Abstract
p62/SQSTM1 is a selective autophagy receptor that drives ubiquitinated cargos towards autophagic degradation. This receptor is also a stress-induced scaffold protein that helps cells to cope with oxidative stress through activation of the Nrf2 pathway. Functional disorders of p62 are closely associated with multiple neurodegenerative diseases and cancers. The gene encoding the E3 ubiquitin ligase substrate-binding adapter SPOP is frequently mutated in prostate cancer (PCa), but the molecular mechanisms underlying how SPOP mutations contribute to PCa tumorigenesis remain poorly understood. Here, we report that cytoplasmic SPOP binds and induces the non-degradative ubiquitination of p62 at residue K420 within the UBA domain. This protein modification decreases p62 puncta formation, liquid phase condensation, dimerization, and ubiquitin-binding capacity, thereby suppressing p62-dependent autophagy. Moreover, we show that SPOP relieves p62-mediated Keap1 sequestration, which ultimately decreases Nrf2-mediated transcriptional activation of antioxidant genes. We further show that PCa-associated SPOP mutants lose the capacity to ubiquitinate p62 and instead promote autophagy and the redox response in a dominant-negative manner. Thus, our findings indicate oncogenic roles of autophagy and Nrf2 activation in the tumorigenesis of SPOP-mutated PCa.
Similar content being viewed by others
Introduction
Autophagy is a highly conserved pathway among eukaryotic cells, through which cytoplasmic components are degraded and nutrients recycled [1, 2]. Autophagy and the ubiquitin-proteasome system (UPS) are two major protein quality control pathways, and have been long considered independent from each other. However, recent research suggests that these two processes are closely interconnected and mutually regulated through the common ubiquitination signals [3]. Identification of autophagy cargo receptors, which simultaneously bind ubiquitin and ATG8 family proteins, has illustrated molecular links between UPS and autophagy [4]. p62 is the best studied autophagy cargo receptor and plays a critical role in autophagy upon metabolic, proteotoxic, and oxidative stresses. In addition, p62 sequesters the Cullin 3 ubiquitin ligase adaptor Kelch Like ECH Associated Protein 1 (Keap1), thereby leading to increased stabilization and activity of nuclear factor, erythroid 2 like 2 (Nrf2), a key transcription factor in reprogramming oxidative stress responses [5,6,7]. Although the pathophysiological importance of p62 is well-recognized, the dynamic regulation of p62 activity under basal and stress conditions is not fully understood.
Speckle type BTB/POZ protein (SPOP) is a substrate-binding adaptor of the Cullin 3-RING E3 ubiquitin ligase complexes (CRL3) [8]. SPOP mutations are recurrently found in PCa and endometrial cancer (EC), but such mutations are relatively rare in cancers of other types [9, 10]. The majority of PCa or EC-associated SPOP mutations are within the substrate-binding MATH domain, suggesting that there is a change in binding affinity between SPOP and its interacting partners. Multiple oncoproteins, such as SRC-3, AR, ERα, ERG, DEK, BRD2/3/4, PD-L1, SENP7, Caprin1, DAXX, c-MYC, Cyclin E, and Nanog, are ubiquitinated and degraded by the SPOP [11]. Cancer-associated SPOP mutants generally display impaired substrate-binding capacity and oligomerization, which disrupts the function of the entire E3 ubiquitin complexes in a dominant-negative manner [12]. Accumulating evidence indicates that SPOP mutations promote the initiation and progression of PCa due to the dysregulation of substrates; however, the underlying mechanisms are not fully understood [13].
To identify additional SPOP substrates whose dysregulation plays role in SPOP mutation-driven PCa tumorigenesis, we perform an affinity–purification followed by mass spectrometry (AP–MS) to identify novel SPOP interacting partners. In the represent study, we reveal that SPOP binds p62 and acts as a negative regulator of autophagy and Nrf2 pathway.
Results
p62 is a SPOP-interacting protein
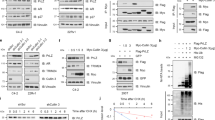
We used FLAG-HA-SPOP as a bait to identify SPOP-binding partners in prostate cancer cell line DU145 via AP-MS method. From this experiment, p62 ranked high on the interaction hit list (Supplementary Table. 1). Given that p62 participates in autophagy and oxidative stress response, we embarked to identify whether SPOP exerts tumor-suppressive roles through its interaction with p62. We first verified the binding of p62 with SPOP via co-immunoprecipitation (co-IP) assays. Ectopically overexpressed SPOP and p62 interacted with each other (Figs. 1A, B). FLAG-SPOP was also capable of immunoprecipitating endogenous p62 and a reported substrate, BRD4 in DU145 cells (Fig. 1C). Importantly, the endogenous SPOP–p62 interaction was observed in DU145 cells (Figs. 1D, E). We also demonstrated that SPOP did not interact with other autophagy cargo receptors examined (Figs. 1E, F). The MATH domain of SPOP is responsible for recruiting substrates [8]. We demonstrated that the deletion of the MATH domain, but not the BTB (for BR-C, ttk and bab) domain, abolished the SPOP–p62 interaction (Figs. 1G, H). SPOP usually binds its substrates via the SPOP-binding consensus (SBC) motif [8]. We sought to determine the protein sequence in p62 that is responsible for its interaction with SPOP. We found that one region (273-PESSS-277 aa) in p62 perfectly matched the SBC motif and is similar to those present in reported SPOP substrates (Fig. 1I). We generated p62 mutants in which the motif is deleted or single residue is mutated to alanine. co-IP assay results showed that SPOP only bound p62-WT and not with any of the mutants that disrupt the SBC motif (Figs. 1J–L). Together, our findings indicate that SPOP interacts with p62 via the SBC motif in p62 and the MATH domain in SPOP.
A Western blot analysis of the whole cell lysates (WCL) and immunoprecipitates from 293 T cells transfected with the indicated plasmids. B Western blot analysis of the WCL and immunoprecipitates from 293 T cells transfected with the indicated plasmids. C Western blot analysis of the WCL and immunoprecipitates from DU145 cells transfected with the indicated plasmids. D Western blot analysis of the WCL and immunoprecipitates from DU145 cells immunoprecipitated with IgG or p62 antibody. E Western blot analysis of the WCL and immunoprecipitates from DU145 cells immunoprecipitated with IgG or SPOP antibody. F Western blot analysis of the WCL and immunoprecipitates from 293 T cells transfected with the indicated plasmids. G Schematic representation of SPOP-WT and deletion mutants. Binding capacity of SPOP to p62 is indicated with the symbol. H Western blot analysis of the WCL and immunoprecipitates from 293 T cells transfected with the indicated plasmids. I Amino acid sequence alignment of the SBC motif in SPOP substrates. NANOG, Puc, and Ci are reported SPOP substrates containing well-characterized SBC motif (Φ-π-S-S/T-S/T; Φ: nonpolar residues, π: polar residues). J Diagram showing wild-type p62 and the SBC-motif-deleted mutant (ΔSBC). K Western blot analysis of the WCL and immunoprecipitates from 293 T cells transfected with the indicated plasmids. L Western blot analysis of the WCL and immunoprecipitates from 293 T cells transfected with the indicated plasmids.
