Abstract
Caspases exert critical functions in diverse cell death pathways, including apoptosis and pyroptosis, but some caspases also have roles in the processing of cytokines into their functional forms during inflammation. The roles of many caspases have been unravelled by the generation of knockout mice, but still very little is known about the overlapping functions of caspases as only a few studies report on double or triple caspase knockout mice. For example, the functions of caspase-12 in cell death and inflammation, on its own or overlapping with the functions of caspase-1 and caspase-11, are only poorly understood. Therefore, we generated a novel mutant mouse strain lacking all three inflammatory caspases, caspases-1, -11 and -12. Analysis under steady state conditions showed no obvious differences between caspase-1/11/12−/− and wildtype (WT) mice. Since caspases-1 and -11 are involved in endotoxic shock, we analysed the response of caspase-1/11/12−/− mice to high-dose LPS injection. Interestingly, we could not detect any differences in responses between caspase-1/11/12−/− mice vs. caspase-1/11 double knockout mice. Furthermore, cell lines generated from caspase-1/11/12−/− mice showed no differences in their apoptotic or necroptotic responses to a diverse set of cytotoxic drugs in vitro when compared to WT cells. Importantly, these drugs also included ER stress-inducing agents, such as thapsigargin and tunicamycin, a form of cell death for which a critical pro-apoptotic function of caspase-12 has previously been reported. Additionally, we found no differences between caspase-1/11/12−/− and WT mice in their in vivo responses to the ER stress-inducing agent, tunicamycin. Collectively, these findings reveal that caspase-12 does not have readily recognisable overlapping roles with caspases-1 and -11 in the inflammatory response induced by LPS and in necroptosis and apoptosis induced by diverse cytotoxic agents, including the ones that elicit ER stress.
Similar content being viewed by others
Introduction
Inflammatory caspases play a crucial role in the innate immune response and are therefore an important part of defence against pathogenic attack [1]. The murine inflammatory caspase locus on chromosome 9 encodes for caspases-1, -11 and -12 [1]. Humans have four inflammatory caspases (encoded by genes located on chromosome 11), namely caspases-1, -4, -5 and -12 [1]. Caspases-4 and -5 in humans are thought to be the orthologues of mouse caspase-11 and probably arose from the duplication of an ancestral gene [1]. The inflammatory caspases have N-terminal CARD (caspase activation and recruitment domains) pro-domains, and because they can proteolytically activate the effector caspases (i.e. caspases -3, -6 and -7) they are classified as initiator caspases [1,2,3]. This places them alongside the CARD-containing caspase-9, which plays a role in the intrinsic apoptotic pathway [4, 5] and the DED-containing caspase-8, which is essential for death receptor-induced apoptosis [6] and the prevention of necroptosis [7, 8]. The CARD and DED pro-domains function as protein–protein interaction motifs serving as activation platforms to convert inactive zymogens into fully active initiator caspases [1, 9]. This results in the assembly of multimeric protein complexes, such as the apoptosome (APAF-1, cytochrome c plus pro-caspase-9) or inflammasome (NLRP1, NLRC4, or ASC plus pro-caspase-1) [10, 11].
CARD-containing caspases, such as caspases-1 and −11, promote inflammation by driving the secretions of functional IL-1β and IL-18 [1]. Upon infection or injury, caspases are required to destroy and rapidly clear the cells. These processes must be tightly regulated because too much or too little cell death or inflammation can lead to failure of clearing an infection, cause auto-inflammatory diseases, or cancer [12, 13].
Caspase-1, formerly known as ICE (Interleukin-1 Converting Enzyme) was discovered through its ability to generate the biologically active form of the pro-inflammatory cytokine IL-1β by cleavage [14, 15]. Caspase-1 becomes activated via two signals: an initial ‘priming’ stimulus provided by the membrane-bound receptors and an ‘activation’ signal produced by cytosolic pattern recognition receptors (PRRs) [16]. Stimulation of membrane receptors by pathogen-associated molecular patterns (PAMPs), such as TLR4 being activated by bacterial LPS or host-derived danger-associated molecular patterns (DAMPs), triggers signalling cascades that promote the transcription of the genes encoding pro-interleukin-1β (pro-IL-1β), intracellular PRRs (i.e. NOD-like receptors, NLRs) and other mediators of inflammation [10]. Upon sensing cytosolic DAMPs or PAMPs, the newly expressed NLRs prime the assembly of a multi-protein scaffold, termed as inflammasome [17,18,19]. This structure recruits and thereby facilitates the activation of caspase-1. This causes proteolytic activation of the cytokines pro-IL-1β and pro-IL-18, and induction of pyroptosis. This lytic form of cell death is driven by caspase-1 or caspase-11mediated proteolytic activation of gasdermin D, which permeabilises the plasma membrane [2, 9, 17, 20].
