Abstract
Stimulator of interferon genes (STING), a major adaptor protein in antiviral innate immune signaling, is considered as one of the most important regulators of antiviral and antitumor immunity. Although STING agonists are now intensively studied in clinical trials as a new class of adjuvants to boost cancer immunotherapy, the tumor-intrinsic role of the STING pathway in shaping the tumor microenvironment remains controversial. Here, we discovered that STING plays a vital role in regulation of myeloid-derived suppressor cell (MDSC) differentiation and antitumor immunity in Epstein-Barr virus (EBV)-associated nasopharyngeal carcinoma (NPC). Mechanistic analyses reveal that STING represses NPC-derived MDSC induction by enhancing SOCS1 expression in both tumor cells and MDSCs. SOCS1 physically interacts with STAT3 through its SH2 domain to prevent STAT3 phosphorylation and dimerization, resulting in reduced MDSC induction via inhibition of GM-CSF and IL-6 production. Notably, reduced tumoral STING expression was found to be significantly associated with a poor prognosis for NPC patients. Our findings reveal a novel mechanism linking STING to tumor microenvironmental cytokine production and MDSC induction.
Similar content being viewed by others
Introduction
The differentiation of myeloid lineage cells is perturbed in the tumor microenvironment, resulting in expansion of tumor-derived myeloid cell populations, primarily myeloid-derived suppressor cells (MDSCs) and tumor-associated macrophages (TAMs), which suppress cytotoxic T-cell functions, contributing to tumor escape from immune surveillance [1,2,3]. This interplay between tumor cells and MDSCs results in tumor growth, relapse, and metastasis [4, 5]. MDSC induction is mainly regulated by the JAK-STAT signaling pathway, which is activated by IL-6 and GM-CSF [6]. However, the function and regulation of these key immune signaling molecules during MDSC formation and expansion in the tumor microenvironment has not yet been fully investigated.
Stimulator of interferon genes (STING; also known as ERIS, MYPS, or MITA), encoded by TMEM173, is a central adaptor protein in DNA virus-related innate immunity [7, 8]. Upon recognition of aberrant DNA species released from a number of invading pathogens, many DNA sensors, including cGAS and DDX41, activate STING to mediate TBK1-dependent IRF3 activation, which induces the production of type I interferons (IFNs) and numerous pro-inflammatory cytokines [8,9,10,11,12,13]. In addition to these important roles in host immunity and autoimmune diseases, STING signaling is usually inactivated in tumor cells and involved in antitumor immunity against various tumors, such as glioma [14, 15], sarcoma [3], melanoma [16,17,18], and colorectal tumors [19, 20]. Priming the STING signaling pathway using STING agonists has shown potent antitumor efficacy in several cancer therapeutic models [21, 22].
Thus far, the majority of STING-centric studies have focused on its function in myeloid populations, especially dendritic cells (DCs) [23, 24]. However, STING is widely expressed in many tumor types, where its tumor-intrinsic role in non-myeloid cells in shaping the tumor microenvironment remains controversial. TRIM29-mediated STING degradation in epithelial cells has been reported to result in immunosuppression, which was proposed as the etiology for chronic EBV infection in nasopharyngeal carcinoma (NPC) cells [25]. On the other hand, Liang et al. investigated radiation-induced tumor-intrinsic STING activation and surprisingly found that, unlike its role in DCs, intra-tumoral STING activation led to MDSC recruitment and immunosuppression [26].
In this study, we used nasopharyngeal carcinoma (NPC), a DNA virus-related tumor, as a model to investigate the functions of STING signaling in tumor cells and its impact on regulating cross-talk between tumor cells and MDSCs in the tumor microenvironment. Using both gain- and loss-of-function approaches, we demonstrate that STING restrains NPC-derived MDSC differentiation by preventing IL-6 and GM-CSF production by NPC cells. This inhibitory function of STING is dependent on cross-talk between TBK1-dependent type I IFN signaling and STAT3 signaling via SOCS1 in the tumor microenvironment. Mechanistically, SOCS1 is induced by the STING-TBK1-IRF3 axis and deactivates STAT3 signaling by physically interacting with the SH2 domain of STAT3 in both NPC cells and MDSCs. Importantly, STING protein expression is decreased in human NPC tissues and is negatively associated with the density of tumor-infiltrating CD33+ cells. Of note, reduced levels of tumoral STING correlate with poor NPC patient survival. Taken together, our findings identify an unrecognized role of the STING-SOCS1-STAT3 axis in MDSC induction, highlighting the potential for targeting innate immune molecules in designing and optimizing cancer therapies in DNA virus-associated malignancies.
Results
STING inhibits tumor-associated MDSC differentiation
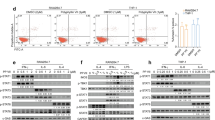
STING signaling, the major innate antiviral signaling pathway in response to DNA viral infection, has been linked to antitumor immunity in recent years [27]. Here, we observed that the protein level of STING but not that of other key antiviral signaling proteins was reduced in NPC cell lines, including CNE2, C666, and TW03, compared with the normal NP cell line NP69 (Fig. 1a). Ectopic expression of STING in TW03 and CNE2 cells had no direct effect on the growth of NPC cells (Supplementary Fig. 1a, b). Interestingly, the percentage of HLA-DR−CD11b+CD33+ MDSCs induced by TW03 and CNE2 cells with forced exogenous STING expression was significantly decreased in an in vitro co-culture system (Fig. 1b). We next generated STING-knockdown TW03 and CNE2 cells using STING-specific small hairpin RNAs (shRNAs) (Supplementary Fig. 1c) and observed that the percentage of HLA-DR−CD11b+CD33+ MDSCs induced by STING-knockdown TW03 and CNE2 cells was increased (Fig. 1c). Furthermore, MDSCs induced by NPC cells with forced exogenous STING expression showed a sharply decreased suppressive ability against CD4+ and CD8+ T-cell proliferation, while MDSCs induced by NPC cells with knockdown of endogenous STING showed an increased suppressive ability against the proliferation of CD4+ and CD8+ T cells (Supplementary Fig. 1d). Notably, among the suppressive molecules, including p-STAT3, PD-L1, Arg-1 and iNOS, the p-STAT3 and PD-L1 levels in NPC-induced MDSCs mediated by CNE2-STING cells were significantly decreased compared with corresponding control cells (Fig. 1d). The opposite p-STAT3 and PD-L1 expression pattern was observed in the NPC-induced MDSCs mediated by CNE2-shSTING cells (Fig. 1e). Previously, we identified IL-6 and GM-CSF release from NPC cells are critical for NPC-induced MDSC differentiation [28]. Here, we found that STING potently repressed IL-6 and GM-CSF secretion from NPC cells (Fig. 1f). These results suggest that STING restrains NPC-induced MDSC differentiation by suppressing IL-6 and GM-CSF secretion.