SPOP promotes the non-degradative ubiquitination of p62
We explored whether SPOP could promote the ubiquitination and degradation of p62. The ectopic overexpression of SPOP-WT or its deletion mutants did not alter the coexpressed p62 levels (Fig. 2A). Stable overexpression of SPOP in DU145 and another prostate cell lines PC-3 did not alter endogenous p62 levels (Figs. 2B). SPOP depletion via short hairpin RNA (shRNA)-mediated knockdown or CRISPR/Cas9-mediated knockout (KO) did not alter endogenous p62 levels (Figs. 2C, D). To confirm the loss of SPOP activity, BRD4, a degradative substrate of SPOP, was stabilized in SPOP-depleted cells (Figs. 2C, D). Although p62 did not show any degradation effect by SPOP, p62 was robustly polyubiquitinated by the coexpression of SPOP-WT, and not by SPOP-ΔBTB or -ΔMATH mutant (Fig. 2E, Supplementary Fig. 1A). Deletion of the SBC motif or point mutations of the SBC motif also prevented SPOP-mediated ubiquitination (Supplementary Figs. 1B, C). Accordingly, SPOP depletion decreased the endogenous p62 ubiquitination (Figs. 2F). These results indicate that SPOP promotes p62 ubiquitination through binding to the SBC motif in p62, but surprisingly, this ubiquitination event does not induce p62 degradation.
A Western blot analysis of the WCL from 293 T cells transfected with FLAG-p62 and increasing doses of Myc-tagged SPOP-WT or deletion mutants. B Western blot analysis of the WCL from PC-3 or DU145 cells stably expressing empty vector (EV) or FLAG-SPOP. C Western blot analysis of the WCL from PC-3 or DU145 cells stably expressing SPOP-specific shRNA or negative control (NC). D Western blot analysis of the WCL from PC-3 or DU145 cells with SPOP KO using CRISPR/Cas9 method. Parental PC-3 and DU145 cells were used as the controls. E 293 T cells were transfected with indicated plasmids. 24 h after transfection, the WCL were prepared and the in vivo ubiquitination assays were performed as described in methods. The polyubiquitinated forms of p62 were detected by Western blot with anti-HA antibody. F 293 T cells were infected with lentivirus expressing SPOP-specific shRNA or NC. 72 h after infection, the stable cell lines were transfected with HA-Ub. 24 h after transfection, the WCL were prepared and the in vivo ubiquitination assays were performed. The polyubiquitinated forms of p62 were detected by Western blot with anti-HA antibody. G 293 T cells were transfected with the indicated plasmids. 24 h after transfection, the WCL were prepared and the in vivo ubiquitination assays were performed. The polyubiquitinated forms of p62 were detected by Western blot with anti-HA antibody. H The distribution of the ubiquitin attachment sites of p62 catalyzed by different E3 ubiquitin ligases. I Western blot analysis of WCL and immunoprecipitates from 293 T cells transfected with Myc-SPOP and FLAG-tagged p62-WT or mutants. J 293 T cells were transfected with the indicated plasmids. 24 h after transfection, the WCL were prepared and the in vivo ubiquitination assays were performed. The polyubiquitinated forms of p62 were detected by Western blot with anti-HA antibody.
Given that SPOP-mediated p62 ubiquitination is non-degradative, we then examined the polyUb chain linkage specificity on p62. We performed ubiquitination assays using a panel of ubiquitin mutants containing single K→R mutations at each of seven lysine residues. We also included a lysine-free ubiquitin mutant wherein all the lysine are replaced with arginine (K-ALL-R). Co-expression of the Ub–K-ALL-R mutant abolished SPOP-mediated p62 ubiquitination, excluding the possibility that SPOP catalyzes multiple mono-ubiquitination on p62 proteins (Fig. 2G). The co-expression of Ub-K11R, -K33R, -K48R or -K63R mutant did not alter p62 ubiquitination (Fig. 2G), suggesting that K11, K33, K48, and K63 in Ub were largely dispensable for SPOP-mediated p62 ubiquitination. By contrast, a moderate reduction in p62 ubiquitination was observed when using the Ub-K6R, -K27R, or -K29R mutant (Fig. 2G). We then chose to use a reciprocal series of Ub mutants. These mutants contained only one lysine present, with the other six lysine mutated to arginine. The expression of Ub-K11O, K33O, K48O, or K63O mutant nearly abolished SPOP-mediated p62 ubiquitination, but remnant ubiquitination was observed with the Ub-K6O, -K27O, or -K29O mutant (Fig. 2G). These results indicate that SPOP catalyzes the synthesis of mixed-linkage polyUb chains on p62 and that K6, K27, and K29 in Ub are involved.
Having established that SPOP promotes the atypical ubiquitination of p62, we next sought to identify the ubiquitin attachment sites on p62. Previous studies reported that specific lysine residues of p62, including K7, K13, K91, K189, and K420, are targeted by E3 ubiquitin ligases TRIM21, Parkin, RNF166, and Keap1, respectively (Fig. 2H) [14,15,16,17,18,19,20]. We constructed single K→R point mutants of p62, and evaluate whether SPOP induces p62 ubiquitination through these lysine residues. Although these p62 mutants interacted with SPOP at a comparable level to p62-WT (Fig. 2I), only p62-K420R mutant was incapable of ubiquitination by SPOP, suggesting that K420 is the primary ubiquitin attachment site catalyzed by SPOP (Fig. 2J). Together, our findings indicate that SPOP promotes the non-degradative ubiquitination on p62, and that K420 within the UBA domain of p62 serves as the predominant ubiquitin attachment site.
