Abstract
In TNF signaling, ubiquitination of RIP1 functions as an early cell-death checkpoint, which prevents the spatial transition of the signaling complex from complex-I to death-inducing complex-II. Here, we report that ankyrin repeat domain 13a (ANKRD13a) acts as a novel component of complex-II to set a higher signal threshold for the cytotoxic potential of TNF. ANKRD13a deficiency is sufficient to turn the response to TNF from survival to death by promoting the formation of complex-II without affecting NF-κB activation. ANKRD13a binds to ubiquitinated-RIP1 via its UIM, and subsequently limits the association of FADD and caspase-8 with RIP1. Moreover, high ANKRD13a expression is inversely correlated with apoptotic phenotypes in ovarian cancer tissues and is associated with poor prognosis. Our work identifies ANKRD13a as a novel gatekeeper of the early cell-death checkpoint, which may function as part of an escape mechanism from cell death in some cancers.
Similar content being viewed by others
Introduction
Tumor necrosis factor (TNF) is a pleiotropic cytokine that elicits a variety of cellular responses, ranging from inflammation and proliferation to programmed cell death (PCD), in the form of apoptosis and necroptosis [1]. This pleiotropic nature primarily depends on the balance of two sequential signaling complexes: a plasma membrane-bound primary complex and a cytoplasmic secondary complex, referred to as complex-I and complex-II, respectively [2, 3]. Ligation of TNF receptor 1 (TNFR1) initiates assembly of complex-I composed of RIP1, cIAP1/2, LUBAC and IKKs, which triggers the induction of prosurvival genes via activation of NF-κB [4,5,6,7,8,9,10]. Subsequently, the RIP1-containing complex dissociates from complex-I and recruits FADD and caspase-8 to form cytotoxic complex-II, which then induces PCD [2, 11]. Therefore, the cell fate decision of life or death in response to TNF has long been thought to result from the balance of complex-I and complex-II.
Despite the ability of TNF to promote the formation of the cytotoxic complex-II, survival is the predominant response in most cell types, indicating that PCD is actively suppressed by specific checkpoint molecules [12, 13]. Historically, NF-κB-mediated transcriptional induction of prosurvival genes has been shown to be a late cell death checkpoint [14,15,16]. It is now believed that an early checkpoint protects cells from RIP1-dependent PCD by regulating its ubiquitination and phosphorylation status [13, 17]. Within a few minutes of TNF stimulation, diverse ubiquitin (Ub) linkages to RIP1 in complex-I have been shown to keep RIP1 in survival mode and thus prevent complex-II formation [1, 10]. Furthermore, IKKα/β, TBK1, and MK2 have also been reported to function as an early checkpoint by phosphorylation of ubiquitinated RIP1 in complex-I [18,19,20,21,22,23]. It has therefore been postulated that ubiquitination of RIP1 is a key node to control the cytotoxic potential of TNF by preventing sequential complex formation. Despite such progresses, how RIP1 ubiquitination functions as an early checkpoint remains to be determined.
The Ub-interacting motif (UIM; also termed the LALAL motif) is a highly conserved sequence motif found in a variety of eukaryotic proteins [24]. It has dual functions in the recognition of monoUb or polyUb chains as well as in facilitating ubiquitination [25,26,27]. The ankyrin repeat domain (ANKRD) 13 family proteins, including ANKRD13a, 13b, and 13d, contain multiple UIMs. Although little is known about its cellular function, recent studies have shown that ANKRD13a interacts to some endocytic proteins through its UIM and suppresses their internalization via as yet unknown mechanisms [28,29,30]. Therefore, it is possible that the ANKRD13 family are involved in the trafficking of ubiquitinated-substrates. Although ubiquitination of the TNFR1 signaling complex, particularly on RIP1, is one of the best characterized PTMs, the ANKRD13 family has not yet been implicated in TNFR1 signaling.
In this study, we explored the role of ANKRD13a in TNFR1 signaling and showed that ANKRD13a is able to suppress PCD by acting as an early checkpoint molecule in a process independent of NF-κB.
Materials and methods
Antibodies and reagents
All commercial antibodies and chemicals were purchased from the following: anti-ANKRD13a (GTX107299) and anti-ANKRD13d (GTX48917) antibodies were from GeneTex International Corporation; anti-phoshpho-IKKα/β (#2697), anti-IκBα (#9242), anti-A20 (#5630), anti-caspase-8 (#9746), anti-cleaved-caspase-8 (#9496), anti-caspase-3 (#9662), anti-phospho-MK2 (#3042), anti-Bid (#2002), anti-PARP (#9541), anti-phospho-RIP1 (#65746), anti-RIP3 (#13526) and goat HRP-conjugated anti-mouse IgG (#7076) antibodies were from Cell Signaling Technology; anti-TNFR1 (sc-8436), anti-TRADD (sc-46653), anti-TRAF2 (sc-876), anti-HOIL (sc-393754), anti-Xp (sc-7270, sc-499) and anti-HA (sc-7392, sc-805) antibodies were from Santa Cruz Biotechnology; anti-actin (A2066) and anti-flag (F3165, F7425, A2220) antibodies, cycloheximide (#C8527) and tebufenozide (31652), flag-agarose (A2220) were from Sigma-Aldrich; anti-PARP (611039, 556362), anti-FADD (610400) and anti-RIP1 (610459) antibodies were from BD Biosciences; anti-MLKL (ab184718), anti-phospho-MLKL (ab187091) antibodies and HRP-conjugated Protein A (ab7456) were from Abcam; anti-TNFR1 (AF-425-PB) and anti-caspase-8 (AF-750) antibodies and recombinant TNF (410-MT) were from R&D system; anti-cIAP1 (07-759) antibody was from EMD Millipore; anti-caspase-10 antibody (M059-3) was from MBL International Corporation; anti-cFLIP (ALX-804-961) antibody was from Enzo life sciences; goat anti-Rabbit IgG antibody (31463) was from ThermoFisher; anti-A20 (AM63) antibody, SB203580 (558389), TPCA-1 (401481) and NEC-1(480065) were from Calbiochem; protein G-sepharose (17-0618-01), protein A-sepharose (17-0780-01) and glutathione sepharose 4B bead (17-0756-01) were from GE healthcare. Recombinant human cIAP1 (#E3-280), cIAP2 (#E3-285), UbcH5a (#E2-616), UBE1 (#E-305) and Ubiquitin (#U-100H) were from Boston Biochem. SM-164 (A8815) was from APExBIO; Z-VAD(OMe)-FMK (HY-16658) was from Med Chem Express.
Construction of expression plasmids
Full-length of RIP1 (671aa), caspase-8 (538aa) and ANKRD13a (590aa) were prepared by amplifying the human normal colon tissue cDNA library in-frame with corresponding primers into pcDNA3.1C or pCMV-tag2B using SolgTM Pfu DNA Polymerase (SolGent). cDNA encoding truncated forms of RIP1 (ΔDD; 1–560aa, KD; 1–289aa, ΔKD; 290–671aa and DD; 561–671aa), caspase-8 (2′; 1–433aa, 3′; 1–237aa, 4′; 1–157aa, 5′; 276–433aa and 6′; 444–538aa) and ANKRD13a (UIM4; 1–568aa, UIM3/4; 1–538aa and UIM2/3/4; 1–502aa) were generated by standard PCR methods. The point-mutant ANKRD13a (M2; A527G/S531A, M3; A557G/S561A and M4; V582G/S586A) and RIP1 (K115R, K163R, K184R, K306R, K316R, K377R, K571R and K115/377R) were created using a QuickchangeTM Site-Directed Mutagenesis kit (Stratagene).