STING inhibits the differentiation of NPC-induced MDSCs. a Protein levels of STING and other signaling proteins in the NPC cell lines CNE2, C666, and TW03 or the normal NP cell line NP69. b Representative flow cytometry plot (left) and quantification (right) of MDSC differentiation assays are shown as the CD33+CD11b+ cells in the HLA-DR− gate induced by medium, NPC-empty vector (EV) or NPC-STING cells co-cultured with CD33+ cells isolated from healthy PBMCs. The percentage of MDSCs was decreased in STING-transfected TW03- and CNE2-mediated MDSC induction. c Representative plot (left) and quantification (right) are shown as the CD33+CD11b+ cells in the HLA-DR− gate induced by medium, NPC-shControl (shCtrl) or NPC-shSTING in different combinations. The percentage of MDSCs was increased in shSTING-transfected TW03- and CNE2-mediated MDSC induction. d Histogram of flow cytometry staining for the p-STAT3 and PD-L1 level in NPC-induced MDSCs mediated by CNE2-EV and CNE2-STING cells. e Representative histogram of p-STAT3 and PD-L1 expression in NPC-induced MDSCs mediated by CNE2-shCtrl and CNE2-shSTING-02 cells. f IL-6 and GM-CSF production were determined by ELISA in the cultural supernatants of NPC-EV, NPC-STING, NPC-shCtrl, and NPC-shSTING-02 cells. All of the experiments were performed at least three times, and quantification data are plotted as the means ± SEM. Statistical analyses were conducted with an unpaired Student’s t-test; *p < 0.05, **p < 0.01, and ***p < 0.001 vs. the corresponding control
To investigate whether STING affects MDSC differentiation in vivo, we measured the proportion of MDSCs in Tmem173+/− mice and observed a significant increase in the proportion of murine MDSCs (CD11b+Gr-1+) in spleens from Tmem173+/− mice (Supplementary Fig. 2). Murine MDSCs consist of two major subsets: granulocytic MDSCs (G-MDSCs) that express Ly6G (CD11b+Ly6G+Ly6C−) and monocytic MDSCs (Mo-MDSCs) that express Ly6C (CD11b+Ly6G−Ly6C+) [29]. We found that the G-MDSC population was significantly increased in spleens from Tmem173+/− mice (Supplementary Fig. 2). Taken together, these findings indicate that STING inhibits MDSC differentiation under physiological conditions.
STING suppresses tumor-induced MDSC differentiation by inhibiting STAT3 signaling
Given the important role of the STAT3 signaling pathway in MDSC differentiation by promoting the production of IL-6 and GM-CSF [30, 31], we next explored whether STING directly regulates STAT3 activation in NPC cells. STAT3 phosphorylation (p-STAT3, both pY705 and pS727) was decreased when STING was overexpressed in CNE2 cells with or without IL-6 stimulation (Fig. 2a), while p-STAT3 (pY705 and pS727) was increased when endogenous STING was knocked down in CNE2 cells (Fig. 2b). STAT3 reporter assays further demonstrated that STING inhibits the transcriptional activity of STAT3 (Fig. 2c, d), suggesting that STING potently inhibits STAT3 activation in NPC cells.
STING downregulates STAT3 signaling during NPC-derived MDSC differentiation. a CNE2 cells were transfected with a Myc-tagged-empty vector (Myc-EV) and a Myc-tagged-STING (Myc-STING) expression vector for at least 24 h, followed by treatment with IL-6 (20 ng/ml) for 30 min. The STAT3 pY705, STAT3 pS727, total STAT3, Myc, and β-actin levels were detected by immunoblot assay. b Immunoblot analysis of the indicated CNE2 cells treated with IL-6 (20 ng/ml) for 30 min before collecting of the lysates. c CNE2 cells were transfected with a STAT3-targeted gene promoter-driven luciferase reporter (STAT3-luc) and TK-Renilla luciferase (TK-luc), together with expression plasmids encoding Myc-EV or Myc-STING, for at least 24 h before treatment with or without IL-6 stimulation for 30 min. Luciferase assays (top) were performed to determine the relative STAT3 luciferase expression (fold), and an immunoblot assay (bottom) was used to detect STING expression. STAT3 (WT) and STAT3 (Y705F) mutants were used as positive and negative controls for STAT3 transcriptional activity, respectively. d Control or STING-knockdown CNE2 cells were transfected with STAT3-luc and TK-luc expression vectors, followed by IL-6 stimulation for 30 min. After 24 h, luciferase assays (top) and an immunoblot assay (bottom) were performed to determine STAT3 activity and STING expression. e ELISA assay of IL-6 and GM-CSF production in the culture supernatants of shCtrl NPC cells or of shSTING-02 NPC cells treated with cryptotanshinone for 48 h. f Representative image (top) and quantification (bottom) of MDSC differentiation assays in which CD33+ cells were co-cultured with NPC-shCtrl or cryptotanshinone-treated shSTING-02 NPC cells for 48 h. CD33+ cells in medium alone were included as a control. All experiments were performed at least three times, and quantification data are plotted as the mean ± SEM. Statistics was conducted with an unpaired Student’s t-test; *p < 0.05, **p < 0.01, and ***p < 0.001 vs. the corresponding control
We further investigated the effect of STING-mediated STAT3 inhibition during the NPC-induced MDSC differentiation process and found that the increased secretion of IL-6 and GM-CSF induced by STING deficiency was reversed by cryptotanshinone [32], a STAT3 inhibitor (Fig. 2e). Moreover, cryptotanshinone reversed the promotion of NPC-induced MDSC differentiation mediated by both wild-type NPC cells and STING-deficient NPC cells (Fig. 2f). Taken together, these results suggest that STING suppresses the differentiation of NPC-induced MDSCs by inhibiting STAT3 signaling.
STING-mediated type I IFN signaling is involved in NPC-induced MDSC differentiation
STING recruits TBK1 and IRF3 to induce type I IFN signaling upon DNA virus infection, and the S358 site at the C-terminal region of STING is critical for STING to activate type I IFN signaling [33]. Therefore, we generated a STING S358A (STING-SA) mutant as an inactive form of STING and found that this mutation completely eliminated STING’s ability to activate type I IFN signaling in NPC cells (Fig. 3a, b). We confirmed that the STING-SA mutant neither activates TBK1 nor inhibits STAT3 phosphorylation (Fig. 3c). Furthermore, ectopically expressed STING-SA did not inhibit IL-6 or GM-CSF production by TW03 and CNE2 cells (Fig. 3d). Consistent with these results, NPC-induced MDSC differentiation was impeded by forced expression of exogenous STING-WT but not STING-SA in NPC cells (Fig. 3e and Supplementary Fig. 3a). Collectively, these data indicate that activation of type I IFN signaling is required for STING to suppress NPC-induced MDSC differentiation.
STING-mediated Type I IFN signaling inhibits NPC-induced MDSC differentiation. a Top: schematic diagram of the STING wild-type (WT) and STING S358A mutant (SA). Bottom: the lysates of NPC cells stably expressing Flag-tagged EV, STING-WT, and STING-SA were subjected to immunoblotting with anti-Flag antibody. b Luciferase assay of the empty vector (EV), STING-WT, and STING-SA NPC cells transfected with IFNβ-luc (top) and ISRE-luc (bottom), together with TK-luc. c The lysates of NPC cells stably expressing Flag-tagged EV, STING-WT, and STING-SA were subjected to immunoblotting with the indicated antibodies. d ELISA assay of IL-6 and GM-CSF levels in the culture supernatants of cells stably expressing EV, STING WT, and STING SA. e Quantification analysis of the percentage of MDSCs induced by NPC cells stably expressing EV, STING WT, and STING SA. f Wild type (WT) and TBK1-KO TW03 cells were transfected with Flag-tagged EV and STING for 48 h. The cell lysates were immunoblotted with the indicated antibodies. g ELISA assay and h MDSC differentiation assay of the supernatants from WT and TBK1-KO TW03 cells transfected with Flag-tagged EV and STING for 48 h. All experiments were performed at least three times, and the quantification data are plotted as the mean ± SEM. Statistical analyses were conducted with an unpaired Student’s t-test; *p < 0.05, **p < 0.01, and ***p < 0.001 vs. the corresponding control. NS not significant
To further explore the impact of STING-mediated type I IFN signaling on NPC-induced MDSC differentiation, we knocked out the TBK1 gene, a major kinase downstream of STING in the type I IFN signaling pathway, in TW03 cells using the CRISPR/Cas9 system (Supplementary Fig. 3b). In these TBK1-KO cells, STING did not inhibit STAT3 phosphorylation (Fig. 3f) or suppress the secretion of IL-6 and GM-CSF (Fig. 3g). The STING-dependent reduction in MDSC differentiation was also abrogated in TBK1-KO cells (Fig. 3h and Supplementary Fig. 3c). Taken together, these findings indicate that STING inhibits MDSC differentiation by activating type I IFN signaling in a TBK1-dependent manner.
