Abstract
Hierarchical organization of intestine relies on the self-renewal and tightly regulated differentiation of intestinal stem cells (ISCs). Although signals like Wnt are known to sustain the continued intestinal renewal by maintaining ISCs activity and lineage commitment, molecular mechanisms underlying ISCs ‘stemness’ and supportive niche have not been well understood. Here, we found that CUL4B-RING ubiquitin ligase (CRL4B) regulates intestinal homeostasis by targeting immunity-related GTPase family M member 1 (IRGM1) for proteasomal degradation. CUL4B was mainly expressed at ISCs zone. Deletion of Cul4b led to reduced self-renewal of ISCs and a decreased lineage differentiation towards secretory progenitors through downregulated Wnt signals. Besides, Cul4b-null mice exhibited impaired Paneth cells number and structure. Mechanistically, CRL4B complex were associated with WD40 proteins and targeted IRGM1 at K270 for ubiquitination and proteosomal degradation. Impaired intestinal function caused by CUL4B deletion was rescued by down-regulation of its substrate IRGM1. Our results identified CUL4B as a novel regulator of ISCs and revealed a new 26 S proteasome degradation mechanism in intestine self-renewal and lineage commitment.

Similar content being viewed by others
Introduction
The mammalian intestine is organized into crypt-villi structures and self-renews with a rapid turnover rate of 4–5 days. Lgr5+ (also called crypt base columnar, CBC) cells at the bottom of crypt act as intestinal stem cells (ISCs) during intestinal homeostasis [1]. ISCs maintain continued self-renewing ability and produce differentiated progeny including rapidly proliferating transit-amplifying (TA) cells, post-mitotic enterocytes, absorptive and secretory lineages [2, 3]. Among them, larger, granule-containing Paneth cells intermingled with ISCs at the bottom crypt function as components of stem cell niche by secreting factors and host defensing [4, 5].
ISC stemness is tightly controlled by various pathways. Wnt signals restricted to the lower crypt form a gradient to fuel the continued renewal by maintaining ISC activity and regeneration [6, 7]. Lgr5, the marker of stem cells, is the direct target gene of Wnt signaling pathway [8]. Deletion of Ctnnb1 and Tcf7l2 (TCF4), the downstream transcription factors of Wnt pathway, in pVillin-CreERT2 mice results in a rapid loss of Lgr5+ cells [9]. Stem cell-specific activation of Wnt signals by Apc mutation confers a growth advantage [10, 11]. Conversely, overexpression of the secreted Wnt inhibitor Dickkopf1 (Dkk1) leads to the loss of crypts [12]. Paneth-cell derived Wnt3a and R-spondin1 are proved to be essential to maintain 3D organoids culture for ex vivo expansion [7]. Besides epithelial signals, reduced mTOR activity by calorie restriction or inhibition of glycolysis in Paneth cells metabolically also boosts ISC function [13]. The growth factors and cytokines secreted by mesenchymal cells also provide vital supporting niche for stem cells [14]. Thus identification of additional regulators of Wnt pathway in ISC niche will definitely contribute to a better understanding of ISC regulating network. Although loss of IRGM1 has been reported to result in disrupted intestinal epithelial function, gut dysbiosis, defects in anti-microbial peptide secretion and stress responses to pathogenic microbes [15, 16], however its role in ISC regulation has not been documented.
Cul4 encodes the scaffold protein of Cullin-Ring family, the biggest E3 ubiquitin ligase complex in mamallian cells [17]. CUL4B assembles DDB1, ROC1 and substrate receptors to form various CUL4B-RING E3 ligase complexes (CRL4B). CRL4B catalyzes polyubquitination of substrates for proteosomal degradation [18]. Although CUL4B possesses high homology with CUL4A, the nuclear localization sequences (NLS) of CUL4B distinguishes it from CUL4A in multiple biological processes [19]. We and other group have found that CUL4B mutation causes human X-linked intellectual disabilities [20, 21]. Disruption of Cul4b in mice results in early embryonic lethality before E9.5 [22, 23]. CUL4B in extraembryonic tissues is essential for early development. Selective deletion of CUL4B in the epiblast shows no significant defects except for the abnormal spermatogenesis [24]. Our recent study reveal the function of CUL4B in adipogenesis [25], but no research has addressed its role in intestine regulation.
We here studied the function of CUL4B in intestinal homeostasis. Our results showed a surprising cytoplasm-specific localization of CUL4B at the ISCs zone. To determine the function of CUL4B in the intestine, we generated conditional knockout mice and in vitro organoids to investigate how CUL4B modulates ISC stemness and cell lineage commitment. We found that CUL4B was required for intestine self-renewal and proliferation. Loss of Cul4b caused scarcity of Lgr5+cells, Paneth cells and secretory lineages by decreasing Wnt signals. We uncovered IRGM1 as the ubiquitination substrate of CRL4B. CRL4B ubiquitylated IRGM1 at K270 and promoted Wnt3 secretion. Our findings suggest that CUL4B is essential in maintaining ISCs stemness and intestinal homeostasis.
Materials and methods
Animals and administration
All animal experiments were approved by the Animal Care and Use Committee of the School of Basic Medical Science of Shandong University (No. LL-201502043 and ECSBMSSDU2019-2-008). Lgr5-EGFP-ires-CreERT2, CAG-CreERT2, and Rosa26-LSL-Cas9-tdTomato mice were from the Jackson Laboratory (stock number 008875 and 004682) or GemPharmatech co. Ltd (stock numbers T002249). pVillin-Cre mice were provided by Dr. Baichun Jiang. Cul4b transgenic mice were generated as described previously [26]. Mice were housed in SPF facilities. The wildtype or Cul4b knockout mice from 8 to16 weeks were used for most experiments. For Cre induction, mice with the genotyping of Lgr5-EGFP-ires-CreERT2 and CAG-CreERT2 were intraperitoneally injected with 100 μl tamoxifen at the concentration of 2 mg/ml for 5 consecutive days.
Tissue histology and immunofluorescence
Small intestines or colons from WT or KO mice were isolated, rinsed with cold PBS, fixed in 4% paraformaldehyde overnight at 4 °C, and embedded in Tissue-Tek® OCT or paraffin. Tissues were cut into 4 μm sections. Unstained paraffin sections were washed twice with xylene dewax, dehydrated in ethanol solution with decreasing concentration (100%, 95%, 90%, 80%, 75%, 70%, 50%, 25%), and stained with hematoxylin-eosin staining (H&E). To assess tissue histology, the crypts and villus length were quantified from crypt/villus units per mouse with triplicates. Positive staining cells with PCNA, Olfm4, or Lyz were quantified from approximately 50 well-oriented crypt/villus units per mouse with triplicates.
Unfixed tissue segments were embedded with Tissue-Tek® OCT and frozen in freezing table. Sections were cut on a Freezing Microtomes. For immunofluorescence, antigen retrieval was achieved by boiling in Citrate Antigen Retrieval Solution. Sections were washed in PBS, then permeabilized (0.2% Triton X-100 in PBS) and incubated in blocking solution (10% v/v goat/donkey serum in PBS) at 37 °C for one hour. Primary antibody treatment was performed overnight at 4 °C in blocking solution. Secondary antibody was incubated one hour at room temperature. Microscopic images were obtained with Laser Confocal Microscope (LSM880, LSM900) and OLYMPUS imaging.
The primary antibodies included the following: anti-CUL4B (Sigma, St. Louis, MO, USA, 1:750), anti-Lyz (Abcam, Cambridge, UK, 1:1000), anti-Ki67 (Abcam, 1:500), anti-PCNA (GeneTex, Alton PkwyIrvine, CA, USA, 1:500), anti-β-catenin (Santa Cruz, CA, USA, 1:1000), anti-GFP (Rockland, USA, 1:500), anti-Olfm4 (Cell signaling, Beverly, MA, USA 1:400), anti-BrdU (Abcam, 1:1000), anti-UEA-1 (Sigma, 1:200), anti-Mucin2 (Santa, 1:100), anti-ChgA (Santa, 1:100), anti-Villin(Abcam, 1:400) and anti-WDR77(Abcam, 1:200). Secondary antibodies included peroxidase-conjugated goat anti-rabbit IgG, goat anti-mouse IgG (Jackson ImmunoResearch, West Grove, PA, USA, 1:200) and donkey anti-goat IgG (Invitrogen, Carlsbad, CA, USA, 1:500).