CRISPR/Cas-9-mediated KO cells generation
For the depletion of ANKRD13a, 13d, A20 and RIP1 in several cells including HeLa cells, HT29 cells, U87 and HEK293 cells, oligos were synthesized and inserted into the px330-puro vector (kind gift from Dr. Macfarlan, NICHD, NIH, USA) through standard protocol to generate gRNA with hCas9 protein. gRNA sequences were designed using the open access software provided at http://crispr.mit.edu/. gRNA sequences were as follows: ANKRD13a—TGTGATATCGACGCGCAGTT; ANKRD13d—CCCTCTGGTAGTCCCGATAC; A20—TGAAGTCCACTTCGGGCCAT; RIP1-GGCACCGCTAAGAAGAATGG). Each targeting plasmids were transfected into HeLa cells using Lipofectamine 2000 reagent according to the manufacturer’s instructions (Invitrogen Life Technologies). After 24 h, cells were exposed with 3 μg/ml puromycin for two days, and clones propagated from single cells were picked out. The depletion of target genes was confirmed by both immunoblotting and genomic DNA sequencing.
Lentiviral shRNA generation and lentiviral transduction
Coding sequences of ANKRD13a was inserted into the pCDH-CMV-MCS-EF1a-Puro (CD510B1) expression vector (SBI system biosciences). hRIP3-pBABE construct was provided by Prof. Jaewhan Song (Yonsei University, Seoul, Rep of Korea). shRNA-expression lentiviral plasmid targeting ANKRD13a were purchased from Mission shRNA (Sigma). For viral packaging, HEK293T cells were transfected with vectors using FuGENE 6 Transfection reagent (Promega). The virus-containing medium was harvested 48 h post-transfection and used for transduction of HeLa cells. Two days after transduction, viral infected cells were grown in puromycin-containing selectivemedium, and selected cells were expanded and examined by immunoblotting for the stable KD and expression of proteins.
Immunoblot analysis and Immunoprecipitation
After treatment as described in the figure legends, the cells were collected and lysed in M2 buffer (20 mM Tris, pH 7.6, 0.5% NP-40, 250 mM NaCl,3 mM EDTA, 3 mM EGTA, 0.5 mM phenylmethyl sulphonyl fluoride (PMSF), 20 mM β-glycerol phosphate, 1 mM sodium vanadate and 1 μg/ml leupeptin). Cell lysates were fractionated by SDS-ployacrylamide gel electrophoresis (SDS–PAGE) and visualized by enhanced chemiluminescence, according to manufacturerʹs instruction (Thermo Fisher). For immunoprecipitation assays, the lysates were mixed and precipitated with relevant antibodies and the protein G or A-sepharose beads for overnight incubation at 4 °C. The beads were washed four times with M2 buffer, and the bound proteins were resolved in 8% SDS–PAGE for immunoblot analysis.
Purification of recombinant proteins
Various form of recombinant human RIP1 (WT, K115R, K377R and K115/377R) proteins from the expression vectors (pcDNA3.1 C and pCMV-tag2B) were expressed in ExpiCHO-STM cells using ExpiCHOTM Expression system according to the manufacturer’s recommendations (Life Tech Corp #A29133). Briefly, ExpiFectamineTM and DNA mixtures were inoculated to CHO cells (6 × 106 cells/ml) with the expression enhancers for 24 h, and 7 days later, cells were collected and lyzed with M2 Buffer (20 mM Tris, pH 7.6, 0.5% NP-40, 250 mM NaCl, 3 mM EDTA, 3 mM EGTA, 0.5 mM PMSF and protease inhibitors cocktail). The soluble fraction was separated by centrifugation and incubated with flag-agarose or anti-Xp antibody with protein A-sepharose beads at 4 °C for 4 h. Each protein-bound bead was washed with M2 buffer and 1x PBS. For purifying recombinant GST-ANKRD13a protein, the cloned human GST-ANKRD13a in pGEX-4T-1 plasmid was expressed in E coli, BL21 (DE3). E. Coli containing expressed GST-ANKRD13a was collected and lyzed with lysis buffer (50 mM Tris, 100 mM NaCl, 5 mM EDTA, 0.03% Triton X-100, 1 μg/ml leupeptin, 1 μg/ml pepstatin, 0.15 mM PMSF, protease inhibitors cocktail, pH 8.0). The soluble fraction was separated by centrifugation and incubated with glutathione sepharose 4B bead at 4 °C for 4 h. The protein-bound beads were washed with lysis buffer and eluted with elution buffer (50 mM Tris, 50 mM NaCl, 10% glycerol, 0.03% Triton X-100, 50 mM l-glutathione reduced, pH 8.0). Eluted GST-ANDKRD13a protein was dialyzed with 1x PBS, and used for in vitro binding assay.
In vitro ubiquitination and binding assay
In vitro ubiquitination assay was performed with E1 (UBE1, 100 nM), E2 (UbcH5a, 1μM), E3 (1μM of cIAP1 and cIAP2) with recombinant RIP1 protein-bound beads in E3 ligation buffer (50 mM Tris, pH 7.5, 2.5 mM MgCl2, 2 mM DTT, 2 mM ATP) at 37 °C for 1 h. After samples were washed with 1x PBS and treated with Laemmli sample buffer (Bio-Rad) and supernatants separated by 6–8% SDS–PAGE. In vitro ubiquitinated-RIP1 was detected by immunoblot with anti-ubiquitin and anti-RIP1 antibodies. For in vitro binding assays, purified recombinant GST-ANKRD13a (1 mg/ml) and RIP1 (1 mg/ml) were incubated in binding buffer (1X PBS, 0.1% NP-40, 0.5 mM DTT, 10% Glycerol and 1 mM PMSF) for 2 h. Samples were then precipitated using flag-agarose bead or anti-Xp antibody at 4 °C for 3 h. The beads were washed with binding buffer, and resolved in 6–8% SDS–PAGE for immunoblot analysis with the relevant antibodies.
Cell viability assay
After treatment as described in the figure legends, cell viability was conducted utilizing Cell Titer-gloLuminescent Cell Viability Assay kit (Promega), which measures cell viability based on adenosine triphosphate (ATP) levels present in live cells. Luminescent measurements were taken on a Tecan Infinite Plate Reader (Mannedorf, Switzerland). Presented data were from representative experiments of at least 3 independent assays.
Human ovarian cancer tissue samples and tissue extracts preparation
Total 52 fresh surgical ovarian cancer specimens were obtained from Biobanks of Chungnam National University Hospital and Pusan National University Hospital of the Korea Biobank Network. After washing with cold saline, the tissue specimens were homogenized in M2 buffer (20 mM Tris, pH 7.6, 0.5% NP-40, 250 mM NaCl,3 mM EDTA, 3 mM EGTA, 0.5 mM PMSF, 20 mM β-glycerol phosphate, 1 mM sodium vanadate and 1 μg/ml leupeptin). Tissue homogenates were incubated on ice for 30 min and centrifuged at 15,000 x g for 15 min at 4 °C. Protein concentration of homogenates was determined for each sample using the Bradford method (Bio-Rad).
Tissue microarray preparation for ANKRD13a
We initially retrieved, from our surgical pathology files, 142 adnexectomy specimens performed at Chungnam National University Hospital from 2002 to 2016. All sections were reviewed for confirmation of the original diagnosis by a pathologist on the study and staged according to the 2010 American Joint Committee on Cancer–TNM classification. Of the 142 samples, 91of ovarian serous carcinoma were qualitatively and quantitatively suitable for immunohistochemistry (IHC). Tissue microarray were constructed using a manual tissue arrayer (Chemicon ATA-100, California, USA) using 3-mm cores. Two representative cores from each donor block were selected with hematoxylin and eosin-stained sections.