STING inhibits tumor-induced MDSC differentiation by enhancing SOCS1 expression
We next asked whether type I IFN directly affects MDSC differentiation. Interestingly, we found that IFN-β, a typical type I IFN, has no functional role in MDSC differentiation (Supplementary Fig. 4), suggesting that the inhibition of NPC-induced MDSC differentiation mediated by STING signaling is independent of type I IFN-activated JAK-STAT signaling. Therefore, we assessed downstream genes induced by STING signaling and observed that the mRNA levels of SOCS1 and other ISG genes, including ISG15, ISG54, and ISG56, were upregulated by STING overexpression (Fig. 4a and Supplementary Fig. 5a) but depressed by STING knockdown in NPC cells (Fig. 4b and Supplementary Fig. 5b). However, unlike ISG15, ISG54 and ISG56, SOCS1 could not be upregulated by IFN-α or IFN-β in CNE2 cells (Supplementary Fig. 5c), suggesting that SOCS1 is regulated directly by the STING-TBK1-IRF3 axis. SOCS1 is a potent-negative regulator of JAK-STAT signaling [34, 35], and we further observed that STING but not STING-SA enhances TBK1 phosphorylation and SOCS1 expression and inhibits STAT3 phosphorylation in TW03 and CNE2 cells (Fig. 4c). Conversely, STING knockdown decreased the levels of p-TBK1 and SOCS1 but increased the level of p-STAT3 in TW03 and CNE2 cells (Fig. 4d). Similar alterations in SOSC1, p-TBK1, and p-STAT3 levels were observed in STING-KO CNE2 cells (Supplementary Fig. 5d).
STING inhibits NPC-induced MDSC differentiation by activating SOCS1. a The mRNA expression levels of SOCS1 were determined in NPC cells stably expressing STING-WT or STING-SA or in control cells using quantitative real-time PCR (qRT-PCR). b qRT-PCR assays to determine the mRNA levels of SOCS1 in STING-knockdown (shSTING-01 and shSTING-02) and control (shCtrl) NPC cells. c, d Lysates of TW03 and CNE2 cells with stable overexpression of STING-WT or STING-SA c or transfected with STING-specific shRNAs d were subjected to immunoblotting with the indicated antibodies. e WT and SOCS1-KO CNE2 cells were transfected with Flag-tagged EV and STING for 48 h. The cell lysates were then immunoblotted with the indicated antibodies. f, g ELISA assay f and MDSC differentiation assay g of WT and SOCS1-KO CNE2 cells transfected with Flag-tagged EV and STING for 48 h. All experiments were performed a minimum of three times, and the quantification data are plotted as the means ± SEM. Statistical analysis was performed with an unpaired Student’s t-test; ***p < 0.001 vs. the corresponding control. NS not significant
We next found that in SOCS1-KO cells, STING no longer inhibited STAT3 phosphorylation (Fig. 4e). In addition, forced STING overexpression did not inhibit the production of IL-6 and GM-CSF when SOCS1 was knocked out (Fig. 4f). Therefore, SOCS1 deficiency abrogates the inhibition of NPC-induced MDSC differentiation caused by forced expression of exogenous STING in NPC cells (Fig. 4g and Supplementary Fig. 5e). Taken together, these data indicate that STING promotes SOCS1 activation to suppress STAT3 phosphorylation, leading to inhibition of IL-6 and GM-CSF secretion and NPC-induced MDSC differentiation.
SOCS1 prevents STAT3 phosphorylation and dimerization via its SH2 domain
To further investigate the mechanism by which SOCS1 regulates STAT3 phosphorylation in NPC, we overexpressed Flag-tagged SOCS1 in NPC cells and found that SOCS1 inhibits STAT3 phosphorylation (pY705 and pS727) in a dose-dependent manner (Fig. 5a). Importantly, ectopic SOCS1 expression still inhibited STAT3 phosphorylation in STING-KO CNE2 cells (Fig. 5b), indicating that SOCS1 exerts its inhibitory function downstream of STING. Indeed, exogenous STAT3 specifically interacted with SOCS1 but not SOCS3 (Fig. 5c). Further endogenous co-immunoprecipitation assessments illustrated that SOCS1 binds STAT3 under physiological conditions in NPC cells (Fig. 5d). To confirm whether SOCS1 could directly bind to STAT3, we prepared His-SOCS1 and GST-STAT3 from bacteria, and in vitro GST-pulldown assays showed that SOCS1 physically associated with STAT3 through direct interaction (Fig. 5e).
SOCS1 prevents STAT3 phosphorylation and dimerization via its SH2 domain. a TW03 and CNE2 cells were transfected with increasing amounts of Flag-SOCS1. Cell lysates were immunoblotted with the indicated antibodies. b Wild-type (WT) and STING-KO CNE2 cells were transfected with Flag-EV and Flag-SOCS1 for 48 h. Cell lysates were subjected to immunoblotting with the indicated antibodies. c 293T cells were transfected with Flag-EV, Flag-SOCS1, and Flag-SOCS3, together with HA-STAT3 expression vectors, for 24 h. The cell extracts were subjected to immunoprecipitation with anti-Flag-beads and immunoblot analysis with anti-HA antibody. d Extracts of CNE2 cells were subjected to immunoprecipitation with anti-SOCS1 antibody and immunoblot analysis with an anti-STAT3 antibody. e His-SOCS1 and GST-STAT3 were purified from bacteria and incubated in reaction buffer in vitro. The proteins were immunoprecipitated with anti-GST beads and immunoblotted with an anti-His antibody. f Top: Schematic diagram of SOCS1 and its SH2 domain mutant. Bottom: Lysates from 293T cells transfected with Myc-EV, Myc-SOCS1, and Myc-SOCS1-SH2 mutant, together with Flag-STAT3, were subjected to immunoprecipitation with anti-Myc-beads and immunoblot analysis with anti-Flag antibody. g 293T cells were transfected with Myc-EV, Myc-STAT3 WT, and ΔSH2 mutant, together with Flag-SOCS1, and the lysates from these cells were immunoprecipitated with anti-Myc beads and immunoblotted with an anti-Flag antibody. h 293T cells were transfected with HA-EV, HA-STAT3, and Flag-STAT3 for 8 h, followed by transfection with increasing amounts of Myc-SOCS1 for 36 h. The lysates were subjected to immunoprecipitation with anti-Flag-beads and immunoblot analysis with an anti-HA antibody. i 293T cells were transfected with HA-STAT3 and Flag-STAT3 for 8 h, followed by transfection with Myc-SOCS1 SH2 for 36 h. The lysates were subjected to immunoprecipitation with anti-Flag-beads and immunoblot analysis with an anti-HA antibody. All of the experiments were performed at least three times
To determine the detailed mechanisms underlying how SOCS1 inhibits STAT3 phosphorylation and activation, we generated a deletion mutant containing the SH2 domain of SOCS1, which is the element responsible for binding and competing for substrates [36]. Subsequent co-immunoprecipitation assays showed that the SOCS1 SH2 domain strongly interacts with STAT3 (Fig. 5f). We next generated three domain-deletion mutants of STAT3 and found that STAT3 interacts with SOCS1 through its C-terminal region, which contains the SH2 and TAD domains (Supplementary Fig. 6a). In addition, SOCS1 failed to interact with STAT3 in the absence of its SH2 domain, which contains the Y705 binding domain (Fig. 5g and Supplementary Fig. 6b). These data suggest that SOCS1 can inhibit STAT3 signaling by physically binding to the SH2 domain of STAT3. Interestingly, the C-terminal domain of STAT3 has been found to be responsible for STAT3 dimerization [37] (Supplementary Fig. 6c); we thus hypothesized that SOCS1 might also disrupt STAT3 dimerization via its SH2 domain. Indeed co-immunoprecipitation experiments revealed that SOCS1 disrupts STAT3 dimerization in a dose-dependent manner (Fig. 5h). In addition, the SH2 domain of SOCS1 alone inhibits STAT3 phosphorylation and dimerization (Fig. 5i). Taken together, these data indicate that SOCS1 disrupts STAT3 phosphorylation and dimerization through a heteromeric SH2-SH2 domain interaction, leading to inactivation of STAT3 signaling.
