Abstract
Mitochondrial cristae are critical for efficient oxidative phosphorylation, however, how cristae architecture is precisely organized remains largely unknown. Here, we discovered that Mic19, a core component of MICOS (mitochondrial contact site and cristae organizing system) complex, can be cleaved at N-terminal by mitochondrial protease OMA1 under certain physiological stresses. Mic19 directly interacts with mitochondrial outer-membrane protein Sam50 (the key subunit of SAM complex) and inner-membrane protein Mic60 (the key component of MICOS complex) to form Sam50–Mic19–Mic60 axis, which dominantly connects SAM and MICOS complexes to assemble MIB (mitochondrial intermembrane space bridging) supercomplex for mediating mitochondrial outer- and inner-membrane contact. OMA1-mediated Mic19 cleavage causes Sam50–Mic19–Mic60 axis disruption, which separates SAM and MICOS and leads to MIB disassembly. Disrupted Sam50–Mic19–Mic60 axis, even in the presence of SAM and MICOS complexes, causes the abnormal mitochondrial morphology, loss of mitochondrial cristae junctions, abnormal cristae distribution and reduced ATP production. Importantly, Sam50 displays punctate distribution at mitochondrial outer membrane, and acts as an anchoring point to guide the formation of mitochondrial cristae junctions. Therefore, we propose that Sam50–Mic19–Mic60 axis-mediated SAM–MICOS complexes integration determines mitochondrial cristae architecture.
Similar content being viewed by others
Introduction
Mitochondria are fully articulated and highly organized organelles which surrounded by two membranes: the outer mitochondrial membrane (OMM) and the inner mitochondrial membrane (IMM) [1]. IMM is consists of the inner boundary membrane (IBM) and the cristae membrane. OMM and IBM are firmly connected by mitochondrial contact sites. The IBM is closely apposed to the OMM, and it protrudes into the matrix of the mitochondria to form cristae [2,3,4]. The connections between IBM and the cristae are the crista junctions (CJs), which are relatively uniform narrow, tubular and slot-like structures [5].
Mitochondrial cristae and crista junctions are important for mitochondrial organization and functions [6]. The formation of cristae and crista junctions requires Mgm1 (known as OPA1 in mammals), the dimeric form of the F1FO-ATP synthase (F1FO), Prohibitins (PHBs), and MICOS complex. Mgm1 (OPA1) cooperates with dimeric F1FO to stabilize the cristae membranes and to thereby generate the sac-like structure [5, 7, 8]. mitochondrial inner membrane protease OMA1 and Yme1L processes OPA1 at the S1 and S2 cleavage site, respectively, and the processing of OPA1 regulates mitochondrial morphology and cristae remodeling [8,9,10,11]. Additionally, OPA1 cleavage at its C-terminal is associated with the loss of cristae in the mouse postprandial model [12]. The assembly of MICOS complex is proposed to limit the fusion process by forming a crista junction [13,14,15]. However, so far, how mitochondrial cristae and crista junctions are formed remained largely unknown.
Eight subunits of MICOS complex in mammalian are described: Mic60/IMMT, Mic19/CHCHD3, Mic10/MINOS1, Mic23/Mic26/APOO, Mic27/APOOL, Mic13/QIL1, Mic25/CHCHD6 and Mic14/CHCHD10 [16,17,18,19,20,21,22,23,24]. In MICOS complex, Mic60, Mic19 and Mic10 play a dominant role in cristae patterning [16, 25]. Additionally, Mic10 and Mic60 show the ability in bending the liposome membrane in vitro [26, 27]. Mic60 or Mic19 ablation can affect the mitochondrial dynamics [16, 17, 28]. Furthermore, Mic60 and Mic19 could bind to Sam50 to sustain the cristae structure [29,30,31]. Sam50 is a key component of the SAM complex, which is essential for membrane integration and assembly of β-barrel proteins into the mitochondrial outer membrane [32, 33]. The SAM complex is assembled by three components: the core subunit Sam50 (Tob55) and two peripheral membrane proteins, Sam35 and Sam37 (Metaxin1 (MTX1) and Metaxin2 (MTX2) in mammals) [32]. Sam50 depletion, but not Metaxin2 depletion, could trigger abnormal mitochondrial morphology and cristae structures [34,35,36], but the mechanism of Sam50 in regulating cristae organization is still obscure. In addition, it remains unclear how the synergistic interactions between SAM and MICOS complex play role in the formation of mitochondrial contact sites and the biogenesis of mitochondrial cristae structure. Recently, some groups reported a larger protein complex, around 2200–2800 kDa, stretching across the inner and out membrane which termed “mitochondrial intermembrane space bridging” (MIB) complex [35, 37, 38], in which MICOS complex interacts with DNAJC11, Sam50, MTX1, and MTX2 to regulate the cristae function [3, 39, 40]. However, the mechanism of MIB complex assembly and the role of MIB complex in mitochondrial morphology and ultrastructure are largely unknown.
In this study, we find that Mic19 can be cleaved at N-terminal by mitochondrial metalloprotease OMA1 upon certain stresses. This feature provides us with excellent tools to prove the key role of the Sam50–Mic19–Mic60 axis in mediating mitochondrial outer- and inner-membrane contact. The cleaved short form of Mic19 (S-Mic19) disrupts the interaction between SAM and MICOS and then causes the abnormal mitochondrial morphology, loss of mitochondrial crista junctions, and impairs ATP production. Overall, we provide a new mode that Sam50–Mic19–Mic60 axis-mediated OMM and IMM contact dominantly regulates the formation of cristae junction.
Results
Mic19 can be cleaved by mitochondrial protease OMA1
Mitochondrial proteases, which are distributed in the different mitochondrial compartment and responsible for the turnover of mitochondrial proteins, regulate the mitochondrial homeostasis. Mic19 knockdown resulted in Sam50 degradation, and Yme1L knockdown partly inhibited Sam50 degradation (Figure S1A); furthermore, Yme1L physically interacted with Sam50 (Figure S1B). These data suggest that Yme1L regulates Sam50 degradation. Unexpectedly, instead of causing the degradation of Mic19, Sam50 knockdown (shSam50) resulted in Mic19 cleavage, which forms a short form of Mic19 (S-Mic19) (Fig. 1a). However, the depletion of Mic60 or Mic25 did not lead to Mic19 cleavage (Figures S1C and S1D).
