Abstract
Argonaute 2 (AGO2), the core component of microRNA (miRNA)-induced silencing complex, plays a compelling role in tumorigenesis and aggressiveness. However, the mechanisms regulating the functions of AGO2 in cancer still remain elusive. Herein, we indentify one intronic circular RNA (circRNA) generated from AGO2 gene (circAGO2) as a novel regulator of AGO2-miRNA complexes and cancer progression. CircAGO2 is up-regulated in gastric cancer, colon cancer, prostate cancer, and neuroblastoma, and is associated with poor prognosis of patients. CircAGO2 promotes the growth, invasion, and metastasis of cancer cells in vitro and in vivo. Mechanistic studies reveal that circAGO2 physically interacts with human antigen R (HuR) protein to facilitate its activation and enrichment on the 3’-untranslated region of target genes, resulting in reduction of AGO2 binding and repression of AGO2/miRNA-mediated gene silencing associated with cancer progression. Pre-clinically, administration of lentivirus-mediated short hairpin RNA targeting circAGO2 inhibits the expression of downstream target genes, and suppresses the tumorigenesis and aggressiveness of xenografts in nude mice. In addition, blocking the interaction between circAGO2 and HuR by cell-penetrating inhibitory peptide represses the tumorigenesis and aggressiveness of cancer cells. Taken together, these results indicate that oncogenic circAGO2 drives cancer progression through facilitating HuR-repressed functions of AGO2-miRNA complexes.
Similar content being viewed by others
Introduction
Argonaute 2 (AGO2), a component of mammalian AGO protein family, is ubiquitously expressed and involved in post-transcriptional gene silencing [1]. As a unique protein of RNA-induced silencing complex (RISC) with endonuclease activity, AGO2 recognizes specific targets through base-pairing of small interfering RNA or microRNA (miRNA) [2], resulting in degradation of mRNA [3] or repression of translation [1]. Recent studies have shown that AGO2 is aberrantly expressed in many types of cancer, such as gastric cancer [4], colorectal cancer [5], prostate cancer [6], and neuroblastoma [7], and regulates the proliferation, migration, and invasion of cancer cells [8], suggesting a compelling role of AGO2 in the development and progression of human cancers [5]. However, the mechanisms regulating the functions of AGO2 in tumorigenesis and aggressiveness still remain elusive.
Circular RNAs (circRNAs) are a class of stable RNA molecules that are covalently closed loops and resistant to the degradation by exonuclease RNase R [9]. Emerging evidence shows that circRNAs exhibit aberrant expression in many types of cancer, including gastric cancer [10], colorectal cancer [11], hepatocellular carcinoma [12], and ovarian cancer [13], and function as an important type of endogenous RNAs regulating the proliferation, migration, invasion, and therapeutic resistance of cancer cells [10, 14]. One action mode of circRNAs is to serve as endogenous sponges of miRNAs to regulate target gene expression. For example, ciRS-7 is able to bind with miRNA-7 to activate oncogenes epidermal growth factor receptor (EGFR) and v-raf-1 murine leukemia viral oncogene homolog 1 (RAF1), resulting in aggressive phenotypes of colorectal cancer cells [15]. In addition, circRNAs may bind and sequester RNA-binding proteins (RBPs) or even base-pair with other RNAs to regulate their activities [16]. Previous studies show that circular RNA of forkhead box O3 (circFoxo3) represses cell cycle progression by binding to cyclin-dependent kinase 2 and cyclin-dependent kinase inhibitor 1 [17]. Therefore, it is crucial to understand the roles of circRNAs in cancer progression.
In this study, we identify one intronic circRNA generated from AGO2 gene (circAGO2) as a novel regulator of AGO2-miRNA complexes and cancer progression. CircAGO2 is up-regulated in human cancer tissues, associated with poor prognosis of patients, and promotes the growth, invasion, and metastasis of cancer cells. Mechanistically, circAGO2 physically interacts with human antigen R (HuR) protein to facilitate its competitive enrichment on the 3’-untranslated region (UTR) of target genes, resulting in reduction of AGO2 binding, repression of AGO2/miRNA-mediated gene silencing, and promotion of tumorigenesis and aggressiveness, suggesting the crucial roles of circAGO2 in cancer progression.
Results
CircAGO2 is up-regulated in human cancer tissues and cell lines
Mining of circular RNA database circBase [18] revealed six potential circRNAs generated from human AGO2 gene (Fig. 1a). Further reverse transcription PCR (RT-PCR) using divergent primers validated two detectable circRNAs in gastric cancer AGS cells (Supplementary Fig. S1a). One circRNA (hsa_circ_0135889), termed as circRNA generated from AGO2 (circAGO2), was highly expressed in the tissues of gastric cancer, colon cancer, prostate cancer, than their adjacent normal tissues (Fig. 1b and Supplementary Fig. S1b). Higher circAGO2 expression was also noted in more aggressive neuroblastoma, when compared to that in ganglioneuroblastoma (Fig. 1b). In addition, circAGO2 was abundant in many cancer cell lines (Supplementary Fig. S1c). The cyclization of 391-nt circAGO2, consisting of partial first intron of AGO2, was validated by RT-PCR with divergent primers and Sanger sequencing (Fig. 1c and Fig. 1d). Subcellular fractionation and RNA fluorescence in situ hybridization (FISH) assays indicated the cytoplasmic and nuclear enrichment and localization of circAGO2 in cancer cells (Fig. 1e, Fig. 1f, and Supplementary Fig. S1d). Moreover, the endogenous or ectopic expression of circAGO2 resistant to efficient RNase R digestion was also detected in cancer cell lines using junction-specific or non-junction probes in Northern blot assay (Fig. 1g, Fig. 1h, Supplementary Figs. S1e, S1f). These results indicated that circAGO2 was up-regulated in cancer tissues and cell lines.
CircAGO2 is up-regulated in human cancer tissues and cell lines. a The flowchart delineating the identification of circAGO2 from cancer cells and tissues. CT, cancer tissues; MNT, matched normal tissues. b Real-time qRT-PCR assay showing the relative levels of circAGO2 (normalized to β-actin) in matched normal tissues (MNT) and cancerous tissues (CT) of gastric cancer, colon cancer, and prostate cancer, and those in ganglioneuroblastoma (GNB) and neuroblastoma (NB). c RT-PCR or PCR assay indicating the detection of circAGO2 using divergent and convergent primers from cDNA or genomic DNA (gDNA) of cancer cell lines AGS and PC-3. d Schematic illustration showing the genomic location of circAGO2 generated from its host gene, and validation by Sanger sequencing. e Real-time qRT-PCR indicating the distribution of circAGO2, GAPDH, and U1 in the cytoplasmic and nuclear fractions of cancer cells (mean ± SD, n = 5). f RNA-FISH assay showing the cytoplasmic and nuclear localization of circAGO2 in cultured AGS cells using a junction-specific antisense probe (green), with the nuclei staining with DAPI (blue). Sense probe with RNase R (3 U/mg) treatment was used as a negative control. The GAPDH and U1 were applied as positive controls. Scale bar: 10 μm. g, h Northern blot using a junction-specific probe indicating the endogenous existence (g) of circAGO2 in cancer cell lines, and those of MKN-45 cells transfected by circAGO2 (h) and treated with RNase R (3 U/mg). Student’s t test compared gene expression levels in b
CircAGO2 promotes the growth, invasion, and metastasis of cancer cells
To explore the roles of circAGO2 in tumorigenesis and aggressiveness, the expression vector and short hairpin RNAs (shRNAs) targeting the junction of circAGO2 were established and stably transfected into cancer cells, resulting in its over-expression or knockdown, respectively (Supplementary Figs. S2a and S2b). Ectopic expression or silencing of circAGO2 increased and decreased the anchorage-independent growth and invasion capabilities of gastric cancer cell lines MKN-45 and AGS, colon cancer HCT-116 cells, and prostate cancer PC-3 cells, respectively (Fig. 2a, Fig. 2b, Supplementary Figs. S2c and S2d). In addition, the growth, volume and weight of subcutaneous xenografts formed by cancer cells were significantly increased and decreased by stable over-expression or knockdown of circAGO2, respectively (Fig. 2c, 2d, and 2e). Immunohistochemical staining of xenografts showed that stable ectopic expression or knockdown of circAGO2 increased and decreased the proliferation index Ki-67 and CD31-positive intratumoral microvessels, respectively (Fig. 2f). In experimental metastasis assay, athymic nude mice treated with tail vein injection of circAGO2 over-expressing MKN-45 cells or circAGO2 knockdown AGS cells displayed more or less lung metastatic colonies, and had less or greater survival probability, respectively (Fig. 2g and Supplementary Fig. S2e). These data indicated that circ-AGO2 exerted an oncogenic role in tumorigenesis and aggressiveness.