Caspase-11 was shown to function as a cytosolic sensor for Gram-negative bacteria, such as directly binding the LPS from Escherichia coli, and like caspase-1, it is involved in the induction of pyroptosis [21,22,23]. Furthermore, caspase-11 has been shown to initiate the canonical NLRP3 inflammasome to support the caspase-1driven release of IL-1β and IL-18 and it also promotes the secretion of the pro-inflammatory factors IL-1α and high-mobility group box 1 protein (HMGB1) [22, 24]. Exactly how caspase-11 triggers inflammasome activation is still unclear and, as mentioned above, in humans caspases-4 and -5 are thought to be the orthologues of murine caspase-11 and perform these functions [22].
Only limited insight is available on the other member of this family, caspase-12. Apart from having two caspase-11like genes, another difference between mice and humans is that due to an early stop codon (TGA), all humans of Asian and Caucasian descent can only express a truncated version of caspase-12 that contains the CARD, but lacks the domains critical for proteolytic activity [25, 26]. The expression of full-length caspase-12 is confined to 20–25% of people of African descent. A role for caspase-12 in inflammation in mice (and possibly other species) appears likely, given its gene is clustered closely with the genes for caspases-1 and −11 within the inflammatory caspase locus [25]. Caspase-12 is only expressed in certain cell types and can be upregulated in response to certain inflammatory stimuli [27]. Caspase-12 has been implicated in apoptosis triggered by ER stress in one study [28], but this was not substantiated by follow-up investigations [27, 29].
To examine the overall roles of all inflammatory caspases, we generated mice deficient for caspase-1/11/12. Analysis of these animals reveals that additional loss of caspase-12 does not further diminish the septic shock response beyond the attenuation afforded by the combined loss of caspases-1 and -11. Moreover, we found no evidence for an overall role for these three inflammatory caspases in the response of cells to diverse inducers of apoptosis or necroptosis in vitro or to inducers of ER stress in vitro and in vivo.
Results
Generation and validation of caspase-1/11/12 triple deficient mice
To determine the overall role of inflammatory caspases-1, -11 and -12 in inflammation and cell death, we generated mice deficient for these three CARD-containing caspases. Therefore, the entire locus spanning the coding regions of caspases-1, −11 and −12 was constitutively deleted in C57BL/6 ES cells (Fig. 1a). The genomic region encompassing caspase-1 including 1.7 kb of genomic sequence upstream of caspase-1 exon 1 (promoter region), caspase-11 and caspase-12 (total of 76.3 kb) was replaced with a FRT-flanked puromycin resistance (PuroR) cassette that was subsequently removed using site-directed Flp-FRT-mediated recombination. This resulted in a constitutive caspase-1/11/12 knockout (ko) allele. Intercrossing caspase-1/11/12+/− mice generated caspase-1/11/12−/− offspring at the expected mendalian frequency. The caspase-1/11/12−/− mice displayed no overt abnormalities, appeared healthy up to at least 18 months of age and were fertile (data not shown).
Generation and validation of caspase-1/11-12 triple knockout mice. a Targeting strategy to generate mice constitutively deficient (KO) for caspases-1, -11 and -12. b Genotyping of caspase-1/11/12−/− mice. DNA from WT mice or H2O served as controls. Band sizes, WT: 245 bp, KO: 413 bp. c Western blot analysis to detect caspases-1 and -11 in BMDMs of the indicated genotypes after 24 h of stimulation with 20 ng/mL LPS. Left panel: blot probed for caspase-1; right panel: blot probed for caspase-11. Both membranes were also probed for HSP70 (loading control) and the left membrane was additionally probed for pro-IL-1ß. The western blot shown is representative of two independent experiments. d Raw Ct values from qRT-PCR analysis of mRNA expression for caspase-12 and HMBS (loading control) in extracts from the lung (left panel) and brain (right panel) from caspase-1/11/12 triple knockout and WT control mice. n.d. indicates no RNA was detected. n = 3. Graphs show the mean ± S.E.M. of triplicate tests. e, f IL-1ß release from LPS-primed BMDMs of the indicated genotypes that had been stimulated with e 5 mM ATP or f 2 μg/mL LPS, that had been transfected into cells using lipofectamine. (Cont., medium alone). All graphs show mean ± S.E.M. of n ≥ 4. Adjusted p values in Supplementary Table 1 a and b
The loss of the three caspase genes was verified by PCR, amplifying a 413 bp band for the mutant allele and a 245 bp band for the WT allele (Fig. 1b). To further validate the knockout mice, we stimulated primary bone marrow-derived macrophages (BMDMs) derived from these animals with LPS and analysed the expression of caspases-1 and -11 by western blotting. This revealed steady state expression of caspase-1 and LPS-induced upregulation of caspase-11 in BMDMs from WT mice, while as predicted, both caspases-1 and -11 were absent in the cells from caspase-1/11/12 triple knockout mice (Fig. 1c). Due to the lack of suitable caspase-12 antibodies, we verified the loss of caspase-12 expression by qRT-PCR (Fig. 1d). As previously reported [27], we detected caspase-12 mRNA in the lungs and brain from unchallenged WT mice but, as expected, this mRNA was absent from the corresponding tissues from the caspase-1/11/12−/− mice (Fig. 1d).