The STING-TBK1-SOCS1 axis suppresses STAT3 activation in MDSCs
We next investigated whether the function and mechanism of STING-mediated signaling in NPC cell lines also occurs in MDSCs. First, we observed that the SOCS1 mRNA level was positively correlated with the STING mRNA level in MDSCs (Fig. 6a). Exogenous STING expression enhanced TBK1 phosphorylation and SOCS1 induction and inhibited STAT3 phosphorylation in MDSCs (Fig. 6b). Importantly, exogenous STING-WT but not STING-SA directly inhibited MDSC differentiation, whereas knockdown of endogenous STING in MDSCs promoted MDSC differentiation in vitro (Fig. 6c and Supplementary Fig. 7). Moreover, exogenous STING-WT but not STING-SA decreased the p-STAT3 and PD-L1 levels in MDSCs (Fig. 6d). The opposite p-STAT3 and PD-L1 expression pattern was observed in STING-knockdown-MDSCs (Fig. 6e). In addition, we found that STING directly impedes MDSC differentiation through regulating the STAT3 pathway in MDSCs, because the STAT3 inhibitor cryptotanshinone reversed the differentiation of MDSCs mediated by STING deficiency (Fig. 6f). Taken together, these data suggest that the STING-TBK1-SOCS1 axis directly suppresses MDSC differentiation by inhibiting STAT3 signaling activation in MDSCs.
STING signaling directly regulates MDSC differentiation. a CD33+ cells were cultured in medium alone or with administration of recombinant lentivirus vectors containing Flag-empty vector (EV), Flag-STING, Flag-STING SA, shControl or shSTING-02 for 48 h. qRT-PCR was performed to determine the mRNA levels of STING and SOCS1 in the CD33+ cells cultured under the different inducement conditions described above. b Lysates of MDSCs treated with medium or infected with lentivirus containing the indicated expression vectors were subjected to immunoblot analysis with the indicated antibodies. c Quantification of MDSC cells (CD33+CD11b+ cells in the HLA-DR− gated population) induced by medium or infected with lentivirus containing the indicated expression vectors. d The quantification of p-STAT3, PD-L1, Arg-1, and iNOS expression in the indicated MDSCs. e Quantification of p-STAT3 and PD-L1 expression in STING-knockdown MDSCs. f A representative image of CD33+ cells treated with medium or lentivirus containing shControl or shSTING-02 for 24 h and then treated with cryptotanshinone. All experiments were performed at least three times, and quantification data are plotted as the mean ± SEM. Statistical analyses were conducted with an unpaired Student’s t-test; ***p < 0.001 vs. the corresponding control
Reduced STING expression is correlated with poor prognosis of NPC patients
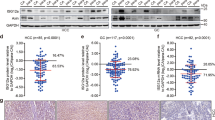
Based on the above observations, we further investigated a possible association between STING expression and MDSC accumulation in NPC tissues. This analysis revealed that STING levels in tumor tissues (T) were lower than in adjacent non-tumor tissues (NTs) from the same NPC patients (Fig. 7a, b, n = 22). Importantly, intra-tumoral STING expression was negatively associated with the number of tumor-infiltrating CD33+ cells (Fig. 7c, n = 61). Moreover, reduced tumoral STING expression was positively correlated with poor patient disease-free survival (DFS) (Fig. 7d, n = 77). Finally, the level of tumoral STING was significantly negatively correlated with NPC patient relapse (p < 0.05) but not sex, age, tumor stage, lymph node metastasis (N) or clinical grade (Supplementary Table 1, n = 77). In sum, these findings suggest that in humans NPC STING signaling impedes the activity of MDSCs, resulting in improved patient outcomes.
STING expression and its clinical relevance in NPC. a Representative images from immunohistochemistry (IHC) assays to examine the expression of STING in pairs of paraffin-embedded non-tumor tissues (NT) and tumor tissues (T) from 22 NPC patients. b The STING level was lower in tumor tissues than in non-tumor tissues. Statistical analyses were conducted with a paired Student’s t-test. Scale bars, 50 μm. c Negative correlations were found between the number of tumor-infiltrating CD33+ cells and tumor STING levels in NPC patients (n = 61). The statistical analysis was performed using Spearman’s correlation and linear regression. R, Spearman’s correlation, is the correlation coefficient. d Disease-free survival (DFS) rates showing that the tumor STING level (n = 77, p = 0.024) was significantly associated with the poor prognosis of NPC patients. e A cartoon demonstrating the role of STING-SOCS1-STAT3 axis in regulation of cross-talk between tumor cells and MDSCs in the NPC microenvironment. The cutoff point of STING for IHC data was 4.7. Cutoff selection was based on X-tile. The data in b are plotted as the mean ± SEM. *p < 0.05, and ***p < 0.001 vs. the corresponding control
Discussion
Although numerous studies have described the myriad functions of STING in antiviral immunity [8, 33, 38,39,40,41,42], increasing evidence indicates that STING signaling is one of most important pathways for remodeling of the tumor microenvironment [21, 43, 44]. STING-mediated antitumor immunity is associated with an enhancement in antigen-presenting cells (APCs) and tumor antigen-specific CD8+ T cells [15, 18, 22]. However, the function of STING in other tumor-infiltrating immune cells and tumor cells within the tumor microenvironment has not been elucidated. In this study, we found that STING expression is negatively associated with the number of tumor-infiltrating CD33+ cells, suggesting that STING might play a role in tumor-associated MDSC differentiation. We then found that STING signaling decreases the percentage of NPC-induced HLA-DR−CD11b+CD33+ MDSCs in vitro, implying that STING has a protective role against tumor progression. Consistent with these observations, another study also observed that CD11b+Gr1+ immature myeloid suppressor cells (mouse MDSCs) are increased in glioma-bearing STINGgt/gt mice [15]. Recently, another study showed that PD-L1 antibody therapy exhibited reduced antitumor efficacy in STINGgt/gt mice, which was associated with an increased MDSC population in the tumor microenvironment [45], consistent with our results.
