Abstract
Inappropriate activation of the p53 transcription factor is thought to contribute to the developmental phenotypes in a range of genetic syndromes. Whether p53 activation drives these developmental phenotypes by triggering apoptosis, cell cycle arrest, or other p53 cellular responses, however, has remained elusive. As p53 hyperactivation in embryonic neural crest cells (NCCs) drives a number of phenotypes, including abnormal craniofacial and neuronal development, we investigate the basis for p53 action in this context. We show that p53-driven developmental defects are associated with the induction of a robust pro-apoptotic transcriptional signature. Intriguingly, however, deleting Puma or Caspase9, which encode key components of the intrinsic apoptotic pathway, does not rescue craniofacial, neuronal or pigmentation defects triggered by p53 hyperactivation in NCCs. Immunostaining analyses for two key apoptosis markers confirm that deleting Puma or Caspase9 does indeed impair p53-hyperactivation-induced apoptosis in NCCs. Furthermore, we demonstrate that p53 hyperactivation does not trigger a compensatory dampening of cell cycle progression in NCCs upon inactivation of apoptotic pathways. Together, our results indicate that p53-driven craniofacial, neuronal and pigmentation defects can arise in the absence of apoptosis and cell cycle arrest, suggesting that p53 hyperactivation can act via alternative pathways to trigger developmental phenotypes.
Similar content being viewed by others
Introduction
A broad spectrum of genetic syndromes, characterized by distinct constellations of developmental phenotypes, has been associated with increased activity of the p53 tumor suppressor protein [1]. p53 is a transcription factor normally maintained at low levels in cells but becomes stabilized and activated in response to various cellular stresses [2]. In a number of genetic syndromes, mutations that disrupt critical cellular processes, such as ribosome biogenesis and DNA repair, trigger intracellular stress responses that activate p53. Many of the developmental defects in animal models of these syndromes can be rescued by loss of p53, suggesting that they arise as a direct consequence of inappropriate p53 activation [1]. However, the specific downstream pathways through which p53 acts to drive these developmental defects are not well defined.
A variety of mouse models have been generated to study the role of p53 in driving developmental defects. In these mouse models, p53 is hyperactivated during embryogenesis due to stabilizing mutations in Trp53, the gene encoding p53, or due to inactivating mutations in Mdm2 or Mdm4, which encode the major negative regulators of p53 [1]. These mouse models point to a direct role for p53 hyperactivation in driving diverse developmental phenotypes, including craniofacial, neuronal, pigmentation, cardiovascular and hematopoietic defects. These studies also indicate that p53 can act in different embryonic cell compartments to drive distinct sets of developmental phenotypes. In particular, p53 hyperactivation in the neural crest, which is a multipotent cell population arising from the embryonic neural tube [3], can induce a host of craniofacial, neuronal, pigmentation and other developmental defects that overlap with those observed in human genetic syndromes [4].
Once active, p53 binds DNA and induces hundreds of target genes in a cell type-dependent and context-dependent manner [5]. These target genes encode proteins involved in diverse cellular responses, including apoptosis, cell cycle arrest, DNA repair, and modulating metabolic reprogramming [6, 7]. In mouse models of p53-driven developmental defects, increased apoptosis and/or decreased proliferation is observed in certain cell compartments, suggesting that p53 may drive developmental defects at least in part by compromising tissue homeostasis through effects on cell proliferation and viability [4, 8,9,10,11]. Other studies suggest that p53 activation can affect additional developmental programs, such as cell fate changes through endothelial-to-mesenchymal transition during heart development [12], and increased secretion of growth factors that modulate skin pigmentation [13]. Thus, the role of p53 in driving developmental phenotypes may be more complex than previously appreciated.
Given that p53 inactivation can rescue many of the observed phenotypes in mouse models of various developmental disorders [1], pharmacological strategies that inhibit inappropriate p53 activity might have utility for mitigating developmental phenotypes in humans. However, these strategies should be approached with caution given that p53 inhibition itself could promote developmental phenotypes, such as neural tube defects [14], or could promote tumorigenesis [6]. Delineating the molecular pathways through which p53 hyperactivation acts to drive developmental defects may reveal specific components acting downstream of p53 that could be targeted therapeutically to prevent developmental defects in human genetic syndromes without completely abolishing p53 activity.
Here, we sought to further define the specific pathways through which p53 hyperactivation acts to drive developmental defects. We focused in particular on the craniofacial, neuronal and pigmentation defects caused by p53 hyperactivation in neural crest cells (NCCs) [4]. In mouse models with hyperactive p53, increased apoptosis is observed in NCCs, and this is associated with an overall decrease in the size of the NCC population [4, 15]. These observations raise the possibility that p53-driven neural crest-based developmental defects are primarily caused by NCC insufficiency due to increased NCC apoptosis. Here, we sought to determine whether inhibition of apoptosis could mitigate the manifestation of developmental defects in mouse models where p53 is hyperactivated in NCCs. Intriguingly, we observed that p53-driven craniofacial, neuronal and pigmentation defects induced by p53 hyperactivation in NCCs were not rescued by loss of key components of the intrinsic apoptotic pathway, Puma and Caspase9, despite their loss being sufficient to prevent p53-hyperactivation-induced apoptosis. These findings suggest that apoptosis is dispensable for p53-driven neural crest-based developmental defects.
Methods
Mouse husbandry
All mouse work was approved and performed in compliance with the Stanford University Administrative Panel on Laboratory Animal Care (APLAC). Mice carrying the following previously described alleles were maintained on a mixed 129/Sv-C57BL/6 background: Trp53LSL-25,26 [16], Wnt1-Cre [17], Rosa26LSL-tdTomato [18], Mdm2flox [19], and Mdm4-null [20]. Mice carrying Puma-null [21] and Caspase9-null [22] alleles were maintained on a C57BL/6 background. To generate embryos, a male mouse was housed with 1–2 females overnight, and the day a vaginal plug was observed was considered embryonic day 0.5 (E0.5). For 5′-Bromo-2′-deoxyuridine (BrdU) incorporation experiments, pregnant females were injected with BrdU (Millipore) prepared in phosphate buffered saline (PBS) at a dose of 0.1 mg per gram body weight 10 min before the embryos were harvested. Embryos were harvested from pregnant females that had been euthanized via CO2 exposure, according to APLAC-approved methods. Yolk sac DNA was collected for genotyping, and embryos were imaged in PBS under a dissecting microscope. For fluorescent imaging, a NIGHTSEA Stereo Microscope Fluorescence Adapter (Electron Microscopy Services) was used.