Next, we conducted a functional assay for caspases-1 and -11. Both of these caspases are known to be critical for the production of IL-1β during pyroptosis [22]. BMDMs were primed with LPS, and caspases-1 or -11 were activated by the further addition of ATP or transfection of cells with LPS, respectively (Fig. 1e, f). Substantial levels of IL-1β were detected in the supernatants of stimulated BMDMs from wt mice, but no IL-1β was produced by BMDMs from the caspase-1/11/12 knockout mice (Fig. 1e, f). BMDMs from caspase-1/11 double knockout mice served as a control and, as predicted, these cells also did not produce IL-1β (Fig. 1e, f). Collectively, these findings demonstrate that we have successfully generated a novel strain of mice deficient for all three inflammatory caspases-1/11/12.
Caspase-12 mRNA expression is upregulated to a similar extent in wildtype and caspases-1/11 deficient cells upon treatment with LPS in vitro
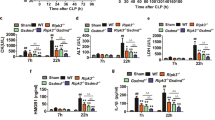
It is possible that the deletion of caspases-1 and -11 may interfere with caspase-12 gene expression. To investigate this possibility and determine the expression of caspases-1, -11 and -12, we treated primary BMDMs with 20 ng/mL or 500 ng/mL LPS and analysed the expression of these genes using qRT-PCR.
Upon LPS stimulation, the expression of caspase-11 increased ~20-fold in BMDMs from WT mice, but was not detected in BMDMs from the caspase-1/11−/− mice (Figure S1). Stimulation with LPS increased the levels of caspase-1 mRNA in wt BMDMs approximately threefold. The caspase-1 gene in the caspase-1/11−/− mice lacks parts of exons 6 and 7 and therefore no functional caspase-1 protein can be produced. However, a shortened caspase-1 transcript that can be recognised by the PCR primers can be generated in cells from these animals, which explains why a qRT-PCR signal was obtained in the BMDMs from caspase-1/11−/− mice (Figure S1).
The levels of caspase-12 mRNA were not significantly different between untreated BMDMs from WT vs. those from the caspase-1/11−/− mice. Upon LPS stimulation, the levels of caspase-12 mRNA rose to a similar extent in BMDMs of either genotype (Figure S1). This reveals that loss of caspases-1/11 does not affect the levels of caspase-12 mRNA expression.
Caspase-1/11/12 −/− mice do not exhibit noticable defects in the haematopoietic system
To study the roles of caspases-1, -11 and -12 in the haematopoietic system, we performed flow cytometric analysis to compare the myeloid and lymphoid cell subset composition between the caspase-1/11/12 knockout and WT mice. In the thymus no differences were found in the frequencies and numbers of double-negative (CD4−CD8−; including all DN1-4 subsets), double-positive (CD4+CD8+) or single-positive (CD4+ or CD8+) T cells (Fig. 2a and S2a). Moreover, the caspase-1/11/12 knockout mice had normal numbers of CD4+ as well as CD8+ mature T cells in their spleens, with normal distributions of naïve (CD62LhiCD44−), activated (CD62LloCD44+) and memory (CD62hiCD44+) subsets found in both the CD4+ and CD8+ T cell lineages (Fig. 2b and S2b). Finally, the caspase-1/11/12 knockout mice had similar numbers of CD4+FOXP3+ T regulatory cells (Treg) as WT controls (Fig. 2c and S2c).
Normal haematopoietic cell subset composition in the bone marrow (both tibia and femur), lymph nodes (axial, brachial and inguinal), thymus and spleen of caspase-1/11/12 triple knockout mice. a (Left) Representative flow cytometry plots to examine T cell development in the thymus. Cell populations during development defined as CD8+, CD4+ and DP for CD4+CD8+ double-positive T cells. Additional analysis on double-negative (CD8−CD4−) progenitor cells, denoted DN, using antibodies against CD25 and CD44. Gating strategy on DN progenitor cells depicted in second flow cytometry plot from the left to further subdivide into DN1–4 stages as follows: DN1 (CD44+CD25−), DN2 (CD44+CD25+), DN3 (CD44−CD25+) and DN4 (CD44−CD25−). a (Right) Percentages for each of these T cell populations in the thymus of caspase-1/11/12 knockout and WT control mice. b (Left) Representative gating strategy for CD4+ T-cell activation. Naïve T cells that have not yet encountered an antigen do not express CD44 on the cell surface (top left quadrant). Upon activation by an antigen, T cells upregulate the CD44 expression (right panels). T cells with upregulated CD62L expression are considered memory T cells, whereas the so-called activated CD4+ T cells circulating in the periphery express low levels of CD62L. b (Right) Quantification of percentages of naïve, memory and activated CD4+ T cells in the spleen and lymph nodes from caspase-1/11/12 triple knockout and WT control mice. c (Left) Representative flow cytometry plots of T-cell populations in the lymphoid organs. Gating on regulatory T cells (Tregs) (CD4+CD25+FOXP3+). c (Right) Treg cell percentages in the thymus, lymph nodes and spleen. d (Left) Representative plots of B cell populations defined as B-1 (IgM+CD5+B220−) or marginal zone (IgM+B220+CD5−) B cells. d (Right) Percentages in lymph nodes and spleen are shown for each of these B-cell populations. e (Left) B-cell development using flow cytometric analysis. Representative plots showing pro-B/pre-B (B220+IgM−), immature B (B220+IgMlo), transitional B (B220+IgMhi) and mature B cells (B220hiIgMlo) in the bone marrow. e (Right) Quantitative analysis of these B cell populations. f (Left) Representation of myeloid cell populations analysed by flow cytometry. Top right represents neutrophils (GR-1+MAC-1+) and bottom right the monocyte population (GR-1−MAC-1+). f (Right) Percentages of these cells found in the bone marrow and lymph nodes. Graphs show means ± S.E.M. Cell numbers are presented in Supplementary Figure 2. n ≥ 5. Symbols represent individual mice. Results tested with two-way ANOVA or unpaired two-tailed Student's t test
We also found no abnormalities in the B-lymphocyte compartment of the caspase-1/11/12 knockout mice. They contained normal frequencies and numbers of pro-B/pre-B (B220+IgM−), immature B (B220+IgMhiIgDlo) and mature B cells (B220 + IgMloIgDhi) in the bone marrow and normal frequencies, and numbers of follicular (B220+IgD+IgMlo), marginal zone and B-1 B cells in the spleen and lymph nodes (Fig. 2d, e and S2d, e).
