Abstract
The cystine/glutamate antiporter SLC7A11 (commonly known as xCT) functions to import cystine for glutathione biosynthesis, thereby protecting cells from oxidative stress and ferroptosis, a regulated form of non-apoptotic cell death driven by the accumulation of lipid-based reactive oxygen species (ROS). p14ARF, a well-established tumor suppressor, promotes ferroptosis by inhibiting NRF2-mediated SLC7A11 transcription. Here, we demonstrate the crucial role of Cullin 2 RING E3 ligase (CRL2)-KLHDC3 E3 ubiquitin ligase complex in regulating p14ARF protein stability. KLHDC3 acts as a CRL2 adaptor that specifically recognizes a C-terminal degron in p14ARF and triggers p14ARF for ubiquitin–proteasomal degradation. This regulation mode is absent in the murine p14ARF homolog, p19arf which lacks the C-terminal degron. We also show that KLHDC3 suppresses ferroptosis in vitro and supports tumor growth in vivo by relieving p14ARF-mediated suppression of SLC7A11 transcription. Overall, these findings reveal that the protein stability and pro-ferroptotic function of p14ARF are controlled by a CRL2 E3 ubiquitin ligase complex, and suggest that suppression of the p14ARF-NRF2-SLC7A11 regulatory pathway by KLHDC3 overexpression likely contributes to cancer progression.
Similar content being viewed by others
Introduction
p14ARF (murine homolog is called p19arf) was originally identified as an alternative reading frame within the Ink4a/arf tumor suppressor locus, which encodes p16Ink4a inhibitor of cyclin-dependent kinases [1]. p14ARF loss has been frequently observed in human cancers [2]. Predisposition to spontaneous tumors in p19arf-null mice and tumor resistance in mice carrying a transgenic copy of the entire Ink4a/arf locus collectively indicated that p19arf is a bona fide tumor suppressor [3, 4]. The tumor-suppressive roles of p14ARF and p19arf are initially considered to be elicited through the Mdm2-p53 pathway [5,6,7]. Oncogenic stresses induced by c-MYC [8], Ras [9], or E2Fs [10], lead to Mdm2 sequestration in the inhibited state by inducing p14ARF (p19arf in mice) expression as a safeguard measure, thereby activating the tumor-suppressive function of p53. However, accumulating evidence showed the additional p53-independent functions of p14ARF by interacting with diverse proteins, besides Mdm2 [11]. We recently showed that NRF2 is a major target of p14ARF in p53-independent ferroptosis, which is a regulated form of non-apoptotic cell death driven by the accumulation of lipid-based ROS, particularly lipid hydroperoxides [12,13,14]. p14ARF inhibits NRF2’s ability to transcriptionally activate its target genes, including SLC7A11 (xCT), which is a component of the cystine/glutamate antiporter that suppresses ROS-induced cell death [15].
E3 ubiquitin ligases select substrates for ubiquitin conjugation. The largest E3 subclass consists of Cullin-RING Ligases (CRLs), which are modular, multi-subunit enzymes consisting of hundreds of distinct CRL complexes with the capacity to recruit numerous substrates. Human cells express eight Cullins (CUL1, 2, 3, 4A, 4B, 5, 7, 9) that each Cullin nucleates a multi-subunit complex except CUL9 [16]. CUL2 is a unique Cullin and expresses in multi-cellular organisms only. The CRL2 complex is composed of CUL2, RBX1, Elongin B (ELOB), Elongin C (ELOC), in combination with an adaptor protein for substrate binding [17]. The founding member of the CRL2 adaptors is VHL. An assembled CRL2-VHL complex suppresses hypoxia-inducible transcription by targeting HIF1/2α for degradation [18]. Human genome encodes 17 putative CRL2 adaptors, most of which have been poorly studied [19].
Through multiple biochemical and cell biology approaches, we showed that KLHDC3 acts as a CRL2 adaptor to specifically recognize a C-terminal degron in p14ARF and triggers p14ARF degradation via the ubiquitin–proteasomal pathway. We also showed that the KLHDC3-p14ARF-NRF2-SLC7A11 regulatory pathway mediates ferroptosis in vitro and tumor growth in vivo.
Materials and methods
Cell culture
293T, H1299, SKOV3, OVCAR-3, SJSA, HeLa, Huh-7, MCF-7, A549, Hep3B, and CT26 cells were obtained from the American Type Culture Collection. L02 cells were obtained from the Shanghai Cell Bank. HCCLM3 cells were obtained from the Liver Cancer Institute of Zhongshan Hospital, Fudan University. The cell line authenticity and mycoplasma infection are routinely checked by DNA fingerprinting and PCR. All these cell lines were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% FBS. The antibodies, chemicals, and Kits used in this study are listed in Supplementary Tables 1, 2.
CRISPR-Cas9 mediated gene KO cell lines
The guide oligos targeting KLHDC3 or NFE2L2 (NRF2) gene were subcloned into pX459 plasmid. Cells were plated and transfected with pX459 constructs overnight. After 24 h,1 μg/ml puromycin was used to screen cells for 3 days. Living cells were seeded in 96-well plate by limited dilution to isolate monoclonal cell line. The knockout cell clones are screened by western blot (WB) and validated by Sanger sequencing. The sequences of gene-specific sgRNAs are listed in Supplementary Table 3.
Protein complex purification
The epitope-tagging strategy to isolate the CUL2 or KLHDC3-containing protein complex from H1299 cells was performed essentially as previously described with some modifications [12]. In brief, to obtain a FLAG-HA (FH)-CUL2 or KLHDC3 expressing cell line, H1299 cells were transfected with FH-CUL2 or -KLHDC3 constructs and selected for 2 weeks in 1 mg/ml G418. FH-CUL2 or KLHDC3 protein levels were detected by WB. The stable cell lines were chosen to expand for protein complex purification. For purification, cells were lysed in BC100 buffer (20 mM Tris-Cl, pH 7.9, 100 mM NaCl, 0.2 mM EDTA, 20% glycerol) containing 0.2% Triton X-100 and fresh protease inhibitor on ice for 2 h. The homogenate was centrifuged for 30 min at 12,000 rpm at 4 °C. Cleared lysates were filtered through 0.45 μM spin filters (Millipore) and immunoprecipitated by anti-FLAG antibody-conjugated M2 agarose (Sigma). The bound proteins eluted with the FLAG peptide were further affinity purified by anti-HA antibody-conjugated agarose (Sigma). The final elutes from the HA-beads with HA peptides were resolved by SDS-PAGE on a 4–20% gradient gel (Bio-Rad) for Coomassie Blue (CB) staining. Gel bands were cut out from the gels and subjected to mass-spectrometric sequencing.
Real-time reverse transcription PCR (RT-qPCR)
Total RNA was isolated from cells using the TRIzol reagent (Tiangen), and cDNA was reversed-transcribed using the Superscript kit (TOYOBO) following the manufacturer’s instructions. Amplification was performed using the SYBR Green PCR mix Kit (TOYOBO). All quantitation was normalized to the level of endogenous control GAPDH. The sequences of primers for RT- qPCR are listed in Supplementary Table 3.
