Abstract
Autophagy is a cell biological process that promotes resilience in the face of environmental perturbations. Given that infectious agents represent a major type of environmental threat, it follows that the autophagy pathway is central to the outcome of host–microbe interactions. Detailed molecular studies have revealed intricate ways in which autophagy suppresses or enhances the fitness of infectious agents, particularly intracellular pathogens such as viruses that require the host cell machinery for replication. Findings in animal models have reinforced the importance of these events that occur within individual cells and have extended the role of autophagy to extracellular microbes and immunity at the whole organism level. These functions impact adaptation to bacteria that are part of the gut microbiota, which has implications for the etiology of chronic disorders such as inflammatory bowel disease. Despite major advances in how autophagy regulates inflammatory reactions toward microbes, many challenges remain, including distinguishing autophagy from closely related pathways such as LC3-associated phagocytosis. Here, we review the role of autophagy in microbial pathogenesis at the level of organismal biology. In addition to providing an overview of the prominent function of autophagy proteins in host–microbe interactions, we highlight how observations at the cellular level are informing pathogenesis studies and offer our perspective on the future directions of the field.
Similar content being viewed by others
Facts
Removal of intracellular microbes and inhibition of inflammatory cytokine production are autophagy functions that work in concert to mediate the return to homeostasis.
Autophagy promotes the replication of certain RNA viruses, yet other viruses block autophagy proteins to evade destruction or antigen presentation.
Successful bacterial pathogens have evolved mechanisms to evade autophagic degradation, but other functions of autophagy proteins can still contribute to the outcome of an infection at the whole organism level.
Defense against eukaryotic pathogens in particular is associated with non-autophagy functions of autophagy proteins.
Open questions
Which functions attributed to autophagy proteins represent autophagy versus related pathways such as LC3-associated phagocytosis?
For which pathogens and strains of pathogens are the detailed molecular mechanisms of autophagy-microbe interactions identified in vitro most relevant?
Is targeting autophagy proteins a viable strategy for treating infectious disease in humans?
Introduction
The field of microbial pathogenesis seeks to understand how infectious agents contribute to disease events and lies at the intersection of cellular and molecular microbiology, immunology, and physiology. Cell biology has the potential to bridge these spheres of knowledge. From rewiring of the cellular machinery by pathogens to multicellular communication in lymph nodes, certain cellular pathways are pervasive in host–microbe interactions. One such cell biological process that has captured the attention of many in the field is macroautophagy, more commonly referred to as autophagy.
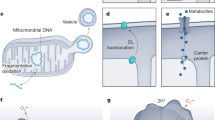
Autophagy is a process by which substrates in the cytosol are sequestered in a double-membrane-bound vesicle termed the autophagosome and transported to the lysosome. This process is necessary for eliminating the unwanted material, such as damaged organelles and protein aggregates. Also, macromolecules (e.g., proteins) are broken down into their constituents (e.g., amino acids) by lysosomal hydrolases and exported back to the cytosol where they can replenish the nutrient pool. These functions of autophagy are mediated by conserved autophagy-related proteins (ATGs) that can be grouped into the following functional units (Fig. 1): (1) the preinitation complex downstream of autophagy-inducing signals, (2) the class 3 phosphatidylinositol kinase (PI3KC3) complex that nucleates the pre-autophagosomal structure, also known as the phagophore, and (3) the ATG16L1 complex that mediates the elongation of the phagophore around the cargo to seal the contents within the autophagosome [1]. Like other vesicle trafficking pathways, RAB GTPases and SNAREs mediate the fusion between the completed autophagosome and endosomes, multivesicular bodies (MVBs), or lysosomes.
The ATG16L1 complex (ATG16L1, ATG5, and ATG12) is generated through a ubiquitin-like pathway by ATG7 and ATG10 that covalently attaches ATG12 to ATG5. Following non-covalent binding between ATG5 and ATG16L1, the complex is recruited to these ER-associated sites by the PI3P-binding protein WIPI2. ATG7 also functions in a second ubiquitin-like pathway by activating and transferring the ubiquitin-like molecule LC3 to ATG3. The ATG16L1 complex then transfers LC3 from ATG3 onto the lipid phosphatidylethanolamine (PE) on the pre-autophagosomal structure. Through its fusogenic properties, LC3 mediates the elongation and closure of the autophagosome. STX17, VAMP8, SNAP29, YKT6 and other membrane integrated SNARE proteins and their cofactors mediate fusion between the autophagosome and endo-lysosomal vesicles. The acidic environment and enzymes mediate the degradation and recycling of cargo molecules such as mitochondria (blue oval), intracellular pathogens (purple hexagon), or protein aggregates (triangle).
Much of our understanding of how these steps are coordinated comes from studies examining the response of yeast and mammalian cells to nutrient starvation. The kinase function of the preinitation complex, which is restricted through inhibitory phosphorylation by mammalian target of rapamycin (mTOR) under nutrient replete conditions, activates the PI3KC3 complex to generate PI3Ps at the endoplasmic reticulum (ER) or sites associated with the ER [2, 3]. The multipass transmembrane molecule ATG9a cycles between vesicular compartments to recruit membrane-building material to this site of the nascent phagophore. ATG16L1 is recruited through interactions with FIP200 (part of the preinitiation complex) and the PI3P-binding protein WIPI2 [4, 5]. ATG16L1 non-covalently binds ATG5−ATG12, which is covalently linked to each other through a ubiquitination-like pathway involving other ATGs. This multimeric complex consisting of ATG16L1-ATG5−ATG12 serves as an E3 ubiquitin ligase-like enzyme that attaches a lipid, phosphatidylethanolamine (PE), to LC3 or homologs of LC3. PE-conjugated LC3 facilitates the efficient closure of the autophagosome and fusion with the lysosome [6, 7]. When autophagy mediates the degradation of mitochondria (mitophagy) or specific macromolecules, these substrates are recruited by autophagy receptors, such as SQSTM1 (also known as p62), that simultaneously interact with LC3 and the cargo, thereby adding specificity to the pathway.
Here, we review how the above ATGs contribute to microbial pathogenesis. We wish to distinguish this review article from previous, excellent ones written on immunity to infection by emphasizing multicellular and whole organism events, especially those that contribute to disease. We will first summarize the established role of autophagy in immunity. Then, we will discuss how autophagy relates to limiting the damage caused by pathogens, dividing the infectious agents into three major classes—viruses, bacteria, and eukaryotes. Infectious agents, including members of the gut microbiota, play a prominent role in chronic inflammatory disorders. Therefore, we will also discuss the role of autophagy in host–microbiota interactions by focusing on inflammatory bowel disease (IBD), a disorder associated with autophagy-related defects. Finally, we will examine the recurring themes presented in this article and offer our perspective on the future directions of the field.
Primer on autophagy and host defense
Autophagy functions as a form of cell autonomous defense when internalized microbes become substrates, a process referred to as xenophagy. Salmonella enterica serovar Typhimurium and Mycobacterium tuberculosis (Mtb), bacteria that cause gastroenteritis and tuberculosis, respectively, are the most extensively examined model pathogens for xenophagy. Host-encoded E3 ubiquitin ligases Parkin, LRSAM1, SMURF1, RNF166, and linear ubiquitin chain assembly complex (LUBAC) tag ubiquitin onto bacteria that escape into the cytoplasm or are exposed to the cytoplasm upon damage to the pathogen-containing vacuole (PcV) [8,9,10,11,12,13,14]. SQSTM1 and NDP52 are examples of autophagy receptors that can crosslink either mitochondria or bacteria to the autophagy machinery through ubiquitin and LC3 binding motifs. Hence, it is possible the mitophagy process was repurposed for xenophagy. NDP52 also binds galectin-8, a cytosolic lectin that recognizes the damaged PcVs [15]. These events are induced by pattern recognition receptors (PRRs) [16]. For example, toll-like receptor 4 (TLR4) recognition of lipopolysaccharide (LPS) activates two downstream kinases to target LC3 to S. Typhimurium: transforming growth factor beta-activated kinase 1 (TAK1) phosphorylates ULK1 in the preinitation complex to initiate autophagy [17], and TANK binding kinase 1 phosphorylates the autophagy receptor OPTN to enhance binding to ubiquitinated bacteria [18]. In another example, detection of cytosolic peptidoglycan by nucleotide-binding oligomerization domain protein-1 (NOD1) and NOD2 induces signaling that is favorable for autophagy and can directly recruit ATG16L1 to the internalized microbe [19,20,21,22,23].
Xenophagy may be a primordial effector function of PRRs that evolved earlier than cytokine production. Stimulator of interferon genes (STING) is best known for mediating antiviral type I interferon (IFN-I) production upon the binding of cyclic dinucleotides that are generated by cyclic GMP-AMP synthease (cGAS) in the presence of cytosolic DNA, such as during viral infection. However, the activation of STING in certain metazoans such as flies and the sea anemone induce autophagy but not interferons [24, 25]. Given that flies do not encode cGAS, STING is likely activated by cyclic dinucleotides that are directly made by bacteria. The observation that PRR-induced autophagy promotes defense against diverse viral and bacterial pathogens in fly, nematodes, zebrafish, and ameba models supports the concept that xenophagy is a conserved defense mechanism [25,26,27,28,29,30].