PCa-associated SPOP mutants are defective in promoting p62 ubiquitination
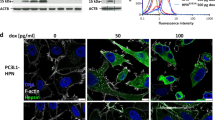
To date, the vast majority of SPOP mutations associated with PCa occur within its MATH domain, which is responsible for selective substrate binding (Fig. 3A) [9, 10]. We postulated that PCa-associated SPOP mutants may be defective in mediating p62 ubiquitination. The p62-binding ability of these SPOP mutants was abolished or severely impaired as compared to SPOP-WT (Fig. 3B). SPOP-mediated p62 ubiquitination is markedly attenuated for these mutants (Fig. 3C). In accordance with previous studies showing that PCa-associated SPOP mutants function as dominant-negative variants to deregulate its substrates [21, 22], we found that co-expression of SPOP-Y87N, -F125V or -F133L mutant markedly reduced the interaction between SPOP-WT and p62 (Fig. 3D), and resulted in the suppression of SPOP-WT-mediated p62 ubiquitination (Fig. 3E), indicating a dominant-negative effect exerted by PCa-associated SPOP mutants. To further analyze the interaction between p62 and SPOP (WT or mutant) in a more physiological level, doxycycline-inducible FLAG-SPOP-WT or -F125V mutant was lentivirally transduced into SPOP KO DU145 cells. Then, doxycycline was added in an attempt to generate similar SPOP levels as those of parental cells. Co-IP assays showed that SPOP-WT, but not the F125V mutant, interacted with endogenous p62 (Supplementary Fig. 2A). Together, these results indicate that PCa-associated SPOP mutants exert dominant-negative effects to downregulate p62 ubiquitination because of impaired p62- binding capacities.
A The distribution of the prostate-associated SPOP mutations occur in the MATH domain of SPOP. B Western blot analysis of WCL and immunoprecipitates from 293 T cells transfected with the indicated plasmids. C 293 T cells were transfected with the indicated plasmids. 24 h after transfection, the WCL were prepared and the in vivo ubiquitination assays were performed. The polyubiquitinated forms of p62 were detected by Western blot with anti-HA antibody. D Western blot analysis of WCL and immunoprecipitates from 293 T cells transfected with the indicated plasmids. E 293 T cells were transfected with the indicated plasmids. 24 h after transfection, the WCL were prepared and the in vivo ubiquitination assays were performed. The polyubiquitinated forms of p62 were detected by Western blot with anti-ubiquitin antibody. F The distribution of the lung and pancreatic cancer-associated p62 mutations occur in the SBC motif. G Western blot analysis of WCL and immunoprecipitates from 293 T cells transfected with the indicated plasmids. H 293 T cells were transfected with the indicated plasmids. 24 h after transfection, the WCL were prepared and the in vivo ubiquitination assays were performed. The polyubiquitinated forms of p62 were detected by Western blot with anti-HA antibody.
Cancer-associated p62 mutants evade SPOP-mediated ubiquitination
Given that p62 interacts with SPOP in an SBC motif-dependent manner, we further explored whether PCa-associated p62 mutations that occur in the SBC motif would attenuate the SPOP–p62 interaction and permit p62 to evade SPOP-mediated ubiquitination. To this end, we examined cancer sequencing data deposited in the cBioPortal database (www.cbioportal.org/). However, we did not identify any SBC motif-localized missense p62 mutations in PCa specimens, but we found a S277G mutation in lung cancer and two mutations (S276Y and S277I) in pancreatic cancer specimens (Fig. 3F). We found that the SPOP-binding capacity of these p62 mutants was completely abolished (Fig. 3G). SPOP-mediated ubiquitination of these p62 mutants was also markedly attenuated (Fig. 3H). Together, our findings indicate that cancer-derived p62 mutations occur in the SBC motif allow p62 to evade SPOP-mediated ubiquitination.
SPOP inhibits p62 phase condensation and dimerization
p62 is a cytoplasmic protein. Our previous study showed that SPOP was either localized exclusively in the nucleus as speckles, or in both the cytoplasm and nuclei [21]. We investigated the subcellular compartment where the SPOP–p62 interaction occurs. Ectopically overexpressed p62 appear as small puncta in the cytoplasm. When p62 and SPOP were co-expressed, p62 was exclusively colocalized with cytoplasmic SPOP but not nuclear SPOP. Our previous study showed that the cytoplasmic retention ability of PCa-associated SPOP mutants is impaired and they are exclusively localized as nuclear puncta in PCa cells [21]. We found that nuclear-localized SPOP-F125V mutant showed no colocalization with cytoplasmic p62. SPOP lacking the nuclear localization sequence (SPOP-ΔNLS) accumulated exclusively in the cytoplasm as puncta and perfectly colocalized with p62. To further probe their interaction, SPOP-ΔNLS showed no colocalization with p62-ΔSBC mutant, and PCa-associated SPOP-ΔNLS mutants (Y87N, F125V, or F133L) showed no colocalization with p62, both suggesting that p62–SPOP interaction is a prerequisite for their colocalization. SPOP did not colocalize with NBR1, a closed paralog of p62, suggesting a selective interaction (Fig. 4A). We also observed a partial co-localization as puncta between endogenous p62 and tet-on inducible SPOP-WT, but not the -F125V mutant (Supplementary Figs. 2B).
A DU145 cells were transfected with indicated plasmids, and then immunostained with HA (SPOP) and FLAG (p62). Scale bar, 10 μm. Enlargements of selected areas were shown. B, C DU145 cells were transfected with the indicated plasmids. 24 h after transfection, cells were subjected to live imaging and FRAP assays. n = 8 for each group from three independent experiments (B). Scale bar, 5 μm. The data were analyzed with Graphpad Prism using nonlinear regression (curve fit) (C). Data were shown as mean ± SD. The p values were calculated using the Two-way ANOVA test. *p < 0.05, ***p < 0.001. D Western blot analysis of the WCL and immunoprecipitates from 293 T cells transfected with the indicated plasmids. E FLAG-p62-WT or mutants were expressed in 293 T cells. HA-Ub was expressed in another set of 293 T cells. 24 h after transfection, the FLAG-p62-WT or mutant-expressing cell lysates were incubated with the same amount of the HA-Ub expressing cell lysates for 3 h. The mixtures were subjected to co-IP with FLAG antibody and then WB analysis with the indicated antibodies.
It is well-established that the p62–Keap1 interaction titrates Keap1 away from Nrf2, stabilizing Nrf2 by decreasing CRL3-Keap1 mediated ubiquitination and allowing for Nrf2-mediated activation of the antioxidant response pathway [23]. Furthermore, Keap1 is degraded through the autophagy pathway in a p62-dependent manner [24]. SPOP and Keap1 are both classified as CRL3 adapters, we were curious whether a similar regulatory pattern might exist for the p62–SPOP interaction. Thus, we generated ATG3, ATG5, ATG7, and p62 KO DU145 cells via CRISPR/Cas9-mediated KO and detected SPOP levels in autophagy-deficient cells. As a positive control, p62 proteins were accumulated in ATG3, ATG5, and ATG7 KO DU145 cells; however, SPOP protein accumulation was not observed (Supplementary Fig. 3A), indicating that SPOP is unlikely an autophagic substrate. Moreover, SPOP protein levels remained unchanged in cells treated with rapamycin or medium lacking amino acids (Supplementary Fig. 3B). As expected, Nrf2 proteins were stabilized in ATG3, ATG5, and ATG7 KO DU145 cells, but destabilized in p62 KO DU145 cells; however, no obvious changes of SPOP and its substrates (BRD4, DEK, and Caprin1) were observed (Supplementary Figs. 3A, C). These results suggested that p62 had no impact on SPOP-mediated substrate degradation, which is distinct from its roles towards Keap1.