Immunohistochemistry (IHC) and histopathological scoring
Immunostaining using a 3,3′-diaminobenzidine peroxidase substrate kit (DAKO Cytomation, Inc., Carpinteria, CA, USA) was performed according to the manufacturer’s protocol. Briefly, paraffin sections (3 µm) were deparaffinized in xylene and rehydrated in graded alcohol. After antigen retrieval by heating in a pressurecooker in 10 mM sodium citrate buffer (pH 6.0) for 4 min, tissue sections were treated with 3% hydrogen peroxide for 10 min to block endogenous peroxidases. The sections were then incubated 30 min at room temperature with anti-ANKRD13a (1:800) antibody diluted with background reducing diluents (S0809; Dako Cytomation). Slides were then incubated with EnVision reagent for 30 min, followed by 3,30-diaminobenzidine chromogen for 5 min, counterstained with Meyer’s hematoxylin and mounted. Exclusion of the primary antibody during immunostaining was used as a negative control. The IHC results were evaluated by two independent pathologists (MKY and JMK), who were blinded to patient’s clinocopathological details. The immunohistochemical staining was scored based on the staining intensity (score 0, no staining intensity; score 1, weak staining intensity; score 2, intermediate staining intensity; score 3, strong staining intensity). In cases of heterogeneous staining within the samples, the respectively higher score was chosen if >50% of the cells showed a higher staining intensity. The scoring results were categorized as low expression group (score 0 and 1) or high expression group (score 2 and 3) to compare with the clinical outcome of relevant ovarian cancer patients.
Publicly available genomic data analysis
The significant association with survival and aberrant expression of ANKRD13a was evaluated in a gene expression data set from Genomic Data Common (GDC) platform from The Cancer Genome Atlas (TCGA) database and a microarray data of GSE17260 from Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/). To evaluate that the repeatability and portability of ANKRD13a gene, we used a data set of Gene Expression Profiling Interactive Analysis 2 (http://gepia2.cancer-pku.cnu; GEPIA2), which is a web server for analyzing the RNA sequencing expression data of 9736 tumors and 8587 normal samples from the TCGA and The Genotype Tissue Expression (GTEx), using a standard processing pipeline. The Cutoff Finder online tool (http://molpath.charite.de/cutoff/) was used to determine the optimal cutoff value of ANKRD13a mRNA expression as described before [31]. Overall survival curves were plotted using the Kaplan–Meier product-limit method, and significant differences between survival curves were determined using the Log-rank test.
Statistical analysis
Data are expressed as the mean ± s.e. from at least three separate experiments performed in triplicate. The difference between groups were analyzed using Student′s t-test using SPSS software ver.13.0. P-value < 0.05 was considered statistically significant.
Results
ANKRD13a deficiency sensitizes TNF-induced apoptosis independent of IKKs/NF-κB
To explore the novel regulators involved in TNFR1-SC, we performed a yeast two-hybrid screen using the Ub-editing enzyme of RIP1, A20 [32, 33] as a bait. As expected, we identified several proteins related to the ubiquitination process such as ubiquitin B/C and ubiquitin conjugating enzyme E21 (Supplementary Tables 1 and 2). Interestingly, two ANKRD13 subtypes including ANKRD13a and ANKRD13d were repeatedly identified in two independent assays. Indeed, the interaction between ANKRD13a and A20 was confirmed in TNF-treated HeLa cells (Supplementary Fig. 1). We first assessed whether ANKRD13a and/or ANKRD13d deficiency could affect TNF signaling by creating a series of HeLa cell lines deficient in various combinations of these genes (Supplementary Table 3). In kinetic analysis, TNF-induced phosphorylation of IKKα/β and p38 in ANKRD13a-KO, ANKRD13d-KO and/or ANKRD13a/d-DKO cells were comparable to those in wild-type (WT) cells (Fig. 1a). Consistently, the transcriptional activity of NF-κB was normal in these cells as measured by TNF-induced A20 induction, suggesting that ANKRD13a and ANKRD13d are not involved in NF-κB and MAPK signaling. In addition, persistent NF-κB activation observed in A20-KO cells was not altered by deficiency of ANKRD13a and ANKRD13d, indicating that A20 functions independently of ANKRD13a/13d.
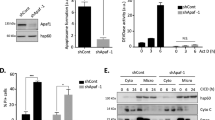
a Wild type (WT) or various combinations of ANKRD13a-, 13d-, and A20-KO HeLa cells were treated with TNF (30 ng/ml) for the indicated times. Whole-cell lysates were immunoblotted with indicated antibodies. b WT and ANKRD13a- KO HeLa cells were treated with TNF for 24 h in the absence or presence of zVAD-fmk (20 μM) and visualized by phase-contrast microscopy. B3 is a magnified view of the boxed area in B2. c, d WT and indicated KO HeLa cell were treated with TNF for 24 h or indicated times. c Cell viability was determined using the Cell Titer-Glo Luminescent Cell Viability Assay kit. Data are presented as the mean ± S.E. (n = 3). *P < 0.05, compared with TNF-treated WT HeLa cells (Student′s t-test). d Whole-cell lysates were immunoblotted with indicated antibodies.
We next examined whether ANKRD13a and/or 13d regulate TNF-induced PCD. As expected, no evident PCD was observed in WT-HeLa cells following treatment with TNF only (Fig. 1b). Surprisingly, significant apoptotic features were observed in TNF-treated ANKRD13a-KO HeLa cells, as evidenced by cell morphology, cell viability and cleavage of caspase-8, PARP and cFLIP; this PCD was blocked by the pan-caspase inhibitor, zVAD-fmk (Fig. 1b–d). Interestingly, TNF-induced apoptosis was only faintly detectable in ANKRD13d-KO HeLa cells, whereas it was more profound in ANKRD13a/d-DKO HeLa cells (Fig. 1c, d), suggesting a synergistic role of ANKRD13a and 13d in regulating apoptosis. Consistent with the results obtained from HeLa cells, significant apoptotic phenotypes were observed in two other ANKRD13a deficient cancer cells (HT-29 and U87) upon TNF treatment (Supplementary Fig. 2), confirming that ANKRD13a indeed protects TNF-induced apoptosis in multiple types of cancer cells. In line with a limited extent of apoptosis in ANKRD13d KO cells, very low expression levels of ANKRD13d mRNA was observed in various cancer cells (Supplementary Fig. 3), suggesting that ANKRD13a may play a predominant role in regulating TNF-induced apoptosis. Furthermore, A20-KO cells showed a marginal degree of apoptosis following TNF exposure, while there were additive effects in A20/ANKRD13a-DKO or A20/ANKRD13a/13d-TKO HeLa cells (Fig. 1c, d), suggesting that A20 is unlikely to play an important role in TNF-induced apoptosis in ANKRD13a-KO cells.