The STAT3 signaling pathway plays an important role in the tumor microenvironment by propagating several levels of cross-talk between tumor cells and immune cells [46]. Constitutively activated STAT3 signaling is frequently observed in tumor cells, leading to upregulation of the MDSC differentiation-associated cytokines IL-6 and GM-CSF [47]. STAT3 activation also directly enhances immunosuppressive activity by upregulating ROS production [48]. Furthermore, STAT3 promotes the production of various immunosuppressive factors in MDSCs, including PD-L1, Arg-1, and iNOS, resulting in alteration of gene expression programs to restrain antitumor immunity [30]. Our results reveal that STING inhibits NPC-derived MDSC induction via TBK1-dependent type I IFN and STAT3 signaling. STING-mediated type I IFN production leads to antitumor immunity in many cancers [23]. However, we found that IFN-β, a typical type I IFN, has no effect on MDSC differentiation in NPC. Notably, it has been reported that chronic IFN-α expression promotes MDSC accumulation to strongly suppress specific CD8+ T cells, leading to a poor immune response against viral infection [49]. This apparent contradiction may be explained by the doses used and times stimulated with type I IFN in different experimental models. Indeed, a high type I IFN dose enhances antiviral immune responses, whereas a low dose has the opposite effect. A similar study demonstrated that chronic but not acute viral infection induces MDSC expansion [50]. These reports illustrate the complexity of type I IFN signaling with regard to MDSC differentiation, which should be further elucidated in future studies.
Here, our data suggest that genes downstream of STING-mediated type I IFN signaling might regulate tumor-associated MDSC differentiation in NPC. Indeed, we observed that SOCS1, which is upregulated by STING-TBK1 signaling, physically interacts with STAT3 at both SH2 domains to inhibit STAT3 phosphorylation at both S727 and Y705. Importantly, this interaction also disrupted STAT3 dimerization, leading to further inhibition of STAT3 signaling. This mechanism underlies the observed STAT3 inhibition by the STING-TBK1 axis, which occurs in a SOCS1-dependent manner.
Currently, the immune function of STING in non-myeloid cells is controversial. On the one hand, Xing et al. identified that TRIM29-mediated STING degradation in epithelial cells results in immunosuppression [25]. On the other hand, Liang et al. found that intra-tumoral STING activation led to MDSC recruitment and immunosuppression [26]; however, they did not investigate the function of STING in induction of tumor-derived cytokines and MDSC differentiation in the tumor host. Here, our studies provide strong evidence supporting a model in which there is no functional difference between myeloid cells and tumor cells for STING-mediated immune-regulation: tumor-intrinsic STING activation also promotes antitumor immunity by suppressing immunosuppressive cytokine production within the tumor microenvironment.
Based on our findings, we propose a working model to illustrate how STING regulates MDSC differentiation in NPC (Fig. 7e). In EBV-positive NPC tumor microenvironments, STING serves as an antitumor protein to activate TBK1-IRF3 signaling to upregulate SOCS1 in tumor cells and myeloid cells. SOCS1 then interacts with STAT3 to inhibit its phosphorylation and dimerization. Released IL-6 and GM-CSF from those cells eventually leads to induction of MDSC differentiation to promote MDSC-mediated immunosuppression and tumor progression.
In conclusion, our study unveils a previously unrecognized function of the STING-SOCS1-STAT3 axis in regulating MDSC differentiation in virus-associated NPC. Given the critical roles of type I IFN signaling and STAT3 signaling in tumor immunity, a more comprehensive understanding of the cross-talk between STING and STAT3 signaling in the pathogenesis of NPC may provide an opportunity to develop novel therapeutic strategies by adjusting the tumor microenvironment.
Materials and methods
Sample collection and mice
In total, 77 fresh tumor specimens, including 22 pairs of non-tumor and tumor tissues from NPC patients at the first diagnosis, were collected at the Sun Yat-sen University Cancer Center, Guangzhou, China between 2011 and 2013 (Supplementary Table 1). This study was conducted in accordance with the Helsinki Declaration, and all patients and healthy donors signed a consent form approved by the Research Ethics Committee of the Sun Yat-sen University Cancer Center (GZR2013-040).
Female 6- to 7-week-old C57BL/6 mice were purchased from the Guangdong Medical Laboratory Animal Center and maintained in pathogen-free conditions at the Animal Experiment Center of Sun Yat-Sen University. Tmem173+/− mice were a gift from Professor Hui Zhang (Zhongshan Medical School, Sun Yat-sen University). All procedures were approved by the Animal Care and Use Committee of Sun Yat-Sen University and conformed to the legal mandates and national guidelines for the care and maintenance of laboratory animals.
Cell culture and reagents
HEK293T, TW03, C666, and CNE2 cells were cultured in DMEM (Life Technologies, Beijing, China) or RPMI-1640 medium (Life Technologies) supplemented with 10% fetal bovine serum (Gibco, Grand Island, NY, USA) and 1% l-glutamine (Gibco). The normal NP69 cell line was maintained in keratinocyte-SFM medium (Invitrogen). Human peripheral blood mononuclear cells (PBMCs) were isolated from the blood of healthy donors. The pSTAT3 inhibitor cryptotanshinone was purchased from Selleck (Houston, TX, USA).
Colony formation assay
For colony formation assays, NPC cells were directly plated in 6-well plates at a density of 1000 cells per well in RPMI-1640 containing 10% FBS for 2–3 weeks. Cells were stained with crystal violet (Beyotime Biotechnology, Xiamen, China) and counted using ImageJ software.
Enzyme-linked immunosorbent (ELISA) assay
For ELISA assays, human IL-6 (R&D Systems, Minneapolis, MN, USA), and GM-CSF ELISA kits (eBioscience, San Diego, CA, USA) were used. Briefly, a 96-well plate was coated with 100 µL of coating diluent at 4 °C overnight. Then, 100 µL of a standard or samples from the serum of NPC patients and cells was added to the appropriate wells and incubated for 2 h at room temperature (RT). The wells were washed five times with wash buffer, and an anti-STING, anti-IL-6, or anti-GM-CSF conjugate was added to all wells followed by further incubation at RT. The plate was again washed 5–7 times, and 100 µL of avidin-HRP was added. The plate was then incubated for 30 min at RT, and 100 µL of a substrate solution was added to each well after a wash with wash buffer. The plate was subsequently incubated for 15 min at RT in the dark, after which 50 µL of stop solution was added to each well, and the absorption at 450 nm was detected using a 96-well plate reader (Bio-Rad).
Immunohistochemistry (IHC) assay
A primary rabbit anti-human STING antibody (ProteinTech Group, Wuhan, China) was used to stain all specimens from NPC patients, most of which (n = 61) were also stained using a primary mouse anti-human CD33+ antibody (Abcam, Cambridge, MA) according to the manufacturer’s instructions. Normal goat IgG (Santa Cruz Biotechnology, Dallas, TX, USA)-negative control staining was performed and evaluated. Briefly, paraffin-embedded tissues were sectioned continuously at a thickness of 4 µm, followed by staining with a STING antibody according to the manufacturer’s instructions. The data were scored for STING expression in tumor nests and adjacent tissues in 5 separate ×400 high-power microscopic fields (HPFs). Specimens were scored using a semi-quantitative immunoreactivity scoring system, which ranged from 0 to 12 and was derived from multiplication of the intensity of immunohistochemical staining (0—no staining; 1—weak; 2—moderate; and 3—strong) and the percentage of positive tumor cells [1 (<25%), 2 (25–50%), 3 (50–75%), 4 (>75%)]. The slides were scored independently by two pathologists. Cutoff selection was based on X-tile (Version 3.6.1, Yale University, New Haven, CT, USA).