Mic19 can be cleaved by mitochondrial protease OMA1. a-b Wild-type (WT), Yme1L KO (a) or OMA1 KO (b) HCT116 cells were infected by retrovirus particles containing control or shSam50, and further cultured for 7 days. Cell lysates were analyzed by Western blotting using the indicated antibodies. c HeLa cells stably expressing Human Mic19-Flag were treated with DMSO or CCCP (20 μM, 4 h). Cell lysates were analyzed by Western blotting with anti-Flag, anti-Mic19, or anti-HSP60 antibodies. The black arrowhead indicates the specific protein. d HeLa cells stably expressing Mic19-Flag were infected by lentivirus particles containing control or shOMA1, and cultured for an additional 5 days. Cells were then treated with DMSO or CCCP (20 μM) for 4 h. Mic19-Flag cleavage was examined by Western blotting using anti-Flag antibodies. e 293 T cells were transiently transfected with control (empty vector) or plasmid coding for OMA1E324Q-Flag (which glutamic acid (E) at residue 324 was substituted by glutamine (Q) that leads to OMA1 catalytically dead). After 48 h transfection, the cells were harvested and solubilized with IP lysis buffer, and cell lysates were then immunoprecipitated with anti-Flag M2 affinity gel, followed by Western blotting using the indicated antibodies. f The schematic presentation of human Mic19 and Mic25 domain. ‘DUF737’ indicates the domain of unknown function; ‘CHCH’ indicates the coiled-coil helix-coiled-coil helix domain. The comparison of N-terminal 27–38 amino acids (AA) of Mic19 and N-terminal 29–40AA of Mic25 were showed on the right. g HeLa cells stably expressing Mic19-Flag and the indicated Mic19 mutants (Flag-tagged) were treated with DMSO or CCCP (20 μM, 4 h), and cell lysates were then analyzed by Western blotting using anti-Flag or anti-Tubulin antibodies. “Δ” indicates deletion of residues. h-j HeLa cells expressing Mic25-Flag (h) or the indicated Mic25 mutants (i), and WT or OMA1 KO HCT116 cells expressing Mic25 mutants (j) were treated with DMSO or CCCP (20 μM, 4 h). Cell lysates were then analyzed by Western blotting using the indicated antibodies
Because Mic19 locates at the mitochondrial intermembrane space, we investigated whether Yme1L or OMA1 cleaves Mic19. OMA1 but not Yme1L knockout blocked Mic19 cleavage (Fig. 1a, b), indicating that OMA1 mediates Mic19 cleavage. To determine whether the Mic19 cleavage locates at the N-terminus or C-terminus, Mic19-Flag was transiently overexpressed in 293 T cells, and the cleavage of Mic19-Flag was detected (Figure S1E). Additionally, Mic19-Flag stably overexpressed HeLa cells were treated with CCCP to activate OMA1. Both endogenous S-Mic19 and exogenous S-Mic19-Flag were detected in CCCP-treatment cells but not in control (Fig. 1c), implying that Mic19 can be cleaved at N-terminal upon OMA1 activation. In contrast, in response to OMA1 knockdown or OMA1 knockout, Mic19-Flag failed to be cleaved even with CCCP treatment (Fig. 1d and S1F). In addition, co-immunoprecipitation (co-IP) assay revealed that Mic19 interacts with OMA1-E324Q (OMA1-E324Q is a proteolytic inactive mutant that could keep interaction with its substrates last longer [9]) (Fig. 1e). These results suggested that Mic19 is cleaved at N-terminal by OMA1.
To ascertain the cleavage site of Mic19, we firstly compared the molecular weight (MW) of proteins being displayed in SDS-PAGE, and found that S-Mic19 is about 3 kDa smaller than Mic19. 3 kDa is equivalent to the MW of about 36 amino acids (aa). We then deleted a total of 10aa (positions 31–40aa as in human) of Mic19 to infer the location of the cleavage site. We found the cleavage of the Mic19 variant deleting such 10aa is inhibited after CCCP treatment (Fig. 1f, g), indicating that the cleavage site is located around the region 31–40aa of Mic19. Next, we assessed the sequence conservation of Mic19 cleavage site in vertebrates. We obtained protein sequences of Mic19 and Mic25 (another MICOS subunit) of multiple species representing diverse vertebrate lineages from the GenBank database (https://www.ncbi.nlm.nih.gov/). These sequences were aligned and subsequently used to reconstruct a phylogenetic tree (Figure S1G), which is consistent with the vertebrate species tree [41], suggesting that Mic19 and Mic 25 protein sequences are generally conserved across vertebrates, respectively. The protein alignment further confirmed the sequence conservation of Mic19 (Figure S1H). By contrast, in the aligned region 31–40aa, two amino acids “ID” are strictly conserved in Mic19, whereas the corresponding amino acids are “VN” that are conserved in Mic25 (Figure S1H), suggesting that these two amino acids “ID” may be critical for cleavage of Mic19. Moreover, exogenous Mic25-Flag is un-cleavable upon CCCP treatment (Fig. 1h). Therefore, we mutated “ID” in Mic19 to “VN” (Mic19ID33-34VN) and implemented in a manner similar as the control did (Fig. 1g). No cleavage of Mic19ID33-34VN was detected in response to CCCP treatment (Fig. 1g), indicating that “ID” of Mic19 is the site of cleavage. In addition, co-IP assay revealed that both Mic19-Flag and Mic19ID33-34VN-Flag interacted with OMA1E324Q-Myc (Figures S1I), suggesting that “ID” is the cleavage site but not the binding site by OMA1. To further test whether “ID” are indispensable for the cleavage by OMA1, we replaced the “VN” in Mic25 with “ID” and examined the processing of Mic25VN35-36ID. OMA1-mediated cleavage of Mic25VN35-36ID was displayed (Fig. 1i, j). Therefore, the inter-paralog replacement of two amino acids endows Mic19 obvious resistance and Mic25 more susceptible to OMA1 cleavage. Additionally, the cleavage site of all known OMA1’s substrates including OPA1, OMA1, and Mic19 was aligned and compared, and no conserved sites were found in these substrates (Figure S1J).
Overall, these results suggest that the cleavage site of Mic19 by OMA1 is possibly right after the “ID” at its N terminal.
OMA1 mediates Mic19 cleavage in response to the certain physiological stresses
To investigate the physiological relevance of OMA1-mediated Mic19 cleavage, we produced a postprandial mouse liver model according to the previous report [12]. At 5 h postprandial, the hepatic phosphorylated ribosomal protein S6 was markedly decreased (Figures S2A and S2B), suggesting that mTORC1 activity is decreased; moreover, increased Mic19 cleavage was detected in postprandial liver (Figure S2C), indicating that OMA1-mediated Mic19 cleavage is physiologically relevant and associated with the mTORC1 signaling pathway. Furthermore, we assessed the physiological relevance of OMA1-mediated Mic19 cleavage by performing hypoxia-reperfusion injury (H/R) assay. In response to H/R, the OMA1 self-cleavage were increased, suggesting that OMA1 were activated [9], moreover, H/R-induced Mic19 cleavage (Figures S2D and S2E).
To investigate the physiological role of Mic19 cleavage, we assessed cell viability and apoptosis of Mic19 KO cells expressing wild-type Mic19 or mutant Mic19 (ID33-34VN). Upon H/R, compared with WT Mic19, Mic19 (ID33-34VN) expressed Mic19 KO HeLa cells showed an increased cell viability and decreased TUNEL-positive cells that indicate apoptotic cells (Figures S2F-S2H); moreover, the cleaved caspase-3 was decreased in H/R treated Mic19 KO HeLa cells expressing Mic19 (ID33-34VN) (Figure S2E). These data suggest that OMA1-mediated Mic19 cleavage promotes H/R-induced apoptosis. Interestingly, just a small amount of Mic19 were cleaved at 5 h postprandial or upon H/R (Figures S2C-S2E), but this few Mic19 cleavages promotes cell death (Figures S2F-S2H), indicating that S-Mic19, the product of Mic19 cleavage, gains proapoptotic function, in other words, S-Mic19 has a biological function per se.
Mic19 cleavage disrupts Sam50-Mic19-Mic60 axis and disassembles MIB supercomplex
Mic19 interacts with the mitochondrial outer membrane protein Sam50 and the mitochondrial inner membrane protein Mic60 [17]. We also demonstrated these interactions by mass spectrometry, co-IP and GST-pull down assay (Supplementary Table S1 and Figures S3A-3C). Then, we investigated whether Mic19 cleavage affects the interactions between Mic19 and Sam50 or Mic60. Co-IP assay revealed that Mic60 interacts with Mic19-Flag and S-Mic19-Flag but not with Mic19 (1-35aa)-Flag (N-terminal 35 amino acids of Mic19) (Fig. 2a). In contrast, large amounts of Sam50 were precipitated by Mic19-Flag or Mic19 (1-35aa)-Flag, but little amount of Sam50 was precipitated by S-Mic19-Flag (Fig. 2a). A small amount of Sam50 was still precipitated by S-Mic19-Flag probably because S-Mic19-Flag can interact with endogenous Mic19, which binds to this small amount of Sam50. Indeed, Mic19-Flag could precipitate S-Mic19 (Fig. 2a). These data suggest that Mic19 (1-35aa)-Flag but not S-Mic19-Flag interacts with Sam50. Furthermore, GST pull-down assay verified that Mic19 (1-35aa)-Flag but not S-Mic19-Flag specifically binds to Sam50, and S-Mic19-Flag but not Mic19 (1-35aa)-Flag directly interacted with Mic60 (Fig. 2b–d). Therefore, Mic19-Sam50 interaction was disrupted after Mic19 cleavage.