Given the aforementioned results, we then explored an alternative possibility that SPOP regulates p62 function. p62 puncta have viscous liquid-like properties and are subject to phase separation [25]. Since SPOP and p62 were colocalized as cytoplasmic puncta, we sought to identify whether SPOP affects p62 phase separation. We used fluorescence recovery after photobleaching (FRAP) assays to show the exchange of molecules between droplets and its surrounding solution. We confirmed that the GFP-p62 fluorescence signals underwent recovery after photobleaching in DU145 cells, and the recovery rate by FRAP reflects the motile ability of droplets. Our results were in line with previous studies that p62 puncta are viscous (Fig. 4B) [25, 26]. We then investigated whether SPOP affects p62 phase condensation. The fluorescence signals of p62 recovered faster after photobleaching in the presence of SPOP-WT (or -ΔNLS) than that of p62 alone. By contrast, SPOP-F125V has an opposite impact on p62 recovery compared to SPOP-WT (or -ΔNLS) (Figs. 4B, C), suggesting that SPOP functions as a negative regulator on the gel-like nature of p62 puncta. Moreover, similar results were observed in ATG7 KO DU145 cells, indicating that SPOP modulates the FRAP property of p62 in an autophagy-independent manner (Supplementary Figs. 4A, B). Since SPOP inhibits p62 phase condensation, we postulated that SPOP may decrease p62 dimerization. We examined the interaction affinity between FLAG- and HA-tagged p62 proteins in the absence or presence of SPOP. We found that in the presence of SPOP-WT but not the -F125V mutant, less HA-p62 was immunoprecipitated by FLAG-p62, indicating that SPOP decrease the p62–p62 interaction affinity, and thus inhibiting p62 dimerization (Fig. 4D). The localization of K420 at the Ub-binding UBA domain prompted us to speculate that SPOP-mediated p62 ubiquitination may affect the ubiquitin-binding capacity of p62. p62 mutants that evade SPOP-mediated ubiquitination are more effective at binding ubiquitin conjugates than that of p62-WT (Fig. 4E). Moreover, the ubiquitin conjugates-binding capacity of p62 was likewise increased by the co-expression of SPOP-F125V or -F133L mutant (Supplementary Fig. 4C). Together, our findings indicate that SPOP negatively regulates p62 phase condensation and dimerization, thereby reducing its ubiquitin-binding capacity.
SPOP negatively regulates autophagy through p62
One of the main functions of p62 is to recruit proteins into aggregates (sequestosomes) for autophagic degradation [5,6,7]. We assessed the functional impact of SPOP on rapamycin-induced autophagy. Immunofluorescence analysis revealed a decreased LC3B and p62 puncta formation in SPOP-WT (or-ΔNLS) overexpressed and rapamycin-treated cells. By contrast, over-expression of PCa-associated SPOP mutants led to more LC3B and p62 puncta than that in control cells (Figs. 5A–C). Rapamycin-induced p62 degradation and LC3B-II accumulation were attenuated in SPOP- WT (or-ΔNLS) overexpressed cells but were potentiated in cells overexpressing PCa-associated SPOP mutants (Fig. 5D). Moreover, LC3B and p62 puncta formation, LC3B-II accumulation, and p62 degradation were all more prominent in SPOP KO DU145 cells than that in parental cells (Figs. 5E–H). To provide evidence that SPOP KO promotes autophagic flux, we induced autophagy in cells expressing mCherry-EGFP-LC3B reporter. SPOP KO increased both the autophagosomes and autolysosomes formation (Supplementary Figs. 5A–C). These results suggest that SPOP-WT restricts autophagy, while PCa-associated SPOP mutants promote autophagy in a dominant-negative fashion.
A DU145 cells were transfected with the indicated plasmids, treated with DMSO (left panel) or rapamycin (500 nM) (right panel) for 12 h, then cells were immunostained with HA (SPOP), p62, LC3B, and DAPI. Scale bar, 10 μm. B, C Quantification of the area of LC3B puncta (B) or p62 puncta (C) per cell shown in (A). n = 8. D Western blot analysis of the WCL from DU145 cells stably expressing the indicated plasmids. Cells were treated with DMSO or rapamycin (500 nM) for 12 h before harvesting. E Parental and SPOP KO DU145 cells treated with DMSO or rapamycin (500 nM) for 12 h were immunostained with p62 and LC3B, and DAPI. Scale bar, 10 μm. F, G Quantification of the area of LC3B puncta (F) and p62 puncta (G) per cell shown in (E). Data were shown as means ± SD of the puncta area in eight cells from three independent experiments were quantified. H Western blot analysis of the WCL from parental and SPOP KO DU145 cells treated with DMSO or rapamycin, (500 nM) for 12 h before harvesting. Data were all shown as means ± SD. The p values were calculated using the One-way ANOVA test in (B, C, F, G). *p < 0.05, **p < 0.01, ***p < 0.001.
To determine whether the impact of SPOP on autophagy is dependent on p62 but not the other SPOP substrates, doxycycline-inducible p62-WT or p62 S276A (mutant incapable of binding SPOP) was lentivirally transduced into p62 KO DU145 cells. Then, doxycycline was added to the medium in an attempt to generate similar p62 expression levels as those of parental cells (Supplementary Fig. 6A). SPOP overexpression reduced the rapamycin-induced LC3B, p62 puncta formation in p62 KO DU145 cells when p62-WT was reintroduced. By contrast, SPOP overexpression showed no impact on LC3B, p62 puncta formation in p62 KO DU145 cells when p62 S276A mutant was reintroduced (Supplementary Figs. 6B–D), suggesting that the p62–SPOP interaction was a prerequisite for the impact of SPOP on autophagy.
The removal of misfolded, ubiquitinated proteins is an essential step in protein quality control. The selective autophagic clearance of misfolded, ubiquitinated proteins is called aggrephagy [27]. Given that p62 is a major receptor for aggrephagy, we investigated the effect of SPOP on aggrephagy triggered by puromycin. Stable overexpression of SPOP-WT decreased p62 puncta and ubiquitin-positive protein aggregates in puromycin-treated cells, while stable overexpression of PCa-associated SPOP mutants increased p62 puncta and ubiquitin-positive protein aggregates (Supplementary Figs. 7A–D). Furthermore, more p62 puncta and ubiquitin-positive protein aggregates were observed in SPOP KO DU145 cells than that in parental cells (Supplementary Figs. 7E–H). Finally, we investigated whether upstream autophagy signals would change the SPOP–p62 interaction. As shown in Supplementary Figs. 8A–B, rapamycin treatment reduced the SPOP–p62 interaction and SPOP-mediated p62 ubiquitination. Together, our findings indicate that SPOP functions as a negative regulator of autophagy in a p62-dependent manner, and the negative impact of SPOP on p62 is relieved when autophagy is initiated.