Counterintuitively, autophagy frequently inhibits production of cytokines that are important for host-pathogen interactions, most notably interleukin-1β (IL-1β) and IFN-I [31]. The NLRP3 inflammasome is a multimeric complex that induces an inflammatory form of cell death (pyroptosis) while simultaneously mediating the processing and the secretion of IL-1β [32]. Reactive oxygen species (ROS) and DNA from damaged mitochondria can activate the NLRP3 inflammasome. Mitophagy reduces IL-1β production by removing these inflammasome triggers [32, 33]. Similarly, mitophagy dampens IFN-I responses downstream of mitochondrial antiviral signaling protein (MAVS), an adapter molecule for the cytosolic RNA sensors retinoic acid inducible gene I and melanoma differentiation associated gene 5 (MDA5) [34,35,36,37]. These studies show autophagy can remove mitochondria that serve as a platform for MAVS signaling or directly target MAVS. When TLR3 or TLR4 are activated in macrophages, autophagy inhibits IFN-I production and cell death by removing the signaling adapter TRIF that is recognized by the autophagy receptor TAX1BP1 [38, 39]. Autophagy also inhibits IL-1β production through the caspase-11 inflammasome and IFN-I production through STING by reducing the availability of cytosolic LPS and DNA, respectively [40]. We propose the removal of intracellular microbes through xenophagy and the inhibition of inflammatory cytokine production are examples of autophagy functions working in concert to mediate a return to homeostasis and the swift resolution of infection. In the subsequent sections, we provide examples of how this regulation impacts the course of disease during an infection.
The same ATGs required for autophagy have autophagy-independent functions in immunity [41, 42]. This challenge in studying autophagy is illustrated by the finding that ATG5 protects against Mtb in mice by mediating neutrophil cell death, while other ATGs are dispensable for this function [43]. In contrast, ATGs function together to mediate an alternate form of autophagy where antimicrobial granules, proteins lacking signal sequences such as certain cytokines, or other cargoes relevant to host defense are exocytosed from the cell [41]. The mechanism of secretory autophagy is not completely understood and may involve redirecting the autophagosome to the plasma membrane away from the lysosome by using different SNARE proteins than those involved in degradative autophagy [44]. Certain cargo may be placed within the intermembrane space of the autophagosome rather than the lumen, in which case the fusion of the double-membrane vesicle with the cell surface would lead to the release of soluble material in-between the outer and inner membranes [45]. Alternatively, it seems likely that there are instances such as exosome secretion in which ATGs act in an autophagosome-independent manner to induce trafficking of the multivesicular body and other endo-lysosomal vesicles [41]. For LC3-associated phagocytosis (LAP), ATGs that are part of the PI3KC and ATG16L1 complexes mediate the transport of cargo internalized from the extracellular environment to the endo-lysosomal compartment. LAP is distinguished by the absence of a double-membrane vesicle and its dependence on NADPH oxidase 2 (NOX2) and Rubicon, a protein that recruits the PI3KC complex to phagocytosed microbes, TLR ligands, and apoptotic corpses [46,47,48,49]. Internalized material processed through LAP induces cytokine expression and serves as a source of antigens for presentation on T cells [46, 49, 50]. For simplicity, a general rule is that autophagy targets cytosolic material while LAP is more likely to be relevant for cargo originating from the extracellular environment. ULK1 and ATG9a each have distinct autophagy-independent functions in inhibiting STING [51, 52], and the establishment of the Brucella abortus PcV is dependent on preinitiation and PI3K complex proteins but independent of the LC3 conjugation machinery [53]. Several other examples of non-autophagy roles of ATGs will be discussed below.
Viral pathogens
Autophagy at the whole organism level has been difficult to investigate for many medically important viruses due to the restricted host tropism and unavailability of practical animal models. Despite this limitation, several generalizable themes have emerged that are consistent with both in vitro and in vivo observations. First, viruses have evolved mechanisms to subvert autophagy, as exemplified by picornaviruses, a large family of single-stranded RNA viruses. Mice in which Atg5 is deleted in pancreatic acinar cells display a striking decrease in the viral burden and disease in a model of coxsackievirus-induced pancreatitis [54], likely reflecting multiple stages of the life cycle impacted by ATGs. Upon entry into the host cell, the lipid-modifying enzyme PLA2G16 enables the delivery of the viral genome from the endosome to the cytosol through a pore before the damaged vesicle is targeted by the autophagy machinery for destruction [55]. During the generation of new virions, rather than target them for degradation, ATGs have been shown to be hijacked by multiple picornaviruses (coxsackievirus, poliovirus, and rhinovirus) to generate LC3+ membranes that serve as a replication platform [56,57,58] (Table 1). In the case of coxsackievirus B3 and enterovirus D68, autophagosomes generated through this process are redirected to the plasma membrane in a manner similar to secretory autophagy by viral proteases that cleave the SNARE protein SNAP29, leading to the exocytosis of single-vesicles that cloak the virions [59, 60]. The phosphatidylserine decorating these pseudo-enveloped virions generated through an autophagy-like process facilitates entry into neighboring cells, especially macrophages [61, 62]. Therefore, ATGs aid the replication, egress, and subsequent entry of Picornaviruses. Herpesviruses, which are enveloped DNA viruses, also hijack LC3 for the maturation of viral particles and egress [63,64,65]. However, several herpesviruses related to these encode molecules that block autophagy to evade the immune system, which may be particularly important for chronic infections. Flaviviruses, which include medically important viruses like hepatitis C virus, have also been shown to depend on autophagy at various steps in their life cycles. One study provided evidence that this dependence represents a therapeutic opportunity by showing that inhibiting autophagy reduces Zika virus (ZIKV) replication in human trophoblasts and limits transmission from the pregnant mother to fetus in mice [66].
For other viruses, autophagy can protect the host. Therapeutically enhancing autophagy by inoculating mice with a Beclin-1 peptide decreases viral burden and improves survival during chikungunya virus and West Nile virus infections [67]. Herpesviruses actively block autophagy. Murine gammaherpesvirus 68 (MHV68) and herpes simplex virus type 1 (HSV-1) engineered to lack viral Bcl-2 and ICP34.5, Beclin-1 inhibitors encoded by the respective viruses, display impaired virulence and fail to establish chronic infections [68, 69] (Table 1). Autophagy also has an essential role in protecting against tissue injury and cell death during viral infection. The capsid protein of Sindbis virus is degraded by selective autophagy through autophagy receptors, which prevents virus-induced death of neurons during infection of the central nervous system [70, 71]. In addition, ATG5 is required to control HSV-1 replication in dorsal root ganglionic neurons following intra-vaginal inoculation of mice [72], and for control of ZIKV in the fly brain [25]. ATGs also prevent necrotic cell death in the intestinal epithelium in response to TNFα produced during persistent murine norovirus (MNV) infection [73]. Although we focus on mammalian autophagy in this review, it is worth noting that similar paradigms are observed in plants where autophagy binds viral proteins or regulates cell death during infections, which can be pro- or antiviral depending on the virulence strategies employed by the virus [74].
In many of the above examples, autophagy functions within parenchymal cells such as epithelia and neurons to protect the tissue from damage. In other cases, ATGs function in immune cells to limit immune-mediated injury. Mitophagy is induced following the recognition of cytosolic influenza virus RNA to dampen the NLRP3 inflammasome and prevent an over-exuberant immune response that causes lung pathology [75]. This mechanism in place to restrict tissue injury is a double-edged sword because the human parainfluenza virus type 3 (HPIV3) matrix protein induces mitophagy to inhibit IFN-I production downstream of MAVS, leading to the evasion of antiviral immunity and increased viral replication [76]. In other studies examining the response to influenza virus or MHV68 infection, mice harboring deletions in one of any number of ATGs in myeloid cells were found to display a striking increase in the production of multiple cytokines that impact tissue injury and viral replication through a mechanism that is not easily explained by currently known autophagy functions [77, 78].
Finally, autophagy and LAP have essential roles in adaptive immunity during viral infections. The deletion of Atg5 in CD11c+ cells (i.e., dendritic cells and some macrophages) leads to compromised antigen presentation to T cells in response to HSV-2 and yellow fever virus vaccine challenges [79, 80]. In a model of corneal infection, an ICP34.5 mutant HSV-1 that fails to inhibit Beclin-1 generates a stronger CD4+ T cell response, reinforcing a role for the autophagy machinery in MHC-II antigen presentation [81]. In addition, ATGs in T and B lymphocytes help adapt to changing metabolic needs and stress during differentiation and proliferation [1]. For example, autophagy is upregulated during the contraction phase when clonal expansion has ended and is essential for the persistence of memory CD8+ T cells following the infection by LCMV, MCMV, and influenza virus [82, 83]. T cells from older mice display a decrease in autophagy, potentially contributing to decreases in immunity that occur with aging [83]. To summarize, autophagy or related processes promote the replication of certain RNA viruses or dampen the antiviral response, but other viruses block the function of ATGs to evade direct destruction or antigen presentation. Then, during the resolution of viral infection, autophagy continues to be important in lymphocytes for sustaining the memory of the infection.
