Abstract
Spermatogenesis is an important cellular differentiation process that produces the male gametes and remains active throughout the individual’s lifespan. Sertoli cell-only syndrome (SCO) refers to the dysfunction of the male reproductive system, including infertility. Accurate self-renewal of spermatogonial stem cells (SSCs) is essential to prevent SCO syndrome. This study investigated the role of microtubule-associated serine/threonine kinase family member 4 (MAST4) in spermatogenesis in mice. MAST4 was localized in Sertoli cells before puberty, providing a somatic niche for spermatogenesis in mice and MAST4 expression shifted to Leydig cells and spermatids throughout puberty. Mast4 knockout (KO) testes were reduced in size compared to wild-type testes, and germ cell depletion associated with an increase in apoptosis and subsequent loss of tubular structure were similar to the SCO phenotype. In addition, MAST4 phosphorylated the Ets-related molecule (ERM), specifically the serine 367 residue. The phosphorylation of ERM ultimately controls the transcription of ERM target genes related to SSC self-renewal. The expression of spermatogenesis-associated proteins was significantly decreased whereas Sertoli cell markers were increased in Mast4 KO testes, which was well-founded by RNA-sequencing analysis. Therefore, MAST4 is associated with the fibroblast growth factor 2 (FGF2)/ERM pathway and this association helps us explore the capacity of SSCs to maintain a vertebrate stem cell niche.
Similar content being viewed by others
Introduction
Spermatogenesis is a complex and cyclic process involving spermatogonial stem cell (SSC) differentiation, meiotic cell division, and sperm production [1]. SSCs continuously maintain spermatogenesis via self-renewal to replenish themselves and differentiation that generates sperms [2]. However, it is unclear which molecules control the balance between SSC self-renewal and differentiation.
Sertoli cells are the only somatic cells in the seminiferous tubules and regulate the self-renewal, division and differentiation of SSCs into sperms [3]. Both undifferentiated and differentiating spermatogonia are found at the peripheries of seminiferous tubules, in contact with the basal membrane [4]. Testes undergo significant developmental and structural changes during puberty, including Sertoli cell proliferation and maturation and spermatogenesis onset [5]. The crosstalk between SSCs and Sertoli cells is crucial in spermatogenesis, but how it is controlled in SSCs is unclear [6].
SSCs produce most proteins essential for maintaining themselves, including promyelocytic leukemia zinc finger (PLZF), inhibitor of DNA binding 4 (ID4), and paired box protein 7 (PAX7) [7,8,9]. However, Sertoli cell proteins, namely, ETS-related molecule (ERM) or ETS variant transcription factor 5 and glial cell derived neurotrophic factor (GDNF), are also necessary for SSC maintenance [10,11,12]. ERM is a member of the ETS transcription factor family and expressed in several organs, including thyroid and brain [13, 14]. Mice with Erm targeted disruption show a loss of maintenance of SSC self-renewal, thus experience the progressive germ cell depletion and exhibit Sertoli cell-only (SCO) syndrome [1].
Fibroblast growth factor (FGF) signaling also regulate SSC self-renewal and male germline development. FGF2 is produced by Sertoli cells, Leydig cells and differentiating germ cells in the testes [2]. FGF2 is involved in spermatogonial proliferation [15], while FGF9 is a downstream target of the male sex determining gene Sry, and males with Fgf9 mutations show the male-to-female sex reversal phenotype [16, 17]. Many ETS transcription factors are regulated by FGF signaling in Sertoli cells [18].
Microtubule-associated serine/threonine kinase family member 4 (MAST4) of the protein kinase superfamily contains the following highly conserved domains: the domain of unknown function, the catalytic domain of the protein serine/threonine kinase, and the postsynaptic density protein 95, disks large MAGUK scaffold protein 1 (DLG1) and zona occludens 1 (ZO-1) (PDZ) domain [19]. Although MAST4 has been studied mainly in neurobiology [20, 21], fundamental role of MAST4 in stem cells during vertebrate development is unclear. In this study, Mast4 knockout (KO) mice exhibit the infertility with the reduced number of sperms. Therefore, we aim to examine MAST4 expression pattern in seminiferous tubules during testis development and to determine how MAST4 regulates spermatogenesis. Our results show that Mast4 KO mice exhibit the Sertoli cell-only morphology, which is similar to the phenotype of Erm KO mice. MAST4 positively regulates ERM function via direct MAST–ERM interaction. Specifically, MAST4 phosphorylates serine 367 residue of ERM. The transcription of ERM target genes is activated by either phosphorylated ERM or FGF2. Therefore, MAST4 controls SSC self-renewal in the FGF2/ERM pathway.
Materials and methods
All animal experiments were approved by Yonsei University Health System Institutional Animal Care and Use Committee in accordance with the Guide for the care and use of laboratory animal (National Research Council, USA). The animal study plan for these experiments (2017-0206) was reviewed and approved by this committee. And all experiments were performed in accordance with the guidelines of this committee.
Animals
Mice were housed in a temperature-controlled room (22 °C) under artificial illumination (lights on from 05:00 to 17:00) and 55% relative humidity, and they had ad libitum access to food and water. Mice from each developmental stage (E15.5, PN 1D, PN 1W, PN 3W, PN 6W, PN 13W, PN 12M and PN 20M) were used in this study. All the operational procedures were performed under deep anesthesia.
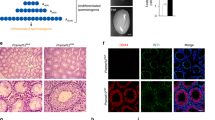
To generate Mast4 KO mice by CRISPR/Cas9-mediated gene targeting, we targeted exon 1 and exon 15 of Mast4 (RefSeq Accession number: 175171); 5′- GGAAACTCTGTCGGAGGAAG-3′ (exon 1) and 5′-GGCACAAAGAGTCCCGCCAG-3′ (exon 15). We then inserted each sequence into the pX330 plasmid, which carried both guide RNA and Cas9 expression units, from Dr. Feng Zhang (Addgene plasmid 42230) [22]. We named these vectors pX330-Mast4-E1 and pX330-Mast4-E15. The pregnant mare serum gonadotropin (5 units) and the human chorionic gonadotropin (5 units) were intraperitoneally injected into female C57BL/6J mice (Charles River Laboratories, Kanagawa, Japan) at 48 h interval, which were then mated with male C57BL/6J mice. The pX330-Mast4-E1 and pX330-Mast4-E15 (circular, 5 ng/μl each) were co-microinjected into 231 zygotes collected from oviducts of the mated female mice. The surviving 225 injected zygotes were transferred into oviducts in pseudopregnant ICR females, and 47 newborns were obtained. We collected genomic DNA from the tails of 31 founder mice that survived. To confirm the indel mutation induced by CRISPR/Cas9, we amplified the genomic region, including the target sites by polymerase chain reaction (PCR) with the primers for exon 1 target (Mast4-1 genotype F: 5′-GTAGGGACTCCACGCTCCAG-3′ and Mast4-1 genotype R: 5′-CCGGACCCTAGTCTCTTCG-3′) and for exon 15 target (Mast4-15 genotype F: 5′-GGGTTCTCTGCGAAAGTCAG-3′ and Mast4-15 genotype R: 5′-ATCCCTGTGTTCCGTTTCAG-3′). The PCR products were sequenced by using BigDye Terminator v3.1 Cycle Sequencing Kit (Thermo Fisher Scientific, MA, USA), Mast4-1 genotype F primer and Mast4-15 genotype F primer. In male founder #38, we found indel mutations in both exon 1 and exon 15 without pX330 random integration. To identify the indel sequence and whether indel mutations in exon 1 and exon 15 occurred on the same chromosome (cis manner), the founder #38 was mated with a wild-type female and the indel mutations in F1 were sequenced. We obtained 17 F1 newborns, and 12 of them carried 71 bp deletion (chr13:103,333,981-103,334,051: GRCm38/mm10) in exon 1 and 3 bp deletion (chr13:102,774,360-102,774,362) in exon 15 in a cis manner. The deletion in exon 1 produces a frameshift so that translation stops at exon 1 (Fig. 1A).