SPOP negatively regulates p62-mediated sequestration of Keap1 and Nrf2-mediated stress response
p62 plays a critical role in the Nrf2-activated antioxidant response by sequestering Keap1 into aggregates, resulting in the inhibition of Keap1-mediated Nrf2 degradation [23]. Reciprocally, Keap1 facilitates the p62-mediated ubiquitin aggregate clearance via autophagy by ubiquitinating p62 at K420 (ref. [20]). Upon establishing the notion that SPOP suppresses p62 phase condensation and dimerization, we aim to examine the impact of SPOP on p62-mediated Keap1 sequestration. Immunofluorescence analysis showed that Keap1 was diffusely localized in the cytoplasm. In accordance with previous studies [14], Keap1 and p62 were colocalized in larger puncta as compared to p62 expression alone. SPOP overexpression had no impact on the cytoplasmic localization of Keap1, but when SPOP, Keap1, and p62 were co-expressed, three proteins co-localized as smaller puncta compared to conditions with only p62 expression. By contrast, PCa-associated SPOP mutants did not co-localize with the Keap1/p62 puncta and Keap1/p62 puncta formation was enhanced. SPOP-WT (or-ΔNLS) did not co-localize with Keap1/p62-S276A puncta and did not affect Keap1/p62-S276A puncta formation (Figs. 6A, B, Suppl. Figs. 9A, C). K420 is the primary ubiquitin attachment site catalyzed by CRL3-SPOP (Fig. 2J). We found that the p62-K420R mutant still had the capacity to co-localize with Keap1. SPOP-WT (or-ΔNLS) was co-localized with the Keap1/p62-K420R puncta but did not affect Keap1/p62-K420R puncta formation (Fig. 6A, B, Suppl. Figs. 9A, C). Moreover, cancer-derived p62 mutants sequestered Keap1 to a similar extent as p62-WT, but these effects cannot be disrupted by SPOP (Supplementary Figs. 9B, C). By taking advantage of the cell lines with tet-on inducible expression of SPOP-WT or -F125V mutant, we showed that endogenous p62, LC3B, and p62/Keap1 puncta formation was enhanced in SPOP-F125V expressing cells compared to SPOP-WT-expressing cells (Supplementary Figs. 2C-G). Together, our findings indicate that SPOP negatively regulates p62-mediated Keap1 sequestration, but this effect is dependent on the SPOP–p62 interaction and SPOP-mediated p62 ubiquitination.
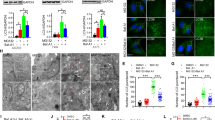
A DU145 cells were transfected with the indicated plasmids, then cells were immunostained with HA (SPOP), FLAG (p62), Myc (Keap1), and DAPI. Scale bar, 10 μm. Enlargements of selected areas were shown. B Quantification of the area of Keap1/p62 double positive puncta per cell shown in (A). n = 8. C Parental and SPOP KO DU145 cells treated with DMSO or AS (100 μM) for 4 h were immunostained with Keap1, p62, and DAPI. Scale bar, 10 μm. D Quantification of the area of Keap1/p62 double positive puncta per cell shown in (C). n = 8. E DU145 cells expressing the indicated plasmids were treated with DMSO or AS (100 μM) for 2.5 h. Cells were lysed in Buffer A as the NP-40-soluble fraction. The insoluble fraction was lysed in 7 M urea-containing Buffer A taken as the NP-40-insoluble fraction. The soluble and insoluble fractions were analyzed by Western blot. D represents DMSO treatment. F Parental and SPOP KO DU145 cells were treated with DMSO or AS (100 μM) for 2.5 h. Cells were lysed in Buffer A as the NP-40-soluble fraction. The insoluble fraction was lysed in 7 M urea-containing Buffer A taken as the NP-40-insoluble fraction. The soluble and insoluble fractions were subjected to Western blot. Data were all shown as means ± SD. The p values were calculated using the One-way ANOVA test in (B, D). *p < 0.05. **p < 0.01, ***p < 0.001, n.s. non-significant.
We investigated whether SPOP had any impact on the Keap1/Nrf2-mediated antioxidant response. Sodium arsenite (AS) was used to elicit oxidative stress. AS treatment led to the formation of Keap1/p62 puncta, and this phenomenon was more evident in SPOP KO DU145 cells than parental cells (Figs. 6C, D). We next prepared detergent-soluble and -insoluble fractions from cells treated with and without AS. SPOP KO DU145 cells and cells overexpressing PCa-associated SPOP mutants had increased amounts of Keap1 in insoluble fractions (Figs. 6E, F), while cells overexpressing SPOP-WT (or-ΔNLS) decreased amounts of Keap1 in insoluble fractions compared to control cells (Fig. 6E).
Stable overexpression of SPOP-WT (or-ΔNLS) led to a decrease in the protein levels of Nrf2 and its target HMOX1. By contrast, stable overexpression of PCa-associated SPOP mutants or SPOP KO elevated the protein level of Nrf2 and HMOX1 (Fig. 7A). Moreover, RT-qPCR measurements showed that SPOP expression negatively regulates the mRNA levels of NQO1 and HMOX1 (Figs. 7B, C). Stable overexpression of SPOP-WT (or-ΔNLS) decreased Nrf2 levels in the nuclei, while overexpression of PCa-associated SPOP mutants or SPOP KO increased the Nrf2 levels in the nuclei (Figs. 7D, E). Consequently, stable overexpression of SPOP-ΔNLS increased the cellular ROS levels in either the basal (Fig. 7F) or AS-treated (Fig. 7G) cells. To determine whether SPOP regulates cellular ROS levels in a Nrf2-dependent manner, we generated Nrf2 KO DU145 cells via CRISPR/Cas9-mediated KO. In contrast to the parental cells, SPOP-ΔNLS had a minimal effect on the ROS levels in the Nrf2 KO cells with or without AS treatment (Figs. 7H–I). Together, our findings indicate that SPOP negatively regulates cellular ROS levels through the p62–Keap1–Nrf2 pathway, which is abnormally enhanced by PCa-associated SPOP mutations.