CircAGO2 promotes the growth, invasion, and metastasis of cancer cells in vitro and in vivo. a, b Representative images (upper panel) and quantification (lower panel) of soft agar (a) and matrigel invasion (b) assays showing the anchorage-independent growth and invasion of MKN-45 and AGS cells stably transfected with empty vector (mock), circAGO2, scramble shRNA (sh-Scb), or sh-circAGO2 #1 (mean ± SD, n = 4). c, d, e Representative images (c), in vivo growth curve (d), and weight at the end points (e) of xenografts formed by subcutaneous injection of MKN-45 and AGS cells stably transfected with mock, circAGO2, sh-Scb, or sh-circAGO2 #1 into the dorsal flanks of nude mice (n = 5 for each group). f Representative images (upper panel) and quantification (lower panel) of immunohistochemical staining revealing the expression of Ki-67 and CD31 within xenografts formed by subcutaneous injection of MKN-45 and AGS cells stably transfected with mock, circAGO2, sh-Scb, or sh-circAGO2 #1. Scale bar: 100 μm. g Representative images (upper panel), HE staining (lower left panel), and quantification (lower right panel) of lung metastatic colonization of nude mice treated with tail vein injection of MKN-45 and AGS cells stably transfected with mock, circAGO2, sh-Scb, or sh-circAGO2 #1 (n = 5 for each group). Scale bar: 100 μm. Student’s t test and analysis of variance compared the difference in a, b, and d–g. *P < 0.05, **P < 0.01, ***P < 0.001 vs. mock or sh-Scb
CircAGO2 interacts with and activates HuR protein in cancer cells
To elucidate the mechanisms responsible for oncogenic roles of circAGO2, we first investigated its effects on the expression of its host gene AGO2. Stable ectopic expression or knockdown of circAGO2 resulted in no changes in the promoter activity, transcript levels, or protein expression of AGO2 in cancer cells (Supplementary Fig. S3a, Fig. S3b, and Fig. S3c). To identify the protein partner of circAGO2, we performed the biotin-labeled linear or circular RNA pull-down followed by a proteomic analysis of RNA-associated protein complex in AGS cells (Fig. 3a), in which the circRNA probe was generated by ligation of biotin-labeled linear circAGO2 transcript in vitro [19]. Mass spectrometry assay revealed 767 differential proteins between circAGO2 and linear transcript pull-down groups (Supplementary Table S1), and over-lapping with established RBP from RBPDB database (http://rbpdb.ccbr.utoronto.ca) and AGO2-binding proteins from BioGRID database (https://thebiogrid.org) indicated four potential circAGO2-interacting partner (Fig. 3b). Further validating RNA pull-down and western blot assays indicated the physical interaction of circAGO2 with HuR, but not with TAR DNA binding protein (TARDBP), ribosomal protein L27A (RPL27A), or RNA binding motif protein 4 (RBM4) (Fig. 3b and Fig. 3c). Consistently, RNA electrophoretic mobility shift assay (EMSA) showed that circAGO2 strongly interacted with endogenous HuR in the lysates of AGS cells or glutathione S-transferase (GST)-tagged recombinant HuR protein (Fig. 3d). In vitro binding assay indicated that RNA recognition motif 1 [RRM1, 19-100 amino acids (aa)], but not RRM2 (103-189 aa), RRM3 (245-326 aa), or hinge (190-244 aa) domain, of GST-tagged or Flag-tagged HuR protein was crucial for its interaction with circAGO2 (Fig. 3e). Mutation of core amino acids (60-63, SLGY) within RRM1 domain analyzed by catRapid [20] and RNABindRPlus [21] abolished the interaction of HuR with circAGO2 (Fig. 3e). The expression levels of HuR were not affected by over-expression or knockdown of circAGO2 (Supplementary Figs. S4a and S4b). Instead, ectopic expression or knockdown of circAGO2 facilitated and attenuated the translocation of HuR from nucleus to cytoplasm in AGS and PC-3 cells, respectively (Fig. 3f, Fig. 3g, and Supplementary Fig. S4c). In dual-luciferase assay with a reporter containing three canonical HuR binding sites [22] locating downstream of Renilla luciferase, ectopic expression or knockdown of circAGO2 or HuR increased and decreased the HuR activity in AGS and PC-3 cells, respectively (Fig. 3h, Supplementary Figs. S4d and S4e). These results suggested that circAGO2 interacted with and activated HuR protein in cancer cells.
CircAGO2 interacts with and activates HuR protein in cancer cells. a Schematic illustration showing the procedure for identifying the circAGO2-interacting protein by biotin-labeled RNA pull-down and mass spectrometry (MS) assays. b Coomassie bright blue staining (left panel), Venn diagram (right upper panel), and western blot (right lower panel) indicating the differential proteins pulled down by linear and circular RNA, their over-lapping analysis with established RBP and AGO2-interacting proteins derived from RBPDB and BioGRID databases, and experimental validation in AGS cells. c Western blot assay showing the HuR protein pulled down by biotin-labeled linear or circular RNA probes from lysates of AGS cells. d RNA EMSA determining the interaction between endogenous (within lysates of AGS cells) or recombinant HuR protein and biotin-labeled RNA probe for circAGO2 (arrowheads), with or without treatment by HuR antibody or competition using an excess of unlabeled circRNA probe. e In vitro binding assay depicting the recovered circAGO2 levels from AGS cells detected by RT-PCR (upper panel) after incubation with full-length, truncation, or mutation forms of Flag-tagged or GST-tagged recombinant HuR protein validated by western blot (lower panel). f Western blot indicating the expression of HuR or histone H3 in total lysates or subcellular fractions of AGS cells stably transfected with empty vector (mock), circAGO2, scramble shRNA (sh-Scb), or sh-circAGO2 #1. g Dual RNA-FISH and immunofluorescence staining assay showing the co-localization of circAGO2 (green) and HuR (red) in AGS cells stably transfected with mock or circAGO2, and treated with RNase R (3 U/mg), with the nuclei staining with DAPI (blue). Scale bar: 10 μm. h Dual-luciferase assay indicating the activity of HuR in AGS cells transfected with mock, HuR, circAGO2, sh-Scb, sh-HuR, or sh-circAGO2#1 (mean ± SD, n = 5). Student’s t test compared the difference in h. *P < 0.05, **P < 0.01 vs. mock or sh-Scb
CircAGO2 facilitates HuR-repressed functions of AGO2-miRNA complexes
Since previous studies implicate the roles of HuR in affecting the functions of AGO2 and miRNAs [23], we next explored the impacts of circAGO2 on AGO2/miRNA-mediated gene silencing in cancer cells. Endogenous interaction between AGO2 and HuR was noted in MKN-45 cells (Fig. 4a), and ectopic expression of circAGO2 restrained their interaction (Fig. 4b). To explore the downstream targets of circAGO2, we observed the differentially expressed genes by RNA sequencing (RNA-seq) in MKN-45 cells stably transfected with circAGO2. There were 624 genes, including 257 up-regulated and 367 down-regulated ones, that showed differential expression (fold change > 2.0, P < 0.05) upon circAGO2 over-expression (Fig. 4c and Supplementary Table S2). Over-lapping analysis with HuR CLIP-seq dataset derived from CLIPdb database [24] revealed 132 target genes with AU-rich elements (ARE) within their 3’-UTR [25] (Fig. 4c). A portion of these genes were associated with cancer progression, including eukaryotic translation initiation factor 4E binding protein 3 (EIF4EBP3) [26], hepatocyte nuclear factor 4 alpha (HNF4A) [27], mitogen-activated protein kinase kinase kinase kinase 1 (MAP4K1) [28], neurogenic locus notch homolog protein 4 (NOTCH4) [29], solute carrier family 2 member 4 (SLC2A4) [30], and solute carrier family 44 member 4 (SLC44A4) [31].