Immunostaining
Embryos were fixed in 4% paraformaldehyde (PFA) at 4 °C overnight and then dehydrated in graded ethanol, cleared in xylene, paraffin-embedded, and sectioned at 5 microns. For immunofluorescence staining, slides were deparaffinized in xylene, rehydrated in graded ethanol and subjected to heat-mediated antigen retrieval in a pressure cooker for 5 min in Tris-EDTA buffer (10 mM Tris, 1 mM EDTA, 0.05% Tween 20, pH 9.0). Slides were permeabilized with tris-buffered saline containing 0.025% Triton X (TBS-TX), blocked in 10% goat serum and 1% bovine serum albumin (BSA) in TBS-TX for 1 h, and incubated overnight at 4 °C in the primary antibody diluted in TBS-TX with 1% BSA. Slides were rinsed in TBS-TX and incubated for 1 h at room temperature in the secondary antibody diluted in TBS-TX with 1% BSA. Slides were incubated in 1μg/ml DAPI for 5 min and were mounted in mounting medium prepared from glycerol, Mowiol 4–88 (Sigma-Aldrich) and DABCO (Sigma-Aldrich). The following primary antibodies and dilutions were used: rabbit anti-p53 CM5 (Leica Biosystems, #NCL-P53-CM5P) 1:1000, mouse anti-BrdU (BD pharmigen, #555627) 1:800, rabbit anti-cleaved Caspase 3 (Cell Signaling, #9661) 1:800. The following secondary antibodies and dilutions were used: Fluorescein goat anti-rabbit (Vector Laboratories, #FI-1000) 1:200 and Alexa Fluor 546 goat anti-mouse (Thermo Fisher Scientific, #A-11003) 1:200. TUNEL staining was performed on 5 micron paraffin sections using the In situ Apoptosis Detection Kit (Abcam, #ab206386), with methyl green counterstaining.
Quantification and statistical analysis
Mouse breeding was performed until at least 3 embryos or pups were obtained per genotype for each phenotype examined. Investigators were blinded to genotype when performing phenotypic analyses and quantifications. No animals were excluded from phenotypic analysis. No method of randomization was applied. Mice of both genders were considered for postnatal phenotypes. For embryonic phenotypes, gender was not determined. Unpaired T-tests were performed when assessing a mutant genotype compared to littermate controls.
Results
Activation of p53 during NCC development induces a robust apoptotic signature
To explore the molecular basis for p53-driven developmental defects, we focused on a previously established model for neural crest-based developmental defects. Specifically, we analyzed RNA-seq data we previously generated from Wnt1-Cre;Trp53LSL-25,26/+ mice, in which a host of embryonic phenotypes, including craniofacial and neuronal defects, are triggered by moderate levels of p53 hyperactivation in embryonic NCCs (Fig. 1A) [4]. These mice carry a conditional knock-in Trp53LSL-25,26 allele, which encodes a mutant p5325,26 protein that is only expressed after NCC-specific Wnt1-Cre-mediated recombination of an upstream transcriptional stop cassette flanked by loxP sites (LSL). The mutant p5325,26 protein contains amino acid substitutions in the first transactivation domain of p53, which compromise its ability to induce most target genes, but lead to increased protein stability, due to impaired binding to its main negative regulator, Mdm2 [16, 23]. When co-expressed with wild-type p53, the mutant p5325,26 protein can bind to and stabilize wild-type p53, leading to a moderate increase in the induction of p53 target genes [4, 11]. By performing RNA-seq on NCCs from Wnt1-Cre;Trp53LSL-25,26/+ embryos and Wnt1-Cre;Trp53+/+ littermate controls, we previously identified a set of genes induced in response to moderate p53 activation, and we showed that this gene set is strongly enriched for genes involved in apoptosis [4].
A Mouse model used to induce moderate levels of p53 activation in NCCs. In wild-type mice, the p53 tetramer is negatively regulated by Mdm2. In Wnt1-Cre;Trp53LSL-25,26/+ mice, which co-express wild-type p53 and p5325,26 in NCCs, p53 stability and target gene induction are increased (likely due to reduced negative regulation by Mdm2), leading to developmental defects. B Heat map of RNA-seq data showing relative expression levels of pro-apoptotic genes expressed at higher levels in NCCs isolated from Wnt1-Cre;Trp53LSL-25,26/+ E10.5 embryos than in Wnt1-Cre;Trp53+/+ littermates. n = 5 embryos per genotype. RNA-seq data were previously published in Bowen et al. [4]. C Model depicting key components of the intrinsic and extrinsic apoptotic pathways, including proteins encoded by genes that are induced (green) or not significantly affected (gray) upon moderate p53 hyperactivation in NCCs.
Prior studies in other cell types indicate that p53 can induce numerous pro-apoptotic genes that encode direct components of the intrinsic or extrinsic apoptotic pathways or that indirectly modulate apoptosis [24]. p53 could also directly or indirectly repress the expression of anti-apoptotic Bcl-2 family members [24]. Given that the transcriptional response to p53 activation can vary based on cell type and context, we sought here to further define the pro-apoptotic signature induced in response to p53 activation specifically in the context of neural crest-based developmental defects. We found that the genes induced in response to moderate p53 activation in NCCs included a number of genes encoding core components of the intrinsic and extrinsic apoptotic pathways. These genes include Puma, Noxa and Bax, which encode pro-apoptotic Bcl-2 family member proteins that regulate mitochondrial outer membrane permeabilization; Apaf1, which encodes an apoptosome component that promotes the activation of caspases at the onset of apoptosis; and Tnfrsf10b, Fas and Eda2r, which encode death receptors that trigger the extrinsic apoptotic pathway upon ligand binding (Fig. 1B, C) [25, 26]. Furthermore, the pro-apoptotic signature included various genes encoding proteins that have been proposed to act as positive regulators of apoptosis in certain contexts, such as Pidd1 [27], Siva1 [28], Aen [29], Dapk1 [30], Phlda3 [31], Ei24 [32], Foxo3 [33], Trp53inp1 [34], Susd6 [35], Jun [36] and Ddit4 [37] (Fig. 1B). Thus, p53 hyperactivation induces a robust pro-apoptotic signature during embryogenesis.