Mic19 cleavage disrupts Sam50-Mic19-Mic60 axis and disassembles the MIB supercomplex. a Cell lysates of 293 T cells expressing control (empty vector), Mic19-Flag, S-Mic19-Flag, and Mic19 (1-35aa)-Flag were used for co-immunoprecipitation (co-IP) assay with anti-Flag M2 resin, and the protein samples were subjected to immunoblot using the indicated antibodies. b–d GST or His fused proteins were expressed in E. coli, cell extracts were used for GST pull-down assay by using the Pierce Glutathione Agarose, and the protein samples were analyzed by Western blotting using antibodies against GST or His. The examination of the direct interaction between Mic19 (1-35AA) and Sam50 (b). The demonstration of the direct interaction between S-Mic19 and Sam50 (c). The detection of direct interaction between Mic19 (1-35AA) or S-Mic19 with Mic60 (371-590AA) (d). e Mitochondria were extracted from WT, Mic19 KO, or Mic19ΔgRNA-Flag or S-Mic19ΔgRNA-Flag expressed Mic19 KO HeLa cells, then non-denatured protein samples from mitochondria were analyzed for MIB, MICOS and SAM complexes by BN-PAGE and Western blotting using indicated antibodies. The bands of the complexes are labeled with corresponding boxes. The MIB complex is approximately 2000kDa, and the MICOS is ~700 kDa. SAM complex is approximately 250 kDa, ~ 125 kDa unidentified complex containing Sam50 is showed. Mitochondrial supercomplex II is detected by anti-SDHA, served as loading control. “Mic19∆gRNA” is a Mic19 mutation (2 nucleotides of Mic19 cDNA were mutated, but the amino acid sequences of Mic19 are not changed) in the sequence of guide RNA (gRNA) against Mic19 gene. Mic19∆gRNA could not be recognized by the Mic19 KO Cas9/CRISPR system. f Control (empty vector), Mic19ΔgRNA-Flag, S-Mic19ΔgRNA-Flag, and Mic19(ID33-34VN)ΔgRNA-Flag were stably expressed in Mic19 KO HeLa cells respectively. Cell lysates were analyzed by Western blot using indicated antibodies. g Cell lysates of WT and Mic19 KO HeLa cells expressing the indicated protein were analyzed for expression of MICOS subunits by Western blotting using the indicated antibodies. h Mitochondria were extracted from indicated cells, BN-PAGE and Western blotting analysis for the related cell lines, the non-denatured protein samples from mitochondria were analyzed for MIB, MICOS and SAM complexes by BN-PAGE and Western blotting. The bands of the complexes are labeled with corresponding boxes
MIB supercomplex could be formed by the interaction of SAM and MICOS complexes by using complexome profiling analysis [3]. We performed BN-PAGE assay to confirm the SAM, MICOS and MIB complexes by using Mic19 KO HeLa cells and the Mic19 cardiac-specific knockout mice heart (Figures S3D and S3E). Next, we examined the effect of Mic19 cleavage on the assembly of these complexes. Mitochondria from WT cells, Mic19 KO cells, Mic19∆gRNA-Flag cells, and S-Mic19∆gRNA-Flag expressed Mic19 KO cells were then analyzed by BN-PAGE. MIB complex was detected in WT (lane 1) and Mic19∆gRNA-Flag expressed Mic19 KO cells (lane 3) but not in Mic19 KO (lane 2) and S-Mic19∆gRNA-Flag expressed Mic19 KO cells (lane 4) (Fig. 2e). Interestingly, MICOS complex (~ 700 kDa) were still maintained in S-Mic19∆gRNA-Flag expressed Mic19 KO cells (Fig. 2e), suggesting that S-Mic19 maintains the ability to assemble MICOS complex. In addition, MICOS subunits Mic60 and Mic10 were recovered by Mic19∆gRNA-Flag, Mic19(ID33-34VN) ∆gRNA-Flag, and S-Mic19∆gRNA-Flag in Mic19 KO cells (Fig. 2f). However, Sam50 was only recovered by Mic19∆gRNA-Flag and Mic19-(ID33-34VN)-∆gRNA-Flag but not S-Mic19∆gRNA-Flag in Mic19 KO cells (Fig. 2f), suggesting that MIB supercomplex is disrupted in S-Mic19∆gRNA-Flag expressed Mic19 KO cells due to the disruption of Sam50-Mic19 interaction. Therefore, these results suggest that Mic19 cleavage disrupts the integrity of MIB supercomplex. Additionally, BN-PAGE assays revealed that S-Mic19∆gRNA-Flag and Myc-Sam50 co-expressed Mic19 KO cells still loss the MIB supercomplex although SAM and MICOS complexes are present (Fig. 2h), further conforming that Mic19-Sam50 interaction is required for the MIB supercomplex. Therefore, Mic19 cleavage disrupts the Sam50-Mic19-Mic60 axis and then result in the disassembly of MIB supercomplex.
Mic60 and Mic25 also bind to Sam50 [29,30,31], indicating that Sam50-Mic60 axis and Sam50-Mic25-Mic60 axis may also be responsible for MIB complex assembly. However, Sam50 is destabilized in S-Mic19∆gRNA-Flag expressed Mic19 KO cells, which maintain the normal level of Mic60 and Mic25 (Fig. 2f, g), suggesting that Mic19-Sam50 but not Mic60-Sam50 or Mic25-Sam50 interaction is responsible for Sam50 stabilization. Therefore, Sam50-Mic19-Mic60 axis dominantly contributes to the MIB supercomplex assembly and the mitochondrial outer- and inner-membrane contact.
Disruption of Sam50-Mic19-Mic60 axis causes the abnormal mitochondrial morphology and the loss of crista junctions
We then investigated the role of Sam50-Mic19-Mic60 axis in the mitochondrial morphology and structure. The certain confocal microscopy images were provided to show the typical mitochondrial shapes of ‘Tubular’, ‘Short Tubular’, ‘Fragmented’, ‘Expanded’, and ‘Large Spherical’ (Figures S4A). Remarkably, almost all Mic19 KO HeLa cells showed the ‘Expanded’ mitochondrial network (Fig. 3a). After re-expression of Mic19∆gRNA-Flag or Mic19-(ID33-34VN) ∆gRNA-Flag in Mic19 KO cells, which restored Sam50-Mic19-Mic60 axis, the normal tubular mitochondria was recovered (Fig. 3a–c). Surprisingly, more ‘large spherical mitochondria’ appeared in S-Mic19∆gRNA- Flag expressed-Mic19 KO cells in which Mic19-Mic60 interaction is restored but Sam50 is not recovered (Figs 2a–f, 3a, b). Because Sam50 depletion causes abnormal spherical mitochondria [35, 36], we overexpressed Myc-Sam50 in Mic19 KO cells or S-Mic19∆gRNA-Flag expressed Mic19 KO cells, and found that these cells expressing Myc-Sam50 still showed the spherical mitochondria (Fig. 3b, c), indicating that Mic19-Sam50 interaction is required for the maintenance of normal mitochondrial morphology. Additionally, “large spherical” mitochondria were also observed in Mic60 depleted MEFs [16]. Therefore, our data suggest that the disruption of Sam50-Mic19-Mic60 axis results in the abnormal spherical mitochondria.