Western blot
Proteins were extracted from mouse tissue and organoids. 25 micrograms of protein were immunoblotted by standard protocols. The primary antibodies included as the following: anti-CUL4B (Sigma), anti-Lgr5 (BBI Life Sciences, Shanghai, China), anti-Histone (GeneTex), anti-α-Tubulin (Proteintech, Wuhan, China), anti-Cytokeratin 20 (GeneTex), anti-GAPDH, anti-pGSK3β(Ser9), anti-p-β-catenin (S33/37/T41), anti-Non-p-β-catenin, anti-Ub (all antibodies from Cell signaling), anti-IRGM1(GeneTex, Cell signaling), anti-PTGES3 (Proteintech), anti-SLC5A1, anti-ABCG2, anti-STX18 (ORIGENE, Rockville, USA), anti-EPCAM (Abways, Shanghai, China), anti-ACTIN, anti-DDB1, anti-β-catenin (Santa Cruz), anti-HA (Rockland) anti-ROC1 and anti-WDR77, anti-WDR1(Abcam). The secondary antibodies included anti-rabbit and anti-mouse horseradish peroxidase (HRP) (Jackson ImmunoResearch, West Grove, PA, USA; 1:10000; 1:5000). The detection reagent Cheminoluminiscent Substrate was provided from the ECL kit (Thermo, USA). The band signals from Western blot results were analyzed by Volume Analysis of Quantity One with volume background subtraction (Bio-Rad, USA).
Lineage tracing of Crypt–Villus units
For BrdU label assay, Lgr5-EGFP-ires-CreERT2; Cul4b-/Y and Lgr5-EGFP-ires-CreERT2; Cul4bfn/Y(KOLgr5) mice were abdominally injected with tamoxifen for 5 consecutive days. Each mouse was injected with 1 mg BrdU. On the 3rd day after injection, small intestines were collected and analyzed by immunofluorescence. For self-renewal analysis, mice were injected with 0.1 mg tamoxifen per gram. SI were collected and analyzed from Lgr5-EGFP-ires-CreERT2; Cul4b-/Y; Rosa26-LSL-Cas9-tdTomato; KI/KI and Lgr5-EGFP-ires-CreERT2; Cul4bfn/Y; Rosa26-LSL-Cas9-tdTomato; KI/KI (KOLgr5-Tom) mice on the 3rd day after tamoxifen injection.
Organoids isolation, culture and passage
The crypt isolation and organoid culture were performed as described previously [11]. Crypts were released from small intestine by incubating with 2 mM EDTA in DPBS (on ice for 30 mins). Crypts were re-suspended with cold Advanced DMEM/F12 medium. Crypts were mixed with Matrigel (Corning, USA), and plated in 24-well plates. The basal differentiation culture medium (Differentiation Medium, DM) included: Advanced DMEM/F12 supplemented with 1×penicillin/streptomycin, 1×HEPES(Invitrogen), 1×Glutamax (Invitrogen), 1×B27 (Invitrogen), and 1 mmol/L N-acetylcysteine (Sigma-Aldrich) with 50 ng/mL recombinant epidermal growth factor (PepRotech, USA), Noggin (conditioned medium, 5% v/v), and R-spondin 1 (conditioned medium, 10% v/v). Expansion medium (EM) included DM plus 15% Wnt3a conditioned medium. The Cre expression was induced with treatment of 10 μM 4-OHT (Sigma-Aldrich) in organoids.
Whole mount staining
Organoids were thoroughly removed from Matrigel with Cell Recovery Solution (Corning) on ice before centrifugation at the speed of 800 rpm. 4% paraformaldehyde was used to fix the organoids overnight at 4 °C. Immunofluorescence proceeded as outlined above. Microscopic observation was performed using a high-speed confocal platform (Andor, Dragonfly 200).
Constructions and transfections
The ORF of mouse Irgm1 (Myc-DDK-tagged) and Cul4b (Myc-DDK-tagged) were purchased from Origene and cloned into the lentiviral expression vector (pCDH-CMV-MCS-EF1-Puro, one kind gift of Dr. Jupeng Yuan, Shandong Cancer Hospital and Institute, China) or the His-Tag lentiviral expression vector (pLent-EF1a-FH-CMV-GFP-P2A-puro purchased from vigene).
For transfection, HEK293T cells were cultured to the concentration of 30%. Transfection was performed in 500 μl opti-DMEM with 20 μl siRNA or control with addition of 30 μl X-tremeGene (Roche, Mannheim, Germany) according to the manufacturer’s instruction. 2 μg constructions for ectopic expression of CUL4B, IRGM1 and HA-Ubiquitin were transfected by PEI transfection reagent (Merck) as indicated respectively. After 48 h incubation, MG132 was added at a final concentration of 10 μM. The cells were then harvested after 3 h incubation.
RNA isolation and qRT-PCR analysis
The RNA was isolated with Trizol reagents (Invitrogen) or by isolation Kit (Qiagen, Dusseldorf, Germany) according to the manufacturer’s instruction. RNA was reversely transcribed into cDNA by reverse transcriptase (Thermo Scientific). Real Time PCR analysis was performed using gene-specific primers as listed in Table S1 and SYBR green master mix (Roche).
RNA-seq and data analysis
RNA was prepared for transcriptome sequencing. Library was prepared and sequenced by Novogene. The sequencing data could be reached in GSE157818. Sequencing results were mapped and differential expression analysis of comparing groups (2–4 biological replicates per group) was performed using the DESeq2 R package (1.16.1). Corrected P-value of 0.05 and absolute fold change of two were set as the threshold for significantly differential expression.
Gene Set Enrichment Analysis (GSEA) analysis was performed by the GSEA analysis tool http://www.broadinstitute.org/gsea/index.jsp. GO, KEGG, Reactome, DO, DisGeNET data sets were used independently. We ranked GSEA database genes by their association with differential genes in crypts from WT (N = 4) and KOIEC (N = 4) groups, organoids from WT (N = 2) and KOIEC (N = 2) and organoids from WT (N = 2) and KOCAG (N = 2) groups.
Differentiated protein/ubiquitylated protein identification and quantification by LC-MS/MS
Crypts from WT (N = 3) and KOIEC (N = 3) were isolated and mixed together. Differentiated proteins and differentiated ubiquitylated proteins were identified by Liquid Chromatography-Mass Spectrometry (LC-MS) in PTM Biolab LLC and analyzed by PTM bioinformatics Team. MS data in PRIDE with the number of PXD-021528. Venn diagram was performed to analyze the overlapping differentiated proteins that both downregulated in ubiquitylation and accumulated at protein level.
Protein stability assay and analysis
HEK293T cells were transfected with Myc-Irgm1 (2 μg). 24 h after transfection, cells were treated with 240 μg/ml cycloheximide (CHX) to inhibit protein synthesis. CHX-treated cells were harvested at different time points (0, 16, 32, 48 h) and processed for immunoblotting with anti-Irgm1 antibody. Anti-ACTIN antibody was used as the internal control. Signals from the Western blots were analyzed by Volume Analysis of Quantity One (Bio-Rad) with background subtraction.
FISH
The small intestine tissue was collected, cleaned, and then immediately put in the fixed fluid (DEPC) for 12 h. The tissue was dehydrated by gradient alcohol with paraffin embedding. The slices were boiled in the retrieval solution for 10–15 mins and naturally cooled. The objective tissue was marked, treated with proteinase K (20 μg/ml) solution and incubated at 37 °C. After washing, the section was incubated in a humidity chamber and hybridized with Lgr5 probe diluted in hybridization solution overnight. After hybridization, the sections were washed in 2×SSC for 10 mins at 37 °C.