Next, we examined the impact of the combined loss of caspases-1, -11 and -12 in myeloid cells. At steady state, the numbers of neutrophils (GR-1+MAC-1+) and monocytes (GR-1loMAC-1+) were found to be normal in the bone marrows and spleens of the caspase-1/11/12 knockout mice (Fig. 2f and S2f). Collectively, these results demonstrate that the combined loss of caspases-1, -11 and -12 does not cause detectable abnormalities in the haematopoietic system of mice under steady state.
The additional loss of caspase-12 does not provide further protection against LPS-induced septic shock beyond the protection afforded by combined loss of caspase-1 and caspase-11
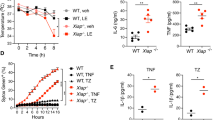
It has previously been shown that mice lacking caspase-11 or both caspases-1 and -11 are resistant to LPS-induced septic shock [22, 30]. Since caspase-12 is a CARD-containing caspase encoded by a gene that is co-located in the inflammatory caspase locus together with the genes encoding caspases-1 and -11, we hypothesised that it might play a role overlapping with the functions of these two caspases in the septic shock response. To test this idea, we injected WT, caspase-1/11 knockout and caspase-1/11/12 knockout mice with intermediate or high doses of LPS (18 or 54 mg/kg body weight, respectively). As previously reported [22], the caspase-1/11 knockout mice survived the septic shock longer than the control WT animals (Fig. 3a, b). Of note, the caspase-1/11/12 knockout mice did not show further extended survival compared with the caspase-1/11 double knockout animals (Fig. 3a, b).
Loss of caspase-12 does not provide additional protection against LPS-induced lethal septic shock above the protection afforded by combined loss of caspases-1 and -11. a Survival of mice of the indicated genotypes after i.p. injection with 18 mg/kg LPS (left panel). Serum levels of IL-18 at 4 h after i.p. injection of 18 mg/kg LPS (Cont. represents no treatment). b Similar presentation of data as in a, but mice were injected i.p. with 54 mg/kg LPS. Symbols represent individual mice. Data are presented as mean ± S.E.M. Adjusted p values are supplied in Supplementary Table 2. Results of the levels of cytokines were analysed by one-way ANOVA
Caspases-1 and -11 exert important functions in the maturation and secretion of several cytokines. It has been postulated that caspase-12, as a CARD-containing caspase being located in the inflammatory caspase locus, may also play a role in cytokine production. To test this hypothesis, we measured the cytokine levels 4 h post injection of high-dose LPS in the blood of mice (Fig. 3b and S3d, e). IL-18 levels were highly elevated in WT mice upon LPS treatment and this was abrogated in the caspase-1/11 double knockout mice. Thus, no further reduction in the levels of IL-18 was possible in the LPS-treated caspase-1/11/12 knockout mice. Generally, for all cytokines tested, the levels were similar between the caspase-1/11/12 triple knockout and the caspase-1/11 double knockout mice, both before and after the treatment with LPS.
At both concentrations of LPS used, there was a trend towards a reduction in IFN-γ in the WT mice compared with the caspase-1/11/12 knockout and caspase-1/11 knockout animals (Figure S3c and Figure S3d). This may be explained by the observation that IFN-γ can be proteolysed by caspase-1 [31]. Furthermore, multiplex analysis suggested an abnormal increase in the levels of chemokine Rantes/CCL5 in the caspase-1/11/12 knockout mice after LPS treatment, but this could not be confirmed in a single cytokine ELISA (Figure S3e). Collectively, these findings demonstrate that caspase-12 does not play a major role in septic shock alongside caspases-1 and -11.