CircAGO2 facilitates HuR-repressed functions of AGO2-miRNA complexes. a, b Co-IP and western blot assays indicating the interaction between AGO2 and HuR in AGS cells (a), and that in MKN-45 cells (b) stably transfected with empty vector (mock) or circAGO2. c Venn diagram (upper panel) showing the over-lapping analysis of RNA-seq results from MKN-45 cells stably transfected with circAGO2 and HuR targets in CLIPdb database. Heatmaps (lower panel) showing 132 differentially expressed genes (fold change > 2.0, P < 0.05) containing ARE within 3’-UTR in MKN-45 cells stably transfected with mock or circAGO2. d Schematic illustration indicating the wild type or mutant (Mut) 3’-UTR of target genes EIF4EBP3, HNF4A, and MAP4K1, and base pairing of corresponding miRNAs. e and f RIP and qRT-PCR assays revealing the endogenous binding of HuR or AGO2 to 3’-UTR of target genes in AGS cells (e) and that in MKN-45 cells (f) transfected with mock, circAGO2, sh-HuR#2, or miRNA mimics (100 nmol/L). g Dual-luciferase assay showing the activity of target gene 3’-UTR reporters with wild type (WT) or mutant (Mut) miRNA binding site in MKN-45 cells transfected with mock, circAGO2, sh-HuR#2, or miRNA mimics (100 nmol/L). h and i Real-time qRT-PCR (h, mean ± SD, n = 5) and western blot (i) assays indicating the transcript and protein levels (normalized to β-actin) of EIF4EBP3, HNF4A and MAP4K1 in MKN-45 cells transfected with mock, circAGO2, negative control miRNA (miRNA-NC), or miRNA mimics (100 nmol/L). Student’s t test and analysis of variance compared the difference in e–h. *P < 0.05, **P < 0.01 vs. IgG, mock, or mock + miRNA-NC. Δ P < 0.01 vs. WT
Notably, analysis using algorithm miRWalk [32] revealed complementary binding sites of miR-224-5p, miR-143-3p, miR-181a-5p, miR-503-5p, or miR-125a-3p at regions adjacent to ARE sites within 3’-UTR of these genes (Fig. 4d and Supplementary Fig. S5a). However, analysis using circInteractome program [33] revealed no potential binding sites of these miRNAs within circAGO2. RNA immunoprecipitation (RIP) assay indicated the endogenous binding of HuR or AGO2 to the 3’-UTR of EIF4EBP3, HNF4A, MAP4K1, NOTCH4, SLC2A4, and SLC44A4 (Fig. 4e). Knockdown of HuR decreased the endogenous enrichment of HuR and AGO2 on the 3’-UTR of target genes, and abolished the increased binding of HuR and decreased enrichment of AGO2 induced by ectopic expression of circAGO2 in MKN-45 cells (Fig. 4f and Supplementary Fig. S5b). Meanwhile, over-expression of miR-224-5p, miR-143-3p, miR-181a-5p, miR-503-5p, or miR-125a-3p increased the enrichment of HuR and AGO2 on 3’-UTR of target genes, which was rescued by circAGO2 over-expression (Fig. 4f, Supplementary Fig. S5b and S5c). Knockdown of HuR or transfection of miRNA mimics suppressed the activity of target gene 3’-UTR reporter, which was abolished by ectopic expression of circAGO2 or mutation of miRNA binding sites (Fig. 4g and Supplementary Fig. S5d). In addition, over-expression of circAGO2 prevented the decrease in the levels of EIF4EBP3, HNF4A, MAP4K1, NOTCH4, SLC2A4, and SLC44A4 in MKN-45 cells transfected with mimics of miR-224-5p, miR-143-3p, miR-181a-5p, miR-503-5p, or miR-125a-3p, respectively (Fig. 4h, Fig. 4i, Supplementary Fig. S5e and S5f). Overall, these findings indicated that circAGO2 facilitated HuR-repressed functions of AGO2-miRNA complexes in cancer cells.
CircAGO2 harbors oncogenic properties through activating HuR
We further investigated the functional interplay between circAGO2 and HuR in regulating the growth and aggressiveness of cancer cells. Knockdown or ectopic expression of HuR abolished the increased and decreased activity of HuR in MKN-45 and AGS cells stably over-expressing or silencing circAGO2, respectively (Supplementary Fig. S6a). Meanwhile, knockdown or ectopic expression of HuR led to a significant decrease or increase in the mRNA stability and expression of target genes EIF4EBP3, HNF4A, MAP4K1, NOTCH4, SLC2A4, and SLC44A4, which was rescued by over-expression or silencing of circAGO2 in MKN-45 and AGS cells, respectively (Supplementary Fig. S6b, S6c, S6d, S6e). In soft agar, scratch, and matrigel invasion assays, cancer cells stably transfected with sh-HuR#2 or HuR presented a decrease or increase in growth, migration, and invasiveness, respectively (Supplementary Fig. S7a, S7b, S7c). Restoration of HuR activity via stable transfection of circAGO2 or sh-circAGO2 #1 prevented the cancer cells from altered biological behaviors induced by knockdown or over-expression of HuR (Supplementary Fig. S7a, S7b, S7c). These data indicated that circAGO2 harbored oncogenic properties through activating HuR in cancer cells.
Therapeutic effects of lentivirus-mediated circAGO2 knockdown in vivo
We further explored the therapeutic efficiencies of circAGO2 knockdown on tumor growth and survival of athymic nude mice bearing xenografts. Intratumoral administration of lentivirus-mediated sh-circAGO2 #1 resulted in significant reduction of the growth, tumor weight, Ki-67 proliferation index, and CD31-positive intratmoral microvessels of subcutaneous xenografts (Fig. 5a and Fig. 5b). The intratumoral expression of circAGO2, EIF4EBP3, HNF4A, MAP4K1, NOTCH4, SLC2A4, and SLC44A4 was significantly down-regulated in xenografts treated with lentivirus-mediated sh-circAGO2 #1 (Fig. 5c and Fig. 5d). In the therapeutic experiments on cancer metastasis, administration of lentivirus-mediated sh-circAGO2 #1 decreased the lung metastatic colonies and increased the survival probability of nude mice (Fig. 5e). These results suggested that lentivirus-mediated circAGO2 knockdown inhibited the tumorigenesis and aggressiveness in vivo.
Therapeutic effects of lentivirus-mediated circAGO2 knockdown in vivo. a The in vivo growth curve (left lower panel), representative images (middle panels), and weight at the end points (right panel) of xenografts formed by subcutaneous injection of AGS cells into the dorsal flanks of athymic nude mice (n = 5 for each group) and received intratumoral injection of lentivirus (LV)-mediated scramble shRNA (sh-Scb) or sh-circAGO2 #1 as indicated (left upper panel). b Representative images (left panel) and quantification (right panel) of immunohistochemical staining revealing the expression of Ki-67 and CD31 within subcutaneous xenografts following intratumoral injection of lentivirus-mediated sh-Scb or sh-circAGO2 #1. Scale bar: 50 μm. c, d Real-time qRT-PCR (c, normalized to β-actin) and western blot (d) assays indicating the expression of target genes EIF4EBP3, HNF4A, MAP4K1, NOTCH4, SLC2A4, and SLC44A4 within xenografts (n = 5 for each group) following intratumoral injection of lentivirus-mediated sh-Scb or sh-circAGO2 #1. e Representative images (upper panel), HE staining (middle panel), quantification of lung metastatic colonization (lower left panel), and survival curves (lower right panel) of nude mice (n = 5 for each group) treated with tail vein injection of AGS cells and subsequent administration of lentivirus-mediated sh-Scb or sh-circAGO2 #1 as indicated (upper panel). Scale bar: 100 μm. Student’s t test and analysis of variance compared the difference in a–c and e. Log-rank test for survival comparison in e. *P < 0.05, **P < 0.01 vs. LV-sh-Scb
Inhibitory peptides suppress cancer progression by blocking circAGO2-HuR interaction
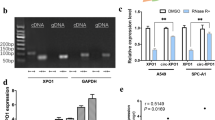
Based on the importance of RRM1 domain (especially 60-63 aa) of HuR protein in interacting with circAGO2 (Fig. 6a), we further designed a cell-penetrating peptide, named as HuR inhibitory peptide with 13 amino acids in length (HIP-13), that might block the circAGO2-HuR interaction. Treatment of AGS cells with fluorescein isothiocyanate (FITC)-labeled HIP-13 resulted in obvious aggregation of peptide within the cytoplasm (Fig. 6b). Biotin-labeled peptide pull-down assay revealed the binding of HIP-13 to endogenous circAGO2 (Fig. 6c). In addition, HIP-13 treatment abolished the endogenous circAGO2-HuR interaction in AGS cells, which was similar to the effects of CMLD-2 (an established inhibitor of HuR activity) [34] (Supplementary Fig. S8a). Administration of HIP-13 or CMLD-2 suppressed the viability, growth, and invasion of AGS cells (Supplementary Fig. S8b, 6d, and 6e). Administration of HIP-13 significantly reduced the growth, tumor weight, Ki-67 proliferation index, and CD31-positive intratumoral microvessels of subcutaneous xenografts formed by injection of AGS cells, with similar effects to those of CMLD-2 (Fig. 6f and Fig. 6g). In experimental metastasis assay, administration of HIP-13 or CMLD-2 via tail vein significantly reduced the lung metastatic colonies and increased the survival probability of athymic nude mice treated with tail vein injection of AGS cells (Fig. 6h and Supplementary Fig. S8c). These data suggested that inhibitory peptide blocking circAGO2-HuR interaction suppressed the growth and aggressiveness of cancer cells in vitro and in vivo.