Protein purification and pull-down assays
pGEX4T3-WDR77 and pRsfduet-IRGM1 were individually expressed in E.coli and purified using glutathione-sepharose (GE Healthcare) or Ni2+-NTA (GE Healthcare). The purified GST-WDR77 and His-IRGM1 were stored in protein buffer containing 25 mM Tris7.6, 150 mM NaCl, and 1 mM DTT. The pull-down assays were performed by mixing GST-WDR77 and His-IRGM1 proteins. The mixture was then incubated with anti-WDR77 antibody (Abcam) and protein A/G sepharose (Santa Cruz) for 2 h at 4 °C. Immunoprecipitates were boiled in sample loading buffer for 5 mins and detected by Western blot.
Ubiquitination assay
HEK293T cells were first transfected with indicated plasmids or siRNAs and treated with MG132 (20 μM). Cells were then harvested with PBS containing 10 mM NEM to prevent deubiquitination. Cells were lysed in cell lysis buffer containing 10 μM Tris 8.0, 150 mM NaCl, and 2% SDS by boiling the samples for 10 mins, followed by sonication (8 s, 1 cycle). Lysed sample supernatants were then incubated with anti-IRGM1 antibody (Cell Signaling Technology), mixed with protein A/G sepharose (Santa Cruz) for 12 h at 4 °C, followed by Western blot.
In vitro ubiquitination assay
His-IRGM1 was expressed in E.coli and purified with Ni2+-NTA (GE Healthcare). His-IRGM1 was incubated with 200 ng E1 (UBE1), 500 ng E2 (UbcH5c), 10 μg His-Ub and 2 mM ATP (Enzo life sciences). The reaction was performed in the absence and presence of Flag-CUL4B co-IP products and incubated at 37 °C for 1 h. Samples were quenched in 6 M guanidinium-HCl (pH=8) containing 5 mM NEM. His-ubiquitinated proteins were pulled down with Ni2+-NTA (GE Healthcare), washed and eluted in sample buffers as described previously [27]. The mixture was then boiled in loading dye at 95 °C for 10 mins to disrupt the protein-protein interactions, followed by Western blot.
Immunoprecipitation
Protein supernatants were incubated with indicated antibodies and protein A/G sepharose (Santa Cruz) for 2 h at 4 °C. Immunoprecipitates were boiled in sample loading dye at 95 °C for 5 mins, followed by Western blot.
Wnt luciferase activity and conditional medium assay
For Wnt luciferase activity, medium was collected from wildtype or knockout organoids. HEK293 STF cells were treated with the medium for 48 h and luciferase was detected. For conditional medium assay, wildtype organoids were digested into single cells and plated at the concentration of 1 × 104 cells per well in 96-well plates. After 24 h, the culture medium was changed with conditional medium collected from either the wildtype or the knockout organoids. The conditional medium was produced by the same number of wildtype or the knockout organoids after for 24 h and 48 h culturing. RNA was collected or cellular ATP activity and RNA levels were measured.
Statistical analysis
Statistical analyses were conducted with Prism software (GraphPad, La Jolla, CA). For pairwise and two independent group comparison two-tailed t-test was used. Data are presented as mean ± SEM and P values determined by Student t-test; *P < 0.05. **P < 0.01. ***P < 0.001. ****P < 0.0001 was considered statistically significant.
Results
CUL4B is highly expressed in mouse intestinal crypts
To investigate the role of CUL4B in intestinal regulation, we firstly determined the expression pattern of CUL4B in intestine. As shown in Fig. 1a, CUL4B was highly expressed at the bottom of crypts where located the stem cell compartment. Interestingly, unlike many cell types in other mammalian tissues where CUL4B was predominantly localized in the nucleus, CUL4B was mainly detected in the cytoplasm of intestine (Supplementary Fig. S1a and Movie S1). Western blot analysis confirmed its cytoplasmic location at crypts as well (Fig. 1b, c). The statistical analysis of immunofluorescent positive staining at each cell position showed that CUL4B expression was gradually decreasing along the crypt-villi axis (Fig. 1d). In accordance with it, co-staining showed the co-express of CUL4B with Lgr5+ ISCs and Lyz+ Paneth cells (Fig. 1e, f). These results above indicate that CUL4B is highly expressed in bottom crypt, suggesting a potential role of CUL4B in intestine regulation.
a The representative images of CUL4B staining on small intestine (SI) tissue. Green, CUL4B, Blue, DAPI. The scale bar is 50 μm (Left) and 20 μm (Right). b Western blots analysis of CUL4B, the stem cell marker (Lgr5), and the villi marker (CK20) in extracts from crypt and villus of the SI. c Western blots analysis of CUL4B, the nuclear marker (Histone protein, H3), and the cytoplasm marker (α-Tubulin) in cytoplasmic and nuclear extracts from crypts of the SI. d The percentage of CUL4B positive staining by statistical quantification of cell position along the crypt-villus from the bottom. e The representative image of co-staining of Lgr5 and CUL4B in the crypts of Lgr5-EGFP-ires-CreERT2 mouse. The white arrowhead, co-expression of Lgr5 and CUL4B. The red arrowhead, exclusive expression of CUL4B. The green arrowhead, exclusive expression of Lgr5. The scale bar is 10 μm. f The representative image of co-staining of Lyz and CUL4B in the crypts. The scale bar is 50 μm. g The measurement of intestine weight of duodenum, jejunum plus ileum, colon of KOIEC (N = 4), and WT mice (N = 4). Error bars represent standard errors. **P < 0.01, *P < 0.05. h The representative images of H&E staining of sections of SI from WT and KOIEC mice. The scale bar is 100 μm(left), 25 μm(right). i, j The Measurement of villus length (i) and crypt length (j) of KOIEC, Tg and WT mice (N = 3). Error bars represent standard errors. ****P < 0.0001.
Loss of CUL4B in intestinal epithelial cells retards intestinal development
To investigate the function of CUL4B in intestine, we generated the mice with intestinal epithelium specific deletion of Cul4b (pVillin-Cre; Cul4bfn/Y, abbreviated as KOIEC) and compared them with the wildtype control (Cul4bfn/Y, abbreviated as WT). Knockout efficiency of KOIEC mice was confirmed at the mRNA and protein levels (Supplementary Fig. S1b, c). KOIEC mice were born at the expected Mendelian ratios. Epithelial deficiency of CUL4B had no effect on mouse body weight but led to decreased intestinal weight, shortened villi, and a reduction of crypt cell number (Fig. 1g–j and Supplementary Fig. S1d). To confirm the role of CUL4B in regulating intestinal development, a previously generated CUL4B overexpression model of transgenic mice were further evaluated (EGFP-CUL4B under the promoter of CMV, abbreviated as Tg) [26]. As expected, we observed enlongated crypts and villi in the transgenic mice (Fig. 1i, j and Supplementary Fig. S1e). Our results suggest that CUL4B promotes crypt-villi formation and intestinal development.
CUL4B is essential to maintain intestinal self-renewal in vivo and vitro
To determine the mechanisms underlying shortened intestine led by CUL4B defiency, we firstly examined intestinal renewal. 5-bromo-2-deoxyuridine (BrdU) labeling assay, which marks cycling crypt cells and their progenies, was performed. High resolution images revealed a lower self-renewal speed in both KOIEC and KOLgr5 mice (Lgr5-EGFP-IRES-CreERT2; Cul4bfn/Y, deteltion of CUL4B in Lgr5+ stem cells, abbreviated as KOLgr5) compared with the WT control (Fig. 2a, b). We then crossed the KOLgr5 with Rosa26-tdTomato mice to generate Lgr5-EGFP-IRES-CreERT2; Cul4bfn/Y; Rosa26-tdTomato mice (abbreviated as KOLgr5-Tom). This strategy allowed for the tracking of Lgr5+cells by tdTomato by one pulse tamoxifen treatment. Three days after tamoxifen administration, the tdTomato-positive descendants extended markedly along the crypt-villus axis to the top in the wildtype, while most tdTomato-positive cells of KOLgr5-Tom mice still localized in the crypt compartment (Fig. 2c). PCNA staining showed a slower proliferation while cell apoptosis was not affected in KOIEC mice (Fig. 2d and Supplementary Fig. S2a). Our findings revealed that CUL4B is indispensable to maintain intestinal renewal in vivo.