Disruption of Sam50-Mic19-Mic60 axis leads to abnormal mitochondrial morphology and loss of crista junctions. a Mic19 KO HeLa cells stably expressing mito-GFP (a mitochondrial marker) were infected with retrovirus particles containing empty vector, Mic19ΔgRNA-Flag, S-Mic19ΔgRNA-Flag, and Mic19(ID33-34VN)ΔgRNA-Flag respectively. Cells were then immunostained with anti-Flag antibody, and mitochondrial morphology was visualized by confocal microscope. Local magnifications of all morphological graphs were shown on the top panel. b Myc-Sam50, or S-Mic19ΔgRNA-Flag plus Myc-Sam50 were expressed in Mic19 KO HeLa cells stably expressing mito-GFP, respectively. Cells were then immunostained with anti-Flag or anti-Myc antibodies, Mitochondrial morphology was visualized by confocal microscope. c Mitochondrial morphology described in “A and B” was counted according to the criteria detailed in“Materials and Methods”. All data represent the means ± SD of three independent experiments (100 cells per independent experiment). Statistical significance was assessed by student’s t-test, ***P < 0.001 and N.S indicates none significance. d Mitochondrial cristae junctions structure in WT, Mic19 KO, and Mic19 KO HeLa cells expressing Mic19-Flag, S-Mic19-Flag, Myc-Sam50 or S-Mic19-Flag plus Myc-Sam50 were analyzed by transmission electron microscope (TEM). The ultrathin section sample was observed using an electron microscope (JEM-1400 plus, Tokyo, Japan) under the condition of an accelerating voltage of 100 kV. e Statistics of the mitochondrial ultrastructure in the cells described in (d). Error bars represent means ± SD of three independent experiments in which 100 mitochondrial cristae and the number of the corresponding CJs were counted. ***P < 0.001 vs WT. f The relative ATP level of WT, Mic19 KO, and Mic19 KO HeLa cells expressing Mic19-Flag, S-Mic19-Flag, Myc-Sam50 or S-Mic19-Flag plus Myc-Sam50 were measured using an ATP assay kit. Error bars represent means ± SD of three independent experiments, ***P < 0.001 vs WT
Defficiency of MICOS complex key subunits such as Mic60 or Mic10 affects mitochondrial cristae conformation [16, 26]. Since Mic60 and Mic10 expression were recovered in S-Mic19∆gRNA-Flag expressed Mic19 KO cells, we speculated that cristae should be normal. However, S-Mic19∆gRNA-Flag expression could not recover the crista junctions in Mic19 KO cells (Fig. 3d). Furthermore, S-Mic19∆gRNA-Flag and Myc-Sam50 co-expressed Mic19 KO cells, which maintained Sam50, Mic60 and Mic10 expression (Fig. 2g), still could not restore crista junctions (CJs) (Fig. 3d, e), suggesting that Sam50-Mic19 interaction regulates Cjs formation. Because mitochondrial energy production is closely related to mitochondrial cristae [42], we then measured ATP production in cells. The disruption of the Sam50-Mic19 axis, even in the presence of SAM and MICOS complexes (Fig. 2h), resulted in significantly reduced ATP production (Fig. 3f). Therefore, Sam50-Mic19-Mic60 axis plays a critical role in the regulation of mitochondrial morphology, crista junctions formation, and ATP production.
Sam50 may act as a mitochondrial outer membrane anchoring point guiding the formation of crista junctions through Sam50-Mic19-Mic60 axis
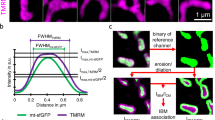
What is the role of Sam50, an outer membrane protein, in the formation of crista junctions? We assessed mitochondrial morphology and structure in short-term depletion of Sam50 (shSam50, 5 days) cells, which maintain the normal level of TOM40 and VDACs [36]. We performed Sam50 knockdown (5 days) in Mic19(ID33-34VN) ∆gRNA-Flag expressed Mic19 KO cells in which Mic19 cleavage is blocked. Upon Sam50 depletion, Mic60 and Mic10 were unaltered no matter whether Mic19 is cleaved or not (Fig. 4a). Confocal microscopy revealed that shSam50 (5 days) cells, shSam50 plus Mic19∆gRNA-Flag or Mic19(ID33-34VN) ∆gRNA-Flag expressed Mic19 KO cells displayed remarkably fragmented mitochondria (Fig. 4b, c). These results suggest that Sam50 deficiency, but not the cleavage of Mic19, directly regulates mitochondrial morphology. Moreover, TEM revealed that most cristae of WT mitochondria connected with inner membrane and crista junctions were regularly arranged (Fig. 4d). However, almost all cristae of shSam50 mitochondria pinched off from the inner membrane, leading to crista junctions collapsed (Fig. 4e), and the similar cristae were also found in shSam50 plus Mic19∆gRNA-Flag or Mic19-(ID33-34VN) ∆gRNA-Flag expressed Mic19 KO cells (Fig. 4e, f). These results indicate that Sam50 depletion is the primary effect leading to cristae deformation. In addition, upon shSam50, the level of MTX1 and MTX2 were remarkably decreased, and Mic19 cleavage was induced (Figure S4B). However, upon shMTX1 or shMTX2, Sam50 remained unchanged, and Mic19 cleavage was not induced (Figure S4B). These data suggest that the loss of Sam50 alone, but not SAM complex, induces Mic19 cleavage. Moreover, mitochondrial cristae are normal in response to MTX2 knockdown [35], indicating that Sam50 but not SAM complex regulates mitochondrial cristae remolding. It should be noted that Mic19 definitely regulates SAM complex stability by directly binding to Sam50, because Mic19 depletion results in a reduced level of Sam50, MTX1 and MTX2, moreover, caused the loss of SAM complex (Fig. 2e and S4C). Thus, we speculated that Sam50 might act as an anchoring point in mitochondria outer membrane to guide the formation of crista junctions. To further illustrate this hypothesis, we performed a super-resolution imaging with Hessian-SIM, which is a new powerful technology in visualizing mitochondrial cristae [43]. In Cos7 cells, mitochondrial cristae membranes were clearly observed by using Hessian-SIM, and Sam50-GFP signal is puncta and localized at mitochondrial outer membrane facing mitochondrial cristae junctions (Fig. 4g, h). In addition, TEM revealed that most mitochondrial cristae junctions were also lost in Sam50 KO MEFs (Figures S4E and S4F). Additionally, unlike Mic19 KO, Sam50 KO does not cause complete loss of cristae junctions, probably because Sam50 is only knockout for 5 days [long time such as 10 days results in cell death (data not shown)] and a small amount of Sam50 still exist (Figure S4D). Overall, Sam50 may serve as an outer membrane anchoring point and connect MICOS complex to anchor the crista junctions at a specific site of the mitochondrial inner membrane.