ANKRD13a protects against TNF-induced dual modes of PCD by suppressing caspase-8 activation
To gain further insight into the role of ANKRD13a, we examined the effects of inhibitors of IKKα/β or p38 (TPCA-1 or SB203580) on TNF-induced apoptosis. While TPCA-1 was not sufficient to switch the TNF response to death in WT cells up to 24 h, it resulted in a remarkable sensitization of TNF response to induce cell death and apoptotic feature in ANKRD13a-KO and 13a/13d-DKO cells, but not in ANKRD13d-KO cells (Fig. 2a, b and Supplementary Fig. 4), suggesting that ANKRD13a deficiency can switch the TNF response from survival to death.
a WT and indicated ANKRD13 genes KO HeLa cell were pre-treated with TPCA-1 (0.5 μM) for 30 min, and treated with TNF for the indicated times. Cell viability was measured as in Fig. 1c. Data are presented as the mean ± S.E. (n = 3). *P < 0.05, compared with TNF/TPCA-treated WT HeLa cells. b WT and indicated ANKRD13 genes KO HeLa cell were treated with TNF for the indicated times in the absence or presence of TPCA-1 (0.5 μM), SB203580 (20 μM) and zVAD-fmk (20 μM). Whole-cell lysates were immunoblotted with indicated antibodies. c WT and indicated ANKRD13 genes KO HeLa cell were treated with CT (30 ng/ml of TNF plus 10 μg/ml of CHX) or CTZ (CT plus 20 μM of zVAD-fmk) for 7 h. Cell viability was measured as in a. *P < 0.05, compared with C/T-treated WT HeLa cells. d, e After RIP3 was stably introduced into WT and indicated ANKRD13 genes KO HeLa cell, cells were treated with CT or CTZ for the indicated times. d Cell viability was measured as in a. Data are presented as the mean ± S.E. (n = 3). *P < 0.05, compared with CT-treated WT HeLa cells. ♯P<0.05, compared with CTZ-treated WT HeLa cells. e Whole-cell lysates were immunoblotted with indicated antibodies. f, g HeLa cells deficient ANKRD13a/13d were reconstituted with ANKRD13a by using lentiviral expression system. Cells were then treated with CT for the indicated times. f Cell viability was measured as in a. Data are presented as the mean ± S.E. (n = 3). *P < 0.05, compared with CT-treated WT HeLa cells. g Whole-cell lysates were immunoblotted with indicated antibodies.
Having shown that the expression levels of cFLIP were substantially decreased in TNF- treated ANKRD13a-KO cells (Fig. 1d), we thus examined the susceptibility of TNF cytotoxicity under conditions of cFLIPL depletion using cycloheximide (CHX) (Fig. 2e; 5th row). Consistently, CHX/TNF (CT)-induced cell death was highly sensitized in ANKRD13a-KO cells compared to WT cells (Fig. 2c), suggesting that antiapoptotic function of ANKRD13a is independent of cFLIP. To examine whether ANKRD13a could also mediate TNF-induced necroptosis, RIP3 was stably expressed in WT and KO cells. In the presence of RIP3, ANKRD13a and 13a/13d deficiency showed marked sensitization to both apoptotic (CT) and necroptotic (CHX/TNF/z-VAD; CTZ) triggers, which was accompanied by increases in PARP1 cleavage and RIP1 phosphorylation, respectively (Fig. 2d, e).
Consistently, reconstitution of ANKRD13a in the ANKRD13a/13d-DKO or ANKRD13a-KO cells protected CT-induced cytotoxicity and caspase-8 activation (Fig. 2f, g and Supplementary Fig. 5), confirming that ANKRD13a predominantly functions to control TNF-mediated PCD. To further confirm the above observation, we generated a tebufenozide (teb)-inducible system to precisely control the ANKRD13a expression in a dose-dependent manner [34]. Consistently, overexpression of ANKRD13a almost completely protected against CT-induced PCD, accompanied by impaired cleavage of caspase-8 depending on the expression of level of ANKRD13a with different dosages of teb (Fig. S6). Thus, all these data support the notion that ANKRD13a prevents TNF-induced PCD by keeping caspase-8 in check.
ANKRD13a is part of TNFR1 complex-II and interferes the complex-II formation
To understand how ANKRD13a protects against caspase-8 activation, we performed a detailed comparative study of the recruitment of signaling components to complex-I and complex-II following TNF stimulation. Treatment of cells with TNF led to no obvious difference in the recruitment and assembly of upstream adaptors including ubiquitinated RIP1 and NF-κB controlling effectors (IKK-β and A20) into complex-I between WT and ANKRD13a-KO HeLa cells (Fig. 3a). Consistently, ANKRD13a was not found in TNFR1-immunoprecipitates, indicating that ANKRD13a is unlikely couples to complex-I.
WT and ANKRD13a KO HeLa cells were treated with TNF for the indicated times. Cell extracts from each samples were subjected to IP with anti-TNFR1 (a) and anti-caspase-8 (b) antibodies. Immunoprecipitates were analyzed by immunoblotting with indicated antibodies. A total of 1% the cell extract volume from each TNF-treated sample was used as input control. (N.S, nonspecific bands for indicated antibodies). c WT and ANKRD13a KO HeLa cells were treated with CT in the absence or presence of z-VAD-fmk (20 μM) for the indicated times. Cell extracts from each samples were subjected to IP with anti-caspase-8 antibody and immunoblotted with indicated antibodies (top panels). A total of 1% WCL from each sample (bottom panels) or CT (3 h) treated sample (top panels, right) was used as input control. d Teb-on ANKRD13a HEK293cells were incubated with or without teb (15 μM) for 24 h, and then further treated with CT for the indicated times. Caspase-8 containing complex-II were purified by IP with anti-caspase-8 antibody, followed by immunoblotting with indicated antibodies. A total of 1% WCL from none treated sample was used as input control.
Next, we examined whether ANKRD13a regulates complex-II formation through the physical interaction. In agreement with a previous study [2], RIP1 and FADD recruitment to caspase-8 was transient in multiple types of cells, peaking at 0.5 to 2 h after TNF stimulation (Fig. 3b and Supplementary Fig. 7). By contrast, substantially prolonged association of the complex-II assembly was observed in pairs of ANKRD13a-KO cells. More importantly, in WT cells, ANKRD13a was detected in caspase-8-immunoprecipitates, suggesting that the recruitment of ANKRD13a into complex-II may affect the caspase-8 activation by limiting the complex-II formation.
To corroborate these data, we investigated the assembly of complex-II at 1 to 3 h after CT treatment, at a time of early stage of apoptosis. At this stage, a certain amount of FADD and full-length RIP1 were associated with caspase-8 in WT cells (Fig. 3c and Supplementary Fig. 8). In ANKRD13a-KO cells, however, this complex-II formation was markedly enhanced with cleaved forms of RIP1 and caspase-8/−10. As expected, pretreatment with z-VAD-fmk in WT cells inhibited the cleavage of RIP1, thereby drastically increased the ubiquitination of RIP1 and the recruitment of ANKRD13a into the complex-II in response to CT (Supplementary Fig. 9). Consistently, we observed that overexpression of ANKRD13a resulted in decreased CT-induced complex-II formation and enhanced association of ANKRD13a with caspase-8 (Fig. 3d), suggesting that ANKRD13a likely functions upstream of caspase-8 in suppressing complex-II formation.