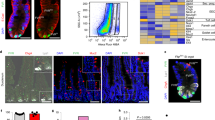
a BrdU label assay shows decreased self-renewal rate in KOIEC mice. The scale bar is 50 μm. b BrdU label assay shows decreased self-renewal rate in KOLgr5 mice 3 days after tamoxifen induction administration. The scale bar is 50 μm. c Lineage tracing of Lgr5+ positive cells and their ascents (tdTomato) in the jejunum of KOLgr5-Tom mice and control 3 days after tamoxifen induction. The scale bar is 50 μm. d The representative images and quantification of PCNA positive staining cells of WT and KOIEC mice (N = 3). The scale bar is 50 μm. ****P < 0.0001. e The representative images of organoids of passage 2 derived from WT and KOIEC mice cultured in differentiation medium (DM). The scale bar is 400 μm. f The average number of buddings per organoid derived from WT and KOIEC mice (N = 3). Error bars represent standard errors. *P < 0.05, ****P < 0.0001. g The representative images of organoids at passage 2 derived from WT and KOIEC mice cultured in expansion medium (EM). The scale bar is 400 μm. h Colony-forming effciency of organoids cultured in expansion medium from KOIEC and WT mice. Error bars represent standard errors. ****P < 0.0001. i Whole-mount staining of Ki67 in intestinal organoids from WT and KOIEC mice. The scale bar is 50 μm. j GSEA analysis for differentially expressed genes between KOIEC and WT crypts. Ribosome and DNA replication pathways were enrichment in WT group. NES, normalized enrichment score; FDR, false discovery rate.
We next generated organoids to evaluate the self-rewal capacity of ISCs ex vivo. CUL4B expression was obviously elevated in organoids cultured in stem cell-enriched expansion medium (EM) than those in differetiation medium (DM) (Supplementary Fig. S2b). Organoids derived from KOIEC mice displayed reduced budding efficiency during passaging (Fig. 2e, f). Decreased size and formation efficiency were also observed in KOIEC organoids cultured in EM in single cell assay (Fig. 2g, h and Supplementary Fig. S2c). Meanwhile immunofluorescent staining of proliferative signal, Ki67 was obviously reduced (Fig. 2i and Supplementary Movie S2 and S3). To investigate the molecular alteration caused by loss of CUL4B, we then performed mRNA sequencing (RNA-seq) of isolated crypts (4KOIEC vs 4WT) and in vitro cultured organoids (2KOIEC vs 2WT). A tamoxifen-induced CUL4B knockout organoid model was also established from CAG-CreERT2; Cul4bfn/Y mice and used for sequencing (2KOCAG vs 2WT) after tamoxifen treatment. Differentially expressed genes (DEGs) in CUL4B-deficient crypts and organoids were identified in comparison with the wildtype respectively (All DEGs were listed in Supplementary Table S2). The gene set enrichment analysis (GSEA) analysis showed that DNA replication and ribosome-associated genes were significantly downregulated in both Cul4b knockout tissues and organoids (Fig. 2j and Supplementary Fig. S2d). Taken together, our data suggest that CUL4B is required for the maintenance of intestinal self-renewal.
CUL4B deficency leads to dysregulated ISC differentiation
We then checked RNA-seq data and performed immunostaining of intestinal cell lineage markers to identify the change of SI cell components after loss of CUL4B. Generally, RNA-seq data suggest Lgr5 was significantly down-regulated and differentiated enterocyte marker Alpi was increased in all three kinds of CUL4B deletion models (Fig. 3a). A decreased presence of ISCs was observed by GFP staining in KOLgr5, Lgr5-FISH and Olfm4 staining in KOIEC intestines (Fig. 3b and Supplementary Fig. S3a, b). Consistently, the ISCs markers such as Lgr5 and Ascl2 were down-regulated in KOIEC crypts (Fig. 3c). For differentiated cell types, immunofluorescent and immunochemical staining showed CUL4B-deficient resulted in the reduction of Paneth cells (Lyz staining, Fig. 3d), Goblet cells (Mucin2 and Alcian Blue staining, Fig. 3f and Supplementary Fig. S3c) and enteroendocrine cells (ChgA+, Fig. 3g) in intestines, and the increased staining of Alkaline phosphatase (Supplementary Fig. S3d). qRT-PCR assay confirmed that Paneth cell marker Lyz was down-regulated (Fig. 3e), whereas the differentiated enterocyte markers such as Alpi, Apoa4, and Krt20 were increased in Cul4b-deficient intestine (Fig. 3h, i). Mucin2+ staining and stem cell markers like Lgr5 and Ascl2 were also decreased in the colon of KOIEC mice (Supplementary Fig. S3e, f). GO analysis revealed that the functions associated with absorption and digestion responsible by enterocytes was strengthened in KOIEC intestine (Supplementary Fig. S3g, h). The peroxisome proliferator-activated receptor (PPAR) signal was also enriched in KOIEC mice by GSEA analysis (Supplementary Fig. S3i). Taken together, these results indicate that CUL4B deficiency in intestinal epithelial cells compromise lineage development. Intestinal CUL4B promotes cell commitment toward secretory progenitors and their terminally differentiated lineages including Paneth cells, Goblet cells and enteroendocrine cells. Meanwhile, terminally differentiated lineage of enterocytes from absorptive progenitors was inhibited (Supplementary Fig. S3j).
a The mRNA expression level of Lgr5 and Alpi isolated from KOIEC intestine crypts relative to WT littermates, KOIEC organoids relative to WT and KOCAG organoids relative to WT. *P < 0.05, ****P < 0.0001. b The representative images of GFP staining in SI of KOLgr5 mice and WT littermates. (N = 3). The scale bar is 50 μm. ****P < 0.0001. c The mRNA expression level of stem cell markers in KOIEC crypts relative to WT littermates. *P < 0.05, **P < 0.01, ****P < 0.0001. d The representative images of Lyz staining in SI of KOIEC mice and WT littermates (N = 3). The scale bar is 50 μm. ****P < 0.0001. e The mRNA expression level of Lyz in KOIEC crypts relative to WT littermates. *P < 0.05. f The representative images and quantification of Mucin2 staining in SI of KOIEC and WT mice (N = 3). The scale bar is 50 μm. ****P < 0.0001. g The representative images and quantification of ChgA staining in SI of KOIEC mice and WT littermates (N = 3). The scale bar is 50 μm. **P < 0.01. h The mRNA level of enterocytes markers Alpi and Apoa4 in KOIEC crypts relative to WT littermates. *P < 0.05, **P < 0.01. i The mRNA level of Krt20 in KOIEC crypts relative to WT littermates. *P < 0.05. Error bars represent standard errors in the figure.
CUL4B regulates intestine through Wnt signals
Wnt signaling is essential for ISC self-renewal, proliferation of progenitors and transit-amplifying (TA) cells, as well as commitment toward secretory progenitors [28]. The fact that loss of CUL4B impaired intestinal self-renewal and secretory lineage differentiation prompted us to check whether CUL4B regulated intestine through Wnt signaling pathway. We examined the effect of CUL4B deficiency on the expression of the Wnt/beta-catenin target genes. qRT-PCR confirmed that lack of CUL4B resulted in a significant decrease of Wnt target genes (Fig. 4a). Western blot showed that decreased β-catenin, p-GSK3β (Ser9) and activating form of p-β-catenin (S33/37/T41) in Cul4b knockout mice. Consistently, they were increased in CUL4B transgenic intestine (Fig. 4b). These data indicate that decreased Wnt signaling could be responsible for the impaired intestinal renewal led by Cul4b deletion. Indeed, rescue attempt with exogenous administration of GSK3β inhibitor, CHIR99021, could efficiently block decreased budding formation and proliferation (Fig. 4c), the reduction of β-catenin (Fig. 4d) and its target genes (Fig. 4e) as well. Furthermore, treating CUL4B overexpression organoids with Wnt inhibitor IWP-2 significantly attenuated increased budding formation and expression of Wnt target genes (Fig. 4f, g). Altogether, these results suggest that decreased Wnt signaling is responsible for the impaired intestinal regulation caused by Cul4b deletion.