Plasmids and transfection
Plasmids encoding expression genes were constructed according to previously described methods [51]. STING-specific shRNA (shSTING, #HSH009286-LVRU6GP) and control (shCtrl, #CSHCTR001-LVRU6GP) vectors were purchased from GeneCopoeia (Guangzhou, China). Flag-SOCS1 was kindly provided by Dr. Xiao Yu. Myc-STAT3 domain mutants and a Myc-STAT3 Y705F plasmid were constructed using standard molecular techniques. Cells were transfected with plasmids using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. For gene-knockdown assays, cells were infected with lentivirus encoding shRNAs.
Lentivirus production and generation of stable expression or knockdown cell lines
For lentivirus production, 293T cells were transfected with corresponding vectors (expression vectors: Δ8.9: VSVG = 5:3:2) for 48 h, and supernatants containing lentiviral particles were collected and stored at 4 °C until use. For generating stable cell lines, lentivirus was tittered by infection of TW03 and CNE2 cells with serial dilutions of lentivirus in the presence of 8 μg/ml polybrene (Abbott Laboratories, Abbott Park, IL, USA). Then, 48 h post infection, lentivirus-infected NPC cells were cultured with 3 μg/ml puromycin (Sigma-Aldrich, St. Louis, MO, USA) for 2 weeks. Puromycin-resistant colonies were then collected and expanded for further analysis under selection conditions.
Generation of knockout cells using CRISPR/Cas9 technology
Plasmids containing STING, TBK1, and SOCS1 sgRNAs for the CRISPR/Cas9 system were constructed as described previously [52]. The sequences are as follows:
GFP sgRNA (control), 5′-GGGCGAGGAGCTGTTCACCG-3′
STING sgRNA 1, 5′-GGTGCCTGATAACCTGAGTA-3′
STING sgRNA 2, 5′-GCGGGCCGACCGCATTTGGG-3′
TBK1 sgRNA 1, 5′-TTTGAACATCCACTGGACGA-3′
TBK1 sgRNA 2, 5′-CATAAGCTTCCTTCGTCCAG-3′
SOCS1 sgRNA 1, 5′-TGCCGGTAATCGGCGTGCGAA-3′
SOCS1 sgRNA 2, 5′-GGGCGAAAAAGCAGTTCCGC-3′.
Fluorescence-activated cell sorting (FACS) analysis
For FACS analysis, single-cell suspensions were stained with corresponding fluorescence-labeled antibodies according to the manufacturer’s instructions. Fluorescence-labeled antibodies against the following proteins, along with matched isotypes, were purchased from eBioscience (San Diego, CA, USA): CD33 (85-45-0338-42); CD11b (85-25-0118-42); CD4 (555346); CD8 (12-0088-42); p-STAT3 (17-9033-41); and PD-L1 (85-11-9969-42). Antibodies targeting iNOS (IC5868F) and Arg-1 (sc-7271AF647) were purchased from R&D Systems (Minneapolis, MN, USA) and Santa Cruz (Dallas, TX, USA), respectively. Antibodies specific for murine MDSC markers, including CD11b (11-0112-85), Gr-1 (12-5931-85), Ly6C (45-5932-82) and Ly6G (12-9668-82), were a kind gift from Professor Jie Zhou (Zhongshan Medical School, Sun Yat-sen University). All data were analyzed using FlowJo software (Treestar).
MDSC differentiation assay and T-cell proliferation assay
CD33+ cells were separated from healthy PBMCs using human CD33 MicroBeads (Miltenyi Biotec, Bergisch Gladbach, Germany) according to the manufacturer’s instructions. Isolated CD33+ cells were co-cultured with NPC cells in 24-well plates using a transwell system (0.4-mm pore, Corning) at a ratio of 1:5 for 48 h. CD33+ cells cultured in medium alone were evaluated as a control. A panel of collected cells was used to analyze MDSC marker expression via staining, and the remaining cells were subjected to the T-cell proliferation assay. Briefly, PBMCs from HDs were labeled with carboxyfluorescein diacetate succinimidyl ester (CSFE, 10 μM), plated in OKT3-coated 96-well plates and added to the induced MDSCs, followed by culture for 3 days. The cells were then stained for CD3, CD4, and CD8 cell surface markers. Flow cytometry was performed using an FC500 flow cytometer, and data were obtained with CXP software (Beckman Coulter, San Jose, CA).
Quantitative RT-PCR analysis
Total RNA from various cells was extracted with Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. Quantification and quality control assessment of total RNA were performed in triplicate with a NanoDrop2000 spectrophotometer (Thermo Scientific). In a 20-μl PCR mixture, 1 mg of total RNA was reverse-transcribed into first-strand cDNA using HiScript® II QRT SuperMix for qPCR (Vazyme, Nanjing, China), and real-time PCR was performed using a 2xRealStar SYBR Mixture (Genestar, Beijing, China). The mRNA expression levels of examined genes were determined using the ΔΔCt method, and RPL13A was used as the reference gene. The data indicate the relative mRNA levels (Fold induction), with the relative quantity of the control cells set to 1. The following specific primers, including the forward (F) and reverse (R) primers (from 5′ to 3′), were used:
RPL13A F: 5′-CCTGGAGGAGAAGAGGAAAGAGA-3′
R: 5′-TTGAGGACCTCTGTGTATTTGTCAA-3′
STING F: 5′-CCAGAGCACACTCTCCGGTA-3′
R: 5′-CGCATTTGGGAGGGAGTAGTA-3′
SOCS1 F: 5′-TTTTCGCCCTTAGCGTGAAGA-3′
R: 5′-GAGGCAGTCGAAGCTCTCG-3′
ISG15 F: 5′-TCCTGGTGAGGAATAACAAGGG-3′
R: 5′-GTCAGCCAGAACAGGTCGTC-3′
ISG54 F: 5′-GGAGGGAGAAAACTCCTTGGA-3′
R: 5′-GGCCAGTAGGTTGCACATTGT-3′
ISG56 F: 5′-TCAGGTCAAGGATAGTCTGGAG-3′
R: 5′-AGGTTGTGTATTCCCACACTGTA-3′
Immunoprecipitation and immunoblot analysis
For endogenous immunoprecipitation, CNE2 cell extracts were prepared and incubated overnight with a SOCS1 antibody plus Protein A/G beads (Thermo, Rockford, IL, USA). For tagged protein immunoprecipitation, 293T or CNE2 cells were transfected with the corresponding expression vectors for 24 h, and then, Flag or Myc was pulled down using Flag-beads or Myc-beads (Sigma-Aldrich, St. louis, MO, USA) and immunoblot analysis was performed.
Horseradish peroxidase (HRP)-FLAG (A8592) and mouse anti-β-actin (A2228) antibodies were purchased from Sigma (St. Louis, MO, USA). Anti-HRP-hemagglutinin (HA, clone 3F10) and anti-c-Myc-HRP (11814150001) antibodies were purchased from Roche Applied Science (Mannheim, Germany). Rabbit anti-STING (13647), anti-STAT3 phosphorylated at Tyr705 (pY705 or pSTAT3, 9131), anti-STAT3 phosphorylated at Ser727 (pS727, 9134), anti-STAT3 (4904), anti-STAT3 phosphorylated at Ser172 (pTBK1, 5483) and anti-TBK1 (3504) antibodies were purchased from Cell Signaling Technology (Danvers, MA, USA). HRP-conjugated 6*His, His-Tag antibody (HRP-66005), rabbit GST Tag antibody (10000-0-AP), anti-SOCS1 antibody (25852-1-AP) was from ProteinTech (Wuhan, China). Mouse anti-c-Myc (9211) and anti-STAT3 (sc-293151) antibodies were purchased from Transgen (Beijing, China) and Santa Cruz (Dallas, TX, USA), respectively.