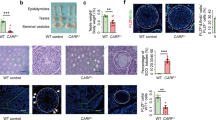
A Mast4 genomic structure is shown. MAST4 has highly conserved kinase domain and PDZ domain. To produce the Mast4 KO mice, 71 base pairs of Mast4 gene are deleted and a frameshift is produced on exon 1 so that translation stops. B At PN 6W, Mast4 KO mice has smaller testes compared to WT mice. C The total sperm number is significantly decreased in Mast4 KO testes (n = 4). D At PN 1D, the migrating germ cells are observed in the developing seminiferous tubules. E MAST4 is expressed in the cytoplasm of the developing Sertoli cells while SOX9 is expressed in the nuclei of the Sertoli cells (arrowhead). F Similarly, strong expression patterns of AMH are localized in the developing Sertoli cells with MAST4 co-localization (arrowhead). G DDX4 is expressed in the cytoplasm of the germ cell (arrowhead), which is located in the outermost layer of seminiferous tubules. H At PN 3 W, the testes have a developed structure with differentiating spermatogonia and Leydig cells. I The expression of MAST4 is shifted to Leydig cells while SOX9 is continuously expressed in Sertoli cells (arrowhead). J MAST4 is expressed in Leydig cells with co-localization of 3β-HSD (arrowhead), while K DDX4 is expressed in spermatogonia located throughout seminiferous tubules (arrowhead). L At PN 6W, WT testes have compact seminiferous tubules with all types of germ cells. M Mast4 KO testis has seminiferous tubules with depleted germ cells or SCO morphology (arrowhead). N MAST4 is expressed in Leydig cells and spermatids although SOX9 is expressed in the nuclei of Sertoli cells in WT testes (arrowhead). O In Mast4 KO testes, SOX9 is strongly expressed in Sertoli cells located at the outermost layer of the SCO tubules along with Sertoli cells in normal seminiferous tubule. P Expression of AMH is not detected in WT pubertal testes (arrow). Q In Mast4 KO testes, AMH is expressed in the SCO tubules (arrowhead). R DDX4 is expressed mainly in differentiating spermatogonia (arrowhead). S In Mast4 KO testes, DDX4 expression is not observed in the SCO tubules (arrow). Scale bars; D–H, L, M, 100 μm; I–K, N-S, 50 μm. SC; Sertoli cell, GC; Germ cell, LC; Leydig cell, SCO; Sertoli cell-only tubule, Sp; Spermatid. **p < 0.01.
Sperm count
To count sperm, both cauda epididymides from PN 6W mice were collected, dissected and placed in 2 ml of Hanks’ Balanced Salt Solution (HBSS; #14025-092, Life Technologies, USA) for 30 min to allow the release of motile cells (swim-out procedure). The total sperm number from suspension sperm was obtained using a hemocytometer.
Immunohistochemistry
Samples were fixed in 4% paraformaldehyde in phosphate buffered saline and then embedded in paraffin using standard procedures. Sections (7-μm thickness) of the specimens were boiled in 10 mM citrate buffer (pH 6.0) for 20 min and cooled at room temperature for 20 min. The specimens were incubated with anti-MAST4 (BS5791, Bioworld Technology, Inc., USA; 1:150), anti-SOX9 (14-9765-80, Invitrogen, OR, USA; 1:200), anti-AMH (SC-166752, Santa Cruz Biotechnology, Inc., USA; 1:100), anti-3β-HSD (SC-515120, Santa Cruz Biotechnology, Inc., USA; 1:100), anti-DDX4 (ab13840, Abcam, UK; 1:100), anti-GATA1 (SC-265, Santa Cruz Biotechnology, Inc., USA; 1:100), anti-ERM (ab102010, Abcam, UK; 1:200), anti-PLZF (SC-28319, Santa Cruz Biotechnology, Inc., USA; 1:100), anti-c-Kit (ab231780, Abcam, UK; 1:100), anti-PCNA (ab29, Abcam, UK; 1:200) antibodies at 4 °C overnight. The specimens were incubated with Alexa Fluor secondary antibodies (Invitrogen, OR, USA) for 2 h at room temperature and were counterstained with TO-PROTM-3 (T3605, Invitrogen, OR, USA; 1:1000) or DAPI (D1306, Invitrogen, OR, USA; 30 nM). For fluorescence detection in double staining of MAST4 and DDX4, TSA Plus Cy3/Fluorescein System (NEL753001KT, PerkinElmer, MA, USA) was used according to the manufacturer’s protocols. The sections were examined using a confocal laser microscope (DMi8, Leica, Germany or C1si, Nikon, Japan).
Section in situ hybridization
The sections were baked at 65 °C, de-waxed in xylene, rehydrated through a graded series of alcohol washes and post-fixed in 4% PFA. The sections were prehybridized in a humid chamber containing 50% formamide in 2 × SSC at 55 °C for 30 min. Digoxigenin (DIG)-labeled RNA probes were pre-warmed at 85 °C and hybridized to the sections overnight at 65 °C. At least ten specimens were examined in each stage. The primer sequences of the Mast4 probe are as follows:
Mast4-F: 5′-CAA AAG GCA AAG AGC CTG TC-3′; R: 5′-TGC GTC TGT GCA TTT CTT TC-3′.
RNA-sequencing analysis
Libraries were prepared for 150 bp paired-end sequences using a TruSeq Stranded mRNA Sample Preparation Kit (Illumina, CA, USA). Namely, mRNA molecules were purified and fragmented from 1 μg of total RNA using oligo (dT) magnetic beads. The fragmented mRNAs were synthesized as single-stranded cDNAs through random hexamer priming. By applying this as a template for second strand synthesis, double-stranded cDNA was prepared. After the sequential process of end repair, A-tailing and adapter ligation, cDNA libraries were amplified with PCR. The quality of these cDNA libraries was evaluated with the Agilent 2100 BioAnalyzer (Agilent, CA, USA), and they were quantified with the KAPA library quantification kit (Kapa Biosystems, MA, USA) according to the manufacturer’s library quantification protocol. Following cluster amplification of denatured templates, sequencing was progressed as paired-end (2 × 150 bp) using Illumina NovaSeq 6000 sequencer (Illumina, CA, USA). Low quality reads were filtered according to the following criteria: reads contain more than 10% skipped bases (marked as “N”s), reads contain more than 40% of bases whose quality scores are less than 20 and reads with an average quality score of less than 20. The whole filtering process was performed using the in-house scripts. Filtered reads were mapped to the reference genome related to the species using the aligner TopHat [23]. The gene expression level was measured with Cufflinks v2.1.1 [24] using the gene annotation database of the species. To improve the accuracy of the measurement, multi-read-correction and frag-bias-correct options were applied. All other options were set to default values.