In vivo ubiquitination assays
293T cells were transfected with HA-ubiquitin and indicated constructs. After 36 h, cells were treated with MG132 (30 μM) for 6 h and then lysed in 1% SDS buffer (Tris [pH 7.5], 0.5 mM EDTA, 1 mM DTT) and boiled for 10 min. For immunoprecipitation, the cell lysates were diluted 10-fold in Tris-HCL buffer and incubated with anti-p14ARF or IgG-conjugated beads (Sigma) for 4 h at 4 °C.The bound beads are then washed four times with BC100 buffer (20 mM Tris-Hcl, pH 7.9,100 mM NaCl,0.2 mM EDTA, 20% glycerol) containing 0.2% Triton X-100. The proteins were eluted with FLAG peptide for 2 h at 4 °C.The ubiquitinated form of p14ARF was detected by WB using anti-HA antibody.
Protein half-life assays
Cells were transfected or treated under indicated conditions. For half-life studies, cycloheximide (20 μg/ml) was added to the media. At indicated time points thereafter, cells were collected and protein abundance was detected by WB.
Cell survival assays
Cell survival rates was determined using Cell Counting Kit-8 (CCK-8) (Beyotime) following the manufacturer’s instructions. Briefly, 1 × 105 cells per well were seeded in a 96-well plate 1 day before treatment. Following treatment with the appropriate drugs where indicated, each well was replaced with fresh medium containing CCK-8 reagent. After incubation for 2 h at 37 °C, the resulting color was measured at OD450 nm using a microplate absorbance reader (Bio-Rad). Each assay was carried out in triplicate.
Liperfluo staining assays
The lipid peroxidation levels were measured as previously described [20]. Briefly, for Liperfluo (Dojindo) staining, cells were incubated in a 60 mm dish containing Erastin (10 μM) for 8 h. Cells were stained with Liperfluo (10 μM) for 30 min at 37 °C, then cells were washed with PBS for 5 min and trypsinized. Cells were then subjected to confocal microscope (LSM880; Carl Zeiss) using the ×40 objective. The blue fluorescence signals in live cells were shown in the figures. The intensity of Liperfluo staining was quantified using Image J software.
GSH measurement assays
GSH levels were detected using the Total Glutathione Assay Kit (Beyotime) following the manufacturer’s instructions. Briefly, 6 × 108 cells per well were seeded in a 12-well plate 1 day before analysis. The culture medium from the wells was carefully removed, and 150 µl reagent S solution was added to remove cell precipitation, and then vortexed. The cells were lysed by freezing and thawing twice. The cells were then centrifuge at 8000 × g for 10 min. The supernatants were used for the assays. Then, 40 µl GSH and GSSG standard solution and samples were loaded into a 96-well plate. Next, 150 µl of total glutathione detection working solution was added to each well and mixed briefly on a plate shaker. Then 50 µl NADPH (0.5 mg/ml) solution was added to each well and the plate was incubated at 37 °C for 1 h and the resulting color was measured at OD412 nm using a microplate absorbance reader (Bio-Rad). Each assay was carried out in triplicate.
GPX4 enzymatic assays
GPX4 enzymatic activity were detected using the Total Glutathione Peroxidase Assay Kit with NADPH (Beyotime) following the manufacturer’s instructions. The cells were harvested and washed with cold PBS. The cells were lysed with 200 µl assay buffer, centrifuged at 10,000 × g for 15 min at 4 °C and the supernatants were transferred to new tubes. Next, Glutathione peroxidase detection buffer, GPX detection working buffe and the samples were loaded into a 96-well plate according to the volume in the table below. The samples were mixed well and incubated at room temperature (RT) for 15 min. 10 μl peroxide solution (30 mM) was added to sample wells and the plate was incubated at 25 °C for 5 min and the resulting color was measured at OD340 nm using a microplate absorbance reader. Each assay was carried out in triplicate.
Immunofluorescence and confocal microscopy
For cultured cell staining, cells on round cover glass were fixed in 4% paraformaldehyde at RT for 15 min. The fixed cells were washed three times in PBS. The Cells were permeabilized with 0.5% Triton X-100 in PBS for 10 min and then in the blocking solution for 30 min at RT. In antibody reaction buffer (PBS plus 1% BSA, 0.3% Triton X-100, pH 7.4), samples were stained with primary antibodies at 4 °C for overnight. After washing with PBS three times (10 min, each), fluorescence-labeled secondary antibodies were incubated for 30 min. The cells were washed three times (10 min, each) after incubation with secondary antibodies, then DAPI (1 µg/ml) was counterstained for 10 min at RT. Cells were then subjected to confocal microscope (LSM880; Carl Zeiss) analysis.
Cell cycle assays
Cells were cultured in six-well plates at a density of 1.2 × 105/well and allowed to adhere to culture plate overnight. Then the medium was replaced with fresh medium containing drugs for the indicated times. The cells were then trypsinized by EDTA-free trypsin and washed twice with cold PBS. Aliquots of the cells were resuspended in 500 µl PI-solution (50 µg/ml PI in PBS, 0.05% Triton X-100) for 40 min at RT. After stained with PI working solution, the cells were washed with PBS three times. Then the cells were suspended in 500 µl PBS. All flow cytometric analyses were performed using a FACSCalibur Flow Cytometer (BD Biosciences).
Apoptosis assays
Annexin V-FITC (Fluorescein isothiocyanate) and propidium iodide (PI) double staining (Dojindo) were used to detect the apoptosis rates. The cells were cultured in six-well plates at a density of 1.2 × 105/well and allowed to adhere to culture plate overnight. Then the medium was replaced with fresh medium containing drugs for another 24 h. The cells were then trypsinized by EDTA-free trypsin and washed twice with cold PBS. Aliquots of the cells were resuspended in 100 µl of binding buffer and stained with 1 µl of annexin V-FITC and 1 µl of PI working solution for 15 min at RT in the dark. All flow cytometric analyses were performed using a FACSCalibur Flow Cytometer.
Mouse tumor implantation
All experimental protocols were approved in advance by the Ethics Review Committee for Animal Experimentation of Fudan University. 4–6-week-old female BALB/c nu/nu mice obtained from SLAC Laboratory Animal Co., Ltd. were bred and maintained in our institutional pathogen-free mouse facilities. Ovarian tumors were established by subcutaneously injecting 5 × 106 SKOV3 cells in 100 μl of PBS buffer into the right flank of 6-week-old nude mice (four mice for each group). At the end of 3 weeks, mice were killed and in vivo solid tumors were dissected and weighed. A portion of tumors was lysed and then subjected to WB with the indicated antibodies.
The Cancer Genome Atlas (TCGA) data analysis
(High-throughput sequencing data were downloaded from the TCGA data portal (https://portal.gdc.cancer.gov/repository). Additional RNA-seq data of normal samples were acquired from GTEx database (https://gtexportal.org/) to achieve balance between tumor and normal data. The raw counts data matrices downloaded from GTEx were normalized to correct for both library size and gene length. The expression data of KLHDC3 was extracted and integrated from TCGA and GTEx datasets, after which the differential analysis of KLHDC3 expression in 29 types of cancers was performed with unpaired student’s t test. The variances of the two groups were thought not to be equal to each other, thus the Welch approximation to the degrees of freedom was used. The statistical significance of difference between tumor and normal samples was characterized by the P value of the student’s t test.