Loss of Puma or Caspase9 does not prevent p53-driven embryonic craniofacial and neuronal defects
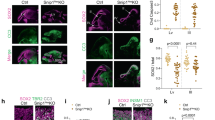
To understand the basis of the p53-triggered developmental phenotypes, we next sought to determine whether inhibiting components of the apoptotic pathway could prevent or diminish p53-driven developmental defects. Puma, which is induced in response to p53 activation in NCCs (Fig. 1B), has been shown to be essential for efficient p53-driven apoptosis in a variety of tissue types [21, 38, 39]. Thus we first asked whether inhibiting Puma-mediated apoptosis could diminish or prevent the neuronal and craniofacial defects we previously reported in Wnt1-Cre;Trp53LSL-25,26/+ embryos with moderate p53 activation in NCCs [4]. To inactivate Puma in our mouse model of p53 hyperactivation, we performed breeding with Puma-null mice, which are viable postnatally and do not display any overt developmental phenotypes [21, 38]. Specifically, we first generated Wnt1-Cre;Puma–/– mice and Trp53LSL-25,26/+;R26LSL-tdTom/LSL-tdTom;Puma+/– mice, which we then interbred to obtain embryos that had moderate p53 activation in NCCs (Wnt1-Cre;Trp53LSL-25,26/+), expressed a tdTomato reporter allele, and were either heterozygous or nullizygous for Puma. To assess neuronal phenotypes, we used a tdTomato reporter allele to visualize the neural crest-derived dorsal root ganglia (DRG) in E11.5 embryos. Consistent with previous reports [4], we found that the DRG in embryos with moderate p53 activation (Wnt1-Cre;Trp53LSL-25,26/+) were notably smaller than the DRG in embryos without p53 hyperactivation (Fig. 2A–D). Interestingly, loss of one or both alleles of Puma did not prevent DRG hypoplasia in embryos with p53 hyperactivation (Fig. 2B, D). While loss of both alleles of Puma slightly increased DRG size in embryos with p53 hyperactivation, the DRG were still approximately two-fold smaller than in littermate controls without p53 hyperactivation (Fig. 2D). Next, to assess craniofacial phenotypes, we focused on the previously reported cleft face phenotype in embryos with moderate p53 activation, which manifests at E11.5 as incomplete fusion of the medial nasal processes [4]. We found that loss of one or both alleles of Puma did not prevent the cleft in the medial nasal process in embryos with p53 hyperactivation (Fig. 2E, F). Thus, Puma is dispensable for the manifestation of p53-driven craniofacial and neuronal defects.
A–C Representative brightfield (i) and fluorescent (ii-iii) images of E11.5 embryos that have moderate p53 activation in NCCs and have lost one or both alleles of Puma or Caspase9. The boxed area in (ii) is shown in (iii). The dorsal root ganglia (DRG) are outlined (iii). Embryos in B are littermates, embryos in C are littermates, and the embryo in A is from a separate litter. All embryos carry a tdTomato reporter allele. Scale bars: 1 mm. D Quantification of DRG size, as measured by the area of fluorescent signal for the fourth DRG in E11.5 embryos carrying a tdTomato reporter allele. n = 3–8 embryos per genotype. Asterisk: p < 0.05. Error bars indicate standard deviation. E–G Representative brightfield images of the dorsal view of E11.5 embryo heads. Arrows indicate a cleft in the medial nasal process. The number of embryos displaying a cleft phenotype is indicated. Scale bars: 0.5 mm.
To corroborate our findings, we next explored the consequences of compromised apoptosis for p53-driven developmental defects by inactivating a more general component of the intrinsic apoptosis pathway, Caspase9. Prior reports indicate that loss of Caspase9 dampens the apoptotic response to a variety of stressors, although this is context-dependent, likely due to redundancy between apoptotic pathway components [40,41,42]. Mice lacking Caspase9 exhibit incompletely penetrant neonatal lethality due to exencephaly but do not display major developmental phenotypes at early embryonic stages [40, 41]. We generated Wnt1-Cre;Caspase9+/– mice and Trp53LSL-25,26/+;R26LSL-tdTom/LSL-tdTom;Caspase9+/– mice, which we then interbred to obtain E11.5 embryos for phenotypic analysis. We found, similar to Puma deficiency, that loss of both Caspase9 alleles in embryos with moderate p53 activation led to a slight increase in DRG size relative to embryos with moderate p53 activation that were heterozygous or wild-type for Caspase9, but did not rescue the DRG hypoplasia phenotype relative to littermate controls without p53 hyperactivation (Fig. 2C, D). Similarly, deletion of Caspase9 did not rescue the facial cleft defect in embryos with moderate p53 activation (Fig. 2G). Together, our results indicate that complete deficiency for either Puma or Caspase9 does not prevent p53-driven craniofacial and neuronal defects.
Puma deficiency does not prevent p53-driven postnatal pigmentation phenotypes
To further define the role for apoptotic factors in p53-driven developmental defects, we sought to determine whether deleting components of the apoptotic pathway could prevent pigmentation defects, which can be assessed in the early postnatal period. Given that Wnt1Cre;Trp53LSL-25,26/+ embryos with moderate p53 activation do not survive postnatally, we used a second mouse model (Wnt1-Cre;Mdm2flox/+;Mdm4+/–) in which p53 is activated to a mild degree in NCCs due to reduced levels of its negative regulators, Mdm2 and Mdm4 (Fig. 3A) [4]. As previously reported, we found that Wnt1-Cre;Mdm2flox/+;Mdm4+/– mice display postnatal hypopigmentation phenotypes, including a white belly spot and limited paw pigmentation at postnatal day 21 (P21) (Fig. 3B) [4]. We next interbred Wnt1-Cre;Puma–/– mice and Mdm2flox/+;Mdm4+/–;Puma+/– mice to generate P21 pups that had mild p53 activation in NCCs (Wnt1-Cre;Mdm2flox/+;Mdm4+/–) and were either heterozygous or nullizygous for Puma. We found that Wnt1-Cre;Mdm2flox/+;Mdm4+/– mice that had lost one or both Puma alleles displayed white belly spots and had lighter paw pigmentation than littermate controls without p53 hyperactivation (Fig. 3C). Quantification of belly spot size in Wnt1-Cre;Mdm2flox/+;Mdm4+/– mice that were wild-type, heterozygous or nullizygous for Puma did not reveal any statistically significant differences between genotypes (Fig. 3D) [4]. These data indicate that Puma is dispensable for the hypopigmentation phenotypes induced by mild p53 activation in NCCs.