Cell culture
TM4 (ATCC® CRL-1715™, USA) and Human Sertoli cell (HSerC; 10HU-149, iXCells Biotechnologies, USA) were cultured in DMEM/F-12 (#11320-033, Life Technologies, USA) supplemented with 2.5% fetal bovine serum and 5% horse serum or in Sertoli cell growth medium (MD-0091, iXCells Biotechnologies, USA), respectively, at 37 °C in a humidified atmosphere with 5% CO2.
LentiCRISPRv2 vector (#52961, Addgene, USA) was digested with BsmBI and ligated with annealed oligonucleotide targeting Mast4 exon 1, 5′-TACCCTGCCGCTGCCGCACC-3′ (LentiCRISPRv2-Mast4 Ex1). The vector without insertion was used as a control. To generate the lentivirus, HEK293T cells were transfected with LentiCRISPRv2-Mast4 Ex1 and packaging vectors (pVSVG and psPAX2) using fugene at 70% confluency. The viral supernatant was harvested at 48 h post-transfection, filtered through 0.45-μm filters and applied to TM4 and human Sertoli cells. The clonal cells were selected with puromycin (A11138-03, Life Technologies, USA) at 24 h post-transfection.
To manipulate FGF signaling, cells were cultured in media containing 50 ng/ml FGF2 for 3, 6, 12, 24 h and 10 μM SU5402 (SML0443, Sigma-Aldrich, USA) for 24 h. SU5402 was dissolved in dimethyl sulfoxide vehicle (DMSO; D2650, Sigma-Aldrich, USA).
Reverse transcription-polymerase chain reaction (RT-PCR) and real time-quantitative polymerase chain reaction (RT-qPCR)
For the RT-PCR, the total RNA was prepared using EasyBlue (Boca Scientific). 2 μg of RNA was reverse-transcribed using M-MLV Reverse Transcriptase (Promega), according to the manufacturer’s instructions. The expression levels of each gene are expressed as normalized ratios against the Gapdh housekeeping gene. The oligonucleotide RT-PCR primers for Mast4, Erm, Cxcl5, Cxcl12, Ccl12, Gapdh are as follows:
Mouse—Mast4-F: 5′- CAA AAG GCA AAG AGC CTG TC-3′; R: 5′- TGC GTC TGT GCA TTT CTT TC-3′; Erm-F: 5′-CCG AGT TGT CGT CCT GTA G-3′; R: 5′-ACT GGC TTT CAG GCA TCA TC-3′; Cxcl5-F: 5′-GAA AGC TAA GCG GAA TGC AC-3′; R: 5′-GGT CCC CAT TTC ATG AGA GA-3′; Cxcl12-F: 5′-TTT CAC TCT CGG TCC ACC TC-3′; R: 5′-TAA TTT CGG GTC AAT GCA CA-3′; Ccl12-F: 5′-TCC TCA GGT ATT GGC TGG AC-3′; R: 5′-GGG AAC TTC AGG GGG AAA TA-3′; Gapdh-F: 5′-ACT CCA CTC ACG GCA AAT TC-3′; R: 5′-CCT TCC ACA ATG CCA AAG TT-3′.
Human – MAST4-F: 5′-CCA GAC GAT ATG GTG CAC TG-3′; R: 5′-GGG AGA GCA AGA TGA AGC TG-3′; ERM-F: 5′-TAC CAT CGG CAA ATG TCA GA-3′; R: 5′-TGG CTG CTG GAG AAA TAA CC-3′.
For the RT-qPCR, the total RNA of the cells was extracted using TRIzol® reagent (#15596-026, Thermo Fisher Scientific, USA). The extracts were reverse transcribed using Maxime RT PreMix (#25081, iNtRON, Korea). RT-qPCR was performed using a StepOnePlus Real-Time PCR System (Applied BioSystems, USA). The amplification program consisted of 40 cycles of denaturation at 95 °C for 15 s and annealing at 61 °C for 30 s. The expression levels of each gene are expressed as normalized ratios against the B2m housekeeping gene. The oligonucleotide RT-qPCR primers for Mast4, Erm, B2m are as follows:
Mouse—Mast4-F: 5′-AGG AAG TCC CGC ATA CCA GG-3′; R: 5′-TCC CAC TCT TCA GCA GGA GC-3′, Erm-F: 5′-GCA GGA GGC TTG GTT AGC TG-3′; R: 5′-CGT GGC TAC AGG ACG ACA AC-3′, B2m-F: 5′-CCT GGT CTT TCT GGT GCT TG-3′; R: 5′-CCG TTC TTC AGC ATT TGG AT-3′.
Human—ERM-F: 5′-TTA CCA GAG GCG AGG TTC CC-3′; R: 5′-ATA GTT CAT GGC TGG CCG GT-3′, B2M-F: 5′-GCC GTG TGA ACC ATG TGA CT-3′; R: 5′-GCT TAC ATG TCT CGA TCC CAC TT-3′.
Nuclear/cytosol fractionation and Western blot
Nuclear and cytoplasmic fractions of TM4 cells were prepared using the NE-PER Nuclear and Cytoplasmic Extraction reagents (Thermo Scientific), following the manufacturer’s protocols. Cell extracts were fractionated by SDS-PAGE transferred to a polyvinylidene difluoride membrane using a transfer apparatus according to the manufacturer’s protocols (Bio-Rad). After incubation with 5% skim milk in TBST (10 mM Tris, pH 7.4, 150 mM NaCl, 0.1% Tween 20) for 60 min, the membrane was incubated with antibodies against anti-MAST4 (PA5-36976; ThermoFisher; 1:500), anti-ERM (ab102010, Abcam, UK; 1:1000), anti-Lamin B (SC-374015, Santa Cruz Biotechnology, Inc., USA; 1:1000) and anti-α-tubulin (T5168, Sigma-Aldrich; 1:3000) at 4 °C overnight. Membranes were washed six times for 10 min and incubated with HRP-conjugated secondary antibodies for 2 h. Blots were washed six times with TBST and developed with the ECL system (RPN2232, GE Healthcare Life Sciences, USA) according to the manufacturer’s protocols.