Recruitment of ANKRD13a into the complex-II depends on RIP1
Given that several types of ubiquitination have been reported in RIP1 and caspase-8 within the complex-II [2, 35, 36], we next examined whether the recruitment of ANKRD13a into complex-II is dependent on RIP1 or caspase-8, respectively. To address this issue, WT or deletion mutants of RIP1 were co-transfected with ANKRD13a, and the ability of endogenous complex-II formation and binding affinities were monitored (Fig. 4a). Consistent with previous reports [37, 38], overexpression of WT and RIP1 mutants (RIP1-ΔKD and -DD) containing a death domain (DD) were able to form a functional complex-II, evidenced by an association with endogenous FADD and caspase-8 (Fig. 4b, left panel) as well as caspase-8 activation in the absence of CrmA (Supplementary Fig. 10). Notably, although RIP1 deletion mutants without a DD (RIP1-ΔDD and RIP1-KD) failed to bind with endogenous FADD or caspase-8, they were able to substantially interact with ANKRD13a. These observations were also confirmed by reverse immunoprecipitation assay using anti-flag antibody (Fig. 4b, right panel), indicating that the involvement of ANKRD13a into complex-II is not caspase-8 or FADD-dependent, but rather by direct interaction with the kinase and intermediate domains of RIP1. We subsequently scrutinized whether the recruitment of ANKRD13a into complex-II requires caspase-8. Consistently, WT and deletion mutants of caspase-8 containing two death effector domains (DEDs) were able to assemble complex-II, which interacted with ANKRD13a (Fig. 4c, d). However, unlike RIP1, deletion mutants of caspase-8 lacking two DEDs did not interact with ANKRD13a, suggesting that ANKRD13a only binds the caspase-8 captured in complex-II. Furthermore, co-immunoprecipitation assay of RIP1-KO HEK293 cells resulted in no detectable interaction between caspase-8 and ANKRD13a, as evident in WT HEK293 cells (Supplementary Fig. 11). Therefore, it is likely that ANKRD13a is recruited to complex-II via a direct interaction with RIP1 rather than caspase-8.
a Schematic diagram of deletion mutants of RIP1 for their expression constructs (left). Summary of binding affinities between ANKRD13a and various truncated forms of RIP1 or activity of complexes-II (FADD-RIP1-caspase-8) formation (right, see the text for detail). b HEK293 cells were co-transfected with plasmids expressing flag-tagged ANKRD13a and Xp-tagged various RIP1 mutants with CrmA to prevent caspase-8-mediated RIP1 cleavage. Endogenous or exogenous interaction between ANKRD13a and RIP1 or association of RIP1-FADD-caspase-8 complex was confirmed in both directions by IP with anti-Xp (left panel) or anti-flag (right panel) antibodies, respectively, and followed by immunoblotting with indicated antibodies. c Schematic diagram of deletion mutants of caspase-8 for their expression constructs (left). Summary of binding affinities between ANKRD13a and various truncated forms of caspase-8 or activity of complexes-II formation (right). d HEK293 cells were co-transfected with plasmids expressing flag-tagged ANKRD13a and Xp-tagged various caspase-8 mutants with CrmA. Endogenous or exogenous interaction between ANKRD13a and caspase-8 or association of caspase-8/−10-FADD-RIP1 complex was determined by IP with anti-Xp antibody and followed by immunoblotting with indicated antibodies. e Deletion mutants of ANKRD13a used in domain mapping experiments. f HEK293 cells were co-transfeced with plasmids expressing Xp-tagged RIP1 and flag-tagged various ANKRD13a mutants with CrmA. ANKRD13a-RIP1 interaction was determined as in b. g Sequence alignment of C-terminal UIMs segment in ANKRD13a. The Ala/Val and Ser residues indicated with a bold in the conserved consensus sequence (Φ-x-x-Ala/Val-x-x-x-Ser-x-x-Ac) were mutated with Gly and Ala. Φ and Ac represent a large hydrophobic and an acidic residue. h HEK293 cells were co-transfeced with plasmids expressing Xp-tagged RIP1 and flag-tagged various ANKRD13a UIM mutants with CrmA. ANKRD13a-RIP1 interaction was determined as in b.
To identify which domain of ANKRD13a was responsible for its interaction with RIP1, we generated serial UIM deletion mutants of ANKRD13a (Fig. 4e). Notably, the deletion of the fourth UIM of ANKRD13a almost completely abolished the interaction with both unmodified and ubiquitinated forms of RIP1 (Fig. 4f). We also established mutants of ANKRD13a in which Ala/Val and Ser residues within each UIM were substituted by Gly and Ala (Fig. 4g). Consistently, only the fourth UIM mutant (M4) markedly decreased interaction with RIP1, whereas the other two UIM mutants (M2 and M3) showed marginal effects in comparison to the WT (Fig. 4h). Such findings thus suggest that ANKRD13a is able to interact with ubiquitinated RIP1 through its 4th UIM.
Polyubiquitination of Lys115 and Lys377 on RIP1 by cIAP1/2 controls its interaction with ANKRD13a and complex-II formation
To identify the potential ubiquitination sites of RIP1 recognized by ANKRD13a, we tested seven K-R mutants, in which were previously known ubiquitination sites within the kinase and intermediate domains of RIP1 by biochemical and proteomic analysis [39, 40] (Fig. 5a). Notably, K115R or K377R mutants of RIP1 drastically reduced their polyubiquitination status as well as the interaction with ANKRD13a in comparison with WT-RIP1 (Fig. 5b), suggesting that K115- or K377-ubiquitination of RIP1 is required for its interaction with ANKRD13a. Several Ub E3 ligases, including cIAP1, cIAP2, TRAF2 and A20 were known to mediate ubiquitination of RIP1 in TNFR1 signaling [7, 8, 32]. To identify the Ub E3 ligase(s) responsible for RIP1 bound to ANKRD13a, the ability of each Ub E3 ligases to ubiquitinate RIP1 in ANKRD13a immunoprecipites were tested. Strikingly, cIAP1/2 markedly enhanced the interaction of ANKRD13a with ubiquitinated-RIP1 (Fig. 5c) raising the possibility that cIAP1/2 may participate in the formation of poly-Ub chains to RIP1 bounded with ANKRD13a. To validate this hypothesis, we performed an in vitro ubiquitination assay and confirmed that cIAP1/2 could directly promote the ubiquitination on the purified RIP1 in the presence of UBE1 and UbcH5a (Fig. 5d). Of note, in vitro binding experiments showed that the direct interaction between ANKRD13a and RIP1 was only marginal, whereas such interaction was drastically enhanced by cIAP1/2 (Fig. 5e), indicating that ANKRD13a interacts with RIP1 in an ubiquitination-dependent manner. In the consequent in vitro binding experiments, the interaction of RIP1K377/115R mutant with ANKRD13a was drastically decreased in comparison with WT-RIP1 (Fig. 5f), indicating that cIAP1/2 is a responsible Ub E3 ligase for RIP1 at both Lys115 and Lys377 residues that regulates the interaction with ANKRD13a.
a Schematic depiction of predicted lysine residues within the kinase and intermediate domains of RIP1. b RIP1-KO HEK293 cells were co-transfected with indicated combination of plasmids with CrmA. After 24 h, cells were treated with TNF (30 ng/ml) for 1 h. The ubiquitination status of RIP1 bound to ANKRD13a was determined by using IP with anti-flag antibody and followed by immunoblotting with indicated antibodies. c HEK293 cells were co-transfected with the indicated combination of plasmids. After IP with anti-HA antibody, co-immunoprecipitated proteins were detected by immunoblotting with indicated antibodies. d In vitro ubiquitination assay was performed with recombinant flag-RIP1, His-cIAP1 or His-cIAP2 and indicated E1/E2 enzymes. RIP1 ubiquitination was monitored by using IP with anti-flag antibody and followed by immunoblotting with anti-Ub antibody. e Purified GST-ANKRD13a was incubated with recombinant RIP1 and in vitro binding assay between ANKRD13a and RIP1 was performed, as described in Methods. f Purified GST-ANKRD13a was incubated with the indicated recombinant RIP1 and E1/E2/E3 enzymes and in vitro interaction between ANKRD13a and each types of RIP1 was examined, as in e. g RIP1-KO HEK293 cells reconstituted with the indicated RIP1 constructs and IκBαSR were treated for 6 h with TNF as indicated. Cell viability was measured as described in Fig. 1c. Data are presented as the mean ± S.E. (n = 3). *P < 0.05, compared with TNF-treated RIP1-KO cells expressing WT-RIP1. #P < 0.05, compared with TNF-treated RIP1-KO cells expressing RIP1K115R. h RIP1-KO HEK293 cells reconstituted with indicated constructs were treated with TNF for 1 h. Endogenous association of RIP1-FADD-caspase-8 complex and the recruitment of ANKRD13a was examined, as in Fig. 3b.