Luciferase reporter assay
For luciferase assays, ISRE-luciferase (ISRE-luc), IFN-β-luc, and TK-luc have been described previously [51). The STAT3 reporter plasmid (STAT3-luc, 11503ES03) was purchased from Yensen (Guangzhou, China). NPC cells were co-transfected with overexpression or knockdown plasmids along with reporter-luc and TK-luc plasmids. Then, the cells were lysed with passive lysis buffer (Promega, Madison, WI, USA) 36–48 h after treatment. The relative luciferase units (RLUs) were calculated by normalizing the reading of reporter-luc to that of TK-luc, and the RLU of the control cells was set to 1. A 500-bp fragment of the STING WT 3′-UTR was cloned downstream of the Gaussia luciferase gene into a pEZX-MT05 vector (GeneCopoeia, Guangzhou, China), and a mutant (MT) 3′-UTR plasmid was subsequently generated via site-directed mutagenesis.
In vitro GST-pulldown binding assay
GST-STAT3 and GST-STAT3 ΔSH2 were cloned into the vectors pGEX-5×, and His-SOCS1 was cloned into pET-28a (+), for bacterial expression. E. coli BL21 were transformed with pGEX-5×-STAT3, pGEX-5×-STAT3 ΔSH2 and pET-28a (+)-SOCS1 vectors. GST-STAT3 and His-SOCS1 protein expression were induced with IPTG and purified using Glutathione Agarose (a kind gift from Dr. Xiya Zhang) and Ni-NTA Agarose beads (Qiagen, #30210) according to standard protocols. Purified GST-STAT3 and His-SOCS1 were incubated in reaction buffer at 4 °C for 1 h and then with GST-beads overnight at 4 °C. Afterwards, the beads were washed four times with low-salt buffer, and bound proteins were eluted and subjected to immunoblot analysis.
Statistical analyses
All analyses were performed using GraphPad Prism 5 (La Jolla, CA, USA) and SPSS 18.0 software (Chicago, IL, USA). Two-tailed Student’s t-test was used for comparison of the numerical data, which are presented as the mean ± standard error of the mean (SEM). Pearson’s chi-squared test was used to analyze the correlation between immunohistochemical variants and patient clinic pathological parameters. The Kaplan–Meier and log-rank test were used for survival analysis, and a χ2 test was also used in some experiments as indicated. Cutoff values were obtained using X-tile (Version 3.6.1, Yale University, New Haven, CT, USA). In this study, *p < 0.05, **p < 0.01, ***p < 0.001, and p < 0.05 were considered significant.
Authenticity of the data
The authenticity of this article has been validated by uploading the key raw data onto the Research Data Deposit (RDD) public platform (www. researchdata.org.cn), with the RDD approval number RDDB2018000483.
References
Gabrilovich DI, Ostrand-Rosenberg S, Bronte V. Coordinated regulation of myeloid cells by tumours. Nat Rev Immunol. 2012;12:253–68.
Kumar V, Patel S, Tcyganov E, Gabrilovich DI. The nature of myeloid-derived suppressor cells in the tumor microenvironment. Trends Immunol. 2016;37:208–20.
Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity’s roles in cancer suppression and promotion. Science. 2011;331:1565–70.
Qu P, Wang LZ, Lin PC. Expansion and functions of myeloid-derived suppressor cells in the tumor microenvironment. Cancer Lett. 2016;380:253–6.
Veglia F, Perego M, Gabrilovich D. Myeloid-derived suppressor cells coming of age. Nat Immunol. 2018;19:108–19.
Gabrilovich DI, Velders MP, Sotomayor EM, Kast WM. Mechanism of immune dysfunction in cancer mediated by immature Gr-1+ myeloid cells. J Immunol. 2001;166:5398–406.
Barber GN. STING-dependent cytosolic DNA sensing pathways. Trends Immunol. 2014;35:88–93.
Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–92.
Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, et al. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478:515–8.
Gao D, Wu J, Wu YT, Du F, Aroh C, Yan N, et al. Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science. 2013;341:903–6.
Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–91.
Woodward JJ, Iavarone AT, Portnoy DA. c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science. 2010;328:1703–5.
Zhang Z, Yuan B, Bao M, Lu N, Kim T, Liu YJ. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat Immunol. 2011;12:959–65.
Fujita M, Scheurer ME, Decker SA, McDonald HA, Kohanbash G, Kastenhuber ER, et al. Role of type 1 IFNs in antiglioma immunosurveillance–using mouse studies to guide examination of novel prognostic markers in humans. Clin Cancer Res. 2010;16:3409–19.
Ohkuri T, Ghosh A, Kosaka A, Zhu J, Ikeura M, David M, et al. STING contributes to antiglioma immunity via triggering type I IFN signals in the tumor microenvironment. Cancer Immunol Res. 2014;2:1199–208.
Diamond MS, Kinder M, Matsushita H, Mashayekhi M, Dunn GP, Archambault JM, et al. Type I interferon is selectively required by dendritic cells for immune rejection of tumors. J Exp Med. 2011;208:1989–2003.
Fuertes MB, Kacha AK, Kline J, Woo SR, Kranz DM, Murphy KM, et al. Host type I IFN signals are required for antitumor CD8 + T cell responses through CD8{alpha} + dendritic cells. J Exp Med. 2011;208:2005–16.
Woo SR, Fuertes MB, Corrales L, Spranger S, Furdyna MJ, Leung MY, et al. STING-dependent cytosolic DNA sensing mediates innate immune recognition of immunogenic tumors. Immunity. 2014;41:830–42.
Xia T, Konno H, Ahn J, Barber GN. Deregulation of STING signaling in colorectal carcinoma constrains DNA damage responses and correlates with tumorigenesis. Cell Rep. 2016;14:282–97.
Zhu Q, Man SM, Gurung P, Liu Z, Vogel P, Lamkanfi M, et al. Cutting edge: STING mediates protection against colorectal tumorigenesis by governing the magnitude of intestinal inflammation. J Immunol. 2014;193:4779–82.
Corrales L, Glickman LH, McWhirter SM, Kanne DB, Sivick KE, Katibah GE, et al. Direct activation of STING in the tumor microenvironment leads to potent and systemic tumor regression and immunity. Cell Rep. 2015;11:1018–30.
Fu J, Kanne DB, Leong M, Glickman LH, McWhirter SM, Lemmens E, et al. STING agonist formulated cancer vaccines can cure established tumors resistant to PD-1 blockade. Sci Transl Med. 2015;7:283ra52.
Curran E, Chen X, Corrales L, Kline DE, Dubensky TW Jr., Duttagupta P, et al. STING pathway activation stimulates potent immunity against acute myeloid leukemia. Cell Rep. 2016;15:2357–66.
Moore E, Clavijo PE, Davis R, Cash H, Van Waes C, Kim Y, et al. Established T cell-inflamed tumors rejected after adaptive resistance was reversed by combination STING activation and PD-1 pathway blockade. Cancer Immunol Res. 2016;4:1061–71.
Xing J, Zhang A, Zhang H, Wang J, Li XC, Zeng MS, et al. TRIM29 promotes DNA virus infections by inhibiting innate immune response. Nat Commun. 2017;8:945.
Liang H, Deng L, Hou Y, Meng X, Huang X, Rao E, et al. Host STING-dependent MDSC mobilization drives extrinsic radiation resistance. Nat Commun. 2017;8:1736.
Corrales L, McWhirter SM, Dubensky TW Jr., Gajewski TF. The host STING pathway at the interface of cancer and immunity. J Clin Invest. 2016;126:2404–11.