A Western blot analysis of WCL from DU145 cells stably expressing the indicated plasmids; Western blot analysis of the WCL from parental and SPOP KO DU145 cells. B RT-qPCR assessment of the indicated gene mRNA levels in DU145 cells expressing EV, SPOP-ΔNLS, or SPOP mutants. The mRNA level of GAPDH was used for normalization. C RT-qPCR assessment of the indicated mRNA expression in parental and SPOP KO DU145 cells. The mRNA level of GAPDH was used for normalization. D Western blot analysis of cytoplasmic (Cyto) and nuclear (Nuc) extracts of DU145 cells stably expressing the indicated plasmids. E Western blot analysis of cytoplasmic and nuclear extracts of parental or SPOP KO DU145 cells with indicated antibodies. F DU145 cells stable expressing EV, SPOP-∆NLS, or mutants were subjected to H2DCFDA staining and then flow cytometry analysis. G DU145 cells stable expressing EV, SPOP-∆NLS or mutants were treated with AS (100 μM) for 4 h. Cells were subjected to H2DCFDA staining and then flow cytometry analysis. H Nrf2 KO DU145 cells stably expressing EV, SPOP-ΔNLS, or SPOP mutants were subjected to H2DCFDA (DCF) staining and then flow cytometry analysis. I Nrf2 KO DU145 cells stably expressing EV, SPOP-ΔNLS, or SPOP mutants were treated with AS (100 μM) for 4 h. Cells were subjected to H2DCFDA staining and then flow cytometry analysis. J Schematic diagram depicts a model where both autophagy and Nrf2 pathway are abnormally elevated in SPOP-mutated prostate cancer cells. Data were all shown as means ± SD. The p values were calculated using the One-way ANOVA test in (B, C, F–I). *p < 0.05, **p < 0.01. ***p < 0.001, n.s. non-significant.
Discussion
Accumulating evidence demonstrated that SPOP-mutated PCa have unique molecular and phenotypical features, but the tumorigenic mechanisms underlying this subtype is still poorly understood. In the current study, we reveal that cytoplasmic SPOP promotes the non-degradative ubiquitination of p62, which leads to the decrease in puncta formation, phase condensation, dimerization, and ubiquitin-binding capacity of p62, thereby suppressing p62-dependent autophagy and Nrf2 activation, two pathways commonly elevated in human cancers. Importantly, these effects were abrogated by PCa-associated SPOP mutations. Besides autophagy and the redox response, p62’s interaction with various binding partners allow it to affect a variety of signaling pathways, such as mTORC1 and NF-κB [28, 29]. Thus, it might be possible that PCa-associated SPOP mutations lead to the aberrant activation of multiple p62-engaged pathways.
In this study, we find that SPOP-mediated p62 ubiquitination negatively regulates p62 puncta formation, and that this effect is distinct from Keap1-mediated p62 ubiquitination, which promotes p62 puncta formation [20]. One possible explanation for these different effects could be that the different linkage types of polyUb chain catalyzed by CRL3-SPOP or CRL3-Keap1 on p62 may determine distinct fates of p62. Our results show that CRL3-SPOP catalyzes non-degradative K6-, K27-, and K29-linked polyUb chains on p62. The linkage type of polyUb chain catalyzed by Keap1 on p62 was not determined in the previous study [20] and should be elucidated for comparison in the future. The UBA domain-mediated polyUb chain binding induces p62 phase separation, but the direct impact of this ubiquitination on p62 phase separation may be more complex. While not entirely analogous, the proteasomal shuttle factor UBQLN2 undergoes phase separation, but upon polyUb binding, the phase separation is eliminated, thereby serving as a switch between droplet and disperse phases [30]. There remains much to be discovered for the regulation and effect of p62 phase separation.
In conclusion, we demonstrate that PCa-associated SPOP mutations elevate levels of autophagy and Nrf2 pathway by directly modulating p62 (Fig. 7J). Elucidation of this oncogenic pathway will guide the targeted therapy towards SPOP-mutated PCa.
Materials and methods
Cell lines, cell culture, transfection, and infection
293 T and prostate cancer cell lines (PC-3 and DU145) were purchased from American Type Culture Collection. 293 T cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% FBS. PC-3 and DU145 cells were cultured in RPMI 1640 medium supplemented with 10% FBS. The cells were maintained in a 37 °C humidified incubator supplied with 5% CO2. For transient transfection, the cells were transfected with PEI (Polyethyleneimine) or Lipofectamine 2000 (Thermo) according to the manufacturer’s instructions. pCDH-FLAG-HA-SPOP-WT or mutant vectors or pLKO-based gene-knockdown lentiviral vectors and packing constructs were transfected into 293 T cells. Virus-containing supernatant was collected 48 h after transfection. DU145 cells and PC-3 cells were infected with virus-containing supernatant in the presence of polybrene (8 μg/ml) and were then selected in growth medium containing 1.5 µg/ml puromycin. Sequences of gene-specific shRNAs are listed in Supplementary Table 2.
Protein complexes purification
The epitope-tagging strategy to isolate SPOP- containing protein complexes from DU145 cells was performed. In brief, to obtain a FLAG-HA-SPOP expressing cell line, DU145 cells were transfected with pCIN4-FLAG-HA-SPOP constructs and selected for two weeks in 1 mg/mL G418. The FH-tagged SPOP protein levels were detected by WB analyses. The stable cell lines were chosen to expand for protein complex purification. For purification, the cells were lysed in BC100 buffer (20 mM Tris-Cl, pH 7.9, 100 mM NaCl, 0.2 mM EDTA, 20% glycerol) containing 0.2% Triton X-100 and fresh protease inhibitor on ice for 2 h. The homogenate was centrifuged for 30 min at 12000 rpm at 4 °C. Cleared lysates were filtered through 0.45 μM spin filters (Millipore, USA) and immunoprecipitated by anti-FLAG antibody-conjugated M2 agarose (Sigma, USA). The bound polypeptides eluted with the FLAG peptide (Sigma) were further affinity purified by anti-HA antibody-conjugated agarose (Sigma). The final elutes from the HA-beads with HA peptides were resolved by SDS-PAGE on a 4–20% gradient gel (Bio-Rad) for Coomassie Blue staining. Gel bands were cut out from the gel and subjected to mass-spectrometric sequencing.
Plasmids and mutagenesis
Expression vectors for wild-type SPOP and mutants were described previously [31]. The cDNAs of p62 were obtained from Genecopia Inc. The cDNAs were subcloned into pCMV-FLAG or Myc vector. SPOP or SQSTM1/p62 mutants were generated by KOD-Plus-Mutagenesis Kit (TOYOBO) following the manufacturer’s instructions.
CRISPR-Cas9 mediated gene knock out stable cell generation
PX459 plasmid was used to clone guide oligos targeting SPOP, SQSTM1, ATG3, ATG5, ATG7 or NFE2L2. PC-3 or DU145 cells were plated and transfected with PX459 plasmids overnight. 24 h after transfection, 1 μg/ml puromycin was used to screen cells for three days. Living cells were seeded in 96-well plate by limited dilution to isolate monoclonal cell line. The knock out cell clones are screened by Western blot and validated by Sanger sequencing. Sequences of gene-specific sgRNAs are listed in Supplementary Table 2.