Statistical analysis
Statistical analysis was performed using GraphPad Prism (GraphPad Software, Inc.) All data are shown as mean values ± SD for experiments performed with at least three replicates. The difference between two groups was analyzed using one-way or two-way ANOVA test unless otherwise specified. * represents p < 0.05; ** represents p < 0.01; *** represents p < 0.001, and **** represents p < 0.0001.
Results
p14ARF protein stability is regulated by a CRL2 E3 ubiquitin ligase (s)
Our previous study showed that p14ARF protein stability was regulated by ULF, a HECT-type E3 ubiquitin ligase [20, 21]. However, we noted that treatment of H1299 cells with proteasome inhibitor MG132 led to a stronger increase in p14ARF protein levels than that in cells depleted of ULF by small interfering RNAs (siRNAs). Moreover, MLN4924, which is a small-molecule inhibitor for CRLs, led to p14ARF accumulation at a level comparable to MG132 treatment (Fig. 1A). However, MG132 or MLN4924 treatment had no overt effect on the corresponding p14ARF mRNA levels (Fig. 1B). These results suggested that p14ARF protein stability is controlled by E3 ubiquitin ligase(s) besides ULF and more probably by a CRL(s). We depleted RBX1 or each Cullin (CUL1, 2, 3, 4A, 4B, or 5), in H1299 (p53 null/p14ARF WT) and SJSA (p53 WT/p14ARF WT) cells and found that only RBX1 or CUL2 depletion led to a marked increase in p14ARF protein levels (Fig. 1C). p14ARF was also elevated in the dominant-negative fragment of CUL2 (CUL2-DN) overexpressed cells, but such impact was not observed when other CULs-DN were overexpressed (Fig. 1D). p14ARF protein levels were monitored after the treatment with the protein synthesis inhibitor cycloheximide (CHX). Without de novo protein synthesis, the half-life of p14ARF is considerably prolonged by CUL2 depletion (Fig. 1E, F). Ablation of CUL2 expression by CRISPR/Cas9-mediated knockout (KO) genome editing resulted in a marked increase in p14ARF protein levels but had no impact on the corresponding mRNAs (Fig. 1G, H). Depletion of two essential CRL2 components (ELOB, ELOC) also led to a marked increase in p14ARF protein levels but not the corresponding mRNAs (Fig. 1I, J). Co-IP assay results showed that CUL2, but not other Cullins, interacted with p14ARF (Fig. 1K). Together, these findings suggest that p14ARF protein stability is directly or indirectly controlled by a CRL2 E3 ubiquitin ligase(s).
A Western blots of indicated proteins in WCLs from H1299 cells treated with DMSO, MG132 (20 μM) or with MLN4924 (100 nM) for 8 h, together with H1299 cells transfected with indicated siRNAs for 48 h. B RT-qPCR assessment of p14ARF mRNA levels in H1299 cells treated with DMSO, MG132 (20 μM) or with MLN4924 (100 nM) for 8 h, together with H1299 cells transfected with indicated siRNAs for 48 h. The mRNA levels of GAPDH were used for normalization. Data are shown as means ± SD (n = 3). C Western blot of indicated proteins in WCLs from H1299 or SJSA cells transfected with indicated siRNAs for 48 h. D Western blot of indicated proteins in WCL from 293T cells transfected with the indicated plasmids. E, F Western blot of indicated proteins in WCLs of H1299 cells transfected with indicated siRNAs for 48 h and then treated with 50 μg/ml CHX and harvested at different time points (E). At each time point, the intensity of p14ARF was normalized to the intensity of Actin and then to the value at 0 h (F). Data are shown as means ± SD (n = 3). G Western blot of indicated proteins in WCLs from parental and CUL2 KO H2199 cells. H RT-qPCR assessment of p14ARF mRNA levels in parental or CUL2 KO H1299 cells. Data are shown as means ± SD (n = 3). I Western blot of indicated proteins in WCLs from H1299 cells transfected with indicated siRNAs for 48 h. J RT-qPCR assessment of p14ARF mRNA levels in H1299 cells transfected with indicated siRNAs for 48 h. Data are shown as means ± SD (n = 3). K Western blot of indicated proteins in WCLs and co-IP samples of anti-FLAG antibody obtained from 293T cells transfected with indicated plasmids. P values are calculated by the One-way ANOVA test in (B, H, J) and Two-way ANOVA test in (F). *p < 0.05; **p < 0.01. ns no significant.
KLHDC3 is the CRL2 adaptor responsible for p14ARF degradation
In CRL2 complex, the substrate recognition adaptor typically containsa BC-box (ELOC binding) motif and a CUL2-box (CUL2 binding) motif [17]. To explore which substrate recognition adaptor (s) is responsible for p14ARF degradation, we isolated the CUL2 protein complex from H1299 cells stably overexpressing FH-CUL2 through tandem affinity purification (Fig. 2A). The proteins present in the complex were determined by mass spectrometry. Besides the core components of the CRL2 complex, six substrate recognition adaptors were identified (Fig. 2B). We found that depletion of KLHDC3, but not the other CRL2 adaptors, led to a marked increase in p14ARF protein at a level comparable to CUL2 or RBX1 depletion in H1299 cells (Fig. 2C). Similar regulation was also observed in other p14ARF-expressing cancer cell lines (Supplementary Fig. 1A). Moreover, KLHDC3 depletion had no obvious impact on p14ARF mRNA levels (Fig. 2D) or p16INK4A protein levels (Fig. 2C). The half-life of p14ARF is considerably prolonged by KLHDC3 depletion (Fig. 2E, F). Ablation of KLHDC3 by KO in H1299 cells also resulted in a marked increase in p14ARF protein levels (Fig. 2G, Supplementary Fig. 2A–D). KLHDC3, but not the other CRL2 adaptors, promoted p14ARF degradation (Fig. 2H), and this effect was completely blocked by MG132 treatment (Fig. 2I). p14ARF was consistently localized to nuclear speckles called nucleoli [21]. Immunofluorescence (IF) analysis showed that the intensity of nucleolar p14ARF was more prominent in KLHDC3 KO cells as compared to parental cells, suggesting that KLHDC3 depletion led to p14ARF accumulation but did not alter its nucleolar localization (Fig. 2J, K). Finally, we showed that KLHDC3, but not the other CRL2 adaptors, augmented p14ARF polyubiquitination (Fig. 2L). Together, these findings suggest that KLHDC3 promotes p14ARF degradation through the ubiquitin–proteasomal pathway.