Li ZL, Ye SB, OuYang LY, Zhang H, Chen YS, He J, et al. COX-2 promotes metastasis in nasopharyngeal carcinoma by mediating interactions between cancer cells and myeloid-derived suppressor cells. Oncoimmunology. 2015;4:e1044712.
Youn JI, Nagaraj S, Collazo M, Gabrilovich DI. Subsets of myeloid-derived suppressor cells in tumor-bearing mice. J Immunol. 2008;181:5791–802.
Yu H, Kortylewski M, Pardoll D. Crosstalk between cancer and immune cells: role of STAT3 in the tumour microenvironment. Nat Rev Immunol. 2007;7:41–51.
Sumimoto H, Imabayashi F, Iwata T, Kawakami Y. The BRAF-MAPK signaling pathway is essential for cancer-immune evasion in human melanoma cells. J Exp Med. 2006;203:1651–6.
Shin DS, Kim HN, Shin KD, Yoon YJ, Kim SJ, Han DC, et al. Cryptotanshinone inhibits constitutive signal transducer and activator of transcription 3 function through blocking the dimerization in DU145 prostate cancer cells. Cancer Res. 2009;69:193–202.
Zhong B, Yang Y, Li S, Wang YY, Li Y, Diao F, et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29:538–50.
Nicholson SE, Hilton DJ. The SOCS proteins: a new family of negative regulators of signal transduction. J Leukoc Biol. 1998;63:665–8.
Sansone P, Bromberg J. Targeting the interleukin-6/Jak/stat pathway in human malignancies. J Clin Oncol. 2012;30:1005–14.
Kubo M, Hanada T, Yoshimura A. Suppressors of cytokine signaling and immunity. Nat Immunol. 2003;4:1169–76.
Yuan ZL, Guan YJ, Chatterjee D, Chin YE. Stat3 dimerization regulated by reversible acetylation of a single lysine residue. Science. 2005;307:269–73.
Herzner AM, Hagmann CA, Goldeck M, Wolter S, Kubler K, Wittmann S, et al. Sequence-specific activation of the DNA sensor cGAS by Y-form DNA structures as found in primary HIV-1 cDNA. Nat Immunol. 2015;16:1025–33.
Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–8.
Wang Q, Liu X, Cui Y, Tang Y, Chen W, Li S, et al. The E3 ubiquitin ligase AMFR and INSIG1 bridge the activation of TBK1 kinase by modifying the adaptor STING. Immunity. 2014;41:919–33.
Yoh SM, Schneider M, Seifried J, Soonthornvacharin S, Akleh RE, Olivieri KC, et al. PQBP1 is a proximal sensor of the cGAS-dependent innate response to HIV-1. Cell . 2015;161:1293–305.
Zhou Q, Lin H, Wang S, Wang S, Ran Y, Liu Y, et al. The ER-associated protein ZDHHC1 is a positive regulator of DNA virus-triggered, MITA/STING-dependent innate immune signaling. Cell Host Microbe. 2014;16:450–61.
Demaria O, De Gassart A, Coso S, Gestermann N, Di Domizio J, Flatz L, et al. STING activation of tumor endothelial cells initiates spontaneous and therapeutic antitumor immunity. Proc Natl Acad Sci USA. 2015;112:15408–13.
Woo SR, Corrales L, Gajewski TF. The STING pathway and the T cell-inflamed tumor microenvironment. Trends Immunol. 2015;36:250–6.
Wang H, Hu S, Chen X, Shi H, Chen C, Sun L, et al. cGAS is essential for the antitumor effect of immune checkpoint blockade. Proc Natl Acad Sci USA. 2017;114:1637–42.
Nefedova Y, Huang M, Kusmartsev S, Bhattacharya R, Cheng P, Salup R, et al. Hyperactivation of STAT3 is involved in abnormal differentiation of dendritic cells in cancer. J Immunol. 2004;172:464–74.
Lechner MG, Liebertz DJ, Epstein AL. Characterization of cytokine-induced myeloid-derived suppressor cells from normal human peripheral blood mononuclear cells. J Immunol. 2010;185:2273–84.
Corzo CA, Cotter MJ, Cheng P, Cheng F, Kusmartsev S, Sotomayor E, et al. Mechanism regulating reactive oxygen species in tumor-induced myeloid-derived suppressor cells. J Immunol. 2009;182:5693–701.
Taleb K, Auffray C, Villefroy P, Pereira A, Hosmalin A, Gaudry M, et al. Chronic type I IFN is sufficient to promote immunosuppression through accumulation of myeloid-derived suppressor cells. J Immunol. 2017;198:1156–63.
Norris BA, Uebelhoer LS, Nakaya HI, Price AA, Grakoui A, Pulendran B. Chronic but not acute virus infection induces sustained expansion of myeloid suppressor cell numbers that inhibit viral-specific T cell immunity. Immunity. 2013;38:309–21.
Cui J, Song Y, Li Y, Zhu Q, Tan P, Qin Y, et al. USP3 inhibits type I interferon signaling by deubiquitinating RIG-I-like receptors. Cell Res. 2014;24:400–16.
Zhang J, Zhang C, Cui J, Ou J, Han J, Qin Y, et al. TRIM45 functions as a tumor suppressor in the brain via its E3 ligase activity by stabilizing p53 through K63-linked ubiquitination. Cell Death Dis. 2017;8:e2831.
Acknowledgements
We thank Professor Hui Zhang and Jie Zhou (Zhongshan Medical School, Sun Yat-sen University) for the STING knockout mice and technical support, respectively. This work was supported by the National Natural Science Foundation of China (grant nos. 31870862, 81773256, 81572982, 81372442, and 81172164), the National Key Basic Research Program of China (2014CB910800, 2015CB859800, and 2013CB910301), the Sci-Tech Key Program of the Guangdong Province Science Foundation (grant no. 2014A020212066), and Science and Technology Planning Project of Guangzhou, China (201804010385 and 201605030012).
Author information
Authors and Affiliations
Contributions
J.C., J.L. and Y.-X.Z. proposed the concept and conceived the entire study. C.-x.Z., S.-b.Y., J.-j.N., T.-t.C., Y.-n.L. and D.-j.H. performed experiments. H.-q.M., Q.-y.C., J.H. and X.-s.Z. selected patients and collected clinical samples. J.C., J.L. and C.-x.Z. wrote the manuscript. Y.-x.Z. helped edit the manuscript. J.C. and J.L. supervised the entire project.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by A. Villunger
Supplementary information
Rights and permissions
About this article
Cite this article
Zhang, Cx., Ye, Sb., Ni, Jj. et al. STING signaling remodels the tumor microenvironment by antagonizing myeloid-derived suppressor cell expansion. Cell Death Differ 26, 2314–2328 (2019). https://doi.org/10.1038/s41418-019-0302-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-019-0302-0
This article is cited by
-
Targeting cGAS/STING signaling-mediated myeloid immune cell dysfunction in TIME
Journal of Biomedical Science (2023)
-
The downregulation of type I IFN signaling in G-MDSCs under tumor conditions promotes their development towards an immunosuppressive phenotype
Cell Death & Disease (2022)
-
Re-engineered BCG overexpressing cyclic di-AMP augments trained immunity and exhibits improved efficacy against bladder cancer
Nature Communications (2022)
-
Extracellular traps from activated vascular smooth muscle cells drive the progression of atherosclerosis
Nature Communications (2022)
-
SPLUNC1 regulates LPS-induced progression of nasopharyngeal carcinoma and proliferation of myeloid-derived suppressor cells
Medical Oncology (2022)