A Mouse model used to induce mild levels of p53 activation in NCCs. Reduced expression levels of Mdm2 and Mdm4 leads to increased p53 target gene induction and causes pigmentation defects. B, C Representative images of the belly spot (white arrow) and light paws (red arrow) observed in P21 mice that have mild p53 activation in NCCs and are wild-type, heterozygous or nullizygous for Puma. Agouti and black littermates without p53 activation are shown for comparison. Scale bar = 0.5 cm. D Quantification of belly spot size in P21 mice that have mild p53 activation and are wild-type (n = 3), heterozygous (n = 6) or nullizygous (n = 3) for Puma. No significant differences were observed between genotypes. Error bars indicate standard deviation.
Puma or Caspase9 nullizygosity diminishes apoptotic pathways associated with Caspase3 activation and DNA fragmentation
We next sought to verify whether loss of Puma or Caspase9 impairs p53-driven apoptosis in NCCs. In the Wnt1-Cre;Trp53LSL-25,26/+ mouse model of moderate p53 activation in NCCs, we previously showed that the p53 protein is stabilized in NCCs, which are present in the dorsal neural tube of E9.5 embryos, and we showed that this increase in p53 stability is associated with an increase in apoptotic cells, as detected by cleaved-Caspase3 (CC3) immunostaining [4]. Here, we first confirmed that loss of Puma or Caspase9 does not prevent p53 from being stabilized in the dorsal neural tube of Wnt1-Cre;Trp53LSL-25,26/+ embryos relative to littermate controls with wild-type p53 (Fig. 4Ai, Bi). We next assessed the apoptotic response using two complementary approaches, namely CC3 immunostaining and TUNEL staining. Caspase3 is the main executioner of apoptosis and immunostaining for the activated form of Caspase3 is a commonly used method to detect apoptotic cells [43]. However, given that apoptosis can proceed via alternative executioner caspases in some contexts [42], we also performed TUNEL staining to identify DNA fragmentation, which is a hallmark of apoptotic cells [44]. Compared to control Trp53+/+;Puma+/– and Trp53+/+;Caspase9+/– embryos without p53 hyperactivation, Wnt1-Cre;Trp53LSL-25,26/+;Puma+/- embryos and Wnt1-Cre;Trp53LSL-25,26/+;Caspase9+/– embryos showed increased apoptosis in the dorsal neural tube based on both CC3 and TUNEL analyses, indicating that apoptosis occurs efficiently in embryos heterozygous for Puma or Caspase9 (Fig. 4Aii-iii, Bii-iii). Of note, apoptosis was more pronounced in Wnt1-Cre;Trp53LSL-25,26/+;Caspase9+/– embryos than in Wnt1-Cre;Trp53LSL-25,26/+;Puma+/– embryos (Fig. 4C). This observation suggests that Puma heterozygosity partially dampens p53-hyperactivation-induced apoptosis, consistent with prior reports that loss of one Puma allele can compromise the apoptotic response [38]. Strikingly, loss of both alleles of Puma reduced the percentage of CC3-positive and TUNEL-positive cells to the levels observed in littermate controls without p53 hyperactivation, and loss of both alleles of Caspase9 led to a complete absence of CC3- and TUNEL-positive cells (Fig. 4C). Together, these data support the idea that loss of Puma or Caspase9 prevents p53-hyperactivation-induced apoptotic pathways associated with Caspase3 activation and DNA fragmentation, and suggest that another mechanism underlies the observed developmental defects in embryos with increased p53 activity.
A, B Representative immunofluorescence staining (green) for p53 (i) and cleaved Caspase3 (CC3) (ii), and TUNEL staining (brown, iii) on transverse sections of the dorsal neural tube of E9.5 embryos that have moderate p53 activation and have lost one or both alleles of Puma or Caspase9. Littermate controls without p53 activation are shown for comparison. Arrows indicate CC3-positive cells (ii) and TUNEL-positive cells (iii) that are observed in the dorsal neural tube. Sections were counterstained with DAPI (i-ii, blue) or methyl green (iii, green). Dotted line outlines the dorsal region of the neural tube. Scale bars: 50 µm. C Quantification of CC3-positive and TUNEL-positive cells in the dorsal neural tube of E9.5 embryos that have moderate p53 activation and have lost one or both alleles of Puma or Caspase9. Quantification in littermate controls without p53 activation is also shown for comparison. The dorsal neural tube was defined as the dorsal-most 20% of the neural tube. Asterisk: p < 0.05. Error bars indicate standard deviation. n = 3–4 per genotype.
Puma or Caspase9 deficiency does not trigger cell-cycle arrest
In some contexts, inhibition of p53-driven apoptosis during embryogenesis can trigger lethality by induction of cell cycle arrest by p53. For example, in Mdm2 null embryos, which display lethality at E3.5 associated with extensive apoptosis, deletion of the pro-apoptotic p53 target gene Bax rescues the lethality observed at E3.5 and allows embryos to survive until E7.5, when p53 activation is associated with cell-cycle arrest [45]. To test whether p53 might drive developmental phenotypes in the neural crest of apoptosis-deficient embryos through similar effects on cell cycle progression, we examined cell cycling by immunostaining for BrdU incorporation. We found that the percentage of BrdU-positive cells in the dorsal neural tube of Wnt1-Cre;Trp53LSL-25,26/+ embryos is similar irrespective of Puma or Caspase9 status (Fig. 5A–C), suggesting that cell cycle arrest is not triggered in these embryos with impaired apoptosis pathways. Thus, hyperactive p53 in the neural crest can drive phenotypes without apparent effects on apoptosis or cell cycle progression.
A, B Immunofluorescence staining (pink) for BrdU on transverse sections of the dorsal neural tube of E9.5 embryos that have moderate p53 activation and have lost one or both alleles of Puma or Caspase9. Littermate controls without p53 activation are shown for comparison. Dotted line outlines the dorsal region of the neural tube. Sections were counterstained with DAPI (blue). Scale bars: 50 µm. C Quantification of BrdU-positive cells in the dorsal neural tube of E9.5 embryos. The dorsal neural tube was defined as the dorsal-most 20% of the neural tube. No significant differences were observed between genotypes. Error bars indicate standard deviation. n = 3–4 per genotype.