Bacterial pathogens
Successful pathogens that depend on intracellular replication have evolved strategies to avoid xenophagy. S. Typhimurium [84, 85], Legionella pneumophila [7], Listeria monocytogenes [86,87,88,89,90], Shigella flexneri [90, 91], and contemporary isolates of Group A Streptococcus [92], encode virulence factors that block the recruitment of ATG16L1 to the PcV, inactivate LC3 and autophagy receptors through enzymatic cleavage, or facilitate escape from the autophagy machinery through motility (Table 1). Despite these evasive tactics, autophagy mutant mice are generally susceptible to infection, suggesting that ATGs are performing other functions in addition to xenophagy. Such complex or tissue-specific roles are difficult to investigate in cell lines. One example where the animal model revealed a role of autophagy that is distinct from the role established in cell culture experiments is in studies of Staphylococcus aureus, a Gram-positive bacterium that causes a range of life-threatening illnesses including pneumonia, endocarditis, and sepsis. While initial tissue culture studies using HeLa cells highlight how S. aureus highjacks autophagy to promote bacterial survival [93], autophagy does not contribute to S. aureus burden during lung and blood stream infections in vivo [94]. Instead, ATG16L1 and other autophagy components are involved in the protection of host cells from the lethal effects mediated by a potent pore-forming toxin (α-toxin) produced by the bacterium [94]. Thus, in this scenario, the autophagy machinery prevents excessive tissue damage and is involved in the “tolerance” or a host resilience response [95]. In another recent example, the ATG16L1 complex, but not other steps of autophagy, was shown to mediate plasma membrane repair in response to listeriolysin O (LLO), a pore-forming toxin produced by L. monocytogenes [86]. Rather than controlling replication in the cytosol, the ATG16L1 complex restricts cell-to-cell spread of bacteria by maintaining the integrity of the plasma membrane [86, 96]. Thus, ATGs may have a general role in countering damage to the plasma membrane during infections (Fig. 2).
In the first example, Listeriolysin O (LLO) produced by Listeria monocytogenes mediates pore formation in the plasma membrane that facilitates cell-to-cell spread of the bacterium. The ATG16L1 complex restricts this spread by promoting plasma membrane repair through an autophagy-independent mechanism involving the exocytosis of lysosomes that confines the damage to surface blebs. In the second example, α-toxin secreted by Staphylococcus aureus binds ADAM10 on the surface of target cells such as the endothelium leading to cell death. The ATG16L1 complex downregulates ADAM10 levels, limiting the availability of the receptor for α-toxin binding and promotes cell survival. In the third example, TNFα produced by immune cells in the gut in response to murine norovirus (MNV) infection is tolerated by intestinal epithelial cells when autophagy is intact. However, upon the disruption of the ATG16L1 complex and inhibition of mitophagy, the accumulation of reactive oxygen species (ROS) licenses signaling through RIPK1 and RIPK3 downstream of the TNFα receptor (TNFR) resulting in the activation of MLKL, the pore-forming executioner molecule of the necroptosis pathway of programmed cell death.
Mouse experiments confirm a membrane repair function of ATG16L1 in the intestinal epithelium during L. monocytogenes infection [86] and also reveal other mechanisms by which the autophagy pathway participates in defense against this agent of food poisoning. ATG5 has a non-xenophagy function in myeloid cells that protects animals from lethal L. monocytogenes infection [97], a process that may be related to a novel IFN-γ effector mechanism observed during protozoan infection (see “Eukaryotic pathogens” section below). Also, a recent study showed LLO specifically induces mitophagy in macrophages via the mitochondria-localized autophagy receptor NLRX1 to rid the cells of ROS generated by damaged mitochondria upon infection, thereby dampening the inflammatory response and promoting intracellular bacterial survival [87]. This elegant study does not preclude other important ways in which ATGs promote intracellular survival of L. monocytogenes through establishment of a protective niche [98], but it demonstrates the power of using recently developed tools to inhibit specific autophagy functions with precision.
Approaches in which pathogens are introduced through artificial routes of infection could miss key autophagy functions at barrier sites. In the gastrointestinal tract, epithelial cells act as sentinels that respond to different bacteria by activating autophagy [99, 100]. Using intestinal epithelial cell (IEC)-specific ATG deficient mice, autophagy downstream of Myd88 (adapter for TLRs and IL-1 receptor) was shown to control extraintestinal dissemination of S. Typhimurium following oral infection. Work with Citrobacter rodentium, a rodent pathogen used as a model for Enteropathogenic Escherichia coli (EPEC) infection of humans, highlights how the contribution of autophagy cannot be automatically extended to similar bacterial pathogens. Although both bacterial species are members of the Enterobacteriaceae family of gut pathogens, C. rodentium is an extracellular bacterium that does not establish the intracellular vacuole characteristic of S. Typhimurium. Mice with decreased expression of Atg16L1 or specific deletion in IECs are hyper-resistant to oral challenge by C. rodentium because of enhanced IFN-I signaling associated with unrestricted MAVS and STING activation [101, 102]. Atg16L1 mutant mice are also resistant to uropathogenic E. coli (UPEC) in a model of urinary tract infections due to increased IL-1β production downstream of the NLRP3 inflammasome in myeloid cells [103, 104]. Observations with these three related pathogens, when introduced via their natural route of infection, underscore the context-specific role of autophagy.
Eukaryotic pathogens
LAP in phagocytic cells appears to be the dominant mechanism by which ATGs promote immunity toward eukaryotic pathogens (Fig. 3). β-glucan moieties on the fungal cell wall triggers the recruitment of LC3 to the internalized fungi following ROS production via NOX2 upon recognition by TLR2 or the C-type lectin receptor Dectin-1 [46, 105]. Inhibiting LAP during the fungal infection can lead to decreased fungicidal activity, altered cytokine production, and reduced antigen presentation [47, 50, 105,106,107]. These functions may be particularly important for Aspergillus fumigatus, an opportunistic pathogen that causes life-threatening pulmonary infections. Germination of A. fumigatus spores exposes β-glucan and induces the release of Ca2+ from ER stores, which activates LAP to reduce the fungal burden and airway inflammation in the mouse model [108, 109]. Chronic granulomatous disease (CGD) patients who carry mutations in NOX2 are susceptible to A. fumigatus and display defective LC3 recruitment to fungi internalized by macrophages [110, 111]. Blocking IL-1β in CGD patients or an animal model improves colitis, one of the consequences of invasive infection, indicating that excess inflammasome activity due to defective autophagy may also be important during aspergillosis [110]. Similar to CGD patients, aspergillosis incidence is higher in hematopoietic stem cell transplant recipients who have a polymorphism in death-associated protein kinase 1 (DAPK1), an IFN-γ inducible gene that is necessary for LAP and proteasomal degradation of NLRP3 [111]. Thus, LAP represents a promising pharmacological target for treating fungal infections.
During LC3-associated phagocytosis (LAP), internalized microbes are recognized by pattern recognition receptors, such as Dectin-1 binding of β-glucan from Aspergillus fumigatus, leading to the recruitment of a PI3KC3 complex. In contrast to autophagy, the PI3KC3 complex assembled during LAP is distinguished by the presence of Rubicon and the concurrent recruitment of the NADPH oxidase 2 (NOX2) complex that generates ROS. LC3 lipidation by the ATG16L1 complex then mediates the maturation of the single-membrane phagosome through fusion with the lysosome and degradation of the contents. Targeting by Autophagy Proteins (TAG) is initiated by IFNγ and involves the decoration of the pathogen-containing vacuole (PcV) by the LC3 homolog GATE-16 during Toxoplasma gondii infection. Although dependent on the ATG16L1 complex, the downstream autophagy factors involved in lysosomal degradation are dispensable. Instead, IFNγ inducible GTPase belonging to the IRG and GBP families are recruited and contribute to the disruption of the PcV membrane.
ATGs may have similar functions during the infection by protozoan parasites. Like fungal infection of macrophages, IFN-γ induced LAP in hepatocytes kills Plasmodium species, the causative agent of malaria [112]. However, the parasite can avoid killing through production of UIS3C, a PcV membrane-associated molecule that binds and inhibits LC3 [113] (Table 1). Similarly, invasion by Toxoplasma gondii activates the epidermal growth factor receptor to prevent LC3 recruitment by inhibiting PKR and eIF2α, known activators of autophagy [114, 115] (Table 1). Conversely, the causative agent of Chagas disease, Trypanosoma cruzi, requires acidification of the PcV for its virulence, which was shown to depend on the host autophagy machinery in fibroblasts [116]. Although the role of ATGs in these examples likely reflects LAP, another IFN-γ inducible non-degradative (lysosome-independent) function of the LC3 conjugation machinery called targeting by AutophaGy proteins (TAG) can mediate parasite killing by recruiting molecules that disrupt membranes (Fig. 3). The ATG16L1 complex restricts T. gondii through TAG by mediating lipidation of LC3 homologs, especially GATE-16 (also known as GABARAPL2), which then recruit interferon-inducible GTPases (IRGs) and guanylate binding proteins (GBPs) to the PcV and compromises the replicative niche [117,118,119,120,121,122,123]. TAG also disrupts the membrane-associated replication complex of MNV [124, 125] and may explain why ATG5 restricts L. monocytogenes in macrophages in vivo [97], suggesting that recruiting IRGs and GBPs by ATGs is a defense response to a broad range of pathogens that target host membranes. The mechanism by which ATGs are directed toward TAG rather than autophagy and how IRGs and GBPs disrupt membranes are two key questions that will need to be addressed. Also, ATG5 is essential for maintaining the intestinal barrier during T. gondii infection by preventing epithelial cell death [126], indicating the role of ATGs in tissue injury and repair during parasitic infection requires additional investigation.