A, B Tandem affinity purification of CUL2-containing protein complex was conducted using H1299 cells stably expressing FH-CUL2. Associated proteins were separated by SDS-PAGE and visualized by CB staining (A). The number of total peptides identified by mass spectrometry analysis are shown in (B). C Western blot of indicated proteins in WCLs from H1299 cells transfected with the indicated siRNAs for 48 h. D RT-qPCR assessment of p14ARF mRNA levels in H1299 cells transfected with the indicated siRNAs for 48 h. Data are shown as means ± SD (n = 3). E, F Western blot of indicated proteins in WCLs of 293T cells transfected with the indicated siRNAs for 48 h and then treated with CHX (50 μg/ml) and harvested at different time points (E). At each time point, the intensity of p14ARF was normalized to the intensity of Actin and then to the value at 0 h (F). Data are shown as means ± SD (n = 3). G Western blot of indicated proteins in WCLs from H2199 cells with KLHDC3 KO through CRISPR/Cas9 methods. Parental H1299 cells were used as the control. H Western blot of indicated proteins in WCLs from 293 T cells transfected with the indicated plasmids. I Western blot of WCL of 293T cells transfected with indicated plasmids for 24 h and treated with DMSO or MG132 (20 μM) for 8 h. J, K Representative IF images of endogenous p14ARF staining in H1299 cells transfected with indicated siRNAs for 48 h. Scale bar, 10 μm. p14ARF staining intensity was quantified using Image J (K). n = 20. L Western blot of the products of in vivo ubiquitination assays from 293 T cells transfected with the indicated plasmids and treated with MG132 (20 µM) for 8 h. P values are calculated by the One-way ANOVA test in (D, K), and Two-way ANOVA test in (F). *p < 0.05; **p < 0.01; ****p < 0.0001. ns no significant.
KLHDC3 interacts with p14ARF and promotes p14ARF degradation in a CRL2 complex-dependent manner
We investigated whether KLHDC3 directly interacts with p14ARF. We isolated the KLHDC3 protein complex from H1299 cells stably expressing FH-KLHDC3 and determined the proteins present in the complex. Three peptides corresponding to p14ARF were detected (Supplementary Fig. 1B, C). Co-IP assay results showed that ectopically overexpressed KLHDC3 interacted with p14ARF (Fig. 3A). Semi-endogenous co-IP assays performed in H1299 cells stably overexpressing KLHDC3 or endogenous co-IP assays both showed that KLHDC3 interacted with endogenous p14ARF (Fig. 3B, C). By contrast, other CRL2 receptors examined showed no interaction with p14ARF (Fig. 3D). To investigate the impact of KLHDC3 on the subcellular localization of p14ARF, FH-KLHDC3 constructs were transfected into H1299 cells. p14ARF staining disappeared in the presence of overexpressed KLHDC3. However, when MG132 was added to block p14ARF degradation, p14ARF was dispersed into the nucleoplasm and colocalized with KLHDC3 (Fig. 3E, Supplementary Fig. 1D), suggesting that KLHDC3 promotes p14ARF translocation from the nucleoli to the nucleoplasm for degradation. CUL2-p14ARF interaction was abolished in KLHDC3 KO cells, suggesting that CUL2-p14ARF interaction is indirect and bridged by KLHDC3 (Fig. 3F). KLHDC3 contains six Kelch domains and a BC-box and CUL2-box motif (Fig. 3G). We generated a series of KLHDC3 deletion mutants and tested their binding capacities to p14ARF. Deletion of any Kelch domain abolished KLHDC3-p14ARF interaction. By contrast, deletion of the C-terminal region harboring the BC-box and CUL2-box had no impact on the KLHDC3-p14ARF interaction, suggesting that Kelch domains are required for their interaction (Fig. 3G, H). However, all mutants were defective in promoting p14ARF degradation (Fig. 3J). To further delineate whether the BC-box and CUL2-box are needed for KLHDC3-mediated p14ARF degradation, we generated two mutants, in which the conserved amino acids were substituted by alanine (Fig. 3I), and found that these mutants lost their capacities to degrade and ubiquitinate p14ARF (Fig. 3J, K). Nucleophosmin (NPM) is a nucleolar protein that keeps p14ARF in the nucleoli to protect p14ARF from degradation [22, 23]. We found that NPM depletion caused p14ARF destabilization. However, co-depletion of NPM and KLHDC3 led to p14ARF stabilization (Fig. 3L). Moreover, NPM depletion increased the interaction between KLHDC3 and p14ARF (Fig. 3M), suggesting that NPM protects p14ARF from KLHDC3-mediated degradation by sequestering it into the nucleoli. Together, these findings suggest that KLHDC3 binds p14ARF and targets p14ARF for degradation in a CRL2 complex-dependent manner.
A Western blot of indicated proteins in WCLs and co-IP samples of anti-FLAG antibody obtained from 293T cells transfected with the indicated plasmids. B Western blot of indicated proteins in WCLs and co-IP samples of anti-FLAG antibody obtained from H1299 cells stably overexpressing empty control (EV) or FH-KLHDC3. C Western blots of WCLs and co-IP samples of anti-KLHDC3 antibody from H1299 cells treated with MG132 (20 µM) for 8 h. D Western blot of indicated proteins in WCLs and co-IP samples of anti-FLAG antibody obtained from 293 T cells transfected with the indicated plasmids. E Representative IF images of staining in H1299 cell transfected with HA-KLHDC3 and treated with DMSO or MG132 (20 μM) for 8 h. Scale bar, 10 μm. F Western blot of indicated proteins in WCLs and co-IP samples of anti-FLAG antibody obtained from parental or KLHDC3 KO H1299 cells transfected with the indicated plasmids. G Schematic representation of KLHDC3 deletion mutants. H Western blot of indicated proteins in WCLs and co-IP samples of anti-FLAG antibody obtained from 293T cells transfected with the indicated plasmids. I The canonical BC-box and Cul2-box sequence of KLHDC3 and the amino acid substitutions corresponding to each mutant are indicated. J Western blot of indicated proteins in WCLs from 293T cells transfected with the indicated plasmids. K Western blot of the products of in vivo ubiquitination assays from 293T cells transfected with the indicated plasmids and treated with MG132 (20 µM) for 8 h. L Western blot of indicated proteins in WCLs from H1299 cells transfected with the indicated siRNAs for 48 h. M Western blot of indicated proteins in WCLs and co-IP samples of anti-FLAG antibody obtained from control or NPM-depleted H1299 cells transfected with the indicated plasmids.
KLHDC3 specifically recognizes a C-terminal degron of p14ARF
We unexpectedly found that KLHDC3 can only promote the degradation of non-tagged p14ARF. The addition of a N-terminal or C-terminal FLAG tag to p14ARF completely abolished KLHDC3-mediated p14ARF degradation (Fig. 4A, B). Co-IP assay results showed that N-FLAG-p14ARF interacted with KLHDC3 at a level comparable to non-tagged p14ARF. By contrast, C-FLAG-p14ARF did not interact with KLHDC3 (Fig. 4A, C). p14ARF is a lysine-less protein and the murine homolog p19arf has a single lysine residue but not required for proteasomal degradation. p19arf was subjected to N-terminal ubiquitination [24]. Thus, the N-terminal tagging may block the N-terminal ubiquitination of p14ARF. The fact that the C-terminal tagging blocked KLHDC3-p14ARF interaction led us to suspect that the C-terminal region of p14ARF may harbor a degron recognized by KLHDC3. Indeed, deletion of the last 10 amino acids of p14ARF totally abolished KLHDC3-p14ARF interaction and KLHDC3-mediated p14ARF degradation (Fig. 4A–C). We then performed an alanine-scanning mutagenesis to determine the key amino acids responsible for KLHDC3-binding. Co-IP assay results showed that the majority of single alanine substitution moderately reduced KLHDC3-p14ARF interaction and KLHDC3-mediated p14ARF degradation. However, R129A mutation completely abolished the KLHDC3-p14ARF interaction and KLHDC3-mediated p14ARF degradation (Fig. 4B, D). The half-life of p14ARF R129A mutant was considerably prolonged than wild-type p14ARF (Fig. 4E, F). Incubation of cell lysates with a C-terminal p14ARF decapeptide before co-IP assay reduced the KLHDC3-p14ARF interaction in a dose-dependent manner (Fig. 4G). To further confirm that the C-terminal sequence of p14ARF acts as a degron, we fused this sequence to the C-terminus of GFP(GFP-p14ARF-C). KLHDC3 interacted with GFP fusion protein and targeting it for ubiquitination and degradation. In contrast, KLHDC3 cannot ubiquitinate and degrade GFP proteins or GFP-p14ARF-C-R129A mutant (Supplementary Fig. 3A–C).