Discussion
A wide range of genetic syndromes have been associated with increased p53 activity. However, the mechanisms by which p53 hyperactivation promotes the phenotypes characteristic of these syndromes are not well defined. Here, we investigate the contribution of apoptosis to p53-driven neural crest-based developmental defects. We demonstrate that p53 hyperactivation in NCCs induces a host of genes encoding components of both the intrinsic and extrinsic apoptotic pathways, as well as numerous reported positive regulators of apoptosis. We show that deletion of Puma or Caspase9, which encode key components of the intrinsic apoptotic pathway, prevents p53 hyperactivation-induced apoptosis in NCCs, as assessed by CC3 and TUNEL staining. Surprisingly, however, we show that this inhibition of Puma- and Caspase9-dependent apoptosis does not rescue the neuronal, craniofacial or pigmentation defects driven by p53 hyperactivation in NCCs. Furthermore, inhibition of apoptosis does not trigger a compensatory dampening of cell cycle progression in NCCs. Our results raise the possibility that p53 can act via alternative pathways to promote developmental defects (Fig. 6).
Hyperactivation of p53 in the neural crest triggers apoptosis and induces developmental defects. Loss of Puma or Caspase9 prevents canonical apoptosis associated with Caspase3 activation and DNA fragmentation but does not prevent developmental defects, suggesting that p53-driven developmental defects can arise via alternative modes of cell death and/or other cellular responses.
Given the well-supported role for p53 as an inducer of apoptosis, a number of studies have sought to determine whether genes encoding key pro-apoptotic factors - including Puma, Noxa and Bax, which are direct p53 target genes, and Bim, which may be indirectly regulated by p53 [24] - are important for inducing the developmental phenotypes observed in mouse models upon p53 hyperactivation. These studies indicate that certain p53-driven embryonic and postnatal phenotypes can indeed be fully or partially rescued by loss of pro-apoptotic factors. Specifically, inactivation of Puma can rescue the p53-dependent hindlimb and tail defects in mice lacking Mysm1, which encodes an epigenetic regulator that represses p53 target genes [46], and combined loss of Puma and Bim can rescue defects in thymocyte development in Rpl22–/– mice, in which impaired ribosome biogenesis triggers p53 activation [47]. Furthermore, loss of Bax can partially rescue p53-driven microcephaly in Aspm- and Kif20b-deficient mice, in which defects in mitotic spindle assembly and cytokinesis, respectively, are associated with p53 hyperactivation [48, 49]. Loss of Puma can also rescue the testes growth defects in Mdm2PND/PND mice, in which an aberrant Mdm2 allele leads to reduced Mdm2 protein levels, resulting in increased p53 activity [13]. Moreover, Puma deficiency can rescue the postnatal growth retardation, hematopoietic defects, and testes defects in Trp53TSD/– mice, which have elevated p53 activity due to phosphorylation mimicking mutations in the first transactivation domain of p53 [50]. These studies collectively indicate that inactivating key pro-apoptotic factors is sufficient to rescue certain p53-driven embryonic and postnatal developmental phenotypes. In contrast, we show here that loss of Puma or Caspase9 does not prevent craniofacial, neuronal and pigmentation defects driven by p53 hyperactivation in embryonic neural crest cells. Taken together, these observations suggest that these central pro-apoptotic factors are essential for some, but not all, p53-driven developmental phenotypes.
Our observation that loss of Puma or Caspase9 does not prevent p53-driven neural crest-based developmental defects raises the possibility that apoptosis, in the absence of Puma or Caspase9, might proceed via alternative apoptotic factors in NCCs. Indeed, redundancy between or compensation by apoptotic pathway members has been observed in other contexts. For example, in mice exposed to gamma irradiation, loss of Puma alone is sufficient to prevent p53-driven apoptosis in certain cell types, whereas combined loss of Puma and Noxa is required to prevent p53-driven apoptosis in other cell types, suggesting that redundancy exists between these two BH3-only proteins [39]. Redundancy may also exist amongst caspases. Indeed, while loss of Caspase9 dampens apoptosis in some contexts [40, 41], in other contexts apoptosis still proceeds in the absence of Caspase9 and is associated with compensatory activation of alternative caspases. Specifically, apoptosis in Caspase9-null hematopoietic cells is associated with activation of Caspase7, Caspase1 and Caspase11 [42], and apoptosis in Caspase9-null liver cells is associated with activation of Caspase2 and Caspase6 [51]. However, in our mouse model of p53 hyperactivation, we show that TUNEL staining in E9.5 NCCs was completely abolished in the absence of Caspase9 and almost completely abolished in the absence of Puma. Given that TUNEL staining detects DNA fragmentation, which is a biochemical hallmark of apoptosis [44], these results strongly suggest that loss of either Caspase9 or Puma is indeed sufficient to prevent canonical apoptotic pathways in NCCs with p53 hyperactivation and argues against a role for redundant or compensatory apoptotic factors in E9.5 NCCs. However, we cannot exclude the possibility that compensatory apoptotic factors act at later stages of neural crest development, as we did not assess apoptosis in embryos beyond E9.5.
In addition to promoting apoptosis, p53 also has a well-supported role in triggering cell cycle arrest. Thus, in cases where inhibiting apoptosis does not prevent p53-driven developmental defects, it is possible that p53 drives these defects by restraining cellular proliferation. For example, decreased cellular proliferation is observed when apoptosis is inhibited upon loss of Bax in Mdm2-null embryos [45]. However, our observation that p53 hyperactivation does not restrain cellular proliferation in the neural crest, in either the presence or absence of Puma or Caspase9, argues against a prominent role for cell cycle arrest in p53-driven neural crest-based developmental defects. Interestingly, deletion of p21, which is a major mediator of p53-driven cell cycle arrest, fails to rescue embryonic or postnatal developmental defects in several mouse models of p53 hyperactivation. These defects include the postnatal growth retardation, hematopoietic defects, and testes defects in Trp53TSD/– mice [50]; the testes defects and hyperpigmentation defects in Mdm2PND/PND mice [13]; the hematopoietic defects in Mdm2–/–;p53515C/515C mice, in which a hypomorphic p53 protein is stabilized in the absence of Mdm2 [52]; the tail and hypopigmentation defects in Rpl24-deficient mice [53], the defects in thymocyte development in Rpl22–/– mice [47], and the hindlimb and tail defects in Mysm1–/– mice [46]. On the other hand, analysis of cells from Fanconi Anemia patients suggests that silencing p21 can mitigate the p53-driven hematopoietic defects associated with this syndrome [54]. Thus, while cell cycle arrest mediated by p21 may contribute to certain p53-driven phenotypes, it is unlikely to play a predominant role in the majority of p53-driven developmental defects, including the neural crest-based developmental defects examined here.