a The mRNA levels of Wnt/β-catenin target genes in KOIEC crypts compared with WT control. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. b The protein level of Wnt/β-catenin pathway was determined by Western blot in SIs organoids from KOIEC and WT mice (Left), Tg and Con (Right). c The representative images in bright field (Upper) and Ki67 staining (Lower) of KOIEC intestinal organoids with or without 3 μM CHIR99021 treatment compared with the WT. The scale bar is 100 μm (Upper) and 15 μm (Lower). d The protein level of Wnt/β-catenin pathway was determined by Western blot in SIs organoids from KOIEC mice with or without 3μM CHIR99021 treatment. e The mRNA levels of Wnt/β-catenin target genes in KOIEC and WT intestinal organoids with or without treatment with CHIR99021 (0.5 μM or 3 μM). *P < 0.05, **P < 0.01,***P < 0.001, ****P < 0.0001, ns, no significance. f The representative images in bright field of Tg intestinal organoids with or without treatment of 3 μM IWP-2 compared with Con. The scale bar is 400 µm. g The mRNA levels of Wnt/β-catenin target genes in Tg intestinal organoids with or without 3 μM IWP-2 treatment compared with Con. *P < 0.05, ***P < 0.001, ****P < 0.0001. Error bars represent standard errors in the figure.
Lack of CUL4B in intestinal epithelial cells impairs their secretory phenotype
To determine the potential roles of CUL4B in regulating ISC niche, we first examined the effect of depletion of CUL4B in Lgr5+ cells by treating KOLgr5 organoids with tamoxifen. With 24 h transient induction, knockout of Cul4b only decreased cell proliferation but did not alter organoid-forming and budding efficiency, implying an indispensable role of CUL4B in non-stem cells (Supplementary Fig. S4a–c). To substantiate this notion, we prepared conditional medium from cultured WT and KOIEC organoid and assessed the effect of CUL4B deletion on their secretory phenotypes by growing single cells of wildtype organoids in the conditional medium (illustrated in Fig. 5a). Compared to the organoids cultured in WT conditional medium, the organoids cultured in KOIEC supernatant grew much more slowly (Fig. 5b, c). qRT-PCR confirmed the down-regulation of Wnt target genes in organoids treated by KOIEC supernatant (Fig. 5d). Furthermore, a Wnt luciferase reporter system was used to detect Wnt activity. The reporter cells treated with supernatant from KOIEC organoids, but not supernatant from KOLgr5 organoids revealed a significantly reduced Wnt activity (Fig. 5e). These data suggest that the function of CUL4B in non-stem cells is also essential to maintain ISC function. Consistently, the addition of recombinant Wnt3a rescued the reduction of budding formation and Wnt pathway in KOIEC organoids (Fig. 5f–h).
a Illustration of supernatant treating assay. The same number of wildtype SI organoids treated with supernatant collected from WT or KOIEC organoids with the same cell number for 24 or 48 h. b, c The representative images (b) and cell numbers (c) of wild type small intestine organoids treated with supernatant collected from WT or KOIEC organoids with the same cell number cultured for 24 or 48 h. The scale bar is 1000 μm (Left), 400 μm (Right). *P < 0.05. d The mRNA level of stem cell markers and Wnt target genes in wildtype SI organoids treated with supernatant collected from WT or KOIEC organoids. The experiment was performed with four replicates. **P < 0.01, ****P < 0.0001, ns, no significance. e The relative luciferase activity of Wnt reporter cells were detected after treatment with supernatant collected from KOIEC and KOLgr5 and WT organoids for 48 h. The same cell number of organoids at the third passage were cultured and seeded. *P < 0.01, ns, no significance. f The representative images of SI organoids from WT and KOIEC mice with or without treatment with recombinant Wnt3a (100 ng/ml). The scale bar is 50 μm. g, h The relative mRNA levels of Wnt/β-catenin target genes and protein level of Wnt pathway in WT and KOIEC SI organoids with or without treatment with recombinant Wnt3a (100 ng/ml). ***P < 0.001, ****P < 0.0001. Error bars represent standard errors in the figure.
Cul4b deficiency impairs Paneth cell development
CUL4B maily expressed at crypt bottom including ISCs and Paneth cells, which located among ISCs and provided the main cell source of secreted Wnt proteins. As the role of CUL4B in non-stem cells is indispensable, we next examined if the Paneth cells were altered by CUL4B deletion. Consistant with the observation in lysosome staining of small intestine tissues, we observed a significant reduction of Paneth cells per bud in KOIEC organoids. In contrast, increased number of Paneth cells per bud was detected in CUL4B transgenic organoids (Supplementary Fig. S5a, b). UEA-1 staining confirmed that depletion of CUL4B from epithelial cells led to the loss of Paneth cells (Supplementary Fig. S5c). Furthemore, the impaired structure of Paneth cells was confirmed by Transmission Electron Microscope (TEM). Abnormal cell morphology including degranulation, disturbed cell junction, increased lysosome and disordered mitochondrion were found in KOIEC intestines (Supplementary Fig. S5d). These data indicate that lack of CUL4B in intestine epithelial leads to abnormal Paneth cell number, inner and inter cell structure.
CRL4B targets intestinal IRGM1 for degradation
CUL4B, as a core component of an E3 ubiquitin ligase complex, associates with DDB1, ROC1, DDB1-and CUL4-associated factor (DCAF) to degrade substrates by 26 S proteasome (Fig. 6a). To reveal the underlying molecular mechanism, we performed proteomics to identify the altered proteins and ubiquitinated proteins in Cul4b-deficient crypts. 210 upregulated and 293 downregulated proteins were identified in Cul4b-deficient mice with the fold change above 1.5 (Supplementary Table S3). The ubiquitinated levels of 251 lysines of 180 proteins were upregulated and 321 lysines of 242 proteins were downregulated with more than 1.5 folds (Supplementary Table S4). Venn analysis of candidate degradation substrates of CUL4B was among the overlapping proteins with upregulated expression and reduced ubiquitinated level (Fig. 6b). Twelve potential substrates were identified, among which immunity-related GTPase subfamily protein (IRGM1) had the biggest change at protein level and most decreased ubiquitinated sites (Fig. 6c). We then performed immunoblot to confirm the expression change of these candidate substrates (Fig. 6d). IRGM1 was significantly upregulated both in isolated crypts and cultured organoids derived from KOIEC and KOLgr5 mice (Fig. 6e-g). Meanwhile, overexpression of CUL4B resulted in a reduction of IRGM1 protein. Importantly, the reduction of IRGM1 caused by the overexpression of CUL4B was efficiently blocked by the administration of MG132, a proteasome inhibitor (Fig. 6h). The results imply that CUL4B degraded IRGM1 via a proteasome-dependent degradation mechanism. To further strengthen this notion, we measured the half-life of IRGM1 protein in CUL4B overexpressed cells. Remarkably, overexpression of CUL4B resulted in a accelarated IRGM1 decay (Fig. 6i, j). These results suggest that CUL4B decreases the stability of IRGM1 protein.
a The structure of CRL4B E3 ligase. b Venn diagram showing the overlap of up-regulated proteins and down-regulated ubiquitylated proteins identified in KOIEC crypts by Mass Spectrometry (MS). c The name list of twelve overlapping proteins. The changing folds and detail modification sites of IRGM1 were indicated. d Western blots analysis of the overlapping proteins identified by proteomics. e–g Western blots were performed to check IRGM1 accumulation in KOIEC crypts (e) KOCAG organoids (f) and KOLgr5 organoids (g) compared to WT. CUL4B, IRGM1, and GAPDH expression were detected. h The IRGM1 level was detected in CUL4B overexpression (Flag-CUL4B) and control (Flag-NC) 293 T cells by Western blot with or without the presence of proteasome inhibitor MG132. The cell lysate was then detected with indicated antibodies. i, j The effect of CUL4B on the IRGM1 stability was detected by Western blot in CUL4B overexpression (Flag-CUL4B) and control (Flag-NC) 293 T cells with CHX chasing. The cell lysate was then detected with indicated antibodies with ACTIN as the control (i). j Signals from immunoblots were analyzed by Quantity One (Bio-Rad) to indicate the half-life of IRGM1 degradation.