Immunoprecipitation
Cells were lysed in a RIPA buffer containing a protease inhibitor cocktail (cOmplete™; #11697498001, Roche, IN, USA). Cell extracts were incubated with the indicated primary antibodies overnight at 4 °C. Antibody-bound proteins were precipitated with Dynabeads™ Protein G (Invitrogen). Samples were separated by SDS-PAGE, followed by electrotransfer to polyvinylidene difluoride membranes (PVDF; Millipore). The membrane was blocked for 1 h at room temperature and incubated overnight at 4 °C with the primary antibody. The primary antibodies used were as follows: Glutathione Sepharose 4B for GST pull-down (17-0756-01; GE Healthcare), anti-GST (sc-138, Santa Cruz; 1:1000), anti-HA (sc-805, Santa Cruz; 1:1000), anti-phosphoserine (P5747, Sigma-Aldrich; 1:1000), anti-β-actin (A5441, Sigma-Aldrich; 1:4000), and anti-Flag (F3165, Sigma-Aldrich; 1:4000). Horseradish peroxidase-conjugated antibodies (Millipore) were used as secondary antibodies. The peroxidase reaction products were visualized with WEST-ZOL (iNtRON, Korea). All signals were detected by Amersham Imager 600 (GE Healthcare Life Sciences, USA).
Luciferase assay
The mouse Cxcl5 (-1127~+122) and Cxcl12 (-1580~+89) promoter were amplified by PCR from the genomic DNA of TM4 cells and isolated by a HiYieldTM Genomic DNA Mini Kit (Real Genomics). For Cxcl5 promoter-F: 5′- GAT CAC GCG TTA AGT CCC ACG GAT GAG TCC -3′; R: 5′- GAT CCT CGA GGA GCA CCA GCT CGG GAT A -3′ and Cxcl12 promoter-F: 5′- GAT CAC GCG TGC GCT TGA TCT CGG ATT ACT -3′; R: 5′- GAT CCT CGA GGA GCT GGA CAG CAA GAG GAC -3′. The amplified PCR fragment was cloned into the MluI and XhoI sites of the pGL3 basic vector (Promega). TM4 cells were transfected with Cxcl5- and Cxcl12-promoter luciferase report plasmids and HA-MAST4 PDZ using polyethylenimine (Polysciences; #24765). Cells were treated with FGF-2 (50 ng/ml for 18 h) (Peprotech). The luciferase activities were analyzed using the Luciferase Assay System Kit (Promega) according to the manufacturer’s protocol. All assays were carried out in triplicate, and all values were normalized for transfection efficiency against β-galactosidase activities.
TUNEL staining
A terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay was performed using an in situ cell apoptosis detection kit (#4810-30-K, Trevigen, Inc., USA) according to the manufacturer’s instructions. The 7-μm thick sections were treated with 20 μg/ml proteinase K in 10 mM Tris–HCl, pH 8.0, for 25 min at room temperature. The samples were incubated with the labeling reaction mixture at 37 °C for 1 h and HRP-streptavidin solution for 5 min at room temperature. A 3, 3′-diaminobenzidine was used as a substrate solution to detect the sites of in situ apoptosis. At least five specimens were examined in each experiment.
Statistical analysis
The graphic results were expressed as the mean ± standard deviation. A GraphPad Prism 7 (GraphPad Software, San Diego, CA, USA) was used to analyze the data. Comparison of two groups was performed using an unpaired two-tailed t-test. Comparison of multiple groups was performed by two-way ANOVA followed by Tukey’s multiple comparisons test. A P value < 0.05 was considered significant.
Results
Dynamic MAST4 expression pattern during spermatogenesis
Undefined reproduction failure has been monitored with the necessary efforts previously in Mast4 KO mice and intensive studies for spermatogenesis are carried out in detail. In this study, Mast4 KO testes showed reduced in size (Fig. 1B) and the total sperm number was dramatically decreased compared to wild-type (WT) testes (Fig. 1C), indicating a characteristic of infertility. To identify the role of MAST4, the temporal and spatial expression pattern of MAST4 was examined in WT testes. At postnatal day 1 (PN 1D) when the developing germ cells are migrating in the developing seminiferous tubules (Fig. 1D), MAST4 was expressed in the cytoplasm of SRY-box transcription factor 9 (SOX9)- and anti-Müllerian hormone (AMH)-expressing Sertoli cells (Fig. 1E, F). On the other hand, MAST4 was not expressed in DEAD-box helicase 4 (DDX4)-expressing gonocytes (Fig. 1G). In addition, at embryonic day 15.5 (E15.5), when the developing and disorganized testicular structures are observed, and at postnatal week 1 (PN 1W), when the more germ cells migrate to the outermost layer of seminiferous tubules, MAST4 was expressed in the cytoplasm in SOX9- and AMH-positive Sertoli cells but not expressed in DDX4-positive germ cells (Supplementary Fig. S1A–H). At PN 3W, when testicular structures comprising differentiating spermatogonia and Leydig cells are fully developed (Fig. 1H), MAST4 expression shifted to Leydig cells being co-localized with 3β-hydroxysteroid dehydrogenase (3β-HSD), although SOX9 was continuously expressed in Sertoli cells (Fig. 1I, J). MAST4 did not show co-localization with DDX4, which was expressed in spermatogonia located throughout the seminiferous tubules (Fig. 1K). At PN 13W, when the adult testicular structures remain mature (Supplementary Fig. S1I), MAST4 was expressed in pubertal elongated spermatids, spermatozoa and Leydig cells. MAST4-positive cells were co-localized with 3β-HSD-expressing Leydig cells but not with DDX4-expressing spermatogonia and spermatocytes (Supplementary Fig. S1J–L). Supplementary Table S1 explains the dynamic co-expression pattern of MAST4 with previously known genes in detail. MAST4 is co-expressed with AMH in the cytoplasm of Sertoli cells from E15.5 to PN 1 W, and with 3β-HSD in Leydig cells from PN 3W to PN 13W, demonstrating that MAST4 expression shifts from Sertoli cells to Leydig cells at PN 3W (Supplementary Table S1).
Mast4 RNA localization was also examined from PN 1D to PN 6W using in situ hybridization. At PN 1D, Mast4 expression was observed at Sertoli cells located in the outermost layer of seminiferous tubules (Supplementary Fig. S2A, B arrowhead). At PN 3 W, Mast4 was expressed in Leydig cells (Supplementary Fig. S2C, D arrowhead) and spermatogonia (Supplementary Fig. S2C, D arrow). At PN 6W, Mast4 was localized in Leydig cells (Supplementary Fig. S2E, F black arrowhead), spermatogonia (Supplementary Fig. S2F black arrow) and spermatids (Supplementary Fig. S2F white arrowhead). Mast4 RNA was expressed in the spermatogonia at PN 3W and PN 6W, and the Mast4 RNA expression pattern was partially different from its protein expression pattern. However the results supported MAST4 expression pattern mostly at testicular developmental stages.