Gut microbiota and IBD
The trillions of bacteria and other infectious agents that are part of the gut microbiota achieve a balanced co-existence with a healthy host. Disruption of this balance has been implicated in the origin of immune and metabolic disorders including IBD, a chronic and relapsing inflammatory condition affecting millions worldwide [127]. Although no single etiologic agent has been identified, decades of observations in patients and animal models suggest a microbial origin of IBD [128]. There has been intense interest in examining the role of autophagy in mediating host–microbe interactions in the gut following the identification of a coding variant in ATG16L1 (T300A) associated with risk of acquiring Crohn’s disease, a type of IBD that commonly affects the small intestine as well as other parts of the gastrointestinal tract [127]. The ATG16L1T300A protein product is susceptible to cleavage by caspase-3 induced by cellular stressors such as TNFα and nutrient deprivation [129, 130]. Consistent with the role of ATG16L1 in autophagy, macrophages and DCs from mice and humans expressing the ATG16L1T300A variant are defective in xenophagy, antigen presentation, and control of IL-1β and IFN-I production [22, 129,130,131]. Animal models provide evidence that an uncontrolled cytokine response is a particularly important function of macrophage ATG16L1 during intestinal inflammation. The anti-inflammatory cytokine IL-10 induces mitophagy in macrophages to promote oxidative phosphorylation and inhibit glycolysis in the presence of LPS [132]. Deleting Atg16l1 in macrophages, therefore, leads to a pro-inflammatory metabolic state associated with damaged mitochondria and NLRP3 activity that exacerbate disease in mouse models of colitis [32, 132, 133].
These findings do not exclude a role for LAP. ATG16L1 has a WD40 C-terminal domain outside the region necessary for complex formation with ATG5-12, and this domain is required for recruitment of LC3 to endosomal membranes for MHC-II antigen presentation [134]. The T300A substitution interferes with binding TMEM59, an interaction that mediates the recruitment of LC3 to internalized S. aureus through a process resembling LAP [135]. Outer membrane vesicles derived from B. fragilis induce LAP in DCs, which can enhance the differentiation of co-cultured T cells into IL-10 producing Tregs. Monocyte-derived DCs from individuals with the ATG16L1T300A variant fail to promote Treg differentiation, and a similar loss of tolerance was observed in an analogous animal model [136]. Thus, the genetic association between ATG16L1 and Crohn’s disease could reflect a defect in autophagy, LAP, or both that leads to an exaggerated cytokine responses toward microbes that are otherwise managed.
ATG16L1 also has an essential role in IECs. Crohn’s disease patients homozygous for the ATG16L1T300A variant display defects in granule production by Paneth cells [137], antimicrobial epithelial cells in the small intestine that maintain barrier function [138]. Mutation of Atg16l1, Atg4b, Atg5, and Atg7 in mice all lead to similar Paneth cell defects [137, 139,140,141,142]. A specific role for the microbial pathogenesis is supported by the observation that an infectious trigger is required for intestinal disease in three independently generated Atg16l1 mutant mice: mice with a hypomorphic allele of Atg16l1, an IEC-specific knockout, or a T300A mutation develop Paneth cell defects and an exacerbated intestinal injury response following infection by MNV [73, 143]. In other situations, MNV infection promotes the differentiation and function of the mucosal immune system similar to beneficial bacteria, and protects against vancomycin resistant Enterococcus and C. rodentium infections by inducing the epithelial regenerative cytokine IL-22 [144, 145]. The anaerobic bacterium Bacteroides ovatus, a common member of the human gut microbiota that has garnered some interest as a probiotic, induces inflammatory T cell differentiation in the intestines of Atg16l1T300A knock-in mice when transferred from Crohn’s disease patients [146]. Hence, viruses and bacteria that are potentially beneficial in other settings evoke inflammatory responses in the gut of Atg16l1 mutant mice. These observations are consistent with a model of IBD in which genetically susceptible individuals lose tolerance to commensal agents.
Mechanistically, the role of ATG16L1 in IECs could be explained by the link between autophagy, organelle homeostasis, and secretion. Unmitigated ER stress upon the inhibition of IKKα signaling in IECs leads to ATG16L1 instability and intestinal inflammation [147]. Individuals with the ATG16L1T300A variant and aged Atg16l1 mutant mice display signs of ER stress in Paneth cells [148, 149], which is associated with a compensatory upregulation of IgA production by B cells [150]. Simultaneous deletion of Atg16l1 and the ER stress transcription factor X-box binding protein-1 (Xbp-1) leads to a dramatic loss of Paneth cells and severe intestinal inflammation [151]. In contrast to colonic IECs that die from apoptosis upon autophagy inhibition [152, 153], Atg16l1 mutant Paneth cells accumulate defective mitochondria and die from necroptosis, a form of programmed necrosis in which mixed lineage kinase domain like protein induces pore formation in the plasma membrane [73, 154] (Fig. 2). The selective effects of ATG mutation on Paneth cells may reflect the vulnerability of cells with a high secretory burden. In addition to resolving organelle stress through autophagy, ATG proteins can compensate for ER-Golgi trafficking defects that occur in Paneth cells during S. Typhimurium infection by mediating exocytosis of the antimicrobial molecule lysozyme [155]. ATGs are necessary for mucus secretion by goblet cells [156], another secretory epithelial cell subset that produces the mucus necessary to prevent invasion by commensal and pathogenic microbes [138]. The loss of ATG16L1 is counterbalanced by a binding partner, the cytokine signaling inhibitor and deubiquitinase A20, and deleting both together leads to severe epithelial cell death and intestinal inflammation [157]. When taken together, these studies show that ATG16L1 has a fundamental role in promoting the viability and function of Paneth cells and other IECs in response to stressful conditions associated with colonization by benign bacteria and viruses. This role of ATG16L1 is linked to organelle homeostasis and could reflect both autophagy and related events such as unconventional protein secretion.
Concluding remarks
A key frontier in infection biology is to apply the molecular level knowledge of autophagy gained from sophisticated cell biological experiments toward elucidating mechanisms of pathogenesis. The pioneering studies that investigated ATG function in simplified models have generated an array of hypotheses, many of which have been supported. In addition, coinciding with the generation of mutant mice, research from this past decade has extended the way in which autophagy intersects host defense. Tissue and cell type-specific activities have become more apparent, and multicellular and multi-organ mechanisms are being discovered.
A few themes have emerged that are consistent across multiple studies. At the individual cell level, ATGs are central players in the battle over membranes. Intracellular pathogens seek to subvert membranes to establish replicative niches, pass through membranes to access various compartments, or to simply avoid degradation. In a pathogen-specific manner, autophagy and related pathways can limit these processes or be hijacked to complete the life cycle. Outside the infected cell, autophagy restricts the damage caused by infections, especially by extracellular pathogens, either by supporting the viability of key cell types or restraining inflammatory cytokine production. Applying the brakes to the inflammatory cascade comes at a cost because it dampens antimicrobial immunity but is essential for preventing chronic immune responses associated with a litany of diseases including IBD. After the initial events associated with infection, autophagy and LAP are critically important for adaptive immunity by mediating antigen presentation and lymphocyte differentiation.
These immune functions of autophagy represent generalities. Mechanisms revealed by examination of one pathogen cannot be extended to others because even closely related microbes use distinct virulence strategies. Similarly, the literature on ATG16L1 highlights the importance of distinguishing the function of ATGs in leukocytes versus parenchymal cell types. It may be particularly interesting to test whether the ability of certain viruses to exploit the autophagy machinery in one cellular compartment is a liability in another cell type or tissue, and how this might determine disease outcomes. Also, the ability to discriminate autophagy from related ATG-dependent pathways remains a challenge, especially when relying on animal models in which genetically inhibiting multiple parts of the pathway is cost prohibitive. Therefore, great care must be taken when considering how autophagy can be targeted for therapeutic purposes. It is clear that additional research is needed before we can harness the benefit of this multistep cellular process to promote immunity toward disease-causing microbes. We suggest that the use of clinical isolates of pathogens, advanced cell culture models, and human specimens can bridge the gap between in vitro studies and the increasing repertoire of animal models.
References
Matsuzawa-Ishimoto Y, Hwang S, Cadwell K. Autophagy and inflammation. Annu Rev Immunol. 2018;36:73–101.
Hamasaki M, Furuta N, Matsuda A, Nezu A, Yamamoto A, Fujita N, et al. Autophagosomes form at ER-mitochondria contact sites. Nature. 2013;495:389–93.
Ge L, Melville D, Zhang M, Schekman R. The ER-Golgi intermediate compartment is a key membrane source for the LC3 lipidation step of autophagosome biogenesis. eLife. 2013;2:e00947.
Dooley HC, Razi M, Polson HE, Girardin SE, Wilson MI, Tooze SA. WIPI2 links LC3 conjugation with PI3P, autophagosome formation, and pathogen clearance by recruiting Atg12-5-16L1. Mol Cell. 2014;55:238–52.
Nishimura T, Kaizuka T, Cadwell K, Sahani MH, Saitoh T, Akira S, et al. FIP200 regulates targeting of Atg16L1 to the isolation membrane. EMBO Rep. 2013;14:284–91.
Tsuboyama K, Koyama-Honda I, Sakamaki Y, Koike M, Morishita H, Mizushima N. The ATG conjugation systems are important for degradation of the inner autophagosomal membrane. Science. 2016;354:1036–41.
Choy A, Dancourt J, Mugo B, O'Connor TJ, Isberg RR, Melia TJ, et al. The Legionella effector RavZ inhibits host autophagy through irreversible Atg8 deconjugation. Science. 2012;338:1072–6.