The combined deletion of caspases-1, -11 and -12 does not confer cells with protection against cytotoxic drugs in vitro
Caspase-12 has been implicated in cell death elicited by diverse cytotoxic agents [27, 28, 32, 33]. For example, loss of caspase-12 was shown to protect mouse embryonic fibroblasts (MEFs) from killing induced by several ER stress-inducing agents [28]. However, this finding was challenged in subsequent studies using B16 [27] and several myeloma-derived cell lines [29]. To examine the overall roles of all three inflammatory caspases in cell death, we generated mouse dermal fibroblasts (MDFs) from caspase-1/11/12 knockout and WT mice, and exposed them to a variety of cytoytoxic agents, including the ER stressors thapsigargin and tunicamycin, the microtubule inhibitor taxol and the DNA-damage-inducing drug etoposide. Furthermore, we generated primary BMDMs from caspase-1/11/12 knockout as well as WT mice and treated them with the combinations of TNF plus a SMAC mimetic (CompA) (T + S) or TNF, SMAC mimetic plus caspase inhibitor QVD-OPH (T + S + Q), to cause apoptosis or necroptosis, respectively [34]. In general, the combined loss of caspases-1/11/12 afforded no significant protection against any of the cytotoxic agents tested in either of the two cell types examined (Fig. 4). These findings reveal that caspases-1, -11 and -12 do not exert significant roles in the intrinsic or death receptor apoptotic pathways or in necroptotic cell death.
Combined loss of caspases-1/11/12 does not confer protection against treatment with a broad range of cytotoxic insults to cells in vitro. a Mouse dermal fibroblasts (MDF) from caspase-1/11/12 triple knockout or WT control mice were treated with thapsigargin (123 nM), taxol (10 μM), etoposide (300 μM), tunicamycin (1 μM), TNF (100 ng/mL) + SMAC mimetic (500 nM) and TNF (100 ng/mL) + SMAC mimetic (500 nM) + caspase inhibitor Q-VD-Oph (10 μM), and cell survival was measured at the indicated time points by flow cytometry. Graphs shown are for one representative experiment with n = 3 cell lines per genotype. Data are normalised to untreated control cells. Cell viability was measured by PI and Annexin V staining, followed by flow cytometric analysis. Data are presented as mean ± S.E.M. and were analysed using two-way ANOVA. b, c Primary BMDMs from caspase-1/11/12 triple knockout and WT control mice were treated with b TNF + SMAC mimetic + caspase inhibitor QVD-OPH (T + S + Q) to induce necroptosis or c TNF + SMAC mimetic (T + S) to induce apoptosis. Cell viability was measured at the indicated time points by Annexin V or Annexin V plus PI staining in the case of T + S + Q treatment, followed by flow cytometric analysis. All data were analysed using one-way ANOVA test. n ≥ 3. Data are presented as mean ± S.E.M.
Caspase-1/11/12 triple deficient mice show a normal ER stress response in vivo
Caspase-12 has been implicated in ER stress-induced cell killing in vivo [28]. To examine the overall roles of the inflammatory caspases in ER stress responses in vivo, we injected caspase-1/11/12−/− mice and WT controls with sublethal doses of tunicamycin. Histological analysis revealed similar damage to the kidneys at 72 h post treatment (Fig. 5a). Untreated caspase-1/11/12 triple knockout mice exhibited normal architecture of their kidneys (Fig. 5a). At 72 h post injection of tunicamycin, the overall architecture was damaged, especially in the outer medullary areas, and there were no differences in severity of damage between the caspase-1/11/12 triple knockout and WT mice (Fig. 5a). TUNEL staining revealed increased numbers of apoptotic cells predominantly in the outer medullary areas (Fig. 5a, b). Computational analysis of TUNEL+ cells revealed that there were no consistent differences between the caspase-1/11/12 knockout vs. control WT mice at 72 h post injection of tunicamycin (Fig. 5b and S4a). Moreover, WT animals treated with vehicle (DMSO) presented negligible numbers of TUNEL+ cells in their kidneys (Figure S4a). Untreated WT and caspase-1/11/12 knockout mice had normal serum levels of ALT (marker of liver damage) (Supplementary Figure 4b), demonstrating that the loss of caspases-1, -11 and -12 had no deleterious impact on these organs. After treatment with tunicamycin for 24 h, caspase-1/11/12 knockout and WT mice showed comparable but minor increases in the serum levels of ALT (Supplementary Figure 4b).
Combined loss of caspases-1, -11 and -12 does not protect mice against ER stress-induced renal damage. a Immunohistochemical staining of kidneys from tunicamycin-treated (1 mg/kg body weight, harvested after 72 h) and -untreated mice to detect TUNEL+ (dead) cells of WT mice are shown in the left panels, and for caspase-1/11/12 triple knockout mice in the right right panels. One representative image shown per genotype and treatment. n ≥ 3. Sections: cortex (1), outer medulla (2) and inner medulla (3). Scale bar represents 1 mm, total magnification of kidney ×20. b Bar graphs showing quantification of results from a n ≥ 3 mice per genotype, two kidney sections per mouse were scored computationally
Finally, we examined the expression of the caspase-1, -11 and -12 genes in the organs examined for ER stress-induced damage, i.e. the kidneys and liver. In the liver and kidneys, unstressed mice express the caspase-1, -11 and -12 genes at low levels (Supplementary Table 5; relative expression compared to actin, depicted as 2−ΔCt values in Supplementary Figure S5).