A Alanine scanning mutagenesis of the C-terminal degron of p14ARF. B Western blot of indicated proteins in WCL from 293 T cells transfected with the indicated plasmids. C Western blot of indicated proteins in WCLs and co-IP samples of anti-Myc antibody obtained from 293T cells transfected with the indicated plasmids. D Western blot of indicated proteins in WCLs and co-IP samples of anti-p14ARF antibody obtained from 293T cells transfected with the indicated plasmids. E, F Western blot of indicated proteins in WCLs of 293T cells transfected with indicated siRNAs for 48 h and then treated with CHX (50 μg/ml) and harvested at different time points (E). At each time point, the intensity of p14ARF was normalized to the intensity of actin and then to the value at 0 h (F). Data are shown as means ± SD (n = 3). P values are calculated by the Two-way ANOVA test. **p < 0.01. G 293T cells were transfected with indicated plasmids. KLHDC3-p14ARF complex was immunoprecipitated by anti-FLAG antibody, and then the bound complex was incubated with the increasing amounts of C-terminal decapeptide of KLHDC3 or the corresponding scramble decapeptide for 12 h. Bound material was subjected to western blot. H Western blot of indicated proteins in WCL from 293T cells transfected with the indicated plasmids. I Western blot of indicated proteins in WCLs and co-IP samples of anti-FLAG antibody obtained from 293T cells transfected with the indicated plasmids. J Western blot of indicated proteins in WCLs from CT26 cells transfected with the indicated siRNAs for 48 h.
Given that p14ARF stability was control by CUL2 (Fig. 1), we further investigated whether this process is dependent on the C-terminal degron of p14ARF. To address this issue, we generated H1299 cells stably overexpressing p14ARF-ΔCter. We found that CUL2 depletion elevated endogenous p14ARF protein levels, but had no impact on p14ARF-ΔCter protein levels (Supplementary Fig. 3D). Moreover, we generated MCF-7 cells (ARF null) stably overexpressing p14ARF-WT or p14ARF-ΔCter and found that CUL2 depletion elevated the protein levels of overexpressed p14ARF, but not 14ARF-ΔCter (Supplementary Fig. 3E). Thus, the genetic and pharmaceutical approaches above collectively suggested that a CRL2 E3 ligase controls p14ARF protein stability in a C-terminal degron-dependent manner.
Although p14ARF and p19arf share similar properties toward tumor suppression, the C-terminal regions between these two proteins are not evolutionarily conserved, and R129 of p14ARF is absent in p19arf (Supplementary Fig. 3F). It is interesting to investigate whether murine homolog klhdc3 regulates p19arf. As shown in Fig. 4H, Klhdc3 efficiently degraded p14ARF. By contrast, p19arf was resistant to klhdc3-mediated degradation. No interaction occurred between p19arf and klhdc3 (Fig. 4I), and klhdc3 depletion in mouse colon carcinoma CT26 cells did not alter p19arf protein levels (Fig. 4J). Together, these findings suggest that KLHDC3 promotes p14ARF degradation by specifically recognizing a C-terminal degron in p14ARF, and this regulation emerges after the human-mouse divergence during evolution.
KLHDC3 regulates in vitro cell growth and apoptosis in a p53-dependent manner
p14ARF interacts with diverse proteins and regulate multiple cellular functions, such as cell growth and apoptosis [11]. We investigated whether KLHDC3 has any impact on the cellular phenotypes through regulating p14ARF. We chose four cell lines with different p53/p14ARF backgrounds, namely, H1299 (p53 null/p14ARF-WT), SKOV3 (p53 null/p14ARF-WT), OVCAR-3 (p53 R248Q/p14ARF-WT), and SJSA (p53-WT/p14ARF-WT) cells. p14ARF proteins were inevitably stabilized by KLHDC3 KO in four cell lines (Fig. 5A–D). KLHDC3 KO had no impact on the protein levels of p53 R258Q mutant in OVCAR-3 cells (Fig. 5B). By contrast, KLHDC3 KO in SJSA cells resulted in an elevation of wild-type p53 levels and its downstream target p21 but did not alter the p53 mRNA levels (Fig. 5D, Supplementary Fig. 4A). Moreover, this effect could be reversed by p14ARF depletion, suggesting that KLHDC3 regulates p53 stability through p14ARF (Fig. 5D). Although p14ARF was strongly stabilized, we unexpectedly found that cell growth (Fig. 5E–G), cell cycle distribution (Fig. 5I–K), and etoposide-induced apoptosis (as assessed by Annexin V/PI staining and Caspase-3 cleavage) (Fig. 5M–O, Supplementary Fig. 4B–I) were marginally affected by KLHDC3 KO in p53 null or mutated cell lines. By contrast, KLHDC3 KO in SJSA cells reduced cell growth, increased G1 phase and etoposide-induced apoptosis, and these effects could be reversed by p14ARF depletion (Fig. 5H, L, P).
A–D Western blot of indicated proteins in WCLs from parental and KLHDC3 KO SKOV3 (A), OVCAR-3 (B), H1299 (C), or SJSA (D) cells. SJSA cell were additionally transfected with indicated siRNAs for 48 h. E–H The relative cell growth of parental and KLHDC3 KO SKOV3 (E), OVCAR-3 (F), H1299 (G), or SJSA (H) were determined by CCK-8 assays. SJSA cell were additionally transfected with the indicated siRNAs. Data are shown as means ± SD (n = 3). I–L The cell cycle distribution of parental and KLHDC3 KO SKOV3 (I), OVCAR-3 (J), H1299 (K), or SJSA (L) cells were determined by flow cytometry. SJSA cells were additionally transfected with the indicated siRNAs for 48 h. Data are shown as means ± SD (n = 3). M–P Parental and KLHDC3 KO SKOV3 (M), OVCAR-3 (N), H1299 (O), or SJSA (P) cells treated with DMSO or etoposide (5 μM for SKOV3, 10 μM for OVCAR-3, 10 μM for H1299, and 2 μM for SJSA) for 24 h, then the cell apoptosis was determined by flow cytometry. SJSA cells were additionally transfected with the indicated siRNAs for 48 h before treatment. Data are shown as means ± SD (n = 3). P values are calculated by the One-way ANOVA test in (I–P) and Two-way ANOVA test in (E–H). *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. ns no significant.
Together, these findings suggest that KLHDC3 affects in vitro cell growth and chemotherapeutic drug-induced apoptosis in cells harboring a functional p53.