Doxycycline inducible expression
pGLV-FLAG- p62 or S276A mutant and virus packing plasmids were co-transfected into 293 T cells. Virus supernatant was collected 48 h after transfection. DU145 cells were infected with viral supernatant in the presence of polybrene (8 μg/ml) and were then tested and then selected in growth media containing 1.5 μg/ml puromycin for 3 days, and then the positive clones were pooled and amplified. Doxycycline was added to stable cells lines for FLAG-p62 induction.
Nuclear/cytoplasmic fractionation
In total, 5 × 106 DU145 cells were collected, washed twice with cold PBS, and lysed in cold hypotonic buffer (10 mM HEPES pH 7.9, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 1 mM DTT) supplemented with complete protease inhibitor cocktail without EDTA (Roche) on ice for 15 min. NP-40 was added to a final concentration of 0.625 % and cells were vortexed vigorously for 10 s. Samples were centrifuged for 30 s at 16000 g and the supernatants were harvested as cytoplasmic fraction. The nuclear pellets were then resuspended in cold hypertonic buffer (20 mM HEPES pH 7.9, 0.4 M NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM DTT) supplemented with a complete protease inhibitor cocktail (Roche). The samples were incubated at 4 °C for 15 min with agitation. The supernatants were collected after centrifugation for 5 min at 16000 g as nuclear proteins. Cytoplasmic and nuclear proteins were frozen in 5x sample buffer at −20 °C until use.
In vivo ubiquitination assays I
293 T cells were transfected with HA-Ub and other indicated constructs. 36 h after transfection, cells were lysed in 1% SDS buffer (Tris pH 7.5, 0.5 mM EDTA, 1 mM DTT) and boiled for 10 min. For immunoprecipitation, the cell lysates were diluted 10-fold in Tris-HCl buffer and incubated with anti-FLAG M2 agarose beads for 4 h at 4 °C. The bound beads are then washed four times with BC100 buffer (20 mM Tris-Cl, pH 7.9, 100 mM NaCl, 0.2 mM EDTA, 20% glycerol) containing 0.2% Triton X-100. The protein was eluted with 3×FLAG peptide for 2 h at 4 °C. The ubiquitinated form of p62 was detected by Western blot using anti-HA or Ub antibody.
In vivo ubiquitination assays II
293 T cells were transfected with (His)6-ubiquitin and other indicated plasmids. 36 h after transfection, cells were lysed in buffer A (6 M guanidine-HCl, 0.1 M Na2HPO4/NaH2PO4, and 10 mM imidazole pH 8.0). After sonication, the WCL was incubated with Ni-NTA beads (QIAGEN) for 3 h at room temperature. Subsequently, the pull-down products were washed twice with buffer A, twice with buffer A/TI (1 volume buffer A and 3 volumes buffer TI), and once with buffer TI (25 mM Tris-HCl, 20 mM imidazole pH 6.8). The pulldown proteins were detected by Western blot using anti-p62 antibody.
RT-qPCR assays
Total RNA was isolated from transiently transfected cells using TRIzol reagent (Thermo), and cDNA was reverse-transcribed using the PrimeScript RT Master Mix Kit (TAKARA) according to the manufacturer’s instructions. PCR amplification was performed using the AceQ Universal SYBR qPCR Master Mix Kit (Vazyme). All quantifications were normalized to the level of endogenous control GAPDH. The primer sequences for the SYBR qPCR used are listed in Supplementary Table. 2.
Immunofluorescence and confocal microscopy
For immunofluorescence, cells were plated on chamber slides, fixed with 4% paraformaldehyde at room temperature for 30 min. After washing with PBS, cells were permeabilized with 0.1% Triton-X100 in PBS for 15 min at room temperature. Cells were then washed with PBST, blocked with 5% donkey serums in PBS for 1 h, and incubated with primary antibodies in PBS at 4 °C for overnight in the dark. After washing with PBST, fluorescence-labeled secondary antibodies were applied and DAPI was counterstained for 1 h at room temperature in the dark. Slides were mounted in ProlongGold (Thermo). Cells were visualized and imaged using a confocal microscope (LSM880, Zeiss) with a 63*/1.4NA Oil PSF Objective. The puncta area was quantified using ImageJ by computing the total puncta area with at least eight cells. The analysis was carried out in triplicate from three different fields.
ROS measurement
Cells were incubated with medium containing 5 μM H2DCFDA for 30 min at 37 °C, then washed twice with PBS. Then, cells were harvested with 0.05 % trypsin-EDTA solution, suspended in a fresh media, and immediately analyzed with flow cytometry. Data were analyzed with Flowjo software.
Fluorescence recovery after photobleaching
DU145 cells or ATG7 KO DU145 cells were transfected with GFP-p62 or mApple-SPOP-WT (or ΔNLS). After the times as indicated, the living cells were subjected to FRAP analysis using a 63*/1.4NA Oil PSF Objective. GFP-p62 puncta were bleached with a 100% laser intensity of 488 nm. Recovery was recorded by fluorescence intensity for the indicated times.
Statistical analysis
The statistical calculations were performed using Graphpad Prism software. All data are shown as mean values ± SD for experiments performed with at least three replicates. The difference between two groups was analyzed using One-way ANOVA test unless otherwise specified. *represents p < 0.05; ** represents p < 0.01; *** represents p < 0.001.
References
Feng YC, He D, Yao ZY, Klionsky DJ. The machinery of macroautophagy. Cell Res. 2014;24:24–41.
Parzych KR, Klionsky DJ. An overview of autophagy: morphology, mechanism, and regulation. Antioxid Redox Sign 2014;20:460–73.
Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol Cell. 2009;34:259–69.
Kirkin V, Rogov VV. A diversity of selective au tophagy receptors determines the specificity of the autophagy pathway. Mol Cell. 2019;76:268–85.
Sanchez-Martin P, Komatsu M. p62/SQSTM1-steering the cell through health and disease. J Cell Sci. 2018;131:jcs222836.
Lamark T, Svenning S, Johansen T. Regulation of selective autophagy: the p62/SQSTM1 paradigm. Signal Mechanisms Autophagy. 2017;61:609–24.
Sanchez-Martin P, Saito T, Komatsu M. p62/SQSTM1: ‘Jack of all trades’ in health and cancer. Febs J. 2019;286:8–23.
Zhuang M, Calabrese MF, Liu J, Waddell MB, Nourse A, Hammel M, et al. Structures of SPOP-substrate complexes: insights into molecular architectures of BTB-Cul3 ubiquitin ligases. Mol Cell. 2009;36:39–50.
Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat JP, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44:685–U107.
Le Gallo M, O’Hara AJ, Rudd ML, Urick ME, Hansen NF, O’Neil NJ, et al. Exome sequencing of serous endometrial tumors identifies recurrent somatic mutations in chromatin-remodeling and ubiquitin ligase complex genes. Nat Genet. 2012;44:1310–5.