Inhibitory peptides suppress cancer progression by blocking the circAGO2-HuR interaction. a Schematic illustration showing the structure and core amino acids (SLGY) of HuR protein essential for interacting with circAGO2. b Confocal images (upper panel) revealing the distribution of synthesized control peptide (CTL Pep) or inhibitory peptide (HIP-13, lower panel) after incubation with AGS cells for 48 h. The nuclei were stained with DAPI (blue), while cellular membrane was stained with Dil (red). Scale bar: 10 μm. c Biotin-labeled peptide pull-down assay indicating the interaction of CTL Pep (30 μmol·L−1) or HIP-13 (15 and 30 μmol·L−1) with circAGO2 of AGS cells. d, e Representative images (upper panel) and quantification (lower panel) of soft agar (d) and matrigel invasion (e) assays showing the anchorage-independent growth and invasion of AGS cells treated with solvent control (DMSO), CMLD-2 (30 μmol·L−1), CTL Pep (30 μmol·L−1), or HIP-13 (30 μmol·L−1) for 48 h. f, g Representative images (f, left lower panel), in vivo growth curve (f, right upper panel), tumor weight at the end points (f, right lower panel), intratumoral expression of Ki-67 and CD31 (g) of xenografts formed by subcutaneous injection of AGS cells into nude mice (n = 5 for each group) that subsequently treated with intratumoral injection of DMSO, CMLD-2 (3 mg·kg−1), CTL Pep (3 mg·kg−1), or HIP-13 (3 mg·kg−1) as indicated (f, left upper panel). Scale bar: 50 μm. h Representative images (upper panel) and quantification (lower left panel) of lung metastatic colonization, and survival curves (lower right panel) of nude mice (n = 5 for each group) treated with tail vein injection of AGS cells and subsequent administration of DMSO, CMLD-2 (3 mg·kg−1), CTL Pep, or HIP-13 (3 mg·kg−1). Student’s t test and analysis of variance compared the difference in d–h. Log-rank test for survival comparison in h. *P < 0.05, **P < 0.01, ***P < 0.001 vs. DMSO or CTL Pep
CircAGO2 and HuR are highly expressed and associated with poor outcome in gastric cancer
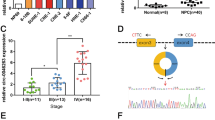
To better understand the clinical relevance of circAGO2 or HuR expression in gastric cancer, fresh tissues were collected from 81 primary cases. Higher levels of circAGO2 (P < 0.0001) or HuR (P = 0.002) were observed in gastric cancer tissues with metastasis, than those without metastasis (Fig. 7a). Kaplan–Meier survival analysis of this series of gastric cancer cases indicated significant difference in overall survival of patients with low or high expression of circAGO2 (P < 1.0 × 10−4) or HuR (P = 1.2 × 10−3, Fig. 7b). Immunohistochemical staining of tissue microarray containing 222 cases indicated high HuR expression in gastric cancer, which was associated with pathological grade (P = 0.010), local invasion (P = 0.004), lymph node metastasis (P = 0.031), or advanced tumor-node-metastasis (TNM) stage (P = 0.002) (Fig. 7c and Supplementary Table S3). The circAGO2 levels were positively correlated with those of target genes EIF4EBP3, HNF4A, MAP4K1, NOTCH4, SLC2A4 and SLC44A4 in our series of 81 gastric cancer cases (Fig. 7d). In addition, Kaplan–Meier survival plots of well-defined gastric cancer cases derived from Kaplan-Meier Plotter (http://kmplot.com/analysis) revealed that higher expression of HuR (P < 1.0 × 10−4 and P = 1.0 × 10−3), EIF4EBP3 (P = 4.3 × 10−3 and P = 2.0 × 10−2), HNF4A (P < 1.0 × 10−4 and P = 2.0 × 10−4), MAP4K1 (P < 1.0 × 10−4 and P = 1.0 × 10−3), NOTCH4 (P < 1.0 × 10−4 and P < 1.0 × 10−4), SLC2A4 (P < 1.0 × 10−4 and P < 1.0 × 10−4), or SLC44A4 (P = 3.0 × 10−2 and P = 8.3 × 10−3) was associated with lower overall (OS) and first progression (FP) survival probability of patients (Fig. 7e and Supplementary Fig. S9). These results indicated that circAGO2 and HuR were highly expressed and associated with poor outcome in gastric cancer.
CircAGO2 and HuR are highly expressed and associated with poor outcome in gastric cancer. a Real-time qRT-PCR assay showing the levels of circAGO2 or HuR (normalized to β-actin) in gastric cancer tissues with differential status of metastasis. b Kaplan-Meier curves indicating overall survival of 81 gastric cancer patients with low or high expression of circAGO2 or HuR (cutoff values = 11.76 and 4.58). c Representative images of immunohistochemical staining showing the nuclear and cytoplasmic expression of HuR in normal counterparts and cancerous tissues of gastric cancer (brown, arrowheads). Scale bar: 100 μm. d The positive correlation between the transcript levels of circAGO2 and its target genes EIF4EBP3, HNF4A, MAP4K1, NOTCH4, SLC2A4, or SLC44A4 in gastric cancer tissues (n = 81). e Kaplan–Meier curves indicating the overall (OS) and first progression (FP) survival of well-defined gastric cancer cases derived from Kaplan-Meier Plotter (http://kmplot.com/analysis) with high or low expression of HuR (cutoff values = 368.00 and 305.00), EIF4EBP3 (cutoff value = 409.00), HNF4A (cutoff value = 72.00), MAP4K1 (cutoff value = 116.00), NOTCH4 (cutoff value = 174.00), SLC2A4 (cutoff value = 4.00), or SLC44A4 (cutoff value = 1110.00). f The mechanisms underlying circAGO2-promoted cancer progression: as an intronic circRNA, circAGO2 interacts with and activates HuR protein to facilitate its binding to 3’-UTR of target genes, resulting in decrease of AGO2 enrichment and repression of AGO2/miRNA-mediated gene silencing associated with cancer progression. Meanwhile, inhibitory peptide HIP-13 is able to block the circAGO2-HuR interaction, resulting in suppression of cancer progression. Student’s t test compared the difference in a. Log-rank test for survival comparison in b, e. Pearson’s correlation coefficient analysis in d
Discussion
CircRNAs are a class of endogenous non-coding RNAs that are characterized by their covalently closed loop structures [9]. Emerging studies reveal the abundance and diversity of circRNAs in mammalian cells [35], including exonic circRNAs (ecircRNA), intronic circRNAs (ciRNAs) [36], or exon-intron circRNAs (EIciRNA) [37]. CircRNAs are widely dysregulated in multiple cancers, and play a tumor suppressive or oncogenic role [38]. For example, circular RNA of itchy E3 ubiquitin protein ligase (circITCH) is under-expressed in esophageal squamous cell carcinoma, while over-expression of circITCH results in up-regulation of host gene and suppression of tumor growth [39]. CircRNAs generated from hippocampus abundant transcript 1 (circHIAT1) and mitochondrial translation optimization 1 (circMTO1) are down-regulated in clear cell renal cell carcinoma and hepatocellular carcinoma, respectively, and suppresses the migration and invasion of cancer cells [14, 40]. On the other hand, circRNA derived from plasmacytoma variant translocation 1 (circPVT1) is up-regulated in gastric cancer tissues and promotes the proliferation of cancer cells [10]. In colorectal cancer, circular RNA of coiled-coil domain containing 66 (circCCDC66) is elevated and associated with poor prognosis, and promotes the growth and invasion of cancer cells [11]. In the current study, we identify circAGO2 (hsa_circ_0135889) as a circRNA significantly up-regulated in multiple cancer tissues, including gastric cancer, colon cancer, prostate cancer, and neuroblastoma. High circAGO2 expression is associated with poor survival of cancer patients. To investigate the protein partner of circAGO2, we applied the biotin-labeled RNA pull-down combined with mass spectrometry analysis. Generally, this method identifies a lot of proteins, while only some of them directly bind to a specific RNA [41]. We demonstrate that circAGO2 physically interacts with HuR protein to facilitate its enrichment on the 3’-UTR of target genes, resulting in reduction of AGO2 binding, repression of AGO2/miRNA-mediated gene silencing, and promotion of tumorigenesis and aggressiveness (Fig. 7f). These findings highlight a novel oncogenic function of circAGO2 in cancer progression.