To investigate whether the ubiquitination of Lys115 and Lys377 on RIP1 is directly involved in TNF-mediated cytotoxicity regulated by ANKRD13a, we re-expressed WT or three mutants of RIP1 with super-repressor form of IκBα (IκBαSR) in RIP1-KO HEK293 cells. Consistent with previous reports [36, 41], TNF-mediated cytotoxicity was more susceptible in RIP1 KO cells reconstituted with mutant RIP1 (K377R) compared with that of WT-RIP1 (Fig. 5g). In addition, the reconstitution of K377/115 R mutant of RIP1 bestowed additional sensitivity to TNF cytotoxicity and complex-II formation than K377R mutant. Of note, compared with WT-RIP1, reconstitution of K377/115 R mutant RIP1 drastically decreased TNF-induced recruitment of ANKRD13a into complex-II, even though it enhances complex-II formation (Fig. 5h), suggesting that the ubiquitination of Lys115 and Lys377 on RIP1serves to interact with ANKRD13a prior to complex-II formation.
Enhanced ANKRD13a levels in ovarian cancers are correlates with poor prognosis
To understand the clinical relevance of our findings in certain types of tumor, we first screened the prognostic association of ANKRD13a expression levels on the Human Protein Atlas, and identified that ovarian cancer (OV) patients with high ANKRD13a expression had significantly poorer overall survival (OS) (https://www.proteinatlas.org, p = 6.77e-4). We then recapitulated both ANKRD13a expression and the OS rate in the cohorts of OV patients from The Cancer Genome Atlas (TCGA) and the Gene Expression Omnibus database (GSE17260). Consistently, the OS rate was significantly lower in OV patients with high ANKRD13a expression (Fig. 6a, b). To confirm the above observations, we performed immunohistochemical staining, and observed that high ANKRD13a immunopositivity was found in 48.4% (47 of 91) of tissue microarray specimens from OV, which mainly detected at the cell periphery (Fig. 6c). Statistical analysis showed that ANKRD13a expression was significantly associated with poor survival in OV (Fig. 6d). We further analyzed the prognostic association of ANKRD13a expression in a pan-cancer manner, which is available in TCGA and GEPIA2 datasets. In both cohort analysis across 28 to 30 cancer types, ANKRD13a expression was significantly correlated with worse prognosis in four types of cancers including OV, adrenocortical cancer (ACC), acute myeloid cancer (LAML) and Mesothelioma (MESO) (Supplementary Figs. 12 and 13), suggesting that the prognostic relevance of ANKRD13a expression may across several cancer types.
a, b Kaplan–Meier survival curve for overall survival rates of patients with ovarian cancer cohort based on the levels of ANKRD13a mRNA expression form the datasets of TCGA (n = 378, P = 0.001) and GSE17260 (n = 110, P = 0.017). Patients were divided into ANKRD13a low and high expression groups with optimal cutoff value. c Representative image of IHC staining of ANKRD13a in ovarian cancer tissues. Scale bars, 100 μm. The IHC staining intensity was divided into low and high expression groups as described in Materials and Methods. Low (C1, C2) and high (C3, C4) staining of ANKRD13 in tissue microarray samples (original magnification, x150; enlarged area of image in bottom, magnification x1800). d Correlation between ANKRD13a staining intensity and overall survival time were determined by the Kaplan–Meier method in 91 collected human ovarian cancer samples (P = 0.012). e Cancer tissues extracts from patients who underwent surgery for malignant ovarian cancers were prepared as described in Materials and Methods, and subjected to immunoblotting with the indicated antibodies. f–h Relative densities of ANKRD13a, cleaved forms of caspase-8/PARP, pMLKL and RIP3 were obtained by densitometry using ImageJ for corresponding actin at each lane. Spearman’s correlation coefficient between the expression levels of cleaved caspase-8 and PARP (f, r = 0.742); ANKRD13a and cleaved caspase-8 (f, r = 0.382); RIP3 and pMLKL (h, r = 0.370) were determined by the scatter plot analysis using SPSS program in 52 collected tissue specimens.
Finally, we examined whether the antiapoptotic role of ANKRD13a discovered in our cellular model was also applicable to OV specimens. By analyzing 52 specimens, relatively high variances in expression of ANKRD13a was observed within the same cancer types, even though actin was expressed consistently as a positive control (Fig. 6e and Supplementary Fig. 14a). However, the expression levels of cleaved caspase-8 and PARP1, were markedly increased in tissue samples with low ANKRD13a expression. In agreement with well-established role of caspase-8, correlation analysis showed that expression of cleaved PARP was strongly correlates with the level of cleaved caspase-8 (Fig. 6f). Importantly, the extent of cleaved caspase-8 had a significant inverse correlation with expression of ANKRD13a but not of ANKRD13d (Fig. 6g and Supplementary Fig. 14b). However, level of p-MLKL was significantly correlated with the expression of RIP3, but not of ANKRD13a (Fig. 6h and Supplementary Fig. 14c), suggesting that RIP3 indeed plays a critical role in necroptotic cell death in human patients in a clinical setting.
Discussion
While the ubiquitination status of RIP1 is known to be critical for cell fate decisions in TNF signaling, the molecular nature of this regulation remained unclear. As several Ub binding motifs, such as UIM, Ub association (UBA), Ub E2 variant (UEV), Cue1-homologous (CUE), and polyubiquitin-associated zinc finger (PAZ), have been identified as ubiquitinated protein binding domains [42,43,44], it remains to be determined whether these proteins contribute to the RIP1 ubiquitination-mediated cell death checkpoint. In this study, we showed that ANKRD13a carrying multiple copies of UIM negatively regulates TNF-induced PCD by interacting with ubiquitinated-RIP1 at the level of complex-II. These observations thus reveal a novel mechanism of RIP1-mediated early cell-death checkpoint control via ANKRD13a in TNFR1 signaling.
It has been established that polyubiquitination of RIP1 at K377 occurs in complex-I, which functions as a negative mechanism in transition to complex-II [3, 7, 8]. In this regard, how ANKRD13a fails to interact with ubiquitinated-RIP1 at K377 in complex-I is a question that remains yet largely unresolved. Previously it was reported that the Ub binding surface in ABIN-1 and NEMO (UBAN) bound to Ub chains with high affinity (Kd: 0.3–3.2 μM) [45]. However, most UIM-containing proteins can be expected to bind Ub chains with dissociation constants in the 150–300 μM range [26]. Given this inherently modest affinity of UIMs, it is possible that K377 ubiquitination of RIP1 within complex-I may not be sufficient to bind ANKRD13a. Furthermore, it is possible that additional site(s) of consecutive ubiquitination of RIP1 may be required for interaction with ANKRD13a in complex-II. Interestingly, it has been reported that RIP1 can undergo K115 ubiquitination exclusively in the necrosome, which stabilizes RIP1 and thus contributes to sensitization to its cytotoxicity [40]. These findings suggest that multiple ubiquitination sites of RIP1 seems to act cooperatively in determining the fate of RIP1. With regard to the functional consequence of ubiquitination site(s) of RIP1, we found that the TNF-induced cytotoxicity was upregulated by the reconstituting RIP1K377/115R mutant rather than an individual mutant form (RIP1K115R or RIP1K377R) of RIP1 in RIP1-deficient cells. Such findings indicate that both ubiquitination sites of RIP1 require for regulating complex-II formation, and suggest that ANKRD13a is likely recruited into complex-II by binding to ubiquitinated-RIP1 at least at two lysine residues (K377 and K115). These observations revealed a novel regulatory mechanism of RIP1 ubiquitination through which ANKRD13a catalyzes the switch regulates a cell fate switch from death to survival.