Wang ZW, Song YZ, Ye MM, Dai XM, Zhu XQ, Wei WY. The diverse roles of SPOP in prostate cancer and kidney cancer. Nat Rev Urol. 2020;17:339–50.
Geng CD, He B, Xu LM, Barbieri CE, Eedunuri VK, Chew SA, et al. Prostate cancer-associated mutations in speckle-type POZ protein (SPOP) regulate steroid receptor coactivator 3 protein turnover. Proc Natl Acad Sci USA. 2019;116:14386–7.
Blattner M, Liu DL, Robinson BD, Huang D, Poliakov A, Gao D, et al. SPOP mutation drives prostate tumorigenesis in vivo through coordinate regulation of PI3K/mTOR and AR signaling. Cancer cell. 2017;31:436–51.
Yang J, Peng H, Xu YM, Xie XD, Hu RG. SQSTM1/p62 (sequestosome 1) senses cellular ubiquitin stress through E2-mediated ubiquitination. Autophagy 2018;14:1072–3.
Pan JA, Sun Y, Jiang YP, Bott AJ, Jaber N, Dou ZX, et al. TRIM21 ubiquitylates SQSTM1/p62 and suppresses protein sequestration to regulate redox homeostasis. Mol Cell. 2016;62:149–51.
Heath RJ, Goel G, Baxt LA, Rush JS, Mohanan V, Paulus GLC, et al. RNF166 determines recruitment of adaptor proteins during antibacterial autophagy. Cell Rep. 2016;17:2183–94.
Jongsma MLM, Berlin I, Wijdeven RHM, Janssen L, Janssen GMC, Garstka MA, et al. An ER-associated pathway defines endosomal architecture for controlled cargo transport. Cell 2016;166:152–66.
Lin Q, Dai Q, Meng HX, Sun AQ, Wei J, Peng K, et al. The HECT E3 ubiquitin ligase NEDD4 interacts with and ubiquitylates SQSTM1 for inclusion body autophagy. J Cell Sci. 2017;130:3839–50.
Song PP, Li SS, Wu H, Gao RZ, Rao GH, Wang DM, et al. Parkin promotes proteasomal degradation of p62: implication of selective vulnerability of neuronal cells in the pathogenesis of Parkinson’s disease. Protein Cell. 2016;7:114–29.
Lee Y, Chou TF, Pittman SK, Keith AL, Razani B, Weihl CC. Keap1/Cullin3 modulates p62/SQSTM1 activity via UBA domain ubiquitination. Cell Rep. 2017;20:1994.
Jin XF, Wang J, Gao K, Zhang PZ, Yao LF, Tang Y, et?al. Dysregulation of INF2-mediated mitochondrial fission in SPOP-mutated prostate cancer. Plos Genet. 2017;13:e1006748.
Jin X, Shi Q, Li Q, Zhou L, Wang J, Jiang L, et al. CRL3-SPOP ubiquitin ligase complex suppresses the growth of diffuse large B-cell lymphoma by negatively regulating the MyD88/NF-kappaB signaling. Leukemia 2020;34:1305–14.
Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat cell Biol. 2010;12:213–U17.
Lau A, Wang XJ, Zhao F, Villeneuve NF, Wu TD, Jiang T, et al. A noncanonical mechanism of Nrf2 activation by autophagy deficiency: direct interaction between Keap1 and p62. Mol Cell Biol. 2010;30:3275–85.
Sun DX, Wu RB, Zheng JX, Li PL, Yu L. Polyubiquitin chain-induced p62 phase separation drives autophagic cargo segregation. Cell Res. 2018;28:405–15.
Yang Y, Willis TL, Button RW, Strang CJ, Fu YH, Wen X, et?al. Cytoplasmic DAXX drives SQSTM1/p62 phase condensation to activate Nrf2-mediated stress response. Nature Communications. 2019;10:3759.
Sun DX, Wu RB, Li PL, Yu L. Phase separation in regulation of aggrephagy. J Mol Biol. 2020;432:160–9.
Linares JF, Duran A, Yajima T, Pasparakis M, Moscat J, Diaz-Meco MT. K63 polyubiquitination and activation of mTOR by the p62-TRAF6 complex in nutrient-activated cells. Mol Cell. 2013;51:283–96.
Duran A, Linares JF, Galvez AS, Wikenheiser K, Flores JM, Diaz-Meco MT, et al. The signaling adaptor p62 is an important NF-kappaB mediator in tumorigenesis. Cancer cell. 2008;13:343–54.
Dao TP, Kolaitis RM, Kim HJ, O’Donovan K, Martyniak B, Colicino E, et al. Ubiquitin modulates liquid-liquid phase separation of UBQLN2 via disruption of multivalent interactions. Mol Cell. 2018;69:965–78. e6
An J, Wang C, Deng Y, Yu L, Huang H. Destruction of full-length androgen receptor by wild-type SPOP, but not prostate-cancer-associated mutants. Cell Rep. 2014;6:657–69.
Acknowledgements
We thank the members of Wang and Gao labs for their technical support. This work was in part supported by the National Natural Science Foundation of China (No. 91954106, 81872109 to K.G.; No. 91957125, 81972396 to C.W.; No. 81872260, 82172938 to P.Z.; No. 31801165 to X.J.), the Natural Science Foundation of Shanghai (No. 18ZR1430100 to K.G.), the Open Research Fund of State Key Laboratory of Genetic Engineering (No. SKLGE-2111 to K.G.). Science and Technology Research Program of Shanghai (19DZ2282100).
Author information
Authors and Affiliations
Contributions
CJW and KG conceived the study. KG, QS, PZZ, QL, ZHL, YD, YJW, YLH, HYH, and XYZ performed the experiments and data analyses. SMZ, YL, KG, and CJW analyzed and interpreted the data. CJW and KG wrote and revised the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by E Baehrecke
Supplementary information
Rights and permissions
About this article
Cite this article
Shi, Q., Jin, X., Zhang, P. et al. SPOP mutations promote p62/SQSTM1-dependent autophagy and Nrf2 activation in prostate cancer. Cell Death Differ 29, 1228–1239 (2022). https://doi.org/10.1038/s41418-021-00913-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-021-00913-w
This article is cited by
-
Therapeutic importance and diagnostic function of circRNAs in urological cancers: from metastasis to drug resistance
Cancer and Metastasis Reviews (2024)
-
Enhanced liquidity of p62 droplets mediated by Smurf1 links Nrf2 activation and autophagy
Cell & Bioscience (2023)
-
Epigenetic and post-translational modifications in autophagy: biological functions and therapeutic targets
Signal Transduction and Targeted Therapy (2023)
-
Targeting PI3K/Akt signaling in prostate cancer therapy
Journal of Cell Communication and Signaling (2023)
-
The SUMOylation and ubiquitination crosstalk in cancer
Journal of Cancer Research and Clinical Oncology (2023)