Although many circRNAs function as endogenous competing RNAs or miRNA sponges, this action model is becoming controversial due to their limited miRNA binding sites [15]. Alternatively, circRNAs can serve as essential regulators of gene transcription, RBP, or protein translation. Previous studies show that intronic circRNA interacts with RNA polymerase II to modulate the transcription of its parent gene in cis [36]. In this study, we identify that the action mode of circAGO2 is different from previously reported intronic circRNAs [36]. CircAGO2 does no affect the transcription or translation of AGO2 in cancer cells. Instead, circAGO2 regulates the expression of target genes in trans through facilitating HuR-repressed functions of AGO2/miRNA complexes. Since knockdown of HuR abolishes the changes in biological features of cancer cells induced by circAGO2, our findings indicate that the cancer promoting functions of circAGO2 are mediated, at least in part, through regulating the HuR activity.
As a RBP, HuR binds to ARE-containing 3’-UTR of mRNA, a major region mediating the regulatory roles of miRNA in post-transcriptional control of gene expression [42]. HuR is mainly localized within nucleus, while its cytoplasmic accumulation is the initial and critical step of its activation [43]. Members of signaling pathways, such as AMP-activated kinase, protein kinase C, and mitogen-activated protein kinase, are able to regulate the nucleocytoplasmic shuttling of HuR [44]. HuR expression is elevated in many types of cancers, and cytoplasmic HuR accumulation is correlated with unfavorable outcome of patients with non-small cell lung cancer, meningioma, esophageal squamous cell carcinoma, or bladder cancer [45]. Previous studies indicate that HuR enhances the mRNA stability and/or translation of genes controlling cell cycle and proliferation, such as cyclin A, cyclin B1, and c-fos [46, 47], whereas HuR inhibits the expression of p16 or c-Myc by recruiting AGO2 to the 3’-UTR [48, 49]. However, the mechanisms regulating these varied roles of HuR in cancer still remain elusive. In this study, our results indicate that high HuR expression is linked to the progression and poor outcome of gastric cancer, and basal HuR activity is associated with AGO2/miRNA-mediated gene silencing. In cultured cancer cells, circAGO2 activates the translocation of HuR from nucleus to cytoplasm, and facilitates its enrichment on the 3’-UTR regions of target genes, resulting in decreased AGO2 binding and repressed functions of AGO2-miRNA complexes. Due to their adjacent binding regions, we believe that circAGO2-facilitated enrichment of HuR on 3’-UTR may exert steric hindrance effects that affect the functions of AGO2-miRNA complexes, which warrants further investigation.
In recent years, pharmacological approaches have been explored to modulate the subcellular localization and RNA-binding activity of HuR. Small molecule MPT0B098, a potent microtubule inhibitor, is able to decrease the HuR translocation from nucleus to cytoplasm, resulting in decreased expression of hypoxia inducible factor 1 alpha [50]. As a low-molecular-weight inhibitor, MS-444 interferes with the RNA binding and trafficking of HuR to suppress the growth properties of glioma cells [51]. In addition, CMLD-2 has been identified as a new disruptor to block the binding of HuR to mRNAs of musashi RNA binding protein 1 (MSI1) and X-linked inhibitor of apoptosis (XIAP) [34], revealing HuR as a promising therapeutic target for cancers. In this study, our evidence shows that a cell-penetrating inhibitory peptide is able to block the circAGO2-HuR interaction, resulting in suppression of the growth and aggressiveness of cancer cells, suggesting a potential approach for the therapeutics of cancers.
In summary, for the first time, we have demonstrated that circAGO2 is up-regulated in cancer tissues, and associated with poor outcome of patients. CircAGO2 interacts with and activates HuR by facilitating its translocation from nucleus to cytoplasm, whereby HuR represses the AGO2/miRNA-mediated gene silencing, resulting in promotion of tumorigenesis and aggressiveness. Lentivirus-mediated circAGO2 knockdown or an inhibitory peptide blocking the circAGO2-HuR interaction is able to suppress the tumorigenesis and aggressiveness of cancer cells. Meanwhile, to facilitate the identification of circRNA-binding protein, quantitative proteomics may be helpful in distinguishing between specific and non-specific interaction partners of circRNA, which warrants further investigation. This study extends our knowledge about the regulation of AGO2/miRNA function by circRNA, and suggests that circAGO2 and HuR may be potential therapeutic targets for human cancers.
Materials and Methods
Cell culture
Human gastric cancer cell line MKN-45 (JCRB0254) was obtained from Japanese Collection of Research Bioresources Cell Bank (Osaka, Japan). Human embryonic kidney HEK293T cells (CRL-1573), and cell lines of gastric [AGS (CRL-1739)], cervical [HeLa (CCL-2)], colon [HCT-116 (CCL-247) and LoVo (CCL-229)], prostate [PC-3 (CRL-1435)] cancer and neuroblastoma [SH-SY5Y (CRL-2266) and SK-N-SH (HTB-11)] were purchased from American Type Culture Collection (Rockville, MD). Cell lines were authenticated by short tandem repeat profiling, and used within 6 months after resuscitation of frozen aliquots. Mycoplasma contamination was regularly examined using Lookout Mycoplasma PCR Detection Kit (Sigma, St. Louis, MO). Cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (Gibco, Grand Island, NY). Cells were incubated in serum-free DMEM for 4 h, and treated with CMLD-2 (Merck Millipore, Temecula, CA) or actinomycin D (ActD, Sigma).
Patient tissue samples
Approval to conduct this study was obtained from the Institutional Review Board of Tongji Medical College (approval number: 2011-S085). All procedures were undertaken in accordance with guidelines set forth by Declaration of Helsinki. Informed written consent was obtained from all patients. Tumor tissues and their adjacent normal tissues were obtained at surgery from patients suffering from gastric cancer, colon cancer, prostate cancer, or neuroblastoma, at Union Hospital of Tongji medical College. Patients with a history of preoperative chemotherapy or radiotherapy were excluded. All fresh specimens were frozen in liquid nitrogen, validated by pathological diagnosis, and stored at -80 °C until use.
RT-PCR and real-time quantitative RT-PCR
Nuclear and cytoplasmic RNA was extracted according to the instruction of RNA Subcellular Isolation Kit (Active Motif, Carlsbad, CA). Total RNA was extracted using RNeasy Mini Kit (Qiagen Inc., Redwood City, CA). For circRNA detection, treatment with RNase R (3 U/mg, Epicenter, Madison, WI) was undertaken at 37 °C for 15 min. Reverse transcription and real-time PCR were performed using Transcriptor First Strand cDNA Synthesis Kit (Roche, Indianapolis, IN), SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA), and primers shown in Supplementary Table S4. The results of transcript levels were analyzed by 2−△△Ct method. To measure mRNA stability, the de novo RNA synthesis was blocked with 5 μg/mL ActD treatment, while half-life of mRNA was examined by transcript levels at indicated time points relative to those before ActD treatment.
Northern blot
The junction or non-junction probe for circAGO2 was synthesized and labeled by digoxigenin (DIG, Supplementary Table S5). For Northern blot, 20 µg of total RNA was separated on 3-(N-morpholino)propanesulfonic acid (MOPS)-buffered 2% (w/v) agarose gel containing 1.2% (v/v) formaldehyde under denaturing conditions for 4 h at 80 V, and transferred to Hybond-N + membrane (Pall Corp., Port Washington, NY). Prehybridization was carried out at 65 °C for 30 min in DIG Easy Hyb solution (Roche). Hybridizations were performed at 65 °C for 16-18 h. Blots were washed stringently, detected by anti-DIG antibody staining, and recorded on X-ray films with chemiluminescence substrate CSPD (Roche).