Irregular seminiferous tubule structures in Mast4 KO mice
To investigate the spermatogenic dysfunction in Mast4 KO testes, the various testis markers were analyzed in PN 6W WT and Mast4 KO mice. WT testes had compact seminiferous tubules with all types of germ cells (Fig. 1L), while Mast4 KO testes had several seminiferous tubules with depleted germ cells or only Sertoli cells, the characteristic SCO phenotype. In addition, relatively wider interstitial space and enlarged seminiferous tubule lumen were observed because of degenerated germ cells in Mast4 KO compared to WT testes (Fig. 1M). SOX9 was expressed in the nuclei of the Sertoli cells in WT mice (Fig. 1N). The number of SOX9-expressing Sertoli cells was increased in the SCO tubules of Mast4 KO mice (Fig. 1O). AMH, an immature Sertoli cell marker, was not detected in WT pubertal testes (Fig. 1P). However, AMH was expressed in the SCO tubules and several normal seminiferous tubules of Mast4 KO testes (Fig. 1Q). DDX4, which was expressed in the spermatogonia of WT testes (Fig. 1R), was not observed in the SCO tubules although DDX4 was expressed in the normal seminiferous tubules in Mast4 KO mice (Fig. 1S). Gata-binding factor 1 (GATA1), a mature Sertoli cell marker, was expressed in Sertoli cells in WT mice, similar to SOX9 (Supplementary Fig. S3A). The number of GATA1-expressing cells was also increased in the SCO tubules of Mast4 KO testes (Supplementary Fig. S3B). RNA-sequencing (RNA-seq) analysis showed that the expression of Sertoli cell markers, such as Amhr2, Gata4, Sox9 and Amh, was increased in the testes of Mast4 KO mice (Supplementary Fig. S4A). Taken together, disrupted development of seminiferous tubules as SCO morphology is observed with increased expression of Sertoli cell markers in Mast4 KO testes.
Synchronous relationship between MAST4 and ERM
Since the SCO phenotype, which is associated with infertility, was observed in Mast4 KO testes, similarly to Erm KO testes and FGF signaling regulates SSC self-renewal through ERM [1, 18, 25], the effects of FGF2 on the relationship between Mast4 and Erm expression was investigated in TM4 cells (mouse Sertoli cells). In the presence of FGF2 (50 ng/ml), RNA expression of Mast4 and Erm was increased (Fig. 2A). In addition, treatment with SU5402 (10 μM), a fibroblast growth factor receptor (FGFR) inhibitor, significantly decreased the expression of Mast4 and Erm (Fig. 2B, C), indicating that FGF2 regulates both Mast4 and Erm in Sertoli cells.
A In the presence of FGF2, expression of Mast4 and Erm is increased in TM4 cells. B After inhibition of the effect of FGF signaling with SU5402, Mast4 expression is significantly decreased (n = 3). C Erm expression is also decreased similarly to Mast4 (n = 3). D In PN 6 W WT, ERM is expressed in Sertoli cells (arrowheads) along with several elongated spermatids. E In PN 6 W Mast4 KO testes, ERM expression pattern is decreased in normal seminiferous tubules (arrowhead) and rarely observed in the SCO tubules (arrow). F In TM4 cell, Erm expression is slightly decreased by MAST4 depletion (n = 3). G) Erm expression is decreased in MAST4-depleted human Sertoli cells (n = 3). H TM4 cells were treated with FGF2 for 6 h. Nuclear/cytoplasmic fractions were analyzed by western blot. Note that both basal and FGF2-induced ERM expression are decreased in MAST4-depleted TM4 cells. Scale bars; 100 μm. **p < 0.01.
Next, ERM expression pattern was also examined in WT and Mast4 KO mice at PN 6W to identify whether MAST4 had any effect on ERM. ERM was expressed in Sertoli cells in WT mice, which was co-localized with SOX9-positive cells, along with several elongated spermatids (Fig. 2D). In contrast, ERM expression was decreased in Mast4 KO mice and rarely observed even in the SCO tubules (Fig. 2E). The decreased Erm expression was also validated at the RNA level in Mast4 KO TM4 cells (Fig. 2F) and MAST4 KO human Sertoli cells (Fig. 2G). The western blots of WT and Mast4 KO TM4 cells after nuclear/cytosol fractionation showed decreased ERM expression in the nucleus of Mast4 KO cells but no change in the cytosol of the cells (Fig. 2H). Taken together, the presence of FGF2 or the absence of MAST4 affects ERM expression.
MAST4–ERM interaction regulates SSC self-renewal-related genes
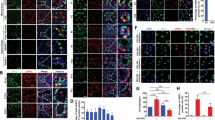
To investigate molecular relationship between MAST4 and ERM, we performed immunoprecipitation (IP) assay and observed the interaction between MAST4 and ERM (Fig. 3A). Considering that MAST4 is a serine/threonine protein kinase and that ERM transactivation is regulated by phosphorylation [26], we further examined whether MAST4 induced ERM phosphorylation. Specifically, IP assay demonstrated that FGF2 increased p-serine of ERM. In fact, MAST4 overexpression alone increased ERM serine phosphorylation, whose level was similar to that induced by FGF2 (Fig. 3B). Since ERM is a transcription factor, its transcriptional activity might be affected. When ERM target genes were examined through luciferase assay, both basal and FGF2-induced transcription of Cxcl5 and Cxcl12 were decreased by MAST4 depletion (Fig. 3C, D). On the other hand, MAST4 PDZ overexpression increased the transcription of Cxcl5 and Cxcl12, even in the absence of FGF2 stimulation (Fig. 3E, F). In the same context, the RNA expression level of Erm, Cxcl5, and Cxcl12 was decreased in Mast4 KO TM4 cell (Fig. 3G) and it was significantly increased with MAST4 PDZ overexpression (Fig. 3H).
A Interaction between MAST4 and ERM is examined. GST-hERM and HA-hMAST4 PDZ were transiently co-transfected into TM4 cells, followed by GST pull-down. Note that MAST4 interacts with ERM. B GST-hERM and HA-hMAST4 PDZ were transiently co-transfected into TM4 cells, followed by FGF2 treatment for 6 h, as indicated. GST-ERM was immunoprecipitated, and the complexes were analyzed by western blot. The bands, which were recognized by p-serine antibody, were later reprobed with GST antibody. Note that ERM serine phosphorylation is increased in the presence of MAST4. C Cxcl5- and D Cxcl12-luciferase reporter and β-gal were co-transfected into the WT and MAST4-depleted TM4 cells. TM4 cells were then treated with 50 ng/ml of FGF2 for 18 h. Note that both basal level and FGF2-induced increase of C Cxcl5 and D Cxcl12 transcription were decreased by Mast4 depletion (n = 3). E Cxcl5- and F Cxcl12-luciferase reporter and β-gal were co-transfected with or without Flag-ERM and HA-hMAST4 PDZ into the WT TM4 cells for 24 h. Both basal level and ERM-induced increase of E Cxcl5 and F Cxcl12 transcription were increased by MAST4 PDZ overexpression (n = 3). The mRNA expression of Erm, Cxcl5 and Cxcl12 is G decreased in MAST4-depleted TM4 cells, but (H) increased in TM4 cells transiently overexpressed with HA-MAST4 PDZ. Luciferase assay was normalized with β-gal activity. *p < 0.05, **p < 0.01, ns; non-significance.