CRL4B functions as an E3 ligase for intestinal IRGM1 at Lys270
We then tested whether IRGM1 was a direct substrate of CRL4B. Firstly, we confirmed the physical association between CRL4B complex and IRGM1. IRGM1 was immunoprecipitated with a substantial amount of CUL4B and DDB1. IRGM1 was coimmunoprecipitated with antibodies against CUL4B (Fig. 7a). The ubiquitination assay showed CRL4B significantly increased the amount of polyubiqutinated IRGM1, whereas knockdown of CUL4B resulted in a significant reduction (Fig. 7b, c) FLAG-CUL4B immunoprecipitated complex was confirmed in Supplementary Fig. S6a. These results support CRL4B as an E3 ligase for IRGM1. Residues Lys270, Lys275 and Lys398 on IRGM1 protein were identified as potential ubiquitination sites by quantitative mass spectrometry. To examine whether CRL4B targets these residues, we mutated Lys270/275/398 to alanine respectively and performed the ubiquitination assay. Our results showed that K270A, but not K275A or K398A mutant significantly reduced the level of polyubiquitinated IRGM1, suggesting that Lys270 was targeted by CRL4B complex (Fig. 7d).
a Immunoprecipitation was performed to detect interactions between CUL4B, DDB1 and IRGM1. Protein extracted from small intestine of mice was immunoprecipitated with the indicated antibodies individually. b The effect of CUL4B on IRGM1 ubiquitination was confirmed by immunoprecipitation and detected by Western blot with indicated antibodies. IRGM1, HA-Ub expression constructions and siRNA (siCUL4B or siNC) were transfected into HEK293T cells as indicated in lane 1, 2, and 3. c In vitro ubiquitination of IRGM1 by the CRL4B complex. Purified His-IRGM1 was incubated with E1, E2, Ub and ATP in the absence and presence of the CRL4B complex (Flag-CUL4B IP products) as indicated in lane 1–4. Ubiquitination of His-IRGM1 was analyzed by Western blot using anti-Ub antibody. d Single lysine mutations on IRGM1 defect ubiquitination by CRL4B. Immunoprecipitation was performed with wildtype and mutant Irgm1 variants together with Flag-CUL4B, HA-Ub with treatment with MG132 for 3 h as indicated in lane 1–5. Western blot was performed and detected with indicated antibodies. Input (3%) was used for Western blotting. e Immunoprecipitation was performed to detect interactions between CUL4B and WDR77. Protein extracted from mice small intestine or HEK293T cells was immunoprecipitated with the indicated antibodies. f, g GST pull down assays identify the direct interaction among IRGM1, DDB1 and WDR77. GST-WDR77, His-DDB1 and His-IRGM1 were expressed in E. coli individually followed by protein purification and immunoprecipitated pull-down. The proteins were detected using indicated antibodies. Input (15%) was used for Western blot. h In vivo ubiquitination assay of IRGM1 by the CRL4B complex with or without WDR77 interfering.
CRL4B uses a variety of DCAFs to assemble different E3 ligases to specifically target substrates. Mass spectrometry identified several CUL4B-interacting proteins including WD40 containing proteins such as WDR1, WDR3, WDR18 and WDR77, among which WDR77 was physically associated with CUL4B, DDB1 and ROC1, as demonstrated by coimmunoprecipitation (Fig. 7e). Importantly, WDR77 was co-expressed with CUL4B at intestinal crypts (Supplementary Fig. S6b). GST-pulldown assay confirmed the direct interaction between WDR77 and IRGM1 as well as DDB1 (Fig. 7f, g). These data suggest that WDR77 functions as a substrate receptor, directly recruits substrate IRGM1, facilitates its ubiquitination, and promotes subsequent proteasomal degradation. However, interfering WDR77 did not lead to significant decrease of ubiquitinated IRGM1 (Fig. 7h). Taken together, our results suggest CRL4B as an E3 ligase for intestinal IRGM1 at Lys270.
CUL4B regulates Paneth cell function and organoid proliferation through IRGM1
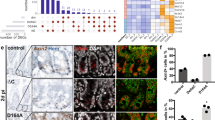
IRGM1 is the autophagy-associated protein susceptible for intestinal inflammation and microbiology infection. Irgm1 KO mice display granules in Paneth cell being finer, irregular in size, and less dense in appearance, selective reduction of Lyz expression and altered antimicrobial peptide (AMP) production [29,30,31]. To check whether CUL4B regulate Paneth cell structure and function through IRGM1, we knocked down Irgm1 by lentivirus in organoids from KOIEC mice to check AMP expression. The reduced number of Paneth cells and transcript levels of lysosome and AMPs in Cul4b KOIEC organoids was rescued by deletion of Irgm1 (Fig. 8a, b). Decreased organoid formation, budding efficiency and cell numbers per bud were rescued after deletion of Irgm1 as well (Fig. 8c–e). Accordingly, proliferation capacity of organoids was also elevated in Cul4b KOIEC organoids by Irgm1 deletion (Fig. 8f). Taken together, our findings suggest that decreased Paneth cell number, morphology and function by lack of CUL4B was mediated through IRGM1 accumulation.
a, b Relative mRNA levels of antimicrobial peptides cryptdin (Defa1), Defa4, Defa5, cathelicidin (Camp), Defa6, and lysozyme (Lyz) in WT + NC, KOIEC infected by lentivirus to knockdown Irgm1 (KOIEC + shIrgm1) and KOIEC control organoids (KOIEC + NC). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. c. The average number of Paneth cells within individual buds (right) from WT + NC, KOIEC + shIrgm1 and KOIEC + NC. ****P < 0.0001. d Representative image the organoids (left) of WT (WT + NC), KOIEC infected by lentivirus to knockdown Irgm1 (KOIEC + shIrgm1) and control (KOIEC + NC). The average budding number per organoid (right) from WT + NC, KOIEC + shIrgm1 and KOIEC + NC. The scale bar is 200 μm. ****P < 0.0001. e The organoids numbers of WT (WT + NC), KOIEC infected by lentivirus to knockdown Irgm1 (KOIEC + shIrgm1) and control (KOIEC + NC). **P < 0.01, ***P < 0.001. f Representative image of Ki67 staining from organoids of WT (WT + NC), KOIEC infected by lentivirus to knockdown Irgm1 (KOIEC + shIrgm1) and control (KOIEC + NC). The scale bar is 15 μm. g Relative mRNA levels of stem cell marker Lgr5 (Left) and Sox9 (Right) in organoids of WT + NC, KOIEC infected by dose-dependent lentivirus to knockdown Irgm1 (KOIEC + shIrgm1) and control (KOIEC + NC). The experiment was performed with triplicates. **P < 0.01, ***P < 0.001, ****P < 0.0001, ns, no significance. h Relative luciferase activity of HEK293 STF Wnt activity reporter lines were detected after 48 h treated with secreted supernatant collected from organoids WT + NC, KOIEC infected by dose-dependent lentivirus to knockdown Irgm1 (KOIEC + shIrgm1) and control (KOIEC + NC). ***P < 0.001, ****P < 0.0001. i Relative mRNA levels of Wnt target gene in organoids of WT (WT + NC), KOIEC infected by lentivirus to knockdown Irgm1 (KOIEC + shIrgm1) and control (KOIEC + NC). The experiment was performed with triplicates. **P < 0.01, ***P < 0.001, ****P < 0.0001, ns, no significance. j Protein level of Wnt/β-catenin pathway was determined by Western blot in SI organoids from KOIEC, KOIEC with Irgm1 ablation (KOIEC + shIrgm1) and WT (WT + NC) mice.