Western blot
Tissue or cellular protein was extracted with 1 × cell lysis buffer (Promega, Madison, WI). Western blot was performed as previously described [52,53,54,55], with antibodies specific for AGO2 (ab186733), TARDBP (ab109535), RPL27A (ab74731), RBM4 (ab222898), HuR (ab200342), HNF4A (ab41898), MAP4K1 (ab33910), NOTCH4 (ab184742), β-actin (ab125402), Flag (ab45766, Abcam Inc, Cambridge, MA), SLC44A4 (A10435, ABclonal, Wuhan, China), GST (sc-33614), histone H3 (sc-10809), EIF4EBP3 (sc-134232), SLC2A4 (sc-53566), and GAPDH (sc-47724, Santa Cruz Biotechnology, Santa Cruz, CA).
Over-expression or knockdown of genes
Human circAGO2 linear sequence (391 bp) was obtained from gastric cancer tissues by PCR (Supplementary Table S5) and inserted into lentiviral vector pLCDH-ciR (Geenseed Biotech Co., Guangzhou, China). Human HuR cDNA (1989 bp) was kindly provided by Dr. Myriam Gorospe [56], and its truncations were amplified with PCR primers (Supplementary Table S5) and subcloned into pCMV-3Tag-1A (Addgene, Cambridge, MA). Mutation of HuR was prepared with GeneTailorTM Site-Directed Mutagenesis System (Invitrogen, Carlsbad, CA), using PCR primers indicated in Supplementary Table S5. Oligonucleotides specific for shRNAs against circAGO2 or HuR (Supplementary Table S6) were inserted into lentiviral vector GV298 (Genechem Co., Ltd, Shanghai, China). After selection for neomycin or puromycin (Invitrogen) resistance, stable cell lines were obtained.
Prediction and measurement of miRNA
The algorithm miRWalk [32] was applied to analyze the potential miRNA targeting sites within 3’-UTR. Confluent cells were transfected with negative control miRNA (miRNA-NC) or miRNA mimics (RiboBio Co. Ltd, Guangzhou, China) using Lipofectamine 2000 (Invitrogen). The miRNA-specific stem-loop primer and Bulge-LoopTM miRNAs qPCR Primer Set (RiboBio Co. Ltd) were used to synthesize the cDNA and measure the levels of mature miRNAs. The results were analyzed by normalizing the miRNA levels to those of U6 snRNA.
Lentivirus construction
The lentiviral vectors were co-transfected with packaging plasmids psPAX2 and pMD2G (Addgene) into HEK293T cells. Infectious lentivirus was harvested at 36 and 60 h after transfection, and filtered through 0.45 μm PVDF filters. Recombinant lentivirus was concentrated 100-fold by ultracentrifugation (2 h at 120,000 g). The lentivirus-containing pellet was dissolved in phosphate buffer saline (PBS) and injected in mice within 48 h.
RNA sequencing assay
Total RNA of cancer cells (1 × 106) was extracted in accordance with the manual of TRIzol® reagent (Life Technologies, Inc., Gaithersburg, MD). Library preparation and transcriptome sequencing on an Illumina HiSeq X Ten platform were carried out at Novogene Bioinformatics Technology Co., Ltd. (Beijing, China) to generate 100-bp paired-end reads. HTSeq v0.6.0 was applied in counting the numbers of read mapping to each gene, and fragments per kilobase of transcript per million fragments mapped (FPKM) of each gene were calculated. Sequencing results have been deposited in GEO database (accession code GSE114055).
Dual-luciferase reporter assay
Human AGO2 promoter (2526 bp) and 3’-UTR of target genes were amplified from genomic DNA using primer sets (Supplementary Table S5) and subcloned into pGL3-Basic (Promega) and psiCHECK2 (Promega), respectively. Human HuR luciferase reporter was established by annealing complementary oligonucleotides containing three canonical HuR binding sites (Supplementary Table S5) and inserting into psiCHECK2 (Promega). Mutation of miRNA binding sites was undertaken with GeneTailorTM Site-Directed Mutagenesis System (Invitrogen) and PCR primers (Supplementary Table S5). Dual-luciferase assay was performed according to the manufacturer’s instructions (Promega). Luciferase activity was measured with a luminometer (Lumat LB9507, Berthold Tech., Bad Wildbad, Germany) [52,53,54]. For AGO2 promoter activity, the luciferase signal was normalized by firefly/Renilla ratio, while the activity of HuR or target gene 3’-UTR reporter was determined by the Renilla/firefly ratio.
Biotin-labeled RNA pull-down and mass spectrometry analysis
Biotin-labeled RNA probe for linear sequence of circAGO2 was in vitro transcribed using Biotin RNA Labeling Mix (Roche) and T7 RNA polymerase, and treated with RNase-free DNase I. After incubation with guide oligonucleotides (Supplementary Table S5), the linear probe was circularized using T4 RNA ligase I, treated with RNase R, and purified by using RNeasy Mini Kit (Qiagen Inc.). RNA pull-down assay was performed at room temperature as previously described [17, 36, 52]. In brief, lysates of 2 × 107 cancer cells were incubated with 3 μg of biotin-labeled linear or circular RNA probe for 2 h, and treated with 35 μl of Streptavidin C1 magnetic beads (Invitrogen) for 1 h. After washed stringently for three times, the retrieved protein was detected by western blot or mass spectrometry analysis at Wuhan Institute of Biotechnology (Wuhan, China).
RNA-FISH assay
Cells were seeded on coverslips, fixed with 4% paraformaldehyde, and incubated with 50% formamide, 2 × SSC, Escherichia coli transfer RNA (0.25 mg/ml), salmon sperm DNA (0.25 mg/ml, Life Technologies, Inc.), bovine serum albumin (2.5 mg/ml, Roche), and DIG-labeled antisense or sense probe for circAGO2 junction (Supplementary Table S5). The probes for GAPDH and U1 were generated by in vitro transcription of PCR products (Supplementary Table S4) using DIG Labeling Kit (MyLab Corporation). Hybridization was carried out in a humidified chamber at 37 °C for 16 h, with or without RNase R (3 U/mg) treatment. The signals of circAGO2 were detected by Fluorescent In Situ Hybridization Kit (RiboBio, Guangzhou, China), while the nuclei were counterstained with 4’,6-diamidino-2-phenylindole (DAPI) [35, 57].
Fluorescence immunocytochemical staining
Cancer cells were grown on coverslips, incubated with 5% milk for 1 h, and treated with antibodies specific for HuR (ab200342, Abcam Inc.; 1:100 dilution) at 4 °C overnight. Then, coverslips were treated with Cy3-conjugated goat anti-rabbit IgG (1:1000 dilution) and DAPI (300 nmol/L) staining. The images were photographed under a Nikon A1Si Laser Scanning Confocal Microscope (Nikon Instruments Inc, Japan).
Co-immunoprecipitation (co-IP) assay
Co-IP was undertaken as previously described [52], with antibodies specific for HuR (ab200342) or AGO2 (ab186733, Abcam Inc.). Briefly, cells were washed with ice-cold PBS, and lysed in 500 μl of co-IP buffer (20 mM Tris-Cl, pH 7.5, 150 mM NaCl, 1 mM ethylenediaminetetraacetic acid, 0.5% NP-40, and 5 μg/ml aprotinin). Equal amounts of protein were incubated with 5 μg of primary antibody and 50 μl of protein A-Sepharose at 4 °C for 4 h. The protein bound by protein A-Sepharose beads (Santa Cruz Biotechnology) was released and detected by western blot.
Cross-linking RIP assay
Cells were ultraviolet light cross-linked at 254 nm (200 J/cm2) in PBS and collected by scraping [52, 53]. RIP assay was performed according to the instructions of Magna RIPTM RNA-Binding Protein Immunoprecipitation Kit (Millipore), with antibodies specific for HuR (ab200342), AGO2 (ab186733) or Flag (ab45766, Abcam Inc.). Co-precipitated RNAs were detected by RT-PCR or real-time quantitative qRT-PCR with specific primers (Supplementary Table S4). Total RNAs (input) and isotype antibody (IgG) were applied as controls.
In vitro binding assay
A series of GST-tagged HuR truncates were kindly provided by Dr. Jan Chemnitz [58], and transformed into E. coli to produce GST-tagged truncated HuR proteins [52, 53]. Biotin-labeled circAGO2 probe was prepared as described above, and incubated with GST-tagged HuR protein. HuR–circRNA complexes were pulled down using GST beads (Sigma). Protein was detected by SDS-PAGE and western blot, while circRNA was measured by RT-PCR with specific primers (Supplementary Table S4).
RNA EMSA assay
Biotin-labeled circAGO2 probe was prepared as described above. RNA EMSA using nuclear extracts or recombinant HuR protein was performed according to the instructions of LightShift Chemiluminescent RNA EMSA Kit (Thermo Fisher Scientific, Inc., Grand Island, NY).