Sam50 may act as an anchoring point for Sam50-Mic19-Mic60 axis to guide the formation of mitochondrial crista junctions. a Knockdown of Sam50 (shSam50, 5 days) in WT, Mic19 KO, and Mic19ΔgRNA-Flag or Mic19 (ID33-34VN)ΔgRNA-Flag expressed Mic19 KO HeLa cells were performed by infecting cells with control or shSam50 lentiviral particles, cell lysates were then analyzed by Western blotting using indicated antibodies. Tubulin was used as loading control. b WT, Mic19ΔgRNA-Flag, or Mic19 (ID33-34VN)ΔgRNA-Flag expressed Mic19 KO HeLa cells were infected with control or shSam50 lentiviral particles. 5 days after infection, shSam50 and control cells were then mixed and grown on the same glass slide, and were fixed and immunostained with anti-HSP60 antibody, mitochondrial morphology was visualized by confocal microscope. shSam50 (shRNAi against Sam50) plasmid, which also encodes a nuclear localization signal fused GFP (nu-GFP), was used to deplete Sam50, and empty vector without nu-GFP was acted as control. Nu-GFP positive cells indicate “shSam50 cells”, and cells without nu-GFP expression indicate“control cells”. c Mitochondrial morphology of indicated cell lines was classified into three types (tubular, short tubular, and fragmented) and counted. Bars represent means ± S.D. of three independent experiments. d-e The mitochondrial cristae junction structures of WT, Mic19ΔgRNA-Flag or Mic19 (ID33-34VN)ΔgRNA-Flag expressed Mic19 KO HeLa cells with or without Sam50 knockdown were analyzed by transmission electron microscope (TEM). f Statistics of the mitochondrial ultrastructure in the indicated cell lines. The mean value and standard deviations (S.D.) were calculated from 3 independent experiments in which 100 mitochondrial cristae and the number of corresponding CJs were counted, ***P < 0.001. g Sam50-GFP transiently expressed COS7 cells were stained with MitoTracker® Red CMXRos dye for labeling mitochondria, and the living cells were visualized and imaged with Hessian-SIM operating at 2D-SIM configuration. A stack of SIM images was processed by Image J software. Boxed regions in the left image are magnified and shown on the right panel. The white arrowhead indicated the co-localization of Sam50-GFP with crista junctions. h Quantification of Sam50-GFP signals in OMM over against to the CJs structure. 300 Sam50-GFP signals were counted, and the relative number of the co-localization of Sam50-GFP with crista junctions were shown
Restoration of Sam50-X-Mic60 axis by overexpression of Mic25 in Mic19 KO cells can reconstruct mitochondrial morphology and crista junctions
Among subunits of the MICOS complex, Mic25 exhibits 36% overall sequence identity and 80% similarity to Mic19 [14]. In addition, Mic60 depletion reduced Mic25, while Mic19 KO decreased Mic60 but not altering Mic25 level (Figure S5A). Moreover, Mic25 is also capable of forming direct interaction with Sam50 and Mic60 (Figures S5B-S5E). Therefore, we investigate whether there is a functional overlap between Mic19 and Mic25. Mic25 KO did not affect the level of other MICOS subunits and mitochondrial morphology (Fig. 5a–e). However, compared with Mic19 KO, Mic25 knockdown (shMic25) plus Mic19 KO converted mitochondrial shape from “expanded” to ‘large spherical’ (Fig. 5b–d), and led to an obvious more reduction of Mic60, Mic10, and Sam50 (Fig. 5e). The similar phenotypes were observed in Mic19 knockdown (shMic19) plus Mic25 KO cells (Fig. 5c, d). These results suggest that Mic25 functionally overlap with Mic19, but Mic19 play a dominant role in mediating Sam50-X-Mic60 axis and maintaining mitochondrial morphology.
Restoration of Sam50-X-Mic60 axis by overexpression of Mic25 in Mic19 KO cells can reconstruct mitochondrial morphology and crista junctions. a–c WT (a), Mic19 KO (b), or Mic25 KO (c) HeLa cells expressing mito-GFP were knocked down for Mic19 (shMic19) or Mic25 (shMic25) respectively, and mitochondrial morphology was analyzed by confocal microscope. d Mitochondrial morphology in cell lines described in “a-c” was classified and counted according to the criteria detailed in “Materials and Methods”, and error bars represent means ± S.D. of three independent experiments, ***P < 0.001. e Cell lysates of the indicated cells described in “a-c” were analyzed by Western blotting using the indicated antibodies. Tubulin protein was used as loading control. f Cell lysates of WT, Mic19 KO, and Mic19-Flag or Mic25-Flag overexpressed Mic19 KO HeLa cells were analyzed for protein levels by Western blotting using the indicated antibodies. g Mic25-Flag was stably overexpressed in Mic19 KO cells expressing mito-GFP, and immunostained with anti-Flag antibody. Mitochondrial morphology in WT, Mic19 KO, and Mic25-Flag overexpressed Mic19 KO HeLa cells was assessed by confocal microscope. h Statistical analysis of mitochondrial morphology in the cells described in “g” was performed according to the criteria detailed in “Materials and Methods”, the mean value and standard deviations (S.D.) were calculated from 3 independent experiments in which 100 cells were scored, ***P < 0.001. i Mitochondrial crista junction structure of WT, Mic19 KO, or Mic25 overexpressed Mic19 KO HeLa cells were analyzed by transmission electron microscope (TEM). j Statistics of the mitochondrial ultrastructure in the indicated cells described in “h”. The mean value and standard deviations (S.D.) were calculated from 3 independent experiments in which 100 mitochondrial cristae and the number of corresponding CJs were counted, ***P < 0.001
Compared with Mic25, Mic19 possess a dominant role in Sam50-X-Mic60 axis, we, therefore, investigated whether Mic19 protein abundance is much higher than that of Mic25. The protein level of endogenous Mic19 and Mic25 are comparable, and Mic25 slightly lower than Mic19 (Figures S5F-S5I). In addition, Mic25 KO cells display normal mitochondrial cristae architecture (Figures S6A and S6B). These data suggest that Sam50-Mic19-Mic60 axis plays a key role in mitochondrial cristae organization, and upon Mic19 downregulation, Mic25 works together with the rest Mic60 and Sam50, and forms Sam50-Mic25-Mic60 axis to partially maintain the function of Sam50-X-Mic60 axis.
We investigated whether overexpressed Mic25 in Mic19 KO cells can form an axis to restore the mitochondrial outer and inner membrane contact. Overexpressed Mic25-Flag (about 10 fold of endogenous Mic25) in Mic19 KO cells recovered the level of other MICOS subunits and the normal mitochondrial morphology (Fig. 5f–h). Additionally, overexpression of Mic25-Flag in WT or Mic19 KO cells resulted in more incorporation of Mic25 or Mic25-Flag into MICOS complex and MIB complex, in contrast, less endogenous Mic19 was assembled into MICOS complex and MIB complex (Figure S6C). Moreover, expression of Mic19-GFP causes the reduced interaction between Mic25-Flag and Mic60 (Figure S6D). These results suggest that Mic25 and Mic19 competitively bind to Mic60, and overexpressed Mic25 could partially replace Mic19 to form a new axis with Sam50 and Mic60 and mediate the mitochondrial outer- and inner- membrane contact. Furthermore, mitochondrial crista junctions was reconstructed and the ATP level was recovered in Mic25 overexpressed Mic19 KO cells (Fig. 5i, j and S6E). Therefore, our data indicate that Mic25 is a compensatory factor for establishing Sam50-X-Mic60 axis, which bridges mitochondrial outer and inner membrane contact and regulates mitochondrial morphology and ultrastructure.
Discussion
We discovered that Mic19 can be cleaved at N-terminal by mitochondrial metalloprotease OMA1. OMA1-mediated Mic19 cleavage provides us a robust tool to explore the key role of the Sam50–Mic19–Mic60 axis in mediating mitochondrial outer and inner membrane contact. The cleavage of Mic19 interrupts the Sam50-Mic19-Mic60 interaction axis, and then results in the disruption of MIB complex. Although Mic60 can also interact with Sam50 [30], Mic60 but not Sam50 can be restored by expression of S-Mic19-Flag in Mic19 KO cells (Fig. 2f), suggesting that Mic60 alone cannot stabilize Sam50. Thus, Mic19 acts as the middle bearings to bridge and stabilize Sam50 (the outer membrane protein) and Mic60 (the inner membrane protein). It has been reported that N-terminal myristoylation of Mic19 may be required for the interaction with the outer membrane protein, and the myristoylation and the CHCH domain are essential for the import and mitochondrial localization of Mic19 [29], thus, S-Mic19-Flag losing its ability to interact with Sam50 may due to the missing of N-terminal myristoylation of Mic19. However, S-Mic19-Flag still maintains the ability to import into mitochondria and assembly MICOS complex (Figs. 2e and 3a).