Dysregulated Wnt signaling caused by CUL4B depletion is rescued by knockdown of Irgm1
To test whether the dysregulation of Wnt signaling in Cul4b-deficient organoids was caused by increased levels of IRGM1, we examined the rescue effect of Irgm1 knockdown on the Wnt activity and its target genes in Cul4b KOIEC organoids. Infection of Irgm1 knockdown lentivirus significantly upregulated the transcription of Wnt target genes (Lgr5 and Sox9) and Wnt luciferase activity in a dose-dependent manner (Fig. 8g, h). Interfering Irgm1 efficiently attenuated the reduction of other Wnt target genes and proteins in Cul4b KOIEC organoids (Fig. 8i, j). Overall, our findings demonstrated a critical role of CUL4B in intestinal development and homeostasis. Mechanistically, we identified IRGM1 as a novel targeting substrate of CRL4B complex. CUL4B ubiquitylates IRGM1 at K270. Impaired intestinal homeostasis caused by CUL4B deletion is mediated by upregulation of IRGM1.
Discussion
In this study, we have demonstrated a critical role of CUL4B in intestinal homeostasis. We observed that ISC self-renewal and differetiation were impaired in the absence of CUL4B. Mechanistically, CUL4B regulates ISCs and their niches by targeting IRGM1 for ubiquitination and degradation, and thereby sustaining Wnt signaling. Our results have important implications in understanding the regulatory network of intestine development and related diseases.
Due to lack of Lgr5+ ISCs, CUL4B KOIEC intestine shows a significant reduction in self-renewal. Constitutive gradient Wnt enriched at crypts was required for ISC proliferation and specific lineage differentiation [7, 32, 33]. The dysregulated Wnt signaling in Cul4b KOIEC mice not only causes shorter crypts/villi length, but also leads to loss of secretory lineage differentiation including Paneth cells, enteroendocrine cells and goblet cells. In contrary, the function of absorptive lineage of enterocytes is accentuated in KOIEC intestine with upregulated Alpi staining and Apoa4 gene expression. The RNA-seq data show that the type 2 diabetes, PPAR pathway and insulin signaling are enriched in KOIEC tissue, which is in accordance with our previous reports that adipocyte specific Cul4b knockout mice are susceptible to obesity [25, 27]. Our results further indicate CUL4B deficiency as a risk factor for metobolic disorders.
Through secreting Wnt3, EGF and Notch that fuel stem cell self-renewal, Paneth cells play critical roles in ISC maintenance as one of the major components of ISC niches [2, 34, 35]. The reduction of organoid-forming and budding efficiency following CUL4B deletion is restricted to ex-vivo cultured organoids derived from KOIEC rather than KOLgr5 intestine. The transient knockout of CUL4B with short-term tamoxifen induction in KOLgr5 organoids leads to reduced size. But in single-cell assay, the expansion medium may contain enough Wnt signals that rescue the decreased organoid formation in KOLgr5 organoids.
Besides, our results of supernatant collection clearly demonstrate that deletion of CUL4B influences ISC stemness by extracellular environment through secretion. The TEM confirms the dysregulated intercellular structure and cell-cell junction between Paneth cell and ISCs in KOIEC mice, that also has been proved as a cause of disrupted Wnt recognization [7, 32]. IFN-gamma, which has been reported to trigger degranulation and extrusion of Paneth cells and release of antimicrobial products, is also enriched in CUL4B KOIEC tissues (data not shown). Taken together, our fingdings suggest the essential role of CUL4B in ISC niche. Increased numbers of granulated Lyz+ cells has been observed in Irgm1 KOIEC mice [31]. Consistantly, the Lyz+ cells were significantly reduced following Cul4b deletion. We perform dose-dependent lentivirus infection of shIrgm1 in KOIEC organoids to clarify that CUL4B regulates Wnt regulation through IRGM1. Total and active β-catenin are rescued in KOIEC organoids after Irgm1 deletion and Wnt recovery is confirmed by the STF reporter system. IRGM1 ablation also has strong rescue effect to re-produce antimicrobial products of Defa1 Defa4, and Defa6 in KOIEC organoids. Collectively, these data strongly suggest a non-redundant role of CUL4B in Paneth cell partially by degrading IRGM1. Besides, IRGM1 has been linked to bacterial immunity, inflammation, and autophagy initiation complexes formation by co-assembling with ULK1 and Beclin1 [15, 29]. Human IRGM1 is a risk allele in Crohn’s disease (CD), one chronic, immune-mediated, inflammatory intestinal disorder. Whether CUL4B functions in host-defense and chronic intestinal inflammation will be determined in our future study.
Our results reveal CRL4B as an E3 ubiquitin ligase to ubiquitylate IRGM1 at K270 and consequently degrade IRGM1. Several candidate WD40 repeat proteins are found by mass spectrometry as potential DCAFs for IRGM1. Although we identify a direct association between WDR77 with DDB1, interfering WDR77 by siRNA in 293 T cells results in no IRGM1 accumulation. In vitro ubiquitylation assay also suggests WDR77 is dispensable for IRGM1 degradation. The other co-factors of CRL4B complex for IRGM1 ubiquitylation need to be further explored.
CUL4B is mainly expressed in the cytoplasm of ISCs zone, which is recapitulated in ex vivo intestinal organoids. Although CUL4B is characterized by the nuclear localization signals at the N terminus, we uncover the specific intestinal cytoplasm location of CUL4B. Our results suggest the cytoplasm location of CUL4B could not be changed in organoids treated with nuclear export inhibitor, Leptomycin B (data not shown). Further experiments are needed to elucidate the mechanism by which CUL4B is excluded from the nucleus in the intestine.
Overall, our findings demonstrate a critical role of CUL4B in intestinal development and homeostasis. A cytoplasm specific expression of CUL4B is found at bottom crypts. Deletion of Cul4b leads to reduced self-renewal and an impaired lineage differentiation towards secretory progenitors of ISCs through Wnt signals. Mechanistically, CRL4B complex ubiquitylate its substrate IRGM1 at K270. These findings advance our understanding of how intestinal homeostasis is regulated and have potential implications in the management of gasteroenterological diseases.
Data availability
Raw and processed data for RNA-seq can be reached from GEO database with the number of GSE157818 and MS data in PRIDE with the number of PXD-021528. All other datasets and code are available upon request from the huhuili@sdu.edu.cn.
References
Muñoz J, Stange DE, Schepers AG, van de Wetering M, Koo BK, Itzkovitz S, et al. The Lgr5 intestinal stem cell signature: robust expression of proposed quiescent ‘+4’ cell markers. EMBO J. 2012;31:3079–91.
Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–7.
Yan KS, Chia LA, Li X, Ootani A, Su J, Lee JY, et al. The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc Natl Acad Sci USA. 2012;109:466–71.
Bevins CL, Salzman NH. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat Rev Microbiol. 2011;9:356–68.
Bel S, Pendse M, Wang Y, Li Y, Ruhn KA, Hassell B, et al. Paneth cells secrete lysozyme via secretory autophagy during bacterial infection of the intestine. Sci (N. Y, NY). 2017;357:1047–52.
Schuijers J, Junker JP, Mokry M, Hatzis P, Koo BK, Sasselli V, et al. Ascl2 acts as an R-spondin/Wnt-responsive switch to control stemness in intestinal crypts. cell stem cell. 2015;16:158–70.
Yan KS, Janda CY, Chang J, Zheng GXY, Larkin KA, Luca VC, et al. Non-equivalence of Wnt and R-spondin ligands during Lgr5(+) intestinal stem-cell self-renewal. Nature. 2017;545:238–42.
Leung C, Tan SH, Barker N. Recent Advances in Lgr5(+) Stem Cell Research. Trends cell Biol. 2018;28:380–91.
Batlle E, Henderson JT, Beghtel H, van den Born MM, Sancho E, Huls G, et al. Beta-catenin and TCF mediate cell positioning in the intestinal epithelium by controlling the expression of EphB/ephrinB. Cell. 2002;111:251–63.
Huels DJ, Bruens L, Hodder MC, Cammareri P, Campbell AD, Ridgway RA, et al. Wnt ligands influence tumour initiation by controlling the number of intestinal stem cells. Nat Commun. 2018;9:1132.