Collectively, these results demonstrate that the inflammatory caspases overall, and hence caspase-12, have no discernible role in tunicamycin-induced cell killing as well as renal or liver damage in vivo.
Discussion
Here, we describe the characterisation of a novel mouse strain lacking the three murine inflammatory caspases-1, -11 and -12. Loss of these three caspases had no impact on the general health of the mice. Moreover, extensive flow cytometric analysis revealed that the caspase-1/11/12 knockout mice had a normal haematopoietic system at steady state.
It is well known that caspases-1 and -11 have critical roles in pyroptotic cell death, the production of IL-1β and IL-18 and in sepsis [18, 22, 30]. We and others have hypothesised that caspase-12, closely related and encoded in the same locus as caspases-1 and -11, may have overlapping functions with caspases-1 and -11. Therefore, we investigated whether the additional loss of caspase-12 might further increase the resistance to LPS-induced septic shock that is afforded by the combined loss of caspases-1 and -11. However, the caspase-1/11/12 knockout mice were no more resistant to LPSinduced septic shock than the caspase-1/11 double knockout animals. The overall condition and body temperature were monitored until the LPStreated mice reached the ethical endpoint. Upon LPS injection, signs of morbidity and poor health appeared later and with reduced severity (at least at early time points) in the caspase-1/11 knockout and the caspase-1/11/12 knockout mice compared with the LPStreated WT animals. Since no significant differences were observed between the caspase-1/11/12 knockout vs. the caspase-1/11 knockout mice, this suggests that caspase-12 does not play a significant role in this septic shock model.
Caspase-12 has also been implicated in ER stress-induced apoptosis [25, 28, 32, 35]. Moreover, caspases-1 and -11 have recently been implicated in the death of certain cancer cells by chemotherapeutic drugs [36]. However, we found that combined loss of caspases-1, -11 and -12 did not protect several types of haematopoietic cells and fibroblasts in vitro against a diverse range of cytotoxic drugs. It has also been reported that caspase-12 deficiency renders mice resistant to tunicamycin-induced renal damage in vivo that is associated with induction of apoptosis in this tissue [28]. However, we did not detect any differences in renal damage and the frequency of apoptotic cells in the kidneys between caspase-1/11/12 knockout and WT mice. These findings may indicate that inflammatory caspases are only critical for cytotoxic drug-induced cell killing in selected cell types. Moreover, it remains possible that we have not yet tested a cell death inducer for which caspases-1/11/12 collectively play critical roles. Activation of the murine inflammatory caspases-1 and -11 requires a two-step process, namely a priming signal and a secondary stimulus, called the activation step [11, 22, 37, 38]. While caspase-1 can be activated by a broad range of stimuli, caspase-11 is only induced in cells and therefore capable of being activated upon sensing Gram-negative bacteria [22, 39]. It is possible that caspase-12 activation may similarly need specific priming and activation signals, that have not been identified yet. In BMDMs as well as in the kidney and liver, the absence of caspase-1/11 had no impact on the levels of caspase-12 mRNA.
The substantial LPSinduced increase in the levels of caspase-12 mRNA suggests a role of caspase-12 in response to bacterial infection and sepsis. Potential regulatory effects of caspase-12 on caspases-1 and -11 (serving as possible effectors) cannot be revealed in our mice since they lack all three inflammatory caspases.
Moreover, similar to caspase-1, the caspase-12 zymogen may have to bind to an adaptor protein to undergo the conformational changes required for activation. An adaptor for caspase-12 has not yet been identified and it is possible that the expression of such an activator of caspase-12 may also need a specific signal. Furthermore, it is possible that caspase-12 acts as a regulator upstream of caspases-1 and -11. Due to an early stop codon in one of the human caspase-12 genes, only a short, truncated version of this protein can be generated in people of Asian and Caucasian descent as well as in most Africans. This non-proteolytic form of human caspase-12 is structurally similar to the so-called CARD-only proteins (COPs), including ICEBERG and COP/pseudo-ICE. These proteins that contain a CARD, but lack the domains for enzymatic activity have previously been shown to have regulatory functions that impact cytokine secretion, NF-κB signalling and certain other signalling pathways [40, 41]. It is therefore possible that caspase-12, particularly the truncated form expressed in most humans, might fulfil a regulatory function or serve as a non-catalytic scaffolding platform in some signalling pathways, similar to the COPs. To rule out the possibility of upstream regulation, a single caspase-12deficient mouse strain would be useful for future studies. Our newly generated caspase-1/11/12 triple knockout mice will serve as a valuable tool in such investigations and we will make them available to the scientific community.
In conclusion, we have generated a strain of caspase-1/11/12 triple knockout mice. The analysis of these animals failed to identify a critical role of caspase-12, overlapping with the functions of caspases-1 and -11, in septic shock induced by LPS or the response of cells to a broad range of cytotoxic insults. Our caspase-1/11/12 triple knockout mice are expected to help identify important functions that are safeguarded by extensive functional overlap between many caspases, such as the ones operating in the fight against pathogens.