Manzanillo PS, Ayres JS, Watson RO, Collins AC, Souza G, Rae CS, et al. The ubiquitin ligase parkin mediates resistance to intracellular pathogens. Nature. 2013;501:512–6.
van Wijk SJL, Fricke F, Herhaus L, Gupta J, Hotte K, Pampaloni F, et al. Linear ubiquitination of cytosolic Salmonella Typhimurium activates NF-kappaB and restricts bacterial proliferation. Nat Microbiol. 2017;2:17066.
Noad J, von der Malsburg A, Pathe C, Michel MA, Komander D, Randow F. LUBAC-synthesized linear ubiquitin chains restrict cytosol-invading bacteria by activating autophagy and NF-kappaB. Nat Microbiol. 2017;2:17063.
Huett A, Heath RJ, Begun J, Sassi SO, Baxt LA, Vyas JM, et al. The LRR and RING domain protein LRSAM1 is an E3 ligase crucial for ubiquitin-dependent autophagy of intracellular Salmonella Typhimurium. Cell Host Microbe. 2012;12:778–90.
Heath RJ, Goel G, Baxt LA, Rush JS, Mohanan V, Paulus GL, et al. RNF166 Determines Recruitment of Adaptor Proteins during Antibacterial Autophagy. Cell Rep. 2016;17:2183–94.
Fiskin E, Bionda T, Dikic I, Behrends C. Global analysis of host and bacterial ubiquitinome in response to salmonella typhimurium infection. Mol Cell. 2016;62:967–81.
Franco LH, Nair VR, Scharn CR, Xavier RJ, Torrealba JR, Shiloh MU, et al. The ubiquitin ligase smurf1 functions in selective autophagy of mycobacterium tuberculosis and anti-tuberculous host defense. Cell Host Microbe. 2017;21:59–72.
Thurston TL, Wandel MP, von Muhlinen N, Foeglein A, Randow F. Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature. 2012;482:414–8.
Delgado MA, Elmaoued RA, Davis AS, Kyei G, Deretic V. Toll-like receptors control autophagy. EMBO J. 2008;27:1110–21.
Liu W, Zhuang J, Jiang Y, Sun J, Prinz RA, Sun J, et al. Toll-like receptor signaling cross-activates the autophagic pathway to restrict Salmonella Typhimurium growth in macrophages. Cell Microbiol. 2019;21:e13095.
Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011;333:228–33.
Irving AT, Mimuro H, Kufer TA, Lo C, Wheeler R, Turner LJ, et al. The immune receptor NOD1 and kinase RIP2 interact with bacterial peptidoglycan on early endosomes to promote autophagy and inflammatory signaling. Cell Host Microbe. 2014;15:623–35.
Travassos LH, Carneiro LA, Ramjeet M, Hussey S, Kim YG, Magalhaes JG, et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol. 2010;11:55–62.
Homer CR, Kabi A, Marina-Garcia N, Sreekumar A, Nesvizhskii AI, Nickerson KP, et al. A dual role for receptor-interacting protein kinase 2 (RIP2) kinase activity in nucleotide-binding oligomerization domain 2 (NOD2)-dependent autophagy. J Biol Chem. 2012;287:25565–76.
Cooney R, Baker J, Brain O, Danis B, Pichulik T, Allan P, et al. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat Med. 2010;16:90–7.
Chauhan S, Mandell MA, Deretic V. IRGM governs the core autophagy machinery to conduct antimicrobial defense. Mol Cell. 2015;58:507–21.
Gui X, Yang H, Li T, Tan X, Shi P, Li M, et al. Autophagy induction via STING trafficking is a primordial function of the cGAS pathway. Nature. 2019;567:262–6.
Liu Y, Gordesky-Gold B, Leney-Greene M, Weinbren NL, Tudor M, Cherry S. Inflammation-Induced, STING-dependent autophagy restricts zika virus infection in the drosophila brain. Cell Host Microbe. 2018;24:57–68 e3.
Jia K, Thomas C, Akbar M, Sun Q, Adams-Huet B, Gilpin C, et al. Autophagy genes protect against Salmonella typhimurium infection and mediate insulin signaling-regulated pathogen resistance. Proc Natl Acad Sci USA. 2009;106:14564–9.
Visvikis O, Ihuegbu N, Labed SA, Luhachack LG, Alves AM, Wollenberg AC, et al. Innate host defense requires TFEB-mediated transcription of cytoprotective and antimicrobial genes. Immunity. 2014;40:896–909.
Moy RH, Gold B, Molleston JM, Schad V, Yanger K, Salzano MV, et al. Antiviral autophagy restrictsRift Valley fever virus infection and is conserved from flies to mammals. Immunity. 2014;40:51–65.
Yano T, Mita S, Ohmori H, Oshima Y, Fujimoto Y, Ueda R, et al. Autophagic control of listeria through intracellular innate immune recognition in drosophila. Nat Immunol. 2008;9:908–16.
Mostowy S, Boucontet L, Mazon Moya MJ, Sirianni A, Boudinot P, Hollinshead M, et al. The zebrafish as a new model for the in vivo study of Shigella flexneri interaction with phagocytes and bacterial autophagy. PLoS Pathog. 2013;9:e1003588.
Cadwell K. Crosstalk between autophagy and inflammatory signalling pathways: balancing defence and homeostasis. Nat Rev Immunol. 2016;16:661–75.
Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264–8.
Zhong Z, Umemura A, Sanchez-Lopez E, Liang S, Shalapour S, Wong J, et al. NF-kappaB restricts inflammasome activation via elimination of damaged mitochondria. Cell. 2016;164:896–910.
Lei Y, Wen H, Yu Y, Taxman DJ, Zhang L, Widman DG, et al. The mitochondrial proteins NLRX1 and TUFM form a complex that regulates type I interferon and autophagy. Immunity. 2012;36:933–46.
Zhao Y, Sun X, Nie X, Sun L, Tang TS, Chen D, et al. COX5B regulates MAVS-mediated antiviral signaling through interaction with ATG5 and repressing ROS production. PLoS Pathog. 2012;8:e1003086.
Mathew R, Khor S, Hackett SR, Rabinowitz JD, Perlman DH, White E. Functional role of autophagy-mediated proteome remodeling in cell survival signaling and innate immunity. Mol Cell. 2014;55:916–30.
Tal MC, Sasai M, Lee HK, Yordy B, Shadel GS, Iwasaki A. Absence of autophagy results in reactive oxygen species-dependent amplification of RLR signaling. Proc Natl Acad Sci USA. 2009;106:2770–5.
Samie M, Lim J, Verschueren E, Baughman JM, Peng I, Wong A, et al. Selective autophagy of the adaptor TRIF regulates innate inflammatory signaling. Nat Immunol. 2018;19:246–54.
Lim J, Park H, Heisler J, Maculins T, Roose-Girma M, Xu M, et al. Autophagy regulates inflammatory programmed cell death via turnover of RHIM-domain proteins. eLife. 2019;8:e44452.
Meunier E, Dick MS, Dreier RF, Schurmann N, Kenzelmann Broz D, Warming S, et al. Caspase-11 activation requires lysis of pathogen-containing vacuoles by IFN-induced GTPases. Nature. 2014;509:366–70.
Cadwell K, Debnath J. Beyond self-eating: the control of nonautophagic functions and signaling pathways by autophagy-related proteins. J Cell Biol. 2018;217:813–22.
Galluzzi L, Green DR. Autophagy-independent functions of the autophagy machinery. Cell. 2019;177:1682–99.
Kimmey JM, Huynh JP, Weiss LA, Park S, Kambal A, Debnath J, et al. Unique role for ATG5 in neutrophil-mediated immunopathology during M. tuberculosis infection. Nature. 2015;528:565–9.
Kimura T, Jia J, Kumar S, Choi SW, Gu Y, Mudd M, et al. Dedicated SNAREs and specialized TRIM cargo receptors mediate secretory autophagy. EMBO J. 2017;36:42–60.
Zhang M, Kenny S, Ge L, Xu K, Schekman R. Translocation of interleukin-1beta into a vesicle intermediate in autophagy-mediated secretion. eLife. 2015;4:e11205.
Sanjuan MA, Dillon CP, Tait SW, Moshiach S, Dorsey F, Connell S, et al. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature. 2007;450:1253–7.
Martinez J, Malireddi RK, Lu Q, Cunha LD, Pelletier S, Gingras S, et al. Molecular characterization of LC3-associated phagocytosis reveals distinct roles for Rubicon, NOX2 and autophagy proteins. Nat Cell Biol. 2015;17:893–906.
Martinez J, Almendinger J, Oberst A, Ness R, Dillon CP, Fitzgerald P, et al. Microtubule-associated protein 1 light chain 3 alpha (LC3)-associated phagocytosis is required for the efficient clearance of dead cells. Proc Natl Acad Sci USA. 2011;108:17396–401.
Henault J, Martinez J, Riggs JM, Tian J, Mehta P, Clarke L, et al. Noncanonical autophagy is required for type I interferon secretion in response to DNA-immune complexes. Immunity. 2012;37:986–97.
Romao S, Gasser N, Becker AC, Guhl B, Bajagic M, Vanoaica D, et al. Autophagy proteins stabilize pathogen-containing phagosomes for prolonged MHC II antigen processing. J Cell Biol. 2013;203:757–66.
Saitoh T, Fujita N, Hayashi T, Takahara K, Satoh T, Lee H, et al. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc Natl Acad Sci USA. 2009;106:20842–6.
Konno H, Konno K, Barber GN. Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling. Cell. 2013;155:688–98.