KLHDC3 suppresses ferroptosis in a p14ARF and SLC7A11-dependent manner
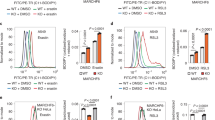
We recently revealed that p14ARF binds NRF2 and inhibits NRF2-mediated SLC7A11 expression to promote p53-independent ferroptosis [12]. Thus, we investigated whether KLHDC3 affects ferroptosis through the p14ARF-NRF2-SLC7A11 regulatory pathway. SKOV3, OVCAR-3, H1299, and SJSA cells have Keap1/NRF2 wild-type backgrounds. Ferroptosis elicited by Erastin or FIN56 was more evident in KLHDC3 KO cells than that in parental cells, and this impact could be eliminated by co-treatment of Ferr-1, a selective inhibitor of ferroptosis (Fig. 6A, Supplementary Fig. 5A, B). Similar results were observed in KLHDC3 KO OVCAR-3, H1299, or SJSA cells (Supplementary Fig. 5C). Reintroduction of KLHDC3-WT but not the mutants (BCM or CULM) into KLHDC3 KO cells (Fig. 6B) or p14ARF depletion (Fig. 6C) restored the cell survival upon erastin treatment. We used Liperfluo staining to monitor lipid peroxidation during ferroptosis and found that Liperfluo signals were more evident in KLHDC3 KO cells, which impact can be eliminated by p14ARF depletion (Fig. 6D, Supplementary Fig. 5D). Similar trends were observed when GPX4 enzymatic activity (Fig. 6E) and cellular GSH levels (Fig. 6F) were measured.
A Parental and KLHDC3 KO SKOV3 cells were treated with DMSO, erastin (4 μM), or erastin (4 μM) plus Ferr-1 (2 μM) for 8 h. Cells were pretreated with Ferr-1 for 4 h before erastin was added. Cell survival was detected by CCK-8 assays. Data are shown as means ± SD (n = 3). B KLHDC3 KO SKOV3 cells stably overexpressing EV, KLHDC3-WT, mutants (BCM or CULM) were treated with erastin (4 μM) for 8 h. Cell survival was detected by CCK-8 assays. Data are shown as means ± SD (n = 3). C Parental and KLHDC3 KO SKOV3 cells were transfected with the indicated siRNAs for 48 h, and then cells were treated with erastin (4 μM) for 8 h. Cell survival was detected by CCK-8 assays. Data are shown as means ± SD (n = 3). D Parental and KLHDC3 KO SKOV3 cells were transfected with the indicated siRNAs for 48 h, and then cells were treated with erastin (10 μM) for 8 h. Liperfluo staining intensity was measured. Data are shown as means ± SD (n = 3). E Parental and KLHDC3 KO SKOV3 cells were transfected with the indicated siRNAs for 48 h. GPX4 activity was measured by the GPX4 enzymatic activity Kit. Data are shown as means ± SD (n = 3). F Parental or KLHDC3 KO SKOV3 cells were transfected with the indicated siRNAs for 48 h. The ratio of reductive GSH to oxidative GSH was measured by the GSH/GSSG quantification Kit. Data are shown as means ± SD (n = 3). G Western blot of indicated proteins in WCLs from parental and KLHDC3 KO SKOV3 cells transfected with indicated siRNAs for 48 h. H RT-qPCR assessment of SLC7A11 mRNA levels in parental or KLHDC3 KO transfected with indicated siRNAs for 48 h. Data are shown as means ± SD (n = 3). I Parental and KLHDC3 KO SKOV3 cells were transfected with the indicated plasmids for 48 h, and then cells were treated with cystine-low (2 µM cystine) medium for indicated times. Cell survival was detected by CCK-8 assays. Data are shown as means ± SD (n = 3). J Tumor image of parental and KLHDC3 KO SKOV3 cells implanted tumors from nude mice. K Weight of implanted tumors formed by parental and KLHDC3 KO SKOV3 cells. n = 4. L Western blot of indicated proteins in WCLs from xenograft tumors (two independent tumors for each group). P values are calculated by the One-way ANOVA test in (A, B, D, E, F, H), Two-way ANOVA test in (C, I), and unpaired two-tailed Student’s t test in (K). *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. ns no significant.
The protein or mRNA levels of SLC7A11 were reduced in KLHDC3 KO cells, which was reversed by p14ARF depletion (Fig. 6G, H) or KLHDC3-WT reintroduction (Supplementary Fig. 6A, B). Such effects were also observed when other NRF2 transcriptional targets (HMOX1 and NOQ1) were detected (Supplementary Fig. 6C). KLHDC3 overexpression decreased the protein levels of p14ARF and increased the protein levels of SLC7A11 in H1299 cells. By contrast, such effect was not observed in NRF2 KO H1299 cells (Supplementary Fig. 6D, E). Moreover, KLHDC3 overexpression had no impact on SLC7A11 protein levels in A549 cells (p14ARF null) which harbor an inactive Keap1 G333C mutant and NRF2 proteins are abnormally accumulated (Supplementary Fig. 6F). These results collectively suggested that KLHDC3 modulates SLC7A11 expression in a NRF2-dependent manner. SJSA cells harbor a wild-type p53 which directly represses SLC7A11 expression in a way distinct from NRF2 [15]. p14ARF depletion completely reversed KLHDC3 KO-induced SLC7A11 downregulation. By contrast, p53 depletion had a minimal impact on SLC7A11 protein levels in KLHDC3 KO SJSA cells. These results suggested that p14ARF seems to play a more important role in the regulation of SLC7A11 than p53, at least in SJSA cells ((Supplementary Fig. 6G). However, given that cellular heterogeneity, the possibility that p53 plays a more important role in the regulation of SLC7A11 than p14ARF in specific cell types or tissues cannot be ruled out.
To further delineate that the impact of KLHDC3 on ferroptosis was dependent on SLC7A11, we stably overexpressed SLC7A11 in KLHDC3 KO cells. The restoration of SLC7A11 expression in KLHDC3 KO cells reversed erastin or low cystine media-induced cell death (Fig. 6I, Supplementary Fig. 5G). Finally, we examined whether KLHDC3 affects p14ARF-mediated tumor growth suppression in xenograft tumor models. We found that p14ARF depletion increased SKOV3 tumor growth, in line with the notion that p14ARF can suppress tumor growth in a p53-independent manner. KLHDC3 KO led to a substantial reduction in tumor growth. More importantly, KLHDC3 KO-induced tumor growth suppression can be largely reversed by p14ARF depletion (Fig. 6J–L), suggesting that p14ARF stabilization is, at least in part, involved in this process. Together, these findings suggest that KLHDC3-mediated ferroptosis suppression acts in a p53-independent manner and relies on the p14ARF-NRF2-SLC7A11 regulatory pathway.
Cell-penetrating p14ARF-derived peptide stabilizes p14ARF and sensitizes cancer cells to ferroptosis
The C-terminal degron was sufficient for p14ARF binding to KLHDC3. Thus, we hypothesized that introduction of the C-terminal degron peptide of p14ARF into cells may competitively inhibit KLHDC3-mediated p14ARF degradation. To efficiently deliver this peptide into cells, we synthesized a fusion peptide, in which the C-terminal degron was connected to the cell-penetrating poly-arginine (R8) sequence. We found that treatment of SKOV3 cells with the fusion peptide elevated p14ARF protein levels but decreased the mRNA and protein levels of SLC7A11 (Supplementary Fig. 6H, I). Moreover, we found that treatment of fusion peptide rendered SKOV3 cells more sensitive to Erastin-induced ferroptosis (Supplementary Fig. 6J). Together, these findings suggest that KLHDC3-mediated p14ARF degradation can be competitively blocked by the C-terminal peptide of p14ARF.