Design and synthesis of inhibitory peptides
The inhibitory peptides for blocking the interaction between circAGO2 and HuR were designed and synthesized (ChinaPeptides Co. Ltd, Shanghai, China). The 11 amino acid long peptide (YGRKKRRQRRR) from Tat protein transduction domain served as a cell-penetrating peptide. Thus, inhibitory peptides were chemically synthesized by linking with biotin-labeled cell-penetrating peptide at N-terminus and conjugating with FITC at C-terminus. The purity of peptides (larger than 95%) was validated by reversed phase-high performance liquid chromatography assay.
Biotin-labeled peptide pull-down assay
Cellular total RNA was isolated using RNeasy Mini Kit (Qiagen Inc.), and incubated with biotin-labeled peptide at 4 °C overnight. Then, incubation of RNA-peptide complex with streptavidin-agarose was undertaken at 4 °C for 2 h. The beads were extensively washed, and circRNAs pulled down were measured by RT-PCR or real-time qRT-PCR.
In vitro cell viability, growth, migration, and invasion assays
The 2-(4,5-dimethyltriazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT; Sigma) colorimetric [52], soft agar [52,53,54], scratch [55], and matrigel invasion [52,53,54] assays were undertaken to measure the in vitro viability, growth, migration, and invasive capabilities of cancer cells.
In vivo growth, metastasis, and therapeutic assays
All animal experiments were approved by the Animal Care Committee of Tongji Medical College (approval number: Y20080290), and undertaken according to NIH Guidelines for the Care and Use of Laboratory Animals. For in vivo tumor growth studies, cancer cells (1 × 106) were subcutaneously injected into the dorsal flanks of blindly randomized four-week-old female BALB/c nude mice (n = 5 per group). The growth and weight of xenografts were detected one month later. In experimental metastasis studies, tail vein injection of cancer cells (0.4 × 106) were performed in blindly randomized four-week-old female BALB/c nude mice (n = 5 per group) [52,53,54]. For in vivo therapeutic studies, cancer cells (1 × 106 or 0.4 × 106) were injected into the dorsal flanks or tail vein of nude mice, respectively. After cancer cell inoculation, mice were blindly randomized and treated by intratumoral or tail vein injection of lentivirus-mediated shRNA (1 × 107 plaque-forming units in 100 μl PBS) or synthesized cell-penetrating peptide (ChinaPeptides, Shanghai, China) as indicated. The tumor volume and survival time of each mouse were monitored and recorded. The animals were imaged using the In-Vivo Xtreme II small animal imaging system (Bruker Corporation, Billerica, MA).
Tissue microarray and immunohistochemistry
Tissue microarray of gastric cancer (n = 222) was obtained from Fanpu Biotech, Inc. (Guilin, China). Immunohistochemical staining and quantitative evaluation were performed as previously described [52,53,54], with antibodies specific for Ki-67 (sc-23900), CD31 (Arigo, ARG52748), or HuR (sc-5261, Santa Cruz Biotechnology) at 4 °C overnight. To assess the reactivity degree, ten different high power fields (400 × ) for each specimen were blindly evaluated. The staining intensity was evaluated on a range from 0 to 3 (0, negative; 1, weakly positive; 2, moderately positive; 3, strongly positive), while percentage of positive cells were evaluated ranging from 0 to 4 (0, negative; 1, positive in 1–25%; 2, positive in 26–50%; 3, positive in 51–75%; 4, positive in 76–100%). Based on the products of staining intensity multiplied by percentage of positive cells, the results of immunohistochemistry were classified into negative (–, 0–1), mildly (+, 2–3), moderately (++, 4–8), and strongly positive (+++, 9–12). The moderate (++) or strong (+++) reactivity was defined as high expression, while negative (−) or mild positive (+) reactivity was defined as low expression.
Statistical analysis
All data were shown as mean ± standard error of the mean (SEM). The cutoff for gene expression was defined by average values. Student’s t test, analysis of variance, and χ2 analysis were used to compare the difference in cancer cells or tissues. Fisher’s exact test was used to analyze the statistical significance of overlap between two gene lists. Pearson’s correlation coefficient assay was used to analyze the expression correlation. Log-rank test was used to assess survival difference. All statistical tests were two-sided.
References
Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute 2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15:185–97.
Shen J, Xia W, Khotskaya YB, Huo L, Nakanishi K, Lim SO, et al. EGFR modulates microRNA maturation in response to hypoxia through phosphorylation of AGO2. Nature. 2013;497:383–7.
Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12:99–110.
Zhang J, Fan XS, Wang CX, Liu B, Li Q, Zhou XJ. Up-regulation of Ago2 expression in gastric carcinoma. Med Oncol. 2013;30:628.
Papachristou DJ, Korpetinou A, Giannopoulou E, Antonacopoulou AG, Papadaki H, Grivas P, et al. Expression of the ribonucleases Drosha, Dicer, and Ago2 in colorectal carcinomas. Virchows Arch. 2011;459:431.
Yoo NJ, Hur SY, Kim MS, Lee JY, Lee SH. Immunohistochemical analysis of RNA-induced silencing complex-related proteins AGO2 and TNRC6A in prostate and esophageal cancers. APMIS. 2010;118:271–6.
Qu H, Zheng L, Song H, Jiao W, Li D, Fang E, et al. microRNA-558 facilitates the expression of hypoxia-inducible factor 2 alpha through binding to 5′-untranslated region in neuroblastoma. Oncotarget. 2016;7:40657–73.
Zhang Y, Wang B, Chen X, Li W, Dong P. AGO2 involves the malignant phenotypes and FAK/PI3K/AKT signaling pathway in hypopharyngeal-derived FaDu cells. Oncotarget. 2017;8:54735–46.
Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32:453–61.
Chen J, Li Y, Zheng Q, Bao C, He J, Chen B, et al. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett. 2017;388:208–19.
Hsiao KY, Lin YC, Gupta SK, Chang N, Yen L, Sun HS, et al. Noncoding effects of circular RNA CCDC66 promote colon cancer growth and metastasis. Cancer Res. 2017;77:2339–50.
Huang XY, Huang ZL, Xu YH, Zheng Q, Chen Z, Song W, et al. Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-100338/miR-141-3p pathway in hepatitis B-related hepatocellular carcinoma. Sci Rep. 2017;7:5428.
Bachmayr-Heyda A, Reiner AT, Auer K, Sukhbaatar N, Aust S, Bachleitner-Hofmann T, et al. Correlation of circular RNA abundance with proliferation-exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci Rep. 2015;5:8057.
Wang K, Sun Y, Tao W, Fei X, Chang C. Androgen receptor (AR) promotes clear cell renal cell carcinoma (ccRCC) migration and invasion via altering the circHIAT1/miR-195-5p/ 29a-3p/29c-3p/CDC42 signals. Cancer Lett. 2017;394:1–12.
Weng W, Wei Q, Toden S, Yoshida K, Nagasaka T, Fujiwara T, et al. Circular RNA ciRS-7-A promising prognostic biomarker and a potential therapeutic target in colorectal cancer. Clin Cancer Res. 2017;23:3918–28.
Wilusz JE, Sharp PA. A circuitous route to noncoding RNA. Science. 2013;340:440–1.
Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846–58.
Glažar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA. 2014;20:1666–70.
Petkovic S, Muller S. RNA circularization strategies in vivo and in vitro. Nucleic Acids Res. 2015;43:2454–65.
Agostini F, Zanzoni A, Klus P, Marchese D, Cirillo D, Tartaglia GG. catRAPID omics: a web server for large-scale prediction of protein-RNA interactions. Bioinformatics. 2013;29:2928–30.
Walia RR, Xue LC, Wilkins K, El-Manzalawy Y, Dobbs D, Honavar V. RNABindRPlus: a predictor that combines machine learning and sequence homology-based methods to improve the reliability of predicted RNA-binding residues in proteins. PLoS One. 2014;9:e97725.
Lopez de Silanes I, Zhan M, Lal A, Yang X, Gorospe M. Identification of a target RNA motif for RNA-binding protein HuR. Proc Natl Acad Sci USA. 2004;101:2987–92.
Balkhi MY, Iwenofu OH, Bakkar N, Ladner KJ, Chandler DS, Houghton PJ, et al. miR-29 acts as a decoy in sarcomas to protect the tumor suppressor A20 mRNA from degradation by HuR. Sci Signal. 2013;6:ra63.