Our observation that p53-driven developmental phenotypes can occur in the absence of apoptosis and cell cycle arrest suggests that p53 can act via alternative pathways to induce developmental phenotypes. One possibility is that, upon inhibition of canonical apoptotic pathways, p53 activation triggers alternative forms of cell death that are not dependent on Puma and Caspase9 and are not associated with Caspase3 activation and DNA fragmentation. Indeed, p53 has been implicated in regulating non-apoptotic forms of cell death, including necroptosis, ferroptosis and autophagic cell death [55]. The molecular pathways involved in cell death are interconnected, and a variety of studies suggest that cellular signals that typically trigger apoptosis can trigger cell death via alternative pathways when apoptosis is inhibited [56]. For example, interdigital cells in developing mouse limb buds, which typically die via apoptosis, are resistant to apoptosis when the Apaf1 apoptotic factor is deleted; however, in the absence of Apaf1, these cells are still eliminated during development, but display morphological features of necrosis instead of apoptosis, as assessed by electron microscopy [57]. Furthermore, in a variety of contexts, treating cells in culture with a pan-caspase inhibitor prevents apoptosis in response to pro-apoptotic stimuli, but does not preserve cell viability, with cells dying via an alternative pathway that exhibits features of necrosis [58,59,60,61]. In addition, disruption of the extrinsic apoptotic pathway during embryogenesis, such as by deletion of Caspase8, can trigger death via a necroptotic pathway mediated by the Ripk1 and Ripk3 kinases and the Mlkl pseudokinase [62]. Furthermore, in other contexts where apoptosis does not occur, the predominant mode of cell death is autophagic cell death [63]. Intriguingly, embryos lacking intrinsic apoptosis altogether through combined deletion of Bax, Bak, and Bok display phenotypes such as cleft face, without any clear evidence of compensation by other forms of cell death [64]. Determining whether p53 activation drives developmental defects by triggering alternative cell death pathways when canonical apoptosis is inhibited is an intriguing area for future research and may shed light on potential therapeutic strategies to prevent p53-driven developmental defects.
In addition to regulating alternative cell death pathways, p53 can also modulate other cellular responses, including migration, invasion and metabolism [7], raising the possibility that these alternative cellular responses could also contribute to p53-driven developmental defects. Intriguingly, our finding that p53 activation can drive developmental defects without requiring apoptosis and cell cycle arrest is consistent with studies suggesting that p53 can suppress tumorigenesis in the absence of apoptosis and cell cycle arrest activity [23, 65, 66]. Together, these studies highlight the complexity of p53 and underscore the importance of further defining the role of alternative cellular responses in both p53-mediated developmental defects and tumor suppression.
References
Bowen ME, Attardi LD. The role of p53 in developmental syndromes. J Mol Cell Biol. 2019;11:200–11.
Vousden KH, Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009;137:413–31.
Bronner ME, Simões-Costa M. The neural crest migrating into the twenty-first century. Curr Top Dev Biol. 2016;116:115–34.
Bowen ME, McClendon J, Long HK, Sorayya A, Van Nostrand JL, Wysocka J, et al. The spatiotemporal pattern and intensity of p53 activation dictates phenotypic diversity in p53-driven developmental syndromes. Dev Cell. 2019;50:212–28.e6.
Fischer M. Census and evaluation of p53 target genes. Oncogene. 2017;36:3943–56.
Kaiser AM, Attardi LD. Deconstructing networks of p53-mediated tumor suppression in vivo. Cell Death Differ. 2018;25:93–103.
Valente LJ, Tarangelo A, Li AM, Naciri M, Raj N, Boutelle AM, et al. p53 deficiency triggers dysregulation of diverse cellular processes in physiological oxygen. J Cell Biol. 2020;219:e201908212.
Hamard P-J, Barthelery N, Hogstad B, Mungamuri SK, Tonnessen CA, Carvajal LA, et al. The C terminus of p53 regulates gene expression by multiple mechanisms in a target- and tissue-specific manner in vivo. Genes Dev. 2013;27:1868–85.
Liu G, Terzian T, Xiong S, Van Pelt C, Audiffred A, Box N, et al. The p53-Mdm2 network in progenitor cell expansion during mouse postnatal development. J Pathol. 2007;213:360–8.
Terzian T, Wang Y, Van Pelt CS, Box NF, Travis EL, Lozano G. Haploinsufficiency of Mdm2 and Mdm4 in tumorigenesis and development. Mol Cell Biol. 2007;27:5479–85.
Van Nostrand JL, Brady CA, Jung H, Fuentes DR, Kozak MM, Johnson TM, et al. Inappropriate p53 activation during development induces features of CHARGE syndrome. Nature. 2014;514:228–32.
Zhang Q, He X, Chen L, Zhang C, Gao X, Yang Z, et al. Synergistic regulation of p53 by Mdm2 and Mdm4 is critical in cardiac endocardial cushion morphogenesis during heart development. J Pathol. 2012;228:416–28.
Pant V, Xiong S, Chau G, Tsai K, Shetty G, Lozano G. Distinct downstream targets manifest p53-dependent pathologies in mice. Oncogene. 2016;35:5713–21.
Delbridge ARD, Kueh AJ, Ke F, Zamudio NM, El-Saafin F, Jansz N, et al. Loss of p53 causes stochastic aberrant X-chromosome inactivation and female-specific neural tube defects. Cell Rep. 2019;27:442–54.e5.
Jones NC, Lynn ML, Gaudenz K, Sakai D, Aoto K, Rey J-P, et al. Prevention of the neurocristopathy Treacher Collins syndrome through inhibition of p53 function. Nat Med. 2008;14:125–33.