In the last dozen years, MICOS and its subunits have been identified to be critical for mitochondrial membranes contact and cristae organization [4, 24, 44]. However, Mic60 or Mic19 deletion always accompanies with the disruption of whole MICOS complex [16, 17]. Therefore, it is hard to verify whether Mic60 or Mic19 alone is sufficient to regulate the cristae structure and mitochondrial membranes contact. Recently, Mic60 was reported to have the ability to bend liposomes in vitro [24], making Mic60 being a dominant player in stabilizing and promoting cristae formation. In addition, Mic10 oligomerization could efficiently bend liposomes in vitro [26]. These results suggest that Mic60 and Mic10 are cristae organization regulating effectors, and MICOS complex itself is supposed to be sufficient for cristae formation and organization. However, depletion of Mic60 or Mic19 not only severely disrupts MICOS subunits but also destabilizes mitochondrial outer membrane Sam50 (Figure S4A), which is closely involved in cristae organization [35, 36]. Therefore, it is questionable whether the abnormal cristae is caused directly by the depletion of Mic60, Mic19 or Sam50. In our study, we proved that Sam50-Mic19-Mic60 axis is critical for MIB assembly and maintenance of normal mitochondrial morphology and ultrastructure. Disrupted Sam50-Mic19-Mic60 axis, even in the presence of SAM and MICOS complex, cause the abnormal mitochondrial morphology, the loss of cristae junctions, and the decrease of ATP production (Fig. 3). Overall, Sam50-Mic19-Mic60 axis, but not MICOS or SAM complex, determines mitochondrial cristae architecture.
The mechanism of formation and maintenance of CJs remains unclear, and most of the previous studies focus on the role of mitochondrial inner membrane proteins, such as OPA1, Mic60, and PHBs, in mitochondrial cristae organization [7, 15, 45]. However, it is rarely reported how mitochondrial outer membrane proteins act on the formation of CJs. We found that CJs fall off from the inner membrane when Sam50 was depleted (5 days) without affecting MICOS core subunits Mic60 and Mic10 (Fig. 4), indicating that outer membrane protein Sam50 is also indispensable for crista junctions formation. Additionally, Sam50-Mic19-Mic60 axis determines the mitochondrial cristae structure (Fig. 3e, f), we put forward a new viewpoint that Sam50 acted as an anchoring point in the mitochondrial outer membrane for the mitochondrial crista junctions. We also used Hessian-SIM to visualize mitochondrial cristae [43]. Mitochondrial cristae membranes were clearly displayed, and Sam50-GFP signal showed puncta localization at mitochondrial outer membrane facing mitochondrial cristae junctions (Fig. 4g, h), indicating that Sam50 acted as an anchoring point in the mitochondrial outer membrane for the mitochondrial crista junctions. Therefore, Sam50 not only functions in the biogenesis of mitochondrial β-barrel proteins, but also cooperates with MICOS to regulate mitochondrial cristae organization.
Taken together, Mic19 stabilizes and links Sam50 and Mic60 to establish Sam50–Mic19–Mic60 axis. The synergistic contact of mitochondrial outer and inner membrane mediated by Sam50–Mic19–Mic60 axis is critical for the establishment and maintenance of mitochondrial cristae architecture.
Materials and methods
Cell culture and reagents
HeLa, HCT116, 293 T, GP2-293, MEFs and COS7 cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% (v/v) fetal bovine serum (PAN, Germany), 100 U/mL Penicillin and 100 mg/mL Streptomycin (Gibco) at 37 °C with 5% (v/v) CO2. Reagents used in this paper were listed as follows: Dimethyl sulfoxide (DMSO, Sigma-Aldrich); Carbonyl cyanide 3-chlorophenylhydrazone (CCCP, Sigma-Aldrich); no-Glucose DMEM (#11966-025, Invitrogen); Puromycin (#A1113803, Invitrogen); Hygromycin B (#V900372, Sigma-Aldrich); Digitonin (#D141, Sigma-Aldrich)
Immunostaining
Cells grown on glass coverslips in a 12-well plate were fixed with 4% paraformaldehyde (dissolved in PBS PH7.4) for 15–20 min at room temperature, and washed with PBS three times, cells were then incubated with 0.1% Triton X-100 (diluted with PBS) for 10 min and washed with PBS buffer once. Next, cells were blocked with PBS plus 10% FBS at room temperature (RT) for 1 h, and incubated with the primary antibody [note: anti-Flag (1:500; Sigma), anti-Myc (1:500; Santa Cruz), and anti-HSP60 (1:1000; Abclonal)] for 2 h at RT and subsequently washed with PBS three times per 5 min. Then, cells were incubated with PBS containing the secondary antibody (1:1000; Jackson ImmunoResearch) for 1 h in the dark at RT, and washed three times with PBS. Finally, coverslips containing cells were mounted, and visualized by a Leica confocal microscope (Leica microsystem, Germany).
Mitochondrial morphology analysis
To examine mitochondrial morphology, cells were randomly selected for quantitative analysis and visually scored into five classifications (‘Tubular’, ‘Short Tubular’, ‘Fragmented’, ‘Expanded’ and ‘Large Spherical’). Representative pictures of different mitochondrial morphology were shown in Figure S4A.
Co-Immunoprecipitation
For the co-immunoprecipitation (co-IP) assay, all procedures were performed at 4 °C. Cells were harvested and then solubilized with IP buffer (150 mM NaCl, 10% glycerol, 20 mM Tris–HCl pH 7.4, 2 mM EDTA, 0.5% Nonidet P-40, 0.5% Triton X-100 and complete protease inhibitor) for 1 h. The insoluble component was removed by centrifugation at 12,000 × g for 10 min, and the supernatant was collected. One-tenth of the supernatant was taken out and used as ‘Input’, and the rest subsequently incubated with anti-FLAG M2 affinity gel (Sigma-Aldrich) at 4 °C overnight. The resin was washed five times with lysis buffer, and then the proteins (IP products) were recovered by boiling the beads in SDS sample buffer and analyzed on SDS-PAGE followed by Western blot analysis.
GST pull-down assay
GST- or His-tagged plasmid was transformed and cultured in E. coli BL21, shaking 200 rpm/min at 37 °C for 20–24 h, then 500 μl of bacteria solution was used to inoculate into 10 ml LB liquid medium, shaking 220 rpm/min at 37 °C about 2–3 h. When the optical density reached OD600 = 0.6, the E. coli were induced with 0.4–1 mM IPTG, shaking 200 rpm/min at 16 °C for 18–24 h. The bacterium was then collected and ultrasonically disrupted in bacteria lysis buffer (1%Triton X-100, 150 mM Tris–HCl, 300 mM NaCl, protease inhibitor mixture pH7.4) on ice. The supernatant was collected by centrifugation at 12,000 × g for 10 min. Next, two protein supernatants to be tested for interaction were subsequently incubated with Pierce Glutathione Agarose beads (Thermo Scientific, Waltham, MA, USA) at 4 °C for 4 h or overnight. Then the beads were collected by centrifugation (1000 × g, 5 min) and washed five times with PBS at 4 °C. The proteins were recovered by boiling the beads in SDS sample buffer and analyzed by SDS–PAGE followed by Western blotting.
Electron microscopy
The procedure for transmission electron microscopy (TEM) was performed according to the previous report [46]. The 100 mM sodium cacodylate buffer was replaced by 100 mM phosphate buffer without CaCl2. The sections were supported on copper grids and then post-stained in uranyl acetate for 10 min and then in lead citrate for 15 min, and the stained sections were imaged onto negatives using a JEM-1400 plus electron microscope operated at 100 kV (Joel Ltd, Tokyo, Japan).