Materials and methods
Generation of caspase-1/11/12 triple knockout mice
The caspase-1/11/12−/− mice were generated by Taconic Bioscience GmbH. The genomic region encompassing caspases-1, -11 and -12 was deleted using the targeting strategy in C57BL/6 ES cells depicted in Fig. 1a. The targeting vector was generated using BAC clones from the C57BL/6J RPCIB-731 BAC library and was transfected into the Taconic C57BL/6N Tac ES cell line. The targeting construct contained a puromycin resistance cassette used for positive selection of recombinant clones as well as a thymidine kinase for negative selection. The positive selection cassette was subsequently excised using Flp-FRT-mediated recombination.
The heterozygous caspase-1/11/12+/− offspring were intercrossed to obtain caspase-1/11/12−/− mice. Genotyping (by PCR) confirmed the absence of the caspase-1/11/12 locus. Primer sets used for genotyping: 23:5′cacacctaaaaacagagtaaaaggc, 24: 5′gggactgtatgaagaatggatcc, 25:5′cagcaccttgaattatgagttgg. PCR programme: 95 °C for 5 min followed by 95 °C (for 30 s), primer annealing at 60 °C (30 s) then 72 °C (1 min). All three steps were repeated for a total of 30 cycles and this was followed by a final single elongatation step at 72 °C.
Endotoxic shock model
Experiments with mice were conducted according to the guidelines and with approval from the Walter and Eliza Hall Institute (Parkville, Victoria, Australia) Animal Ethics Committee. The caspase-1/11 mice used in these (and other following) experiments were derived from a caspase-1/11 double knockout colony published previously [42, 43] and were not littermates from the caspase-1/11/12 triple knockout mice. Mice were injected intraperitoneally (i.p.) with 18 or 54 mg/kg body weight ultrapure LPS (E. coli O111:B4, InvivoGen, catalogue code: tlrl-eblps). Some treated mice were sacrificed after 4 h for heart bleeds and cytokine analysis. Mice for survival studies were taken when showing physical signs of ill health and/or a drop in body temperature below 33 °C. A scoring sheet was followed to assess the overall physical condition of the mice. This included features, such as posture, respiratory rate, dehydration, responsiveness and eye closure. The body temperature and physical condition of each mouse were checked hourly and every 15 min within the first hour until sacrifice. Only males were used for these LPS shock experiments, and all animals were between 6 and 12 weeks of age. The mice used in these experiments were derived from independent colonies, namely the C57BL/6 (WT control), caspase-1/11 double knockout and caspase-1/11/12 triple knockout lines.
In vivo treatment with tunicamycin
C57BL/6 (WT control) and caspase-1/11/12 triple knockout mice were injected with 1 mg/kg body weight tunicamycin (Sigma-Aldrich, product code T776) dissolved in DMSO as per the manufacturer’s instructions and diluted in PBS buffer. Mice were injected i.p. and sacrificed 24 or 72 h post injection. The mice were monitored regularly for signs of discomfort during this period. At sacrifice, kidneys and livers were harvested for histological analysis and TUNEL staining. ALT levels in the sera were measured as described previously [44].
TUNEL staining
After harvesting, the kidneys (and livers) were incubated in formalin for 12 h at room temperature, followed by treatment with 70% EtOH. Preparation of tissue sections and staining with haematoxylin and eosin (H&E) were carried out in-house by the histology department. Dewaxed paraffin sections were incubated in 20 μg/mL proteinase K (Roche, product code 03115879001) for 15 min, followed by three 2-min washes in PBS. Endogenous peroxidase activity was blocked by treatment with 3% H2O2 in methanol for 5 min at room temperature. Following three 2-min washes in PBS, each section was incubated in the TUNEL reaction mix consisting of bio dUTP (0.3 nmol/μL) (Roche, product code 11093070910) + CoCl2 (25 mM) + TdT buffer + TdT (25 U/μL) (Promega, product code M828C) + H2O to a total volume of 50 μL per section for 1 h at 37 °C in a moist chamber. This was followed by further washes with PBS (3 × 2 min). The slides were then incubated with the ABC reagent (Vectastain Elite ABC Kit, Vector labs, product code PK-6100) for 30 min at room temperature (as per the manufacturer’s instructions). After further washes, the sections were incubated in DAB peroxidase substrate mix for 5 min (Vector labs; SK-4100). Finally, slides were washed with H2O, counterstained with haematoxylin and mounted by the in-house histology department. All slides were scored visually by microscopy. Additionally, two organ sections for each genotype and treatment were scanned and computationally analysed. The images were scanned using a 3DHISTEC slide scanner, and quantification of the TUNEL+ cells was performed using FIJI [45] with a custom written scoring macro. For Fig. 5, the image contrast and brightness were adjusted using Adobe Photoshop CS6 version 13.0.4.
Measurement of cytokine levels in the serum of the mice
Blood from mice was taken at death or 4 h after treatment with LPS by cardiac puncture. The levels of cytokines were determined using ELISA (IL-1β from R&D; IL-18 from MBL International Corp.). The levels of all other cytokines and chemokines in the sera from the mice were quantified by using the Bio-Rad Bio-Plex Pro mouse cytokine 23-plex assay (product code M60-009RDPD).