Johnson TM, Hammond EM, Giaccia A, Attardi LD. The p53QS transactivation-deficient mutant shows stress-specific apoptotic activity and induces embryonic lethality. Nat Genet. 2005;37:145–52.
Danielian PS, Muccino D, Rowitch DH, Michael SK, McMahon AP. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr Biol CB. 1998;8:1323–6.
Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–40.
Grier JD, Yan W, Lozano G. Conditional allele of mdm2 which encodes a p53 inhibitor. Genesis. 2002;32:145–7.
Parant J, Chavez-Reyes A, Little NA, Yan W, Reinke V, Jochemsen AG, et al. Rescue of embryonic lethality in Mdm4-null mice by loss of Trp53 suggests a nonoverlapping pathway with MDM2 to regulate p53. Nat Genet. 2001;29:92–5.
Villunger A, Michalak EM, Coultas L, Müllauer F, Böck G, Ausserlechner MJ, et al. p53- and drug-induced apoptotic responses mediated by BH3-only proteins puma and noxa. Science. 2003;302:1036–8.
Simon DJ, Weimer RM, McLaughlin T, Kallop D, Stanger K, Yang J, et al. A caspase cascade regulating developmental axon degeneration. J Neurosci. 2012;32:17540–53.
Brady CA, Jiang D, Mello SS, Johnson TM, Jarvis LA, Kozak MM, et al. Distinct p53 transcriptional programs dictate acute DNA-damage responses and tumor suppression. Cell. 2011;145:571–83.
Aubrey BJ, Kelly GL, Janic A, Herold MJ, Strasser A. How does p53 induce apoptosis and how does this relate to p53-mediated tumour suppression? Cell Death Differ. 2018;25:104–13.
Green DR, Llambi F. Cell death signaling. Cold Spring Harb Perspect Biol. 2015;7:a006080.
Sinha SK, Chaudhary PM. Induction of apoptosis by X-linked ectodermal dysplasia receptor via a Caspase 8-dependent mechanism. J Biol Chem. 2004;279:41873–81.
Tinel A, Tschopp J. The PIDDosome, a protein complex implicated in activation of caspase-2 in response to genotoxic stress. Science. 2004;304:843–6.
Resch U, Schichl YM, Winsauer G, Gudi R, Prasad K, de Martin R. Siva1 is a XIAP-interacting protein that balances NFkappaB and JNK signalling to promote apoptosis. J Cell Sci. 2009;122:2651–61.
Kawase T, Ichikawa H, Ohta T, Nozaki N, Tashiro F, Ohki R, et al. p53 target gene AEN is a nuclear exonuclease required for p53-dependent apoptosis. Oncogene. 2008;27:3797–810.
Singh P, Ravanan P, Talwar P. Death associated protein kinase 1 (DAPK1): a regulator of apoptosis and autophagy. Front Mol Neurosci. 2016;9:46.
Ohki R, Saito K, Chen Y, Kawase T, Hiraoka N, Saigawa R, et al. PHLDA3 is a novel tumor suppressor of pancreatic neuroendocrine tumors. Proc Natl Acad Sci USA. 2014;111:E2404–13.
Mork CN, Faller DV, Spanjaard RA. Loss of putative tumor suppressor EI24/PIG8 confers resistance to etoposide. FEBS Lett. 2007;581:5440–4.
Li Z, Zhao J, Tikhanovich I, Kuravi S, Helzberg J, Dorko K, et al. Serine 574 phosphorylation alters transcriptional programming of FOXO3 by selectively enhancing apoptotic gene expression. Cell Death Differ. 2016;23:583–95.
Ishii K, Ishiai M, Morimoto H, Kanatsu-Shinohara M, Niwa O, Takata M, et al. The Trp53 - Trp53inp1 - Tnfrsf10b pathway regulates the radiation response of mouse spermatogonial stem cells. Stem Cell Rep. 2014;3:676–89.
Tan Y, Huang N, Zhang X, Hu J, Cheng S, Pi L, et al. KIAA0247 suppresses the proliferation, angiogenesis and promote apoptosis of human glioma through inactivation of the AKT and Stat3 signaling pathway. Oncotarget. 2016;7:87100–13.
Ham J, Eilers A, Whitfield J, Neame SJ, Shah B. c-Jun and the transcriptional control of neuronal apoptosis. Biochem Pharm. 2000;60:1015–21.
Li B, Chen R, Chen L, Qiu P, Ai X, Huang E, et al. Effects of DDIT4 in methamphetamine-induced autophagy and apoptosis in dopaminergic neurons. Mol Neurobiol. 2017;54:1642–60.
Jeffers JR, Parganas E, Lee Y, Yang C, Wang J, Brennan J, et al. Puma is an essential mediator of p53-dependent and -independent apoptotic pathways. Cancer Cell. 2003;4:321–8.
Michalak EM, Villunger A, Adams JM, Strasser A. In several cell types tumour suppressor p53 induces apoptosis largely via Puma but Noxa can contribute. Cell Death Differ. 2008;15:1019–29.
Hakem R, Hakem A, Duncan GS, Henderson JT, Woo M, Soengas MS, et al. Differential requirement for caspase 9 in apoptotic pathways in vivo. Cell. 1998;94:339–52.
Kuida K, Haydar TF, Kuan CY, Gu Y, Taya C, Karasuyama H, et al. Reduced apoptosis and cytochrome c-mediated caspase activation in mice lacking caspase 9. Cell. 1998;94:325–37.
Marsden VS, O’Connor L, O’Reilly LA, Silke J, Metcalf D, Ekert PG, et al. Apoptosis initiated by Bcl-2-regulated caspase activation independently of the cytochrome c/Apaf-1/caspase-9 apoptosome. Nature. 2002;419:634–7.
Gown AM, Willingham MC. Improved detection of apoptotic cells in archival paraffin sections: immunohistochemistry using antibodies to cleaved Caspase 3. J Histochem Cytochem. 2002;50:449–54.
Loo DT. In situ detection of apoptosis by the TUNEL assay: an overview of techniques. In: Didenko VV, editor. DNA damage detect situ ex vivo vivo methods protoc [Internet]. Totowa, NJ: Humana Press; 2011. p. 3–13. https://doi.org/10.1007/978-1-60327-409-8_1.