Drost J, van Jaarsveld RH, Ponsioen B, Zimberlin C, van Boxtel R, Buijs A, et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature. 2015;521:43–7.
Kuhnert F, Davis CR, Wang HT, Chu P, Lee M, Yuan J, et al. Essential requirement for Wnt signaling in proliferation of adult small intestine and colon revealed by adenoviral expression of Dickkopf-1. Proc Natl Acad Sci USA. 2004;101:266–71.
Yilmaz OH, Katajisto P, Lamming DW, Gultekin Y, Bauer-Rowe KE, Sengupta S, et al. mTORC1 in the Paneth cell niche couples intestinal stem-cell function to calorie intake. Nature. 2012;486:490–5.
Jardé T, Chan WH, Rossello FJ, Kaur Kahlon T, Theocharous M, Kurian Arackal T, et al. Mesenchymal Niche-derived Neuregulin-1 drives intestinal stem cell proliferation and regeneration of damaged epithelium. Cell Stem Cell. 2020;27:646–62. e647
Mehto S, Jena KK, Nath P, Chauhan S, Kolapalli SP, Das SK, et al. The Crohn’s disease risk factor IRGM Limits NLRP3 inflammasome activation by impeding its assembly and by mediating its selective autophagy. Mol Cell. 2019;73:429–45. e427
Kim BH, Shenoy AR, Kumar P, Bradfield CJ, MacMicking JD. IFN-inducible GTPases in host cell defense. Cell Host Microbe. 2012;12:432–44.
Jackson S, Xiong Y. CRL4s: the CUL4-RING E3 ubiquitin ligases. Trends Biochem Sci. 2009;34:562–70.
Sarikas A, Hartmann T, Pan ZQ. The cullin protein family. Genome Biol. 2011;12:220.
Zou Y, Mi J, Cui J, Lu D, Zhang X, Guo C, et al. Characterization of nuclear localization signal in the N terminus of CUL4B and its essential role in cyclin E degradation and cell cycle progression. J Biol Chem. 2009;284:33320–32.
Tarpey PS, Raymond FL, O’Meara S, Edkins S, Teague J, Butler A, et al. Mutations in CUL4B, which encodes a ubiquitin E3 ligase subunit, cause an X-linked mental retardation syndrome associated with aggressive outbursts, seizures, relative macrocephaly, central obesity, hypogonadism, pes cavus, and tremor. Am J Hum Genet. 2007;80:345–52.
Zou Y, Liu Q, Chen B, Zhang X, Guo C, Zhou H, et al. Mutation in CUL4B, which encodes a member of cullin-RING ubiquitin ligase complex, causes X-linked mental retardation. Am J Hum Genet. 2007;80:561–6.
Jiang B, Zhao W, Yuan J, Qian Y, Sun W, Zou Y, et al. Lack of Cul4b, an E3 ubiquitin ligase component, leads to embryonic lethality and abnormal placental development. PloS one. 2012;7:e37070.
Liu L, Yin Y, Li Y, Prevedel L, Lacy EH, Ma L, et al. Essential role of the CUL4B ubiquitin ligase in extra-embryonic tissue development during mouse embryogenesis. Cell Res. 2012;22:1258–69.
Lin CY, Chen CY, Yu CH, Yu IS, Lin SR, Wu JT, et al. Human X-linked intellectual disability factor CUL4B is required for post-meiotic sperm development and male fertility. Sci Rep. 2016;6:20227.
Li P, Song Y, Zan W, Qin L, Han S, Jiang B, et al. Lack of CUL4B in adipocytes promotes PPARγ-mediated adipose tissue expansion and insulin sensitivity. Diabetes. 2017;66:300–13.
Yuan J, Jiang B, Zhang A, Qian Y, Tan H, Gao J, et al. Accelerated hepatocellular carcinoma development in CUL4B transgenic mice. Oncotarget. 2015;6:15209–21.
Dou H, Duan Y, Zhang X, Yu Q, Di Q, Song Y, et al. Aryl hydrocarbon receptor (AhR) regulates adipocyte differentiation by assembling CRL4B ubiquitin ligase to target PPARgamma for proteasomal degradation. J Biol Chem. 2019;294:18504–15.
Beumer J, Clevers H. Cell fate specification and differentiation in the adult mammalian intestine. Nat Rev Mol cell Biol. 2021;22:39–53.
Chauhan S, Mandell MA, Deretic V. IRGM governs the core autophagy machinery to conduct antimicrobial defense. Mol cell. 2015;58:507–21.
Rogala AR, Schoenborn AA, Fee BE, Cantillana VA, Joyce MJ, Gharaibeh RZ, et al. Environmental factors regulate Paneth cell phenotype and host susceptibility to intestinal inflammation in Irgm1-deficient mice. Dis Models Mech. 2018,11:dmm031070.
Liu B, Gulati AS, Cantillana V, Henry SC, Schmidt EA, Daniell X, et al. Irgm1-deficient mice exhibit Paneth cell abnormalities and increased susceptibility to acute intestinal inflammation. Am J Physiol Gastrointest Liver Physiol. 2013;305:G573–84.
Clevers H, Loh KM, Nusse R. Stem cell signaling. An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science (N. Y, NY). 2014;346:1248012.
Farin HF, Jordens I, Mosa MH, Basak O, Korving J, Tauriello DV, et al. Visualization of a short-range Wnt gradient in the intestinal stem-cell niche. Nature. 2016;530:340–3.
Sato T, van Es JH, Snippert HJ, Stange DE, Vries RG, van den Born M, et al. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469:415–8.
Mei X, Gu M, Li M. Plasticity of Paneth cells and their ability to regulate intestinal stem cells. Stem cell Res Ther. 2020;11:349.
Acknowledgements
We thank Professor Hans Clevers for kindly providing the cell lines to produce recombinant R-spondin1, Noggin and Wnt3a. We thank Professor Jingxin Li and Bo Han for clinical support. We thank Translational Medicine Core Facility of Shandong University for consultation and instrument availability that supported this work.
Funding
This work was supported by the grants from: the National Key R&D Program of China 2019YFA0111400 (MOST, HH),the National Natural Science Foundation of China 31872810 and 81330050 (NSFC, YG), the National Natural Science Foundation of China 31501179, 32122031 and 31970779 (NSFC, HH), the National Natural Science Foundation of China 31771260 (NSFC, CS), the Funds for Youth Interdisciplinary and Innovation Research Group of Shandong University 2020QNQT003 (Shandong University, HH), the Key Research and Development Program of Shandong Province 2016ZDJS07A08 (YG) and the Shandong Provincial Natural Science Foundation ZR2020MH221 (JS).
Author information
Authors and Affiliations
Contributions
HH and YG conceived the project and designed all the experiments. YF performed most experiments, XH, HD and XZ performed immunoprecipitation, GST-pull down and in vitro ubiquitylation. JL, JW, BG, XM, HW, and YS helped to set up organoids and experiments. HH, YY, and HG analyzed the RNA-seq and MS data. HH and YF wrote the manuscript. YG, YZ, CS, and G.S. revised the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics statement
All animal experiments were approved by the Animal Care and Use Committee of the School of Basic Medical Science of Shandong University (No. LL-201502043 and ECSBMSSDU2019-2-008).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by R. Damgaard
Supplementary information
41418_2022_954_MOESM5_ESM.xlsx
Table S2 The differential genes changed in KOIEC mice (N=4), in KOCAG organoids (N=2) or in KOIEC organoids (N=2) compared with wildtype intestine.
41418_2022_954_MOESM7_ESM.xlsx
Table S4 The differential ubiquitylated proteins changed in KO<IEC> mice with fold change of above 1.5 compared with wildtype.
41418_2022_954_MOESM10_ESM.mov
Movie S2 The differential ubiquitylated proteins changed in KOIEC mice with fold change of above 1.5 compared with wildtype.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Fan, Y., Huo, X., Guo, B. et al. Cullin 4b-RING ubiquitin ligase targets IRGM1 to regulate Wnt signaling and intestinal homeostasis. Cell Death Differ 29, 1673–1688 (2022). https://doi.org/10.1038/s41418-022-00954-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-022-00954-9