Mitochondria isolation and BN-PAGE analysis
Homogenization of cells or mouse tissues (liver and heart) and solubilization of mitochondria for BN-PAGE were performed according to the previous protocol [47]. The procedure for isolation of mitochondria of HeLa cells was optimized. A 100-mm dish of ~90% confluent cells were digested and collected in a 2.0 ml EP tube, then were centrifuged at 500 × g for 5 min. Cell pellet was re-suspended in cell homogenization buffer (83 mM sucrose, 6.6 mM imidazole/HCl, pH 7.0), ultrasonically disrupted cell membranes on ice. The nuclei and unbroken cells were removed by centrifugation at 700 × g for 10 min, and the supernatant subsequently was centrifuged at 10,000 × g for 5 min to collect the pellet containing the crude mitochondria. The pellet was further re-suspended and centrifuged at 10,000 × g for 5 min to collect the pellet containing the isolated mitochondria. The mitochondrial pellet was re-suspended in solubilization buffer A (50 mM sodium chloride, 50 mM Imidazole/HCL, 2 mM 6-aminohexanoic acid and 1 mM EDTA PH7.0 at 4 °C) and lysed by the addition of ~2% digitonin for 30 min on ice. The non-denatured protein samples were prepared by adding proportionate glycerol and Coomassie G250 dye, and then separated on 4–14% blue native polyacrylamide gels. The gel was transferred to PVDF and immunoblotted with the indicated antibodies, respectively.
Measurement of ATP Production
Cellular ATP levels were measured using an ATP assay kit (Celltiter-Glo Luminescent Cell Viability Assay, Promega) according to the manufacturer’s instructions. Luminescence was measured using Micro Plate Reader and the values were normalized to the protein concentration assessed by BCA Kit (#P0011, Beyotime Biotechnology, China).
Statistical methods
Densitometry was performed using ImageJ software for the quantitative analysis of the bands on the Western blots. Data were presented as the mean ± S.D. Student’s t-test was used to calculate P-values. Statistical significance is displayed as *p < 0.05, **p < 0.01, ***p < 0.001. Evolutionary analysis was previously described in detail elsewhere [48,49,50]. Briefly, protein sequences were aligned using the MUSCLE program [51]. The phylogenetic tree was reconstructed by the maximum-likelihood and Bayesian methods using PHYML [52] and MRBAYES [53] programs, respectively.
References
Mannella CA. Structure and dynamics of the mitochondrial inner membrane cristae. Biochim Biophys Acta. 2006;1763:542–8.
Demongeot J, Glade N, Hansen O, Moreira A. An open issue: the inner mitochondrial membrane (IMM) as a free boundary problem. Biochimie. 2007;89:1049–57.
Huynen MA, Muhlmeister M, Gotthardt K, Guerrero-Castillo S, Brandt U. Evolution and structural organization of the mitochondrial contact site (MICOS) complex and the mitochondrial intermembrane space bridging (MIB) complex. Biochim Biophys Acta. 2016;1863:91–101.
van der Laan M, Horvath SE, Pfanner N. Mitochondrial contact site and cristae organizing system. Curr Opin Cell Biol. 2016;41:33–42.
Rabl R, Soubannier V, Scholz R, Vogel F, Mendl N, Vasiljev-Neumeyer A, et al. Formation of cristae and crista junctions in mitochondria depends on antagonism between Fcj1 and Su e/g. J Cell Biol. 2009;185:1047–63.
Zick M, Rabl R, Reichert AS. Cristae formation-linking ultrastructure and function of mitochondria. Biochim Biophys Acta. 2009;1793:5–19.
Davies KM, Anselmi C, Wittig I, Faraldo-Gomez JD, Kuhlbrandt W. Structure of the yeast F1Fo-ATP synthase dimer and its role in shaping the mitochondrial cristae. eLife. 2012;109:13602–7.
Patten DA, Wong J, Khacho M, Soubannier V, Mailloux RJ, Pilon-Larose K, et al. OPA1-dependent cristae modulation is essential for cellular adaptation to metabolic demand. EMBO J. 2014;33:2676–91.
Zhang K, Li H, Song Z. Membrane depolarization activates the mitochondrial protease OMA1 by stimulating self-cleavage. EMBO Rep. 2014;15:576–85.
Ehses S, Raschke I, Mancuso G, Bernacchia A, Geimer S, Tondera D, et al. Regulation of OPA1 processing and mitochondrial fusion by m-AAA protease isoenzymes and OMA1. J Cell Biol. 2009;187:1023–36.
Song Z, Chen H, Fiket M, Alexander C, Chan DC. OPA1 processing controls mitochondrial fusion and is regulated by mRNA splicing, membrane potential, and Yme1L. J Cell Biol. 2007;178:749–55.
Sood A, Jeyaraju DV, Prudent J, Caron A, Lemieux P, McBride HM, et al. A Mitofusin-2-dependent inactivating cleavage of Opa1 links changes in mitochondria cristae and ER contacts in the postprandial liver. Proc Natl Acad Sci USA. 2014;111:16017–22.
Hoppins S, Collins SR, Cassidy-Stone A, Hummel E, Devay RM, Lackner LL, et al. A mitochondrial-focused genetic interaction map reveals a scaffold-like complex required for inner membrane organization in mitochondria. J Cell Biol. 2011;195:323–40.
Zerbes RM, van der Klei IJ, Veenhuis M, Pfanner N, van der Laan M, Bohnert M. Mitofilin complexes: conserved organizers of mitochondrial membrane architecture. Biol Chem. 2012;393:1247–61.
Harner ME, Unger AK, Geerts WJ, Mari M, Izawa T, Stenger M, et al. An evidence based hypothesis on the existence of two pathways of mitochondrial crista formation. Elife 2016, 5.
Li H, Ruan Y, Zhang K, Jian F, Hu C, Miao L, et al. Mic60/Mitofilin determines MICOS assembly essential for mitochondrial dynamics and mtDNA nucleoid organization. Cell Death Differ. 2016;23:380–92.
Darshi M, Mendiola VL, Mackey MR, Murphy AN, Koller A, Perkins GA, et al. ChChd3, an inner mitochondrial membrane protein, is essential for maintaining crista integrity and mitochondrial function. J Biol Chem. 2011;286:2918–32.
Alkhaja AK, Jans DC, Nikolov M, Vukotic M, Lytovchenko O, Ludewig F, et al. MINOS1 is a conserved component of mitofilin complexes and required for mitochondrial function and cristae organization. Mol Biol Cell. 2012;23:247–57.
Koob S, Barrera M, Anand R, Reichert AS. The non-glycosylated isoform of MIC26 is a constituent of the mammalian MICOS complex and promotes formation of crista junctions. Biochim Biophys Acta. 2015;1853:1551–63.
Weber TA, Koob S, Heide H, Wittig I, Head B, van der Bliek A, et al. APOOL is a cardiolipin-binding constituent of the Mitofilin/MINOS protein complex determining cristae morphology in mammalian mitochondria. PLoS ONE. 2013;8:e63683.
Guarani V, McNeill EM, Paulo JA, Huttlin EL, Frohlich F, Gygi SP, et al. QIL1 is a novel mitochondrial protein required for MICOS complex stability and cristae morphology. eLife 2015, 4:e06265.
An J, Shi J, He Q, Lui K, Liu Y, Huang Y, et al. CHCM1/CHCHD6, novel mitochondrial protein linked to regulation of mitofilin and mitochondrial cristae morphology. J Biol Chem. 2012;287:7411–26.