Nevertheless, the results of this study also raised several points that remain to be addressed. We were unable to map the RIP1 ubiquitination sites(s) bound to ANKRD13a by K-GG peptide using mass spectrometry due to the difficulty in purifying the protein from co-overexpression systems (data not shown). Further proteomic profiling of ubiquitination events is needed to clarify the dynamic interaction of ANKRD13a with ubiquitinated-RIP1 during the spatial transition of complex-II from complex-I. More critically, it is currently unclear how the ubiquitination-dependent interaction of ANKRD13a and RIP1 functions to antagonize the recruitment of FADD and caspase-8 into complex-II. Given that UIMs are particularly prevalent in proteins that function in the pathways of receptor internalization [29, 30, 46], one possibility is that the interaction of ANKRD13a with the detached RIP1 from the receptor proximal complex-I may change the static orientation of RIP1 to suppress access to FADD in complex-II. Further structural analysis of spatial compartmentalization of ANKRD13a is necessary to elucidate the precise mechanisms by which ANKRD13a limits RIP1 cytotoxicty. In addition, a pan-cancer cohort studies demonstrate that ANKRD13a expression is the most clinically relevant prognostic indicators in OV. In this sense, how ANKRD13a is involved in the pathophysiology of OV is also a question that remains to be further studied. A large body of preclinical and clinical data supports the concept that TNF plays an important role in tumor development and progression to bridge inflammation and carcinogenesis [47, 48]. Furthermore, enhanced level of cIAP expression was commonly observed in several cancer types and also considered to be a poor prognostic indicator in OV [49, 50]. Thus, it is possible that RIP1 ubiquitination caused by cIAP may result in sufficient association with upregulated ANKRD13a, leading to escape mechanisms of OV cells from TNF-mediated cytotoxicity. Future investigation of an integrated network analysis on ANKRD13a and RIP1-associated genes is required to elucidate their pathophysiological relevance in certain types of cancers.
Data availability
All data needed to evaluate the conclusion in this article are included in the paper and/or its supplementary information. The datasets used and/or analyzed in this study are available from the corresponding author on reasonable request.
References
Walczak H. TNF and ubiquitin at the crossroads of gene activation, cell death, inflammation, and cancer. Immunol Rev. 2011;244:9–28.
Micheau O, Tschopp J. Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell. 2013;114:181–90.
Ting AT, Bertrand MJM. More to Life than NF-kappaB in TNFR1 Signaling. Trends Immunol. 2016;37:535–45.
Kanayama A, Seth RB, Sun L, Ea CK, Hong M, Shaito A, et al. TAB2 and TAB3 activate the NF-kappaB pathway through binding to polyubiquitin chains. Mol Cell. 2004;15:535–48.
Ea CK, Deng L, Xia ZP, Pineda G, Chen ZJ. Activation of IKK by TNFalpha requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol Cell. 2006;22:245–57.
Wu CJ, Conze DB, Li T, Srinivasula SM, Ashwell JD. Sensing of Lys 63-linked polyubiquitination by NEMO is a key event in NF-kappaB activation. Nat Cell Biol. 2006;8:398–406.
Bertrand MJ, Milutinovic S, Dickson KM, Ho WC, Boudreault A, Durkin J, et al. cIAP1 and cIAP2 facilitate cancer cell survival by functioning as E3 ligases that promote RIP1 ubiquitination. Mol Cell. 2008;30:689–700.
Yin Q, Lamothe B, Darnay BG, Wu H. Structural basis for the lack of E2 interaction in the RING domain of TRAF2. Biochemistry. 2009;48:10558–67.
Gerlach B, Cordier SM, Schmukle AC, Emmerich CH, Rieser E, Haas TL, et al. Linear ubiquitination prevents inflammation and regulates immune signalling. Nature. 2011;471:591–6.
Silke J. The regulation of TNF signalling: what a tangled web we weave. Curr Opin Immunol. 2011;23:620–6.
Schneider-Brachert W, Tchikov V, Neumeyer J, Jakob M, Winoto-Morbach S, Held-Feindt J, et al. Compartmentalization of TNF receptor 1 signaling: internalized TNF receptosomes as death signaling vesicles. Immunity. 2004;21:415–28.
Brenner D, Blaser H, Mak TW. Regulation of tumour necrosis factor signalling: live or let die. Nat Rev Immunol. 2015;15:362–74.
Annibaldi A, Meier P. Checkpoints in TNF-induced cell death: implications in inflammation and cancer. Trends Mol Med. 2018;24:49–65.
Lee EG, Boone DL, Chai S, Libby SL, Chien M, Lodolce JP, et al. Failure to regulate TNF-induced NF-kappaB and cell death responses in A20-deficient mice. Science. 2000;289:2350–4.
Micheau O, Lens S, Gaide O, Alevizopoulos K, Tschopp J. NF-kappaB signals induce the expression of c-FLIP. Mol Cell Biol. 2001;21:5299–305.
Chen G, Goeddel DV. TNF-R1 signaling: a beautiful pathway. Science. 2002;296:1634–5.
Peltzer N, Darding M, Walczak H. Holding RIPK1 on the ubiquitin leash in TNFR1 signaling. Trends Cell Biol. 2016;26:445–61.
Dondelinger Y, Jouan-Lanhouet S, Divert T, Theatre E, Bertin J, Gough PJ, et al. NF-kappaB-independent role of IKKalpha/IKKbeta in preventing ripk1 kinase-dependent apoptotic and necroptotic cell death during TNF signaling. Mol Cell. 2015;60:63–76.
Dondelinger Y, Delanghe T, Rojas-Rivera D, Priem D, Delvaeye T, Bruggeman I, et al. MK2 phosphorylation of RIPK1 regulates TNF-mediated cell death. Nat Cell Biol. 2017;19:1237–47.
Geng J, Ito Y, Shi L, Amin P, Chu J, Ouchida AT, et al. Regulation of RIPK1 activation by TAK1-mediated phosphorylation dictates apoptosis and necroptosis. Nat Commun. 2017;8:359.
Jaco I, Annibaldi A, Lalaoui N, Wilson R, Tenev T, Laurien L, et al. MK2 Phosphorylates RIPK1 to prevent TNF-induced cell death. Mol Cell. 2017;66:698–710.
Menon MB, Gropengießer J, Fischer J, Novikova L, Deuretzbacher A, Lafera J, et al. p38(MAPK)/MK2-dependent phosphorylation controls cytotoxic RIPK1 signalling in inflammation and infection. Nat Cell Biol. 2017;19:1248–59.
Lafont E, Draber P, Rieser E, Reichert M, Kupka S, de Miguel D, et al. TBK1 and IKKepsilon prevent TNF-induced cell death by RIPK1 phosphorylation. Nat Cell Biol. 2018;20:1389–99.
Hofmann K, Falquet LA. ubiquitin-interacting motif conserved in components of the proteasomal and lysosomal protein degradation systems. Trends Biochem Sci. 2001;26:347–50.