KLHDC3 expression is elevated in ovarian cancer
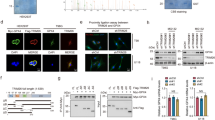
To assess whether KLHDC3 expression is altered in human cancer tissues, we analyzed the RNA-sequencing (RNA-seq) data in the TCGA cohorts. KLHDC3 mRNA expression was significantly overexpressed in the majority of cancer types examined (Fig. 7A). In this study, we focused on ovarian cancer, and we had demonstrated that p14ARF stability was controlled by KLHDC3 in two ovarian cancer cell lines (SKOV3 and OVCAR-3). We then analyzed the mRNA expression of KLHDC3 in several published microarray datasets of ovarian cancer. Similar to the result from the TCGA cohort, KLHDC3 expression was significantly elevated in ovarian cancer tissues as compared to normal ovarian tissues from multiple studies (Fig. 7B). However, KLHDC3 expression had no significant correlation with ovarian cancer prognosis (Fig. 7C). These findings, together with the results from in vivo ovarian cancer cell growth (Fig. 6J–L), suggest that KLHDC3 may act as an oncogene in ovarian cancer.
A The relative mRNA expression of KLHDC3 in various normal and cancer tissues from the TCGA cohorts. Significantly increased and decreased expression in tumor comparing to matched normal tissue indicated as red and green fonts at the top, respectively. B The relative mRNA expression of KLHDC3 in normal ovarian tissues and ovarian cancer tissues from the GEO datasets. C Kaplan–Meier survival curves for patients with high vs. low mRNA expression of KLHDC3 in the ovarian cancer TCGA cohort. High- and low-expression groups were defined by the median KLHDC3 mRNA expression value of the study population. The data was analyzed using GEPIA2 (http://gepia2.cancer-pku.cn/). D Schematic of the proposed mechanism through which CRL2-KLHDC3 complex suppresses ferroptosis through the NRF2-p14ARF-SLC7A11 regulatory pathway. P values are calculated using by the unpaired two-tailed Student’s t test. *p < 0.05; ***p < 0.001.
Discussion
Although cysteine is the least abundant amino acid in cells, it plays critical roles in maintaining cellular redox balance and promoting cell survival and growth. Insufficient cystine uptake induces oxidative stress and provokes ferroptosis [25]. SLC7A11, together with SLC3A2, encode the heterodimeric amino acid transport system, which mediates cystine-glutamate exchange [25]. SLC7A11 is frequently overexpressed in various cancers. Accumulating evidence suggested that SLC7A11 acts as a key factor in modulating ferroptosis in human cancers [26]. Several well-known oncogenes and tumor suppressors, including NRF2, K-Ras, BAP1, p53, and p14ARF, had been reportedly to regulate ferroptosis partly through modulation of SLC7A11 expression [12, 15, 27,28,29]. However, the precise mechanism by which SLC7A11 expression is regulated in human cancers are still not fully understood. In this study, we found that CRL2-KLHDC3 complex relieves p14ARF-mediated SLC7A11 transcription suppression (Fig. 7D). Moreover, KLHDC3 depletion elevated SLC7A11 expression in p53-null, -MUT, and -WT cell lines, suggesting that the impact of KLHDC3 on SLC7A11 expression is largely p53-independent.
The turnover of p14ARF is relative short in most cell lines, suggesting that p14ARF protein is robustly degraded. ULF, SIVA, and MKRN1 were reported as E3 ubiquitin ligases for p14ARF [30,31,32]. Conversely, USP10, a deubiquitinating enzyme, caused p14ARF stabilization by removing the ubiquitin chains from p14ARF [33]. Our results showed that KLHDC3 KD or KO led to a dramatic increase in p14ARF protein levels in all p14ARF-expressing cell lines we examined, suggesting that CRL2-KLHDC3 complex may be the major E3 ubiquitin ligase controlling p14ARF protein stability. Our complex purification results suggested that KLHDC3 may interact with USP10 (Supplementary Fig. 1C). It would be interesting to investigate whether USP10 works cooperatively with KLHDC3 in fine-tuning p14ARF protein levels in response to external stimuli.
One interesting issue raised by the present study is the fact that KLHDC3-mediated p14ARF degradation is not conserved in mice. Addition of a C-terminal FLAG blocked p14ARF binding to KLHDC3, implying that an exposed C-terminal degron is essential for KLHDC3-p14ARF binding. However, p19arf has an extra C-terminal sequence that is not present in p14ARF. This sequence difference may explain why p19arf does not interact with klhdc3. Thus, p14ARF and p19arf may be subjected to different post-translational modification regulation during evolution due to sequence variations. Our study also suggested the potential impact of epitope tagging on protein stability and functions, which should be considered in biological studies concerning p14ARF. Recently, an elegant study by Koren et al. discovered that a subset of CRL2 E3 ubiquitin complexes recognize the C-terminal degron. Specifically, KLHDC3 recognizes substrates through a specific interaction with exposed C-terminal -RG, -KG, or -QG motif [34]. These findings were largely consistent with our results, but we additionally found that R129 at −4 position of p14ARF C-terminus is extremely critical for KLHDC3-binding. This feature may serve as a guide to identify more substrates for CRL2-KLHDC3 complex.
In summary, we identified that KLHDC3 recognizes a C-terminal degron in p14ARF and then targets p14ARF for degradation in a CRL2 complex-dependent manner. We provided evidence that KLHDC3 exerts its pro-tumorigenic functions by protecting cancer cells from ferroptosis partially through the regulation of the p14ARF-NRF2-SLC7A11 regulatory pathway. Finally, we defined a cell-penetrating p14ARF-derived peptide that stabilizes p14ARF and sensitize cancer cells to ferroptosis. Our findings suggest that inhibition of the KLHDC3-p14ARF interaction by peptides or small molecules may represent a promising therapeutic strategy for targeting KLHDC3-overrepresented cancers.
References
Gil J, Peters G. Regulation of the INK4b-ARF-INK4a tumour suppressor locus: all for one or one for all. Nat Rev Mol cell Biol. 2006;7:667–77.
Sherr CJ. Tumor surveillance via the ARF-p53 pathway. Genes Dev. 1998;12:2984–91.
Kamijo T, Zindy F, Roussel MF, Quelle DE, Downing JR, Ashmun RA, et al. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell. 1997;91:649–59.
Matheu A, Pantoja C, Efeyan A, Criado LM, Martin-Caballero J, Flores JM, et al. Increased gene dosage of Ink4a/Arf results in cancer resistance and normal aging. Genes Dev. 2004;18:2736–46.
Kamijo T, Weber JD, Zambetti G, Zindy F, Roussel MF, Sherr CJ. Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. Proc Natl Acad Sci USA. 1998;95:8292–7.