Flow cytometric analysis
Mice were euthanised and organs were harvested into sterile BSS supplemented with 5% foetal calf serum (FCS, Sigma-Aldrich, 12003C). For the detection of cell surface markers, the following rat or hamster monoclonal antibodies that had been conjugated to FITC, APC, R-PE or biotin (made in-house or purchased from eBioscience) were used: TER119 (TER119), MAC-1 (M1/70), NK1.1 (PK136), B220 (14.8 or RA3-6B2), CD3 (145-2C11), CD4 (GK1.5, H129 or YTA3.2.1), CD8 (53-6.7 or YTS169), TCRβ (H57-597), CD44 (IM781), CD25 (PC61), IgD (11-26C), IgM (5.1), CD62L (MEL-14), CD5 (53-7.3) and GR-1 (RB6-8C5). Streptavidin conjugates to PE-Cy7 or APC (BioLegend) were used for the detection of biotin-conjugated antibodies. Cells were washed in BSS supplemented with 2% FCS and analysed using a LSR-II flow cytometer (BD Biosciences). Dead cells (PI+) were excluded from analysis by staining with propidium iodide (PI, 5 μg/mL; Sigma-Aldrich). Intracellular staining for FoxP3 (clone FJK-16s) was performed using the eBioscience FoxP3/transcription factor staining buffer set. All data were analysed using FlowJo 9.9.4 software.
Cell death assays
Three independent MDF cell lines were generated from caspase-1/11/12 triple knockout and WT mice each. The fibroblasts were isolated from the dermis of mice and subsequently immortalised by transfection with an expression vector encoding the SV40 large T antigen. Primary BMDMs were generated (see below) and plated on non-coated 96-well plates over night with 1 × 104 cells seeded into each well containing 100 μL Dulbecco’s modified Eagle’s medium (DMEM) containing 10% FCS (Sigma-Aldrich, product code 12003C), 100 U/mL penicillin, 100 μg/mL streptomycin (complete medium) and supplemented with 20% L929 cell conditioned medium (a source of M-CSF). For immortalised MDFs, 1–2 × 104 cells per well were plated out on regular 96-well coated tissue culture plates in complete medium the night before the treatment. All cell types were treated with the drugs as indicated. The treatments used in these assays included TNF (100 ng/mL, made in-house), SMAC mimetic (CompA from Tetralogic, 500 nM), the caspase inhibitor QVD-Oph (10 μM, MP Biomedicals, OPH109), thapsigargin (Sigma, product code T9033), taxol (paclitaxel from Sigma-Aldrich, product code T7402), tunicamycin (Sigma, product code T7765) and etoposide (Sigma, product code T7402). At the indicated time points, cells were harvested and stained with 1–5 μg/mL propidium iodide alone or in combination with Annexin V-FITC (produced in-house). Cell survival was measured in a BD FACS Calibur flow cytometer. All cell survival data are presented in a manner normalised to untreated control cells unless data for untreated cells are shown separately. Untreated cells were cultured in complete medium.
In vitro assays with BMDMs
Primary BMDMs were generated from single-cell suspensions of bone marrow that had been flushed with PBS containing 2% FCS from mouse tibiae and femora and cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% FCS (Sigma-Aldrich, 12003C), 100 U/mL penicillin and 100 μg/mL streptomycin (complete medium) and supplemented with 20% L929 conditioned medium (a source of M-CSF) for 6 days before plating out on treated tissue culture plates. Approximately 1 × 106 cells were plated per well on coated 12-well tissue culture plates in 1 mL of complete medium plus M-CSF. On day 7, the cells were stimulated with 20 ng/mL ultrapure LPS (LPS from E. coli O111:B4, InvivoGen, catalogue code: tlrl-eblps) for 24 h before harvesting the supernatants and cell pellets separately. For qRT-PCR analysis of the expression of caspases-1, -11 and -12, BMDMs were treated with 20 ng/mL or 500 ng/mL LPS for 3 h. The cell pellet was subsequently washed twice with ice-cold PBS and lysed in 1 mL TRIzol (Thermo Fisher Scientific product code 1559-6018).
Western blot analysis
Cell pellets were lysed in Onyx buffer (20 mM Tris/HCL pH 7.4, 135 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, 1% Triton X-100, 10% glycerol in H2O) with protease inhibitor cocktail (Roche, 4693159001) added fresh, and samples of varying concentration (between 10 and 30 μg, depending on protein abundance) were loaded onto NuPAGE Novex 10% Bis–Tris protein gels (Thermo Fisher Scientific) for size fractioning and run in MES buffer. Transfers were carried out on the iBlot 2 dry blotting system (Invitrogen, IB21001) onto nitrocellulose membranes (Invitrogen, IB23001 or IB23001). Membranes were blocked in 5% skim milk powder in PBS with 0.1% Tween-20 and then probed with monoclonal antibodies against caspase-1 (clone 1H11; made in-house; see below, and available from Enzo LifeSciences), caspase-11 (4E11; made in house; see below available from Adipogene and Enzo LifeSciences) or HSP