Yang YC, Di C, Hu B, Zhou M, Liu Y, Song N, et al. CLIPdb: a CLIP-seq database for protein-RNA interactions. BMC Genom. 2015;16:51.
Fallmann J, Sedlyarov V, Tanzer A, Kovarik P, Hofacker IL. AREsite2: an enhanced database for the comprehensive investigation of AU/GU/U-rich elements. Nucleic Acids Res. 2016;44:D90–D95.
Zhang JH, Seigneur EM, Pandey M, Loshakov A, Dagur PK, Connelly PS, et al. The EIF4EBP3 translational repressor is a marker of CDC73 tumor suppressor haploinsufficiency in a parathyroid cancer syndrome. Cell Death Dis. 2012;3:266.
Tentu S, Nandarapu K, Muthuraj P, Venkitasamy K, Venkatraman G, Rayala SK. DHQZ-17, a potent inhibitor of the transcription factor HNF4A, suppresses tumorigenicity of head and neck squamous cell carcinoma in vivo. J Cell Physiol. 2018;233:2613–28.
Yang HS, Matthews CP, Clair T, Wang Q, Baker AR, Li CC, et al. Tumorigenesis suppressor Pdcd4 down-regulates mitogen-activated protein kinase kinase kinase kinase 1 expression to suppress colon carcinoma cell invasion. Mol Cell Biol. 2006;26:1297–306.
Fukusumi T, Guo TW, Sakai A, Ando M, Ren S, Haft S, et al. The NOTCH4–HEY1 pathway induces epithelial–mesenchymal transition in head and neck squamous cell carcinoma. Clin Cancer Res. 2018;24:619–33.
Chang YC, Chi LH, Chang WM, Su CY, Lin YF, Chen CL. et al. Glucose transporter 4 promotes head and neck squamous cell carcinoma metastasis through the TRIM24-DDX58 axis. J Hematol Oncol. 2017;10:11
Mattie M, Raitano A, Morrison K, Morrison K, An Z, Capo L, et al. The discovery and preclinical development of ASG-5ME, an antibody–drug conjugate targeting SLC44A4-positive epithelial tumors including pancreatic and prostate cancer. Mol Cancer Ther. 2016;15:2679–87.
Dweep H, Gretz N. miRWalk2.0: a comprehensive atlas of microRNA-target interactions. Nat Methods. 2015;12:697.
Dudekula DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M. CircInteractome: A web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016;13:34–42.
Wu X, Lan L, Wilson DM, Marquez RT, Tsao WC, Gao P, et al. Identification and validation of novel small molecule disruptors of HuR-mRNA interaction. ACS Chem Biol. 2015;10:1476–84.
Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–8.
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, et al. Circular intronic long noncoding RNAs. Mol Cell. 2013;51:792–806.
Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256–64.
Qu S, Liu Z, Yang X, Zhou J, Yu H, Zhang R, et al. The emerging functions and roles of circular RNAs in cancer. Cancer Lett. 2018;414:301–9.
Li F, Zhang L, Li W, Deng J, Zheng J, An M, et al. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/β-catenin pathway. Oncotarget. 2015;6:6001–13.
Han D, Li J, Wang H, Su X, Hou J, Gu Y, et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66:1151–64.
Schmidt C, Kramer K, Urlaub H. Investigation of protein-RNA interactions by mass spectrometry-techniques and applications. J Proteom. 2012;75:3478–94.
Fan XC, Steitz JA. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 1998;17:3448–60.
Melling N, Taskin B, Hube-Magg C, Kluth M, Minner S, Koop C, et al. Cytoplasmic accumulation of ELAVL1 is an independent predictor of biochemical recurrence associated with genomic instability in prostate cancer. Prostate. 2016;76:259–72.
Doller A, Pfeilschifter J, Eberhardt W. Signalling pathways regulating nucleo-cytoplasmic shuttling of the mRNA-binding protein HuR. Cell Signal. 2008;20:2165–73.
Kotta-Loizou I, Giaginis C, Theocharis S. Clinical significance of HuR expression in human malignancy. Med Oncol. 2014;31:161.
Wang W, Caldwell MC, Lin S, Furneaux H, Gorospe M. HuR regulates cyclin A and cyclin B1 mRNA stability during cell proliferation. EMBO J. 2000;19:2340–50.
Wang W, Yang X, Cristofalo VJ, Holbrook NJ, Gorospe M. Loss of HuR is linked to reduced expression of proliferative genes during replicative senescence. Mol Cell Biol. 2001;21:5889–98.
Chang N, Yi J, Guo G, Liu X, Shang Y, Tong T, et al. HuR uses AUF1 as a cofactor to promote p16INK4 mRNA decay. Mol Cell Biol. 2010;30:3875–86.
Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, Gorospe M. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 2009;23:1743–8.
Cheng YC, Liou JP, Kuo CC, Lai WY, Shih KH, Chang CY, et al. MPT0B098, a novel microtubule inhibitor that destabilizes the hypoxia-inducible factor-1α mRNA through decreasing nuclear-cytoplasmic translocation of RNA-binding protein HuR. Mol Cancer Ther. 2013;12:1202–12.
Bolognani F, Gallani AI, Sokol L, Baskin DS, Meisner-Kober N. mRNA stability alterations mediated by HuR are necessary to sustain the fast growth of glioma cells. J Neurooncol. 2012;106:531–42.
Li D, Wang X, Mei H, Fang E, Ye L, Song H, et al. Long noncoding RNA pancEts-1 promotes neuroblastoma progression through hnRNPK-mediated β-catenin stabilization. Cancer Res. 2018;78:1169–83.
Zhao X, Li D, Pu J, Mei H, Yang D, Xiang X, et al. CTCF cooperates with noncoding RNA MYCNOS to promote neuroblastoma progression through facilitating MYCN expression. Oncogene. 2016;35:3565–76.
Zhang H, Pu J, Qi T, Qi M, Yang C, Li S, et al. MicroRNA-145 inhibits the growth, invasion, metastasis and angiogenesis of neuroblastoma cells through targeting hypoxia-inducible factor 2 alpha. Oncogene. 2014;33:387–97.
Li D, Mei H, Pu J, Xiang X, Zhao X, Qu H, et al. Intelectin 1 suppresses the growth, invasion and metastasis of neuroblastoma cells through up-regulation of N-myc downstream regulated gene 2. Mol Cancer. 2015;14:47.
Abdelmohsen K, Pullmann R, Lal A, Kim HH, Galban S, Yang X, et al. Phosphorylation of HuR by Chk2 regulates SIRT1 expression. Mol Cell. 2007;25:543–57.
Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–8.
Pieper D, Schirmer S, Prechtel AT, Kehlenbach RH, Hauber J, Chemnitz J. Functional characterization of the HuR:CD83 mRNA interaction. PLoS One. 2011;6:e23290.
Acknowledgements
We appreciate Drs. Myriam Gorospe and Jan Chemnitz for providing vector. This work was granted by the National Natural Science Foundation of China (81272779, 81372667, 81472363, 81402301, 81402408, 81572423, 81672500, 81773094, 81772967, 81874085, 81874066, 81802925), Fundamental Research Funds for the Central Universities (2012QN224, 2013ZHYX003), and Natural Science Foundation of Hubei Province (2014CFA012).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Edited by R. Johnstone
Rights and permissions
About this article
Cite this article
Chen, Y., Yang, F., Fang, E. et al. Circular RNA circAGO2 drives cancer progression through facilitating HuR-repressed functions of AGO2-miRNA complexes. Cell Death Differ 26, 1346–1364 (2019). https://doi.org/10.1038/s41418-018-0220-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-018-0220-6
This article is cited by
-
Emerging roles of circular RNAs in tumorigenesis, progression, and treatment of gastric cancer
Journal of Translational Medicine (2024)
-
TGF-β-activated circRYK drives glioblastoma progression by increasing VLDLR mRNA expression and stability in a ceRNA- and RBP-dependent manner
Journal of Experimental & Clinical Cancer Research (2024)
-
A protein-encoding CCDC7 circular RNA inhibits the progression of prostate cancer by up-regulating FLRT3
npj Precision Oncology (2024)
-
m6A-Mediated Upregulation of lncRNA CHASERR Promotes the Progression of Glioma by Modulating the miR-6893-3p/TRIM14 Axis
Molecular Neurobiology (2024)
-
Circular RNA hsa_circ_0067842 facilitates tumor metastasis and immune escape in breast cancer through HuR/CMTM6/PD-L1 axis
Biology Direct (2023)