Starr T, Child R, Wehrly TD, Hansen B, Hwang S, Lopez-Otin C, et al. Selective subversion of autophagy complexes facilitates completion of the Brucella intracellular cycle. Cell Host Microbe. 2012;11:33–45.
Alirezaei M, Flynn CT, Wood MR, Whitton JL. Pancreatic acinar cell-specific autophagy disruption reduces coxsackievirus replication and pathogenesis in vivo. Cell Host Microbe. 2012;11:298–305.
Staring J, von Castelmur E, Blomen VA, van den Hengel LG, Brockmann M, Baggen J, et al. PLA2G16 represents a switch between entry and clearance of Picornaviridae. Nature. 2017;541:412–6.
Wong J, Zhang J, Si X, Gao G, Mao I, McManus BM, et al. Autophagosome supports coxsackievirus B3 replication in host cells. J Virol. 2008;82:9143–53.
Jackson WT, Giddings TH Jr., Taylor MP, Mulinyawe S, Rabinovitch M, Kopito RR, et al. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 2005;3:e156.
Klein KA, Jackson WT. Human rhinovirus 2 induces the autophagic pathway and replicates more efficiently in autophagic cells. J Virol. 2011;85:9651–4.
Mohamud Y, Shi J, Qu J, Poon T, Xue YC, Deng H, et al. Enteroviral infection inhibits autophagic flux via disruption of the SNARE complex to enhance viral replication. Cell Rep. 2018;22:3292–303.
Corona AK, Saulsbery HM, Corona Velazquez AF, Jackson WT. Enteroviruses remodel autophagic trafficking through regulation of host SNARE proteins to promote virus replication and cell exit. Cell Rep. 2018;22:3304–14.
Chen YH, Du W, Hagemeijer MC, Takvorian PM, Pau C, Cali A, et al. Phosphatidylserine vesicles enable efficient en bloc transmission of enteroviruses. Cell. 2015;160:619–30.
Bird SW, Maynard ND, Covert MW, Kirkegaard K. Nonlytic viral spread enhanced by autophagy components. Proc Natl Acad Sci USA. 2014;111:13081–6.
Nowag H, Guhl B, Thriene K, Romao S, Ziegler U, Dengjel J, et al. Macroautophagy proteins assist epstein barr virus production and get incorporated into the virus particles. EBioMedicine. 2014;1:116–25.
Buckingham EM, Jarosinski KW, Jackson W, Carpenter JE, Grose C. Exocytosis of Varicella-Zoster Virus virions involves a convergence of endosomal and autophagy pathways. J Virol. 2016;90:8673–85.
Taisne C, Lussignol M, Hernandez E, Moris A, Mouna L, Esclatine A. Human cytomegalovirus hijacks the autophagic machinery and LC3 homologs in order to optimize cytoplasmic envelopment of mature infectious particles. Sci Rep. 2019;9:4560.
Cao B, Parnell LA, Diamond MS, Mysorekar IU. Inhibition of autophagy limits vertical transmission of Zika virus in pregnant mice. J Exp Med. 2017;214:2303–13.
Shoji-Kawata S, Sumpter R, Leveno M, Campbell GR, Zou Z, Kinch L, et al. Identification of a candidate therapeutic autophagy-inducing peptide. Nature. 2013;494:201–6.
E X, Hwang S, Oh S, Lee JS, Jeong JH, Gwack Y, et al. Viral Bcl-2-mediated evasion of autophagy aids chronic infection of gammaherpesvirus 68. PLoS Pathog. 2009;5:e1000609.
Orvedahl A, Alexander D, Talloczy Z, Sun Q, Wei Y, Zhang W, et al. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe. 2007;1:23–35.
Orvedahl A, MacPherson S, Sumpter R Jr., Talloczy Z, Zou Z, Levine B. Autophagy protects against Sindbis virus infection of the central nervous system. Cell Host Microbe. 2010;7:115–27.
Sumpter R Jr., Sirasanagandla S, Fernandez AF, Wei Y, Dong X, Franco L, et al. Fanconi anemia proteins function in mitophagy and immunity. Cell. 2016;165:867–81.
Yordy B, Iijima N, Huttner A, Leib D, Iwasaki A. A neuron-specific role for autophagy in antiviral defense against herpes simplex virus. Cell Host Microbe. 2012;12:334–45.
Matsuzawa-Ishimoto Y, Shono Y, Gomez LE, Hubbard-Lucey VM, Cammer M, Neil J, et al. Autophagy protein ATG16L1 prevents necroptosis in the intestinal epithelium. J Exp Med. 2017;214:3687–3705.
Kushwaha NK, Hafren A, Hofius D. Autophagy-virus interplay in plants: from antiviral recognition to proviral manipulation. Mol Plant Pathol. 2019;20:1211–6.
Lupfer C, Thomas PG, Anand PK, Vogel P, Milasta S, Martinez J, et al. Receptor interacting protein kinase 2-mediated mitophagy regulates inflammasome activation during virus infection. Nat Immunol. 2013;14:480–8.
Ding B, Zhang L, Li Z, Zhong Y, Tang Q, Qin Y, et al. The matrix protein of human parainfluenza virus Type 3 induces mitophagy that suppresses interferon responses. Cell Host Microbe. 2017;21:538–47 e4.
Park S, Buck MD, Desai C, Zhang X, Loginicheva E, Martinez J, et al. Autophagy genes enhance murine gammaherpesvirus 68 reactivation from latency by preventing virus-induced systemic inflammation. Cell Host Microbe. 2016;19:91–101.
Lu Q, Yokoyama CC, Williams JW, Baldridge MT, Jin X, DesRochers B, et al. Homeostatic control of innate lung inflammation by Vici syndrome gene Epg5 and additional autophagy genes promotes influenza pathogenesis. Cell Host Microbe. 2016;19:102–13.
Lee HK, Mattei LM, Steinberg BE, Alberts P, Lee YH, Chervonsky A, et al. In vivo requirement for Atg5 in antigen presentation by dendritic cells. Immunity. 2010;32:227–39.
Ravindran R, Khan N, Nakaya HI, Li S, Loebbermann J, Maddur MS, et al. Vaccine activation of the nutrient sensor GCN2 in dendritic cells enhances antigen presentation. Science. 2014;343:313–7.
Leib DA, Alexander DE, Cox D, Yin J, Ferguson TA. Interaction of ICP34.5 with Beclin 1 modulates herpes simplex virus type 1 pathogenesis through control of CD4+ T-cell responses. J Virol. 2009;83:12164–71.
Xu X, Araki K, Li S, Han JH, Ye L, Tan WG, et al. Autophagy is essential for effector CD8(+) T cell survival and memory formation. Nat Immunol. 2014;15:1152–61.
Puleston DJ, Zhang H, Powell TJ, Lipina E, Sims S, Panse I, et al. Autophagy is a critical regulator of memory CD8(+) T cell formation. eLife. 2014;3:03706.
Xu Y, Zhou P, Cheng S, Lu Q, Nowak K, Hopp AK, et al. A bacterial effector reveals the V-ATPase-ATG16L1 axis that initiates xenophagy. Cell. 2019;178:552–66 e20.
Sorbara MT, Foerster EG, Tsalikis J, Abdel-Nour M, Mangiapane J, Sirluck-Schroeder I, et al. Complement C3 drives autophagy-dependent restriction of cyto-invasive bacteria. Cell Host Microbe. 2018;23:644–52 e5.
Tan JMJ, Mellouk N, Osborne SE, Ammendolia DA, Dyer DN, Li R, et al. An ATG16L1-dependent pathway promotes plasma membrane repair and limits Listeria monocytogenes cell-to-cell spread. Nat Microbiol. 2018;3:1472–85.
Zhang Y, Yao Y, Qiu X, Wang G, Hu Z, Chen S, et al. Listeria hijacks host mitophagy through a novel mitophagy receptor to evade killing. Nat Immunol. 2019;20:433–46.
Yoshikawa Y, Ogawa M, Hain T, Yoshida M, Fukumatsu M, Kim M, et al. Listeria monocytogenes ActA-mediated escape from autophagic recognition. Nat Cell Biol. 2009;11:1233–40.
Mitchell G, Cheng MI, Chen C, Nguyen BN, Whiteley AT, Kianian S, et al. Listeria monocytogenes triggers noncanonical autophagy upon phagocytosis, but avoids subsequent growth-restricting xenophagy. Proc Natl Acad Sci USA. 2018;115:E210–E7.
Mostowy S, Bonazzi M, Hamon MA, Tham TN, Mallet A, Lelek M, et al. Entrapment of intracytosolic bacteria by septin cage-like structures. Cell Host Microbe. 2010;8:433–44.
Ogawa M, Yoshimori T, Suzuki T, Sagara H, Mizushima N, Sasakawa C. Escape of intracellular Shigella from autophagy. Science. 2005;307:727–31.
Barnett TC, Liebl D, Seymour LM, Gillen CM, Lim JY, Larock CN, et al. The globally disseminated M1T1 clone of group A Streptococcus evades autophagy for intracellular replication. Cell Host Microbe. 2013;14:675–82.
Schnaith A, Kashkar H, Leggio SA, Addicks K, Kronke M, Krut O. Staphylococcus aureus subvert autophagy for induction of caspase-independent host cell death. J Biol Chem. 2007;282:2695–706.
Maurer K, Reyes-Robles T, Alonzo F 3rd, Durbin J, Torres VJ, Cadwell K. Autophagy mediates tolerance to Staphylococcus aureus alpha-toxin. Cell Host Microbe. 2015;17:429–40.