Specifically, we identified ERM serine sites which might be phosphorylated by MAST4 and involved in MAST4-induced ERM regulation and subsequent target gene expression. After mutating the well-known serine residues of ERM to alanine, we checked mRNA expression level of ERM target genes, which was induced by transient overexpression of ERM mutants in the presence of MAST4. Interestingly, ERM target genes were not induced when ERM serine 367 residue was substituted to alanine (ERM S367A) (Fig. 4A). To determine whether ERM serine 367 residue participated in MAST4-induced ERM phosphorylation, we performed IP assay to examine the interaction between MAST4 and ERM or ERM S367A mutant and the level of ERM S367A mutant serine phosphorylation. Compared to ERM WT, both MAST4-ERM interaction and MAST4-induced ERM serine phosphorylation level were decreased by S367A mutation (Fig. 4B). On the basis of a previous study reporting how ERM S367A mutation decreased the ERM-mediated transcription [26], we examined the transcription of ERM target genes induced by ERM WT or S367A mutant through luciferase assay. Compared to ERM WT, ERM S367A mutation slightly decreased both basal level and MAST4-induced increase of Cxcl5 and Cxcl12 transcription (Fig. 4C, D). These results suggest that MAST4 interacts with ERM and induces serine 367 residue phosphorylation, eventually regulating ERM target genes related to SSC self-renewal.
A The mRNA expression of Mast4, Erm, Cxcl5, Cxcl12 and Ccl12 was examined using TM4 cells transfected with the WT and various phosphorylation inactive mutants of ERM for 24 h in the presence of HA-hMAST4 PDZ. Note that ERM S367A mutant decreased mRNA expression of Cxcl5, Cxcl12 and Ccl12. B HA-hMAST4 PDZ, Flag-hERM WT, and Flag-hERM S367A were transiently co-transfected into TM4 cells, followed by FGF2 treatment for 6 h, as indicated. Flag-ERM was immunoprecipitated, and the complexes were analyzed by western blot. The bands, which were recognized by p-serine antibody, were later reprobed with Flag antibody. C Cxcl5- and D) Cxcl12-luciferase reporter and β-gal were co-transfected with Flag-ERM WT or Flag-ERM S367A into the WT TM4 cells for 24 h in the absence or presence of HA-hMAST4 PDZ. Note that both basal level and MAST4-induced increase of Cxcl5 and Cxcl12 transcriptional activity were slightly decreased by substitution of serine 367 of ERM by alanine (n = 3). Luciferase assay was normalized with β-gal activity. *p < 0.05, **p < 0.01, ns; non-significance.
MAST4-induced regulation of spermatogonial stem cell self-renewal
The mechanisms by which Mast4 KO leads to SSC self-renewal were examined in PN 6W WT and Mast4 KO testes. PLZF, a marker for undifferentiated spermatogonia, was expressed in the spermatogonia sparsely in the seminiferous tubules of WT mice (Fig. 5A arrowheads). However, PLZF expression was significantly decreased in both the SCO tubules and normal seminiferous tubules of Mast4 KO mice (Fig. 5B). c-Kit was observed in 8–16 or more connected differentiating spermatogonia located in the outermost layer of the seminiferous tubules of WT mice (Fig. 5C), while c-Kit expression was decreased in the outermost layer of the seminiferous tubules in Mast4 KO mice. Interestingly, the c-Kit-positive spermatogonia in Mast4 KO mice remained scattered without being merged together, compared to WT mice (Fig. 5D). In addition, proliferating cell nuclear antigen (PCNA) was expressed mostly in the outermost layer of the seminiferous tubules of WT mice (Fig. 5E). However, PCNA was not expressed in the SCO tubules of Mast4 KO mice, although its expression remained similar to that in the normal seminiferous tubules (Fig. 5F). Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) staining detected a few apoptotic cells were detected in several spermatogonia of the seminiferous tubule in WT mice (Fig. 5G arrowhead) and a remarkably number of apoptotic cells in the outermost layer of the seminiferous tubules of Mast4 KO mice (Fig. 5H arrowheads). Table 1 summarizes a comparison of the expression patterns of several markers used in this study between WT and Mast4 KO mice. These results were also supported by RNA-Seq analysis of WT and Mast4 KO testes at PN 6W. Among the undifferentiated spermatogonia markers, Id4 and Gfra1 were not significant in Mast4 KO mice compared to WT mice. Bcl6b and Zbtb16 (also known as Plzf), were decreased in Mast4 KO mice (Supplementary Fig. S4B). These results indicate that spermatogonia and proliferating cell marker expression is decreased and apoptotic cell marker expression is increased in Mast4 KO testes, demonstrating that the MAST4 affects not only SSC self-renewal but also spermatogonial differentiation, sperm production, and cell coordination [27].
A PLZF is expressed in undifferentiated spermatogonia sparsely located at the outermost layer of the seminiferous tubules (arrowheads). B In Mast4 KO testes, PLZF expression is slightly decreased in normal seminiferous tubules, although its expression is not observed in the SCO tubules. C Expression of c-Kit is observed in differentiating spermatogonia located mainly in the outermost layer of seminiferous tubules. D In Mast4 KO testes, c-Kit expression is decreased in the outermost layer of seminiferous tubules. E PCNA is expressed in the overall outermost layer of seminiferous tubules (arrowhead). F In Mast4 KO testes, PCNA is not expressed in the SCO tubules (arrow), although its expression remained similar to normal seminiferous tubules (arrowhead). G TUNEL staining detects a few apoptotic cells in spermatogonia in WT testes (arrowhead). H In Mast4 KO testes, apoptotic cells are increased in the outermost layer of seminiferous tubules (arrowheads). Scale bars; 100 μm. Spg; Spermatogonia.
Discussion
Infertility is a fundamentally important issue in reproductive biology and the mechanisms underlying the spermatogenesis signaling network is main key to understand the infertility.
MAST4 and its assoiciated testis markers are harmoniously co-expressed where the functional intensity is required during spermatogenesis. In this study, MAST4 was localized in seminiferous epithelia and Leydig cells stage-dependently. From E15.5 to PN 1W, MAST4 was localized in Sertoli cells and co-expressed with SOX9 and AMH. SOX9 plays a crucial role in testis development and adult testis maintenance [28, 29]. AMH is one of the first genes to be switched on in Sertoli cells during fetal development in both mice and humans, and is expressed continuously as long as Sertoli cells are immature [30, 31]. However, MAST4 was not co-expressed with DDX4. DDX4 is expressed in the cytoplasm of spermatogonia at the pre-pubertal stage and continuously expressed in spermatocytes and round spermatids at the post-pubertal stage [32]. DDX4 is essential for the regulation of germ cell proliferation and differentiation [33]. After PN 3W, MAST4 expression shifts to spermatids and Leydig cells, which produce testosterone. Overall, MAST4 controls the functional activities of Sertoli cells before the pubertal stage, which regulates SSC self-renewal acting as a pivotal role in the stem cell niche. After puberty, MAST4 regulates testis maturation, indicating that MAST4 plays an indispensable role in securing the spermatogenesis.