Riezman H. Cell biology: the ubiquitin connection. Nature. 2002;416:381–3.
Fisher RD, Wang B, Alam SL, Higginson DS, Robinson H, Sundquist WI, et al. Structure and ubiquitin binding of the ubiquitin-interacting motif. J Biol Chem. 2003;278:28976–84.
Polo S, Confalonieri S, Salcini AE, Di Fiore PP. EH and UIM: endocytosis and more. Sci STKE. 2003:re17.
Satpathy S, Wagner SA, Beli P, Gupta R, Kristiansen TA, Malinova D, et al. Systems-wide analysis of BCR signalosomes and downstream phosphorylation and ubiquitylation. Mol Syst Biol. 2015;11:810.
Tanno H, Yamaguchi T, Goto E, Ishido S, Komada M. The Ankrd 13 family of UIM-bearing proteins regulates EGF receptor endocytosis from the plasma membrane. Mol Biol Cell. 2012;23:1343–53.
Burana D, Yoshihara H, Tanno H, Yamamoto A, Saeki Y, Tanaka K, et al. The Ankrd13 family of ubiquitin-interacting motif-bearing proteins regulates valosin-containing protein/p97 protein-mediated lysosomal trafficking of caveolin 1. J Biol Chem. 2016;291:6218–31.
Budczies J, Klauschen F, Sinn BV, Győrffy B, Schmitt WD, Darb-Esfahani S, et al. Cutoff Finder: a comprehensive and straightforward Web application enabling rapid biomarker cutoff optimization. PLoS One. 2012;7:e51862.
Wertz IE, O’Rourke KM, Zhou H, Eby M, Aravind L, Seshagiri S, et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature 2004;430:694–9.
Heyninck K, Beyaert R. A20 inhibits NF-kappaB activation by dual ubiquitin-editing functions. Trends Biochem Sci. 2005;30:1–4.
Lee S, Sohn KC, Choi DK, Won M, Park KA, Ju SK, et al. Ecdysone receptor-based singular gene switches for regulated transgene expression in cells and adult rodent tissues. Mol Ther-Nucl Acids. 2016;5:e36710.
Jin Z, Li Y, Pitti R, Lawrence D, Pham VC, Lill JR, et al. Cullin3-based polyubiquitination and p62-dependent aggregation of caspase-8 mediate extrinsic apoptosis signaling. Cell. 2009;137:721–35.
O’Donnell MA, Legarda-Addison D, Skountzos P, Yeh WC, Ting AT. Ubiquitination of RIP1 regulates an NF-kappaB-independent cell-death switch in TNF signaling. Curr Biol. 2007;17:418–24.
Jang TH, Zheng C, Li J, Richards C, Hsiao YS, Walz T, et al. Structural study of the RIPoptosome core reveals a helical assembly for kinase recruitment. Biochemistry 2014;53:5424–31.
Meng H, Liu Z, Li X, Wang H, Jin T, Wu G, et al. Death-domain dimerization-mediated activation of RIPK1 controls necroptosis and RIPK1-dependent apoptosis. Proc Natl Acad Sci USA. 2018;115:E2001–E9.
Mollah S, Wertz IE, Phung Q, Arnott D, Dixit VM, Lill JR. Targeted mass spectrometric strategy for global mapping of ubiquitination on proteins. Rapid Commun Mass Spectrom. 2007;21:3357–64.
de Almagro MC, Goncharov T, Izrael-Tomasevic A, Duttler S, Kist M, Varfolomeev E, et al. Coordinated ubiquitination and phosphorylation of RIP1 regulates necroptotic cell death. Cell Death Differ. 2017;24:26–37.
Zhang X, Zhang H, Xu C, Li X, Li M, Wu X, et al. Ubiquitination of RIPK1 suppresses programmed cell death by regulating RIPK1 kinase activation during embryogenesis. Nat Commun. 2019;10:4158.
Hicke L, Schubert HL, Hill CP. Ubiquitin-binding domains. Nat Rev Mol Cell Biol. 2005;6:610–21.
Hurley JH, Lee S, Prag G. Ubiquitin-binding domains. Biochem J. 2006;399:361–72.
Dikic I, Wakatsuki S, Walters KJ. Ubiquitin-binding domains-from structures to functions. Nat Rev Mol Cell Biol. 2009;10:659–71.
Hadian K, Griesbach RA, Dornauer S, Wanger TM, Nagel D, Metlitzky M, et al. NF-kappaB essential modulator (NEMO) interaction with linear and lys-63 ubiquitin chains contributes to NF-kappaB activation. J Biol Chem. 2011;286:26107–17.
Polo S, Sigismund S, Faretta M, Guidi M, Capua MR, Bossi G, et al. A single motif responsible for ubiquitin recognition and monoubiquitination in endocytic proteins. Nature. 2002;416:451–5.
Wang X, Lin Y. Tumor necrosis factor and cancer, buddies or foes? Acta Pharm Sin. 2008;29:1275–88.
Laha D, Grant R, Mishra P, Nilubol N. The role of tumor necrosis factor in manipulating the immunological response of tumor microenvironment. Front Immunol. 2021;12:656908.
Psyrri A, Yu Z, Bamias A, Weinberger PM, Markakis S, Kowalski D, et al. Evaluation of the prognostic value of cellular inhibitor of apoptosis protein in epithelial ovarian cancer using automated quantitative protein analysis. Cancer Epidemiol Biomark Prev. 2006;15:1179–83.
Gadducci A, Cosio S, Tana R, Genazzani AR. Serum and tissue biomarkers as predictive and prognostic variables in epithelial ovarian cancer. Crit Rev Oncol Hematol. 2009;69:12–27.
Funding
This work as supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No. 2020R1A2C2005317; No. 2017R1A5A2015385; No. 2018R1C1B6005332).
Author information
Authors and Affiliations
Contributions
MW and KAP participated in the design of the study, carried out bench experiments and analyzes data. EJ helped carrying out bench experiments related to this study. SK, MY and JMK carried out immunohistochemical experiments and statistical analysis. YK and HY provided clinical ovarian cancer tissues. SYK and SWN carried out the pan-cancer cohort analysis. JL and JO carried out yeast two-hybrid screening to identify novel A20-interacting proteins. EGL helped the experiments for in vitro ubiquitination assay. HR carried out the experiments for the construction of teb-inducible expression plasmid. H-MS helped drafting the manuscript by providing critical intellectual input. GMH. designed this study and wrote the manuscript with comments from the coauthors, and all authors collaborated on the work. This work as supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No. 2020R1A2C2005317; No. 2017R1A5A2015385; No. 2018R1C1B6005332).
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
All studies in human subjects were approved by the Institutional Review Board of Chungnam National University Hospital (approval No. 2016-12-027-001).
Informed consent
The written informed consents were obtained from each patient by research team before surgical operation for malignant ovarian cancers, according to the Declaration of Helsinki.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by M. Piacentini
Supplementary information
Rights and permissions
About this article
Cite this article
Won, M., Park, K.A., Kim, S. et al. ANKRD13a controls early cell-death checkpoint by interacting with RIP1 independent of NF-κB. Cell Death Differ 29, 1152–1163 (2022). https://doi.org/10.1038/s41418-021-00906-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-021-00906-9
This article is cited by
-
The double-edged functions of necroptosis
Cell Death & Disease (2023)
-
The resurrection of RIP kinase 1 as an early cell death checkpoint regulator—a potential target for therapy in the necroptosis era
Experimental & Molecular Medicine (2022)