Maurer K, Torres VJ, Cadwell K. Autophagy is a key tolerance mechanism during Staphylococcus aureus infection. Autophagy. 2015;11:1184–6.
Meyer-Morse N, Robbins JR, Rae CS, Mochegova SN, Swanson MS, Zhao Z, et al. Listeriolysin O is necessary and sufficient to induce autophagy during Listeria monocytogenes infection. PLoS One. 2010;5:e8610.
Zhao Z, Fux B, Goodwin M, Dunay IR, Strong D, Miller BC, et al. Autophagosome-independent essential function for the autophagy protein Atg5 in cellular immunity to intracellular pathogens. Cell Host Microbe. 2008;4:458–69.
Birmingham CL, Canadien V, Kaniuk NA, Steinberg BE, Higgins DE, Brumell JH. Listeriolysin O allows Listeria monocytogenes replication in macrophage vacuoles. Nature. 2008;451:350–4.
Benjamin JL, Sumpter R Jr., Levine B, Hooper LV. Intestinal epithelial autophagy is essential for host defense against invasive bacteria. Cell Host Microbe. 2013;13:723–34.
Conway KL, Kuballa P, Song JH, Patel KK, Castoreno AB, Yilmaz OH, et al. Atg16l1 is required for autophagy in intestinal epithelial cells and protection of mice from Salmonella infection. Gastroenterology. 2013;145:1347–57.
Marchiando AM, Ramanan D, Ding Y, Gomez LE, Hubbard-Lucey VM, Maurer K, et al. A deficiency in the autophagy gene Atg16L1 enhances resistance to enteric bacterial infection. Cell Host Microbe. 2013;14:216–24.
Martin PK, Marchiando A, Xu R, Rudensky E, Yeung F, Schuster SL, et al. Autophagy proteins suppress protective type I interferon signalling in response to the murine gut microbiota. Nat Microbiol. 2018;3:1131–41.
Symington JW, Wang C, Twentyman J, Owusu-Boaitey N, Schwendener R, Nunez G, et al. ATG16L1 deficiency in macrophages drives clearance of uropathogenic E. coli in an IL-1beta-dependent manner. Mucosal Immunol. 2015;8:1388–99.
Wang C, Mendonsa GR, Symington JW, Zhang Q, Cadwell K, Virgin HW, et al. Atg16L1 deficiency confers protection from uropathogenic Escherichia coli infection in vivo. Proc Natl Acad Sci USA. 2012;109:11008–13.
Tam JM, Mansour MK, Khan NS, Seward M, Puranam S, Tanne A, et al. Dectin-1-dependent LC3 recruitment to phagosomes enhances fungicidal activity in macrophages. J Infect Dis. 2014;210:1844–54.
Nicola AM, Albuquerque P, Martinez LR, Dal-Rosso RA, Saylor C, De Jesus M, et al. Macrophage autophagy in immunity to Cryptococcus neoformans and Candida albicans. Infect Immun. 2012;80:3065–76.
Shroff A, Sequeira R, Patel V, Reddy KVR. Knockout of autophagy gene, ATG5 in mice vaginal cells abrogates cytokine response and pathogen clearance during vaginal infection of Candida albicans. Cell Immunol 2018;324:59–73.
Akoumianaki T, Kyrmizi I, Valsecchi I, Gresnigt MS, Samonis G, Drakos E, et al. Aspergillus cell wall melanin blocks LC3-associated phagocytosis to promote pathogenicity. Cell Host Microbe. 2016;19:79–90.
Kyrmizi I, Ferreira H, Carvalho A, Figueroa JAL, Zarmpas P, Cunha C, et al. Calcium sequestration by fungal melanin inhibits calcium-calmodulin signalling to prevent LC3-associated phagocytosis. Nat Microbiol. 2018;3:791–803.
de Luca A, Smeekens SP, Casagrande A, Iannitti R, Conway KL, Gresnigt MS, et al. IL-1 receptor blockade restores autophagy and reduces inflammation in chronic granulomatous disease in mice and in humans. Proc Natl Acad Sci USA. 2014;111:3526–31.
Oikonomou V, Moretti S, Renga G, Galosi C, Borghi M, Pariano M, et al. Noncanonical fungal autophagy inhibits inflammation in response to IFN-gamma via DAPK1. Cell Host Microbe. 2016;20:744–57.
Boonhok R, Rachaphaew N, Duangmanee A, Chobson P, Pattaradilokrat S, Utaisincharoen P, et al. LAP-like process as an immune mechanism downstream of IFN-gamma in control of the human malaria Plasmodium vivax liver stage. Proc Natl Acad Sci USA. 2016;113:E3519–28.
Real E, Rodrigues L, Cabal GG, Enguita FJ, Mancio-Silva L, Mello-Vieira J, et al. Plasmodium UIS3 sequesters host LC3 to avoid elimination by autophagy in hepatocytes. Nat Microbiol. 2018;3:17–25.
Muniz-Feliciano L, Van Grol J, Portillo JA, Liew L, Liu B, Carlin CR, et al. Toxoplasma gondii-induced activation of EGFR prevents autophagy protein-mediated killing of the parasite. PLoS Pathog. 2013;9:e1003809.
Portillo JC, Muniz-Feliciano L, Lopez Corcino Y, Lee SJ, Van Grol J, Parsons SJ, et al. Toxoplasma gondii induces FAK-Src-STAT3 signaling during infection of host cells that prevents parasite targeting by autophagy. PLoS Pathog. 2017;13:e1006671.
Romano PS, Arboit MA, Vazquez CL, Colombo MI. The autophagic pathway is a key component in the lysosomal dependent entry of Trypanosoma cruzi into the host cell. Autophagy. 2009;5:6–18.
Zhao Z, Fux B, Goodwin M, Dunay IR, Strong D, Miller BC, et al. Autophagosome-independent essential function for the autophagy protein Atg5 in cellular immunity to intracellular pathogens. Cell Host amp; Microbe. 2008;4:458–69.
Choi J, Park S, Biering SB, Selleck E, Liu CY, Zhang X, et al. The parasitophorous vacuole membrane of Toxoplasma gondii is targeted for disruption by ubiquitin-like conjugation systems of autophagy. Immunity. 2014;40:924–35.
Haldar AK, Piro AS, Pilla DM, Yamamoto M, Coers J. The E2-like conjugation enzyme Atg3 promotes binding of IRG and Gbp proteins to chlamydia- and toxoplasma-containing vacuoles and host resistance. PLoS One. 2014;9:e86684.
Ohshima J, Lee Y, Sasai M, Saitoh T, Su Ma J, Kamiyama N, et al. Role of mouse and human autophagy proteins in IFN-γ-induced cell-autonomous responses against Toxoplasma gondii. J Immunol. 2014;192:3328–35.
Selleck EM, Orchard RC, Lassen KG, Beatty WL, Xavier RJ, Levine B, et al. A noncanonical autophagy pathway restricts toxoplasma gondii growth in a strain-specific manner in IFN-γ-activated human cells. mBio. 2015;6:e01157–e15.
Park S, Choi J, Biering SB, Dominici E, Williams LE, Hwang S. Targeting by AutophaGy proteins (TAG): targeting of IFNG-inducible GTPases to membranes by the LC3 conjugation system of autophagy. Autophagy. 2016;12:1153–67.
Sasai M, Sakaguchi N, Ma JS, Nakamura S, Kawabata T, Bando H, et al. Essential role for GABARAP autophagy proteins in interferon-inducible GTPase-mediated host defense. Nat Immunol. 2017;18:899–910.
Biering SB, Choi J, Halstrom RA, Brown HM, Beatty WL, Lee S, et al. Viral replication complexes are targeted by LC3-guided interferon-inducible GTPases. Cell Host Microbe. 2017;22:74–85 e7.
Hwang S, Maloney NS, Bruinsma MW, Goel G, Duan E, Zhang L, et al. Nondegradative role of Atg5-Atg12/ Atg16L1 autophagy protein complex in antiviral activity of interferon gamma. Cell Host Microbe. 2012;11:397–409.
Burger E, Araujo A, Lopez-Yglesias A, Rajala MW, Geng L, Levine B, et al. Loss of paneth cell autophagy causes acute susceptibility to toxoplasma gondii-mediated inflammation. Cell Host Microbe. 2018;23:177–90 e4.
Huttenhower C, Kostic AD, Xavier RJ. Inflammatory bowel disease as a model for translating the microbiome. Immunity. 2014;40:843–54.
Wong SY, Cadwell K. There was collusion: microbes in inflammatory bowel disease. PLoS Pathog. 2018;14:e1007215.
Murthy A, Li Y, Peng I, Reichelt M, Katakam AK, Noubade R, et al. A Crohn's disease variant in Atg16l1 enhances its degradation by caspase 3. Nature. 2014;506:456–62.
Lassen KG, Kuballa P, Conway KL, Patel KK, Becker CE, Peloquin JM, et al. Atg16L1 T300A variant decreases selective autophagy resulting in altered cytokine signaling and decreased antibacterial defense. Proc Natl Acad Sci USA. 2014;111:7741–6.
Gao P, Liu H, Huang H, Zhang Q, Strober W, Zhang F. The inflammatory bowel disease-associated autophagy gene Atg16L1T300A acts as a dominant negative variant in mice. J Immunol. 2017;198:2457–67.