Chavez-Reyes A, Parant JM, Amelse LL, de Oca Luna RM, Korsmeyer SJ, Lozano G. Switching mechanisms of cell death in mdm2- and mdm4-null mice by deletion of p53 downstream targets. Cancer Res. 2003;63:8664–9.
Belle JI, Petrov JC, Langlais D, Robert F, Cencic R, Shen S, et al. Repression of p53-target gene Bbc3/PUMA by MYSM1 is essential for the survival of hematopoietic multipotent progenitors and contributes to stem cell maintenance. Cell Death Differ. 2016;23:759–75.
Stadanlick JE, Zhang Z, Lee S-Y, Hemann M, Biery M, Carleton MO, et al. Developmental arrest of T cells in Rpl22-deficient mice is dependent upon multiple p53 effectors. J Immunol. 2011;187:664–75.
Little JN, Dwyer ND. p53 deletion rescues lethal microcephaly in a mouse model with neural stem cell abscission defects. Hum Mol Genet. 2019;28:434–47.
Williams SE, Garcia I, Crowther AJ, Li S, Stewart A, Liu H, et al. Aspm sustains postnatal cerebellar neurogenesis and medulloblastoma growth in mice. Development. 2015;142:3921–32.
Liu D, Ou L, Clemenson GD, Chao C, Lutske ME, Zambetti GP, et al. Puma is required for p53-induced depletion of adult stem cells. Nat Cell Biol. 2010;12:993–8.
Zheng TS, Hunot S, Kuida K, Momoi T, Srinivasan A, Nicholson DW, et al. Deficiency in caspase-9 or caspase-3 induces compensatory caspase activation. Nat Med. 2000;6:1241–7.
Abbas HA, Maccio DR, Coskun S, Jackson JG, Hazen AL, Sills TM, et al. Mdm2 is required for survival of hematopoietic stem cells/progenitors via dampening of ROS-induced p53 activity. Cell Stem Cell. 2010;7:606–17.
Barkić M, Crnomarković S, Grabusić K, Bogetić I, Panić L, Tamarut S, et al. The p53 tumor suppressor causes congenital malformations in Rpl24-deficient mice and promotes their survival. Mol Cell Biol. 2009;29:2489–504.
Ceccaldi R, Parmar K, Mouly E, Delord M, Kim JM, Regairaz M, et al. Bone marrow failure in fanconi anemia is triggered by an exacerbated p53/p21 DNA damage response that impairs hematopoietic stem and progenitor cells. Cell Stem Cell. 2012;11:36–49.
Ranjan A, Iwakuma T. Non-canonical cell death induced by p53. Int J Mol Sci. 2016;17:2068.
Kroemer G, Martin SJ. Caspase-independent cell death. Nat Med. 2005;11:725–30.
Chautan M, Chazal G, Cecconi F, Gruss P, Golstein P. Interdigital cell death can occur through a necrotic and caspase-independent pathway. Curr Biol. 1999;9:967–S1.
Carter BZ, Kornblau SM, Tsao T, Wang R-Y, Schober WD, Milella M, et al. Caspase-independent cell death in AML: caspase inhibition in vitro with pan-caspase inhibitors or in vivo by XIAP or Survivin does not affect cell survival or prognosis. Blood. 2003;102:4179–86.
Hirsch T, Marchetti P, Susin SA, Dallaporta B, Zamzami N, Marzo I, et al. The apoptosis-necrosis paradox. Apoptogenic proteases activated after mitochondrial permeability transition determine the mode of cell death. Oncogene. 1997;15:1573–81.
McCarthy NJ, Whyte MK, Gilbert CS, Evan GI. Inhibition of Ced-3/ICE-related proteases does not prevent cell death induced by oncogenes, DNA damage, or the Bcl-2 homologue Bak. J Cell Biol. 1997;136:215–27.
Xiang J, Chao DT, Korsmeyer SJ. BAX-induced cell death may not require interleukin 1 beta-converting enzyme-like proteases. Proc Natl Acad Sci USA. 1996;93:14559–63.
Shan B, Pan H, Najafov A, Yuan J. Necroptosis in development and diseases. Genes Dev. 2018;32:327–40.
Nassour J, Radford R, Correia A, Fusté JM, Schoell B, Jauch A, et al. Autophagic cell death restricts chromosomal instability during replicative crisis. Nature. 2019;565:659–63.
Ke FFS, Vanyai HK, Cowan AD, Delbridge ARD, Whitehead L, Grabow S, et al. Embryogenesis and adult life in the absence of intrinsic apoptosis effectors BAX, BAK, and BOK. Cell. 2018;173:1217–30.e17.
Li T, Kon N, Jiang L, Tan M, Ludwig T, Zhao Y, et al. Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell. 2012;149:1269–83.
Valente LJ, Gray DHD, Michalak EM, Pinon-Hofbauer J, Egle A, Scott CL, et al. p53 efficiently suppresses tumor development in the complete absence of its cell-cycle inhibitory and proapoptotic effectors p21, Puma, and Noxa. Cell Rep. 2013;3:1339–45.
Acknowledgements
We thank Dr. David Simon and Dr. Mark Tessier-Lavigne for insightful discussions and for providing mice carrying Casapse9-null and Puma-null alleles. This work was supported by a March of Dimes Foundation grant no. 6-FY15-189 (to LDA), R35 grant CA197591 (to LDA), and Jane Coffin Childs Fund Postdoctoral Fellowship (to MEB).
Author information
Authors and Affiliations
Contributions
Conceptualization, MEB and LDA; Investigation, MEB, ASM and AS; Writing – Original Draft, MEB and LDA; Funding Acquisition, MEB and LDA; Supervision, LDA.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics statement
All mouse work was approved and performed in compliance with the Stanford University Administrative Panel on Laboratory Animal Care (APLAC).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by G. Melino
Rights and permissions
About this article
Cite this article
Bowen, M.E., Mulligan, A.S., Sorayya, A. et al. Puma- and Caspase9-mediated apoptosis is dispensable for p53-driven neural crest-based developmental defects. Cell Death Differ 28, 2083–2094 (2021). https://doi.org/10.1038/s41418-021-00738-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-021-00738-7
This article is cited by
-
Apoptotic cell death in disease—Current understanding of the NCCD 2023
Cell Death & Differentiation (2023)