC, JZ, YJ, MC and YW assisted with the experiments. MC and JY provided technical assistance. ZC, XL and LC contributed to the data analysis and manuscript writing. YC was involved in the conception of the research, experimental design, data analysis, and manuscript preparation.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
All animal experiments were performed in compliance with the guide for the care and use of laboratory animals and were approved by the institutional biomedical research ethics committee of Soochow University.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by Y. Shi
Rights and permissions
About this article
Cite this article
Zhang, Y.S., Xin, D.E., Wang, Z. et al. Acetylation licenses Th1 cell polarization to constrain Listeria monocytogenes infection. Cell Death Differ 29, 2303–2315 (2022). https://doi.org/10.1038/s41418-022-01017-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-022-01017-9
This article is cited by
-
Single-cell deconvolution algorithms analysis unveils autocrine IL11-mediated resistance to docetaxel in prostate cancer via activation of the JAK1/STAT4 pathway
Journal of Experimental & Clinical Cancer Research (2024)