Ip WKE, Hoshi N, Shouval DS, Snapper S, Medzhitov R. Anti-inflammatory effect of IL-10 mediated by metabolic reprogramming of macrophages. Science. 2017;356:513–9.
Zhang H, Zheng L, McGovern DP, Hamill AM, Ichikawa R, Kanazawa Y, et al. Myeloid ATG16L1 facilitates host-bacteria interactions in maintaining intestinal homeostasis. J Immunol. 2017;198:2133–46.
Fletcher K, Ulferts R, Jacquin E, Veith T, Gammoh N, Arasteh JM, et al. The WD40 domain of ATG16L1 is required for its non-canonical role in lipidation of LC3 at single membranes. EMBO J. 2018;37:e97840.
Boada-Romero E, Serramito-Gomez I, Sacristan MP, Boone DL, Xavier RJ, Pimentel-Muinos FX. The T300A Crohn's disease risk polymorphism impairs function of the WD40 domain of ATG16L1. Nat Commun. 2016;7:11821.
Chu H, Khosravi A, Kusumawardhani IP, Kwon AH, Vasconcelos AC, Cunha LD, et al. Gene-microbiota interactions contribute to the pathogenesis of inflammatory bowel disease. Science. 2016;352:1116–20.
Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–63.
Ramanan D, Cadwell K. Intrinsic defense mechanisms of the intestinal epithelium. Cell Host Microbe. 2016;19:434–41.
Cabrera S, Fernandez AF, Marino G, Aguirre A, Suarez MF, Espanol Y, et al. ATG4B/autophagin-1 regulates intestinal homeostasis and protects mice from experimental colitis. Autophagy. 2013;9:1188–200.
Cadwell K, Patel KK, Komatsu M, Virgin HWt, Stappenbeck TS. A common role for Atg16L1, Atg5 and Atg7 in small intestinal Paneth cells and Crohn disease. Autophagy. 2009;5:250–2.
Adolph TE, Tomczak MF, Niederreiter L, Ko HJ, Bock J, Martinez-Naves E, et al. Paneth cells as a site of origin for intestinal inflammation. Nature. 2013;503:272–6.
Yang L, Liu C, Zhao W, He C, Ding J, Dai R, et al. Impaired autophagy in intestinal epithelial cells alters gut microbiota and host immune responses. Appl Environ Microbiol. 2018;84:e00880–18.
Cadwell K, Patel KK, Maloney NS, Liu TC, Ng AC, Storer CE, et al. Virus-plus-susceptibility gene interaction determines Crohn's disease gene Atg16L1 phenotypes in intestine. Cell. 2010;141:1135–45.
Kernbauer E, Ding Y, Cadwell K. An enteric virus can replace the beneficial function of commensal bacteria. Nature. 2014;516:94–8.
Abt MC, Buffie CG, Susac B, Becattini S, Carter RA, Leiner I, et al. TLR-7 activation enhances IL-22-mediated colonization resistance against vancomycin-resistant enterococcus. Sci Transl Med. 2016;8:327ra25.
Lavoie S, Conway KL, Lassen KG, Jijon HB, Pan H, Chun E, et al. The Crohn's disease polymorphism, ATG16L1 T300A, alters the gut microbiota and enhances the local Th1/Th17 response. eLife. 2019;8:e39982.
Diamanti MA, Gupta J, Bennecke M, De Oliveira T, Ramakrishnan M, Braczynski AK, et al. IKKalpha controls ATG16L1 degradation to prevent ER stress during inflammation. J Exp Med. 2017;214:423–37.
Tschurtschenthaler M, Adolph TE, Ashcroft JW, Niederreiter L, Bharti R, Saveljeva S, et al. Defective ATG16L1-mediated removal of IRE1alpha drives Crohn's disease-like ileitis. J Exp Med. 2017;214:401–22.
Deuring JJ, Fuhler GM, Konstantinov SR, Peppelenbosch MP, Kuipers EJ, de Haar C, et al. Genomic ATG16L1 risk allele-restricted Paneth cell ER stress in quiescent Crohn's disease. Gut. 2014;63:1081–91.
Grootjans J, Krupka N, Hosomi S, Matute JD, Hanley T, Saveljeva S, et al. Epithelial endoplasmic reticulum stress orchestrates a protective IgA response. Science. 2019;363:993–8.
Adolph TE, Tomczak MF, Niederreiter L, Ko HJ, Bock J, Martinez-Naves E, et al. Paneth cells as a site of origin for intestinal inflammation. Nature. 2013;503:272–6.
Pott J, Kabat AM, Maloy KJ. Intestinal epithelial cell autophagy is required to protect against TNF-induced apoptosis during chronic colitis in Mice. Cell Host Microbe. 2018;23:191–202 e4.
Zhu X, Messer JS, Wang Y, Lin F, Cham CM, Chang J, et al. Cytosolic HMGB1 controls the cellular autophagy/apoptosis checkpoint during inflammation. J Clin Invest. 2015;125:1098–110.
Aden K, Tran F, Ito G, Sheibani-Tezerji R, Lipinski S, Kuiper JW, et al. ATG16L1 orchestrates interleukin-22 signaling in the intestinal epithelium via cGAS-STING. J Exp Med. 2018;215:2868–86.
Bel S, Pendse M, Wang Y, Li Y, Ruhn KA, Hassell B, et al. Paneth cells secrete lysozyme via secretory autophagy during bacterial infection of the intestine. Science. 2017;357:1047–52.
Patel KK, Miyoshi H, Beatty WL, Head RD, Malvin NP, Cadwell K, et al. Autophagy proteins control goblet cell function by potentiating reactive oxygen species production. EMBO J. 2013;32:3130–44.
Slowicka K, Serramito-Gomez I, Boada-Romero E, Martens A, Sze M, Petta I, et al. Physical and functional interaction between A20 and ATG16L1-WD40 domain in the control of intestinal homeostasis. Nat Commun. 2019;10:1834.
Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–39.
Liang C, Feng P, Ku B, Dotan I, Canaani D, Oh BH, et al. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol. 2006;8:688–99.
Gannage M, Dormann D, Albrecht R, Dengjel J, Torossi T, Ramer PC, et al. Matrix protein 2 of influenza A virus blocks autophagosome fusion with lysosomes. Cell Host Microbe. 2009;6:367–80.
Beale R, Wise H, Stuart A, Ravenhill BJ, Digard P, Randow F. A LC3-interacting motif in the influenza A virus M2 protein is required to subvert autophagy and maintain virion stability. Cell Host Microbe. 2014;15:239–47.
Shi J, Wong J, Piesik P, Fung G, Zhang J, Jagdeo J, et al. Cleavage of sequestosome 1/p62 by an enteroviral protease results in disrupted selective autophagy and impaired NFKB signaling. Autophagy. 2013;9:1591–603.
Taylor MP, Kirkegaard K. Modification of cellular autophagy protein LC3 by poliovirus. J Virol. 2007;81:12543–53.
Kyei GB, Dinkins C, Davis AS, Roberts E, Singh SB, Dong C, et al. Autophagy pathway intersects with HIV-1 biosynthesis and regulates viral yields in macrophages. J Cell Biol. 2009;186:255–68.
Gregoire IP, Richetta C, Meyniel-Schicklin L, Borel S, Pradezynski F, Diaz O, et al. IRGM is a common target of RNA viruses that subvert the autophagy network. PLoS Pathog. 2011;7:e1002422.
Acknowledgements
We apologize for omitting many great studies in this review due to lack of space. The selected studies are used as examples of the great complexity and breadth of autophagy studies. The authors laboratories are supported by US National Institute of Health grants R01 AI121244 (KC and VJT), R01 HL123340 (KC), R01 DK093668 (KC), R01 DK103788 (KC), R01 AI130945 (KC), R01 HL125816 (KC), R01AI099394 (VJT) and R01AI105129 (VJT), T32AI007180 and F31HL137304 (MDK) and pilot awards from the NYU CTSA grant UL1TR001445 from the National Center for Advancing Translational Sciences (KC) and NYU Cancer Center grant P30CA016087 (KC). The laboratories have also received recent support from the Faculty Scholar grant from the Howard Hughes Medical Institute (KC), Merieux Institute (KC), Kenneth Rainin Foundation (KC), Crohn’s & Colitis Foundation, and Stony Wold-Herbert Fund (KC). KC and VJT are Burroughs Wellcome Fund Investigators in the Pathogenesis of Infectious Diseases.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
VJT is an inventor on patents and patent applications filed by New York University, which are currently under commercial license to Janssen Biotech Inc. KC has consulted for or received an honorarium from Puretech Health, Genentech, and AbbVie, Inc., has received research support from Puretech Health and Pfizer, Inc, and has a provisional patent, US Patent Appln. No. 15/625,934.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by F. Pentimalli
Rights and permissions
About this article
Cite this article
Keller, M.D., Torres, V.J. & Cadwell, K. Autophagy and microbial pathogenesis. Cell Death Differ 27, 872–886 (2020). https://doi.org/10.1038/s41418-019-0481-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-019-0481-8
This article is cited by
-
SCD1, autophagy and cancer: implications for therapy
Journal of Experimental & Clinical Cancer Research (2021)
-
Emerging roles of the HECT-type E3 ubiquitin ligases in hematological malignancies
Discover Oncology (2021)
-
Mechanisms controlling bacterial infection in myeloid cells under hypoxic conditions
Cellular and Molecular Life Sciences (2021)
-
Autophagy participates in, well, just about everything
Cell Death & Differentiation (2020)