Pomerantz J, Schreiber-Agus N, Liegeois NJ, Silverman A, Alland L, Chin L, et al. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2’s inhibition of p53. Cell. 1998;92:713–23.
Zhang Y, Xiong Y, Yarbrough WG. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell. 1998;92:725–34.
Zindy F, Eischen CM, Randle DH, Kamijo T, Cleveland JL, Sherr CJ, et al. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 1998;12:2424–33.
Palmero I, Pantoja C, Serrano M. p19ARF links the tumour suppressor p53 to Ras. Nature. 1998;395:125–6.
Bates S, Phillips AC, Clark PA, Stott F, Peters G, Ludwig RL, et al. p14ARF links the tumour suppressors RB and p53. Nature. 1998;395:124–5.
Sherr CJ. Divorcing ARF and p53: an unsettled case. Nat Rev Cancer. 2006;6:663–73.
Chen D, Tavana O, Chu B, Erber L, Chen Y, Baer R, et al. NRF2 is a major target of ARF in p53-independent tumor suppression. Mol Cell. 2017;68:224–32.e4.
Stockwell BR, Jiang X, Gu W. Emerging mechanisms and disease relevance of ferroptosis. Trends Cell Biol. 2020;30:478–90.
Jiang X, Stockwell BR, Conrad M. Ferroptosis: mechanisms, biology and role in disease. Nat Rev Mol Cell Biol. 2021;22:266–82.
Jiang L, Kon N, Li T, Wang SJ, Su T, Hibshoosh H, et al. Ferroptosis as a p53-mediated activity during tumour suppression. Nature. 2015;520:57–62.
Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20.
Liu X, Zurlo G, Zhang Q. The roles of Cullin-2 E3 ubiquitin ligase complex in cancer. Adv Exp Med Biol. 2020;1217:173–86.
Ohh M, Park CW, Ivan M, Hoffman MA, Kim TY, Huang LE, et al. Ubiquitination of hypoxia-inducible factor requires direct binding to the beta-domain of the von Hippel-Lindau protein. Nat Cell Biol. 2000;2:423–7.
Mahrour N, Redwine WB, Florens L, Swanson SK, Martin-Brown S, Bradford WD, et al. Characterization of Cullin-box sequences that direct recruitment of Cul2-Rbx1 and Cul5-Rbx2 modules to Elongin BC-based ubiquitin ligases. J Biol Chem. 2008;283:8005–13.
Chen D, Shan J, Zhu WG, Qin J, Gu W. Transcription-independent ARF regulation in oncogenic stress-mediated p53 responses. Nature. 2010;464:624–7.
Chen D, Kon N, Zhong J, Zhang P, Yu L, Gu W. Differential effects on ARF stability by normal versus oncogenic levels of c-Myc expression. Mol Cell. 2013;51:46–56.
Colombo E, Bonetti P, Lazzerini Denchi E, Martinelli P, Zamponi R, Marine JC, et al. Nucleophosmin is required for DNA integrity and p19Arf protein stability. Mol Cell Biol. 2005;25:8874–86.
Korgaonkar C, Hagen J, Tompkins V, Frazier AA, Allamargot C, Quelle FW, et al. Nucleophosmin (B23) targets ARF to nucleoli and inhibits its function. Mol Cell Biol. 2005;25:1258–71.
Kuo ML, den Besten W, Bertwistle D, Roussel MF, Sherr CJ. N-terminal polyubiquitination and degradation of the Arf tumor suppressor. Genes Dev. 2004;18:1862–74.
Daher B, Vucetic M, Pouyssegur J. Cysteine depletion, a key action to challenge cancer cells to ferroptotic cell death. Front Oncol. 2020;10:723.
Koppula P, Zhang Y, Zhuang L, Gan B. Amino acid transporter SLC7A11/xCT at the crossroads of regulating redox homeostasis and nutrient dependency of cancer. Cancer Commun. 2018;38:12.
Fan Z, Wirth AK, Chen D, Wruck CJ, Rauh M, Buchfelder M, et al. Nrf2-Keap1 pathway promotes cell proliferation and diminishes ferroptosis. Oncogenesis. 2017;6:e371.
Lim JKM, Delaidelli A, Minaker SW, Zhang HF, Colovic M, Yang H, et al. Cystine/glutamate antiporter xCT (SLC7A11) facilitates oncogenic RAS transformation by preserving intracellular redox balance. Proc Natl Acad Sci USA. 2019;116:9433–42.
Zhang Y, Shi J, Liu X, Feng L, Gong Z, Koppula P, et al. BAP1 links metabolic regulation of ferroptosis to tumour suppression. Nat Cell Biol. 2018;20:1181–92.
Yang WS, SriRamaratnam R, Welsch ME, Shimada K, Skouta R, Viswanathan VS, et al. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156:317–31.
Ko A, Shin JY, Seo J, Lee KD, Lee EW, Lee MS, et al. Acceleration of gastric tumorigenesis through MKRN1-mediated posttranslational regulation of p14ARF. J Natl Cancer Inst. 2012;104:1660–72.
Wang X, Zha M, Zhao X, Jiang P, Du W, Tam AY, et al. Siva1 inhibits p53 function by acting as an ARF E3 ubiquitin ligase. Nat Commun. 2013;4:1551.
Ko A, Han SY, Choi CH, Cho H, Lee MS, Kim SY, et al. Oncogene-induced senescence mediated by c-Myc requires USP10 dependent deubiquitination and stabilization of p14ARF. Cell Death Differ. 2018;25:1050–62.
Koren I, Timms RT, Kula T, Xu Q, Li MZ, Elledge SJ. The eukaryotic proteome is shaped by E3 ubiquitin ligases targeting C-terminal degrons. Cell 2018;173:1622–35.e14.
Acknowledgements
We thank Dr. Lin Huang for excellent technical assistance in mass spectrometry analysis
Funding
This work was in part supported by the National Natural Science Foundation of China (nos. 81872260, 82172938 to PZ; nos. 91957125, 81972396, to CW; nos. 91954106, 81872109 to KG).
Author information
Authors and Affiliations
Contributions
PZZ, CJW, and GW conceived the study. PZZ, KG, QS, XYZ, LZ, YJC, DYJ, ZL and HRS performed the experiments and data analyses. PZZ, KG, YL, and LZ analyzed and interpreted the data. CJW and PZZ wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
Our studies did not include human participants or human tissue. The animal studies were approved by ethics Review Committee for Animal Experimentation of Fudan University.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by E Baehrecke
Supplementary information
Rights and permissions
About this article
Cite this article
Zhang, P., Gao, K., Zhang, L. et al. CRL2-KLHDC3 E3 ubiquitin ligase complex suppresses ferroptosis through promoting p14ARF degradation. Cell Death Differ 29, 758–771 (2022). https://doi.org/10.1038/s41418-021-00890-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-021-00890-0
This article is cited by
-
Ferroptotic stress facilitates smooth muscle cell dedifferentiation in arterial remodelling by disrupting mitochondrial homeostasis
Cell Death & Differentiation (2023)
-
E3 ubiquitin ligases in cancer stem cells: key regulators of cancer hallmarks and novel therapeutic opportunities
Cellular Oncology (2023)
-
Role of ferroptosis on tumor progression and immunotherapy
Cell Death Discovery (2022)