Mast4 KO testes show to undergo germ cell depletion and finally attain the SCO morphology. PLZF is critical for maintaining SSCs in a low proliferative state and regulates their cell cycle status [34]. On the basis of the decreased expression of PLZF and c-Kit in the present study, it can be suggested that MAST4 depletion lowers the self-maintenance capacity of SSCs and blocks spermatogonial differentiation. In humans and monkeys suffering from SCO syndrome, undifferentiated spermatogonia markers are decreased in a similar pattern as RNA-Seq analysis results in this study [35, 36]. In addition, the total sperm number is significantly decreased in Mast4 KO mice, indicating that Mast4 KO mice also exhibit spermatogenic dysfunction. Although the SCO tubules without germ cells are markedly atrophied, several normal seminiferous tubules show active spermatogenesis and produce sperms, because of the expression of several genes, including DDX4 and PCNA. On the Basis of these results, MAST4 is closely associated with spermatogenesis and could regulate the functions of Sertoli cells in SSC self-renewal, Leydig cells in testosterone production and spermatids which give rise to sperms.
MAST4 is involved in the FGF2 signaling pathway. FGFs induce many kinases, including mitogen-activated protein kinase/extracellular signal-regulated kinase (MAPK/ERK) and phosphatidylinositol 3-kinase/protein kinase B (PI3K/AKT), and are involved in activating their signaling pathways [37]. Currently, FGF2 treatment increases Mast4 and Erm expression, while FGF2 inhibition decreases their expression level. MAST4 binds to ERM and positively regulates its transcriptional activity during induction of the target genes related to SSC self-renewal. However, ERM expression is significantly decreased in all SCO tubules of Mast4 KO testes, suggesting that SSC self-renewal is inhibited. In addition, the transcription and mRNA expression of ERM target genes (Cxcl5 and Cxcl12) are downregulated in Mast4 KO cells. It has been reported that FGF2 activates ERM expression [18] and our results show that FGF2 might induce ERM through MAST4 to promote SSC self-renewal.
The relationship between Mast4 KO and aging was also examined because germ cells were depleted and the SCO morphology was induced in aged mice testes (Supplementary Fig. S5A–F) as well as in Mast4 KO adult mice testes. However, the expression patterns of the key molecules of spermatogenesis are different between Mast4 KO and aged mice testes. MAST4 shows gradually fainted expression as aging progresses (Supplementary Fig. S5G-I). Likewise, the expression of SOX9, GATA1 and ERM is gradually decreased and is not observed in testes at postnatal 20 months (PN 20M) (Supplementary Fig. S5J–R). This is clearly in contrast to the increased expression of SOX9 and GATA1 in the SCO tubules of Mast4 KO testes, although ERM expression is consistent with Mast4 KO testes. Taken together, despite sharing a common point of the SCO phenotype with Mast4 KO testes, aged mice is thought to have unraveled reasons rather than a failure of SSC self-renewal.
In human SCO syndrome, aberrant gene expression by Sertoli cells might decrease their niche function and contribute to SSC deficiency and dysfunction in infertile men [35]. Furthermore, Sertoli cells express abnormally low levels of GDNF, FGF8 and bone morphogenic protein 4 (BMP4), which are growth factors well characterized as regulators of SSCs and progenitors of spermatogonia in mice [38,39,40]. SCO syndrome is a complex phenomenon associated with multiple deficiencies in Sertoli cell gene expression. We propose the necessity of further studies to gain insights into the accurate understanding of the function of Sertoli cells in SCO syndrome. The development of advanced therapies could put forward for maintaining active SSCs during SCO syndrome progress.
In summary, the current study has showed that MAST4 is tightly related to spermatogenesis (Fig. 6). In the first place, FGF signaling plays a central role in regulating SSC self-renewal during spermatogenesis. Especially, the ERM transcription factor mediates the FGF2 signaling pathway. In the second place, MAST4 phosphorylates ERM and regulates ERM transcriptional activity. The transcription of ERM target genes (Cxcl5, Cxcl12, Ccl12, Ccl7, and Bcl6b) coordinates SSC self-renewal [1, 18, 25]. To sum it up, Mast4 KO mice undergo a drastic decrease in germ cells caused by the failure of SSC self-renewal through the FGF2/ERM pathway.
The FGF signaling pathway regulates SSC self-renewal during spermatogenesis and ERM transcription factor mediates the FGF2 signaling pathway. In addition, MAST4 interacts with ERM directly and regulates its transcriptional activities. ERM activates the transcription of its target genes and coordinates spermatogonial stem cell self-renewal. Mast4 KO testes show a loss of maintenance of SSC self-renewal and thus present progressive germ cell depletion and an SCO morphology, indicating that MAST4 is involved in the FGF2/ERM pathway in SSC self-renewal.
References
Chen C, Ouyang W, Grigura V, Zhou Q, Carnes K, Lim H, et al. ERM is required for transcriptional control of the spermatogonial stem cell niche. Nature. 2005;436:1030–4.
Chen S-R, Liu Y-X. Regulation of spermatogonial stem cell self-renewal and spermatocyte meiosis by Sertoli cell signaling. Reproduction. 2015;149:R159–67.
Pui HP, Saga Y. Gonocytes-to-spermatogonia transition initiates prior to birth in murine testes and it requires FGF signaling. Mech Dev. 2017;144:125–39.
Niedenberger BA, Busada JT, Geyer CB. Marker expression reveals heterogeneity of spermatogonia in the neonatal mouse testis. Reproduction. 2015;149:329.
Guo J, Nie X, Giebler M, Mlcochova H, Wang Y, Grow EJ, et al. The dynamic transcriptional cell atlas of testis development during human puberty. Cell Stem Cell. 2020;26:262–76. e264.
Sharma M, Braun RE. Cyclical expression of GDNF is required for spermatogonial stem cell homeostasis. Development. 2018;145:dev151555.
Lovelace DL, Gao Z, Mutoji K, Song YC, Ruan J, Hermann BP. The regulatory repertoire of PLZF and SALL4 in undifferentiated spermatogonia. Development. 2016;143:1893–906.
Sun F, Xu Q, Zhao D, Chen CD. Id4 marks spermatogonial stem cells in the mouse testis. Sci Rep. 2015;5:17594.
Aloisio GM, Nakada Y, Saatcioglu HD, Peña CG, Baker MD, Tarnawa ED, et al. PAX7 expression defines germline stem cells in the adult testis. J Clin Investig. 2014;124:3929–44.
Spinnler K, Köhn F, Schwarzer U, Mayerhofer A. Glial cell line-derived neurotrophic factor is constitutively produced by human testicular peritubular cells and may contribute to the spermatogonial stem cell niche in man. Hum Reprod. 2010;25:2181–7.
Chen L-Y, Willis WD, Eddy EM. Targeting the Gdnf Gene in peritubular myoid cells disrupts undifferentiated spermatogonial cell development. Proc Natl Acad Sci. 2016;113:1829–34.