Genin EC, Plutino M, Bannwarth S, Villa E, Cisneros-Barroso E, Roy M, et al. CHCHD10 mutations promote loss of mitochondrial cristae junctions with impaired mitochondrial genome maintenance and inhibition of apoptosis. EMBO Mol Med. 2016;8:58–72.
Rampelt H, Zerbes RM, van der Laan M, Pfanner N. Role of the mitochondrial contact site and cristae organizing system in membrane architecture and dynamics. Biochim Biophys Acta. 2017;1864:737–46.
Bohnert M, Zerbes RM, Davies KM, Muhleip AW, Rampelt H, Horvath SE, et al. Central role of Mic10 in the mitochondrial contact site and cristae organizing system. Cell Metab. 2015;21:747–55.
Barbot M, Jans DC, Schulz C, Denkert N, Kroppen B, Hoppert M, et al. Mic10 oligomerizes to bend mitochondrial inner membranes at cristae junctions. Cell Metab. 2015;21:756–63.
Hessenberger M, Zerbes RM, Rampelt H, Kunz S, Xavier AH, Purfurst B, et al. Regulated membrane remodeling by Mic60 controls formation of mitochondrial crista junctions. Nat Commun. 2017;8:15258.
Cho B, Cho HM, Jo Y, Kim HD, Song M, Moon C, et al. Constriction of the mitochondrial inner compartment is a priming event for mitochondrial division. Nat Commun. 2017;8:15754.
Darshi M, Trinh KN, Murphy AN, Taylor SS. Targeting and import mechanism of coiled-coil helix coiled-coil helix domain-containing protein 3 (ChChd3) into the mitochondrial intermembrane space. J Biol Chem. 2012;287:39480–91.
Ding C, Wu Z, Huang L, Wang Y, Xue J, Chen S, et al. Mitofilin and CHCHD6 physically interact with Sam50 to sustain cristae structure. Sci Rep. 2015;5:16064.
Sastri M, Darshi M, Mackey M, Ramachandra R, Ju S, Phan S, et al. Sub-mitochondrial localization of the genetic-tagged mitochondrial intermembrane space-bridging components Mic19, Mic60 and Sam50. J Cell Sci. 2017;130:3248–60.
Wiedemann N, Kozjak V, Chacinska A, Schonfisch B, Rospert S, Ryan MT, et al. Machinery for protein sorting and assembly in the mitochondrial outer membrane. Nature. 2003;424:565–71.
Qiu J, Wenz LS, Zerbes RM, Oeljeklaus S, Bohnert M, Stroud DA, et al. Coupling of mitochondrial import and export translocases by receptor-mediated supercomplex formation. Cell. 2013;154:596–608.
Capala ME, Pruis M, Vellenga E, Schuringa JJ. Depletion of SAM50 specifically targets BCR-ABL-expressing leukemic stem and progenitor cells by interfering with mitochondrial functions. Stem Cells Dev. 2016;25:427–37.
Ott C, Ross K, Straub S, Thiede B, Gotz M, Goosmann C, et al. Sam50 functions in mitochondrial intermembrane space bridging and biogenesis of respiratory complexes. Mol Cell Biol. 2012;32:1173–88.
Jian F, Chen D, Chen L, Yan C, Lu B, Zhu Y, et al. Sam50 Regulates PINK1-Parkin-Mediated Mitophagy by Controlling PINK1 Stability and Mitochondrial Morphology. Cell Rep. 2018;23:2989–3005.
Ott C, Dorsch E, Fraunholz M, Straub S, Kozjak-Pavlovic V. Detailed analysis of the human mitochondrial contact site complex indicate a hierarchy of subunits. PLoS ONE. 2015;10:e0120213.
Kozjak-Pavlovic V. The MICOS complex of human mitochondria. Cell Tissue Res. 2017;367:83–93.
Xie J, Marusich MF, Souda P, Whitelegge J, Capaldi RA. The mitochondrial inner membrane protein mitofilin exists as a complex with SAM50, metaxins 1 and 2, coiled-coil-helix coiled-coil-helix domain-containing protein 3 and 6 and DnaJC11. FEBS Lett. 2007;581:3545–9.
Ioakeimidis F, Ott C, Kozjak-Pavlovic V, Violitzi F, Rinotas V, Makrinou E, et al. A splicing mutation in the novel mitochondrial protein DNAJC11 causes motor neuron pathology associated with cristae disorganization, and lymphoid abnormalities in mice. PLoS ONE. 2014;9:e104237.
Li D, Zhang J. Diet shapes the evolution of the vertebrate bitter taste receptor gene repertoire. Mol Biol Evol. 2014;31:303–9.
Tao M, You CP, Zhao RR, Liu SJ, Zhang ZH, Zhang C, et al. Animal mitochondria: evolution, function, and disease. Curr Mol Med. 2014;14:115–24.
Huang X, Fan J, Li L, Liu H, Wu R, Wu Y, et al. Fast, long-term, super-resolution imaging with Hessian structured illumination microscopy. Nat Biotechnol. 2018;36:451–9.
van der Laan M, Bohnert M, Wiedemann N, Pfanner N. Role of MINOS in mitochondrial membrane architecture and biogenesis. Trends Cell Biol. 2012;22:185–92.
Hering T, Kojer K, Birth N, Hallitsch J, Taanman JW, Orth M. Mitochondrial cristae remodelling is associated with disrupted OPA1 oligomerisation in the Huntington’s disease R6/2 fragment model. Exp Neurol. 2017;288:167–75.
McCaffery EMPaJM. Conventional and immunoelectron microscopy of mitochondria. Methods Mol Biol. 2007;372:467–83.
Wittig I, Braun HP, Schagger H. Blue native PAGE. Nat Protoc. 2006;1:418–28.
Feng P, Zheng J, Rossiter SJ, Wang D, Zhao H. Massive losses of taste receptor genes in toothed and baleen whales. Genome Biol Evol. 2014;6:1254–65.
Wu J, Jiao H, Simmons NB, Lu Q, Zhao H. Testing the sensory trade-off hypothesis in New World bats. Proc Biol Sci. 2018;285:20181523
Jiao H, Wang Y, Zhang L, Jiang P, Zhao H. Lineage-specific duplication and adaptive evolution of bitter taste receptor genes in bats. Mol Biol Evol. 2018;27:4475–88.
Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinf. 2004;5:113.
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–21.
Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–4.
Acknowledgements
This work is supported by National Natural Science Foundation of China (91854107 and 31671393), and the Fundamental Research Funds for the Central Universities (2042017kf0197 and 2042017kf0242).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by L. Scorrano
Supplementary information
Rights and permissions
About this article
Cite this article
Tang, J., Zhang, K., Dong, J. et al. Sam50–Mic19–Mic60 axis determines mitochondrial cristae architecture by mediating mitochondrial outer and inner membrane contact. Cell Death Differ 27, 146–160 (2020). https://doi.org/10.1038/s41418-019-0345-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-019-0345-2
This article is cited by
-
Vitamin D ameliorates age-induced nonalcoholic fatty liver disease by increasing the mitochondrial contact site and cristae organizing system (MICOS) 60 level
Experimental & Molecular Medicine (2024)
-
Mic19 depletion impairs endoplasmic reticulum-mitochondrial contacts and mitochondrial lipid metabolism and triggers liver disease
Nature Communications (2024)
-
TRAP1 inhibits MARCH5-mediated MIC60 degradation to alleviate mitochondrial dysfunction and apoptosis of cardiomyocytes under diabetic conditions
Cell Death & Differentiation (2023)
-
Adipocyte IRE1α promotes PGC1α mRNA decay and restrains adaptive thermogenesis
Nature Metabolism (2022)
-
Mechanical instability generated by Myosin 19 contributes to mitochondria cristae architecture and OXPHOS
Nature Communications (2022)