Sada A, Hasegawa K, Pin PH, Saga Y. NANOS2 acts downstream of glial cell line‐derived neurotrophic factor signaling to suppress differentiation of spermatogonial stem cells. Stem Cells. 2012;30:280–91.
Puli OR, Danysh BP, McBeath E, Sinha DK, Hoang NM, Powell RT, et al. The transcription factor ETV5 mediates BRAFV600E-induced proliferation and TWIST1 expression in papillary thyroid cancer cells. Neoplasia. 2018;20:1121–34.
Fontanet PA, Ríos AS, Alsina FC, Paratcha G, Ledda F. Pea3 transcription factors, Etv4 and Etv5, are required for proper hippocampal dendrite development and plasticity. Cereb Cortex. 2018;28:236–49.
Zhang Y, Wang S, Wang X, Liao S, Wu Y, Han C. Endogenously produced FGF2 is essential for the survival and proliferation of cultured mouse spermatogonial stem cells. Cell Res. 2012;22:773–6.
Gustin SE, Stringer JM, Hogg K, Sinclair AH, Western PS. FGF9, activin and TGFbeta promote testicular characteristics in an XX gonad organ culture model. Reproduction. 2016;152:529–43.
Yu M, Wang J, Liu W, Qin J, Zhou Q, Wang Y, et al. Effects of tamoxifen on the sex determination gene and the activation of sex reversal in the developing gonad of mice. Toxicology. 2014;321:89–95.
Ishii K, Kanatsu-Shinohara M, Toyokuni S, Shinohara T. FGF2 mediates mouse spermatogonial stem cell self-renewal via upregulation of Etv5 and Bcl6b through MAP2K1 activation. Development. 2012;139:1734–43.
Sun L, Gu S, Li X, Sun Y, Zheng D, Yu K, et al. Identification of a novel human MAST4 gene, a new member of human microtubule associated serine/threonine kinase family. Mol Biol. 2006;40:724–31.
Garland P, Quraishe S, French P, O’Connor V. Expression of the MAST family of serine/threonine kinases. Brain Res. 2008;1195:12–19.
Gongol B, Marin TL, Jeppson JD, Mayagoitia K, Shin S, Sanchez N, et al. Cellular hormetic response to 27-hydroxycholesterol promotes neuroprotection through AICD induction of MAST4 abundance and kinase activity. Sci Rep. 2017;7:1–11.
Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–23.
Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–11.
Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, Van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511.
Yoon KA, Chae YM, Cho JY. FGF2 stimulates SDF‐1 expression through the Erm transcription factor in Sertoli cells. J Cell Physiol. 2009;220:245–56.
Baert J-L, Beaudoin C, Coutte L, De Launoit Y. ERM transactivation is up-regulated by the repression of DNA binding after the PKA phosphorylation of a consensus site at the edge of the ETS domain. J Biol Chem. 2002;277:1002–12.
De Rooij DG, Griswold MD. Questions about spermatogonia posed and answered since 2000. J Androl. 2012;33:1085–95.
Lavery R, Lardenois A, Ranc-Jianmotamedi F, Pauper E, Gregoire EP, Vigier C, et al. XY Sox9 embryonic loss-of-function mouse mutants show complete sex reversal and produce partially fertile XY oocytes. Dev Biol. 2011;354:111–22.
Rahmoun M, Lavery R, Laurent-Chaballier S, Bellora N, Philip GK, Rossitto M, et al. In mammalian foetal testes, SOX9 regulates expression of its target genes by binding to genomic regions with conserved signatures. Nucleic acids Res. 2017;45:7191–211.
Rey RA, Grinspon RP. Normal male sexual differentiation and aetiology of disorders of sex development. Best Pract Res Clin Endocrinol Metab. 2011;25:221–38.
Sharpe RM, McKinnell C, Kivlin C, Fisher JS. Proliferation and functional maturation of Sertoli cells, and their relevance to disorders of testis function in adulthood. Reproduction. 2003;125:769–84.
Kim J, Jung H, Yoon M. VASA (DDX4) is a putative marker for spermatogonia, spermatocytes and round spermatids in stallions. Reprod Domest Anim. 2015;50:1032–8.
Hickford DE, Frankenberg S, Pask AJ, Shaw G, Renfree MB. DDX4 (VASA) is conserved in germ cell development in marsupials and monotremes. Biol Reprod. 2011;85:733–43.
Sharma M, Srivastava A, Fairfield HE, Bergstrom D, Flynn WF, Braun RE. Identification of EOMES-expressing spermatogonial stem cells and their regulation by PLZF. Elife. 2019;8:e43352.
Paduch DA, Hilz S, Grimson A, Schlegel PN, Jedlicka AE, Wright WW. Aberrant gene expression by Sertoli cells in infertile men with Sertoli cell-only syndrome. PloS One. 2019;14:e0216586.
Lau X, Munusamy P, Ng MJ, Sangrithi M. Single-cell RNA sequencing of the cynomolgus macaque testis reveals conserved transcriptional profiles during mammalian spermatogenesis. Dev Cell. 2020;54:548–66.
Park O-J, Kim H-J, Woo K-M, Baek J-H, Ryoo H-M. FGF2-activated ERK mitogen-activated protein kinase enhances Runx2 acetylation and stabilization. J Biol Chem. 2010;285:3568–74.
Parker N, Falk H, Singh D, Fidaleo A, Smith B, Lopez MS, et al. Responses to glial cell line-derived neurotrophic factor change in mice as spermatogonial stem cells form progenitor spermatogonia which replicate and give rise to more differentiated progeny. Biol Reprod. 2014;91:92. 91–99
Hasegawa K, Saga Y. FGF8-FGFR1 signaling acts as a niche factor for maintaining undifferentiated spermatogonia in the mouse. Biol Reprod. 2014;91:145. 141–8
Yang Y, Feng Y, Feng X, Liao S, Wang X, Gan H, et al. BMP4 cooperates with retinoic acid to induce the expression of differentiation markers in cultured mouse spermatogonia. Stem Cells Int. 2016;2016:9536192.
Acknowledgements
We are grateful to Prof. G Yamada and C Tickle for critical reading of this manuscript. This research was supported in part by the National Research Foundation of Korea (NRF) Grant funded by the Korea Government (MSIP) (NRF-2019R1A2C3005294, NRF-2017M3A9B3061833 and NRF-2016R1A5A2008630).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by G. Melino
Supplementary information
Rights and permissions
About this article
Cite this article
Lee, SJ., Park, J., Lee, DJ. et al. Mast4 knockout shows the regulation of spermatogonial stem cell self-renewal via the FGF2/ERM pathway. Cell Death Differ 28, 1441–1454 (2021). https://doi.org/10.1038/s41418-020-00670-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-020-00670-2
This article is cited by
-
RNF20 is required for male fertility through regulation of H2B ubiquitination in the Sertoli cells
Cell & Bioscience (2023)
-
Mast4 determines the cell fate of MSCs for bone and cartilage development
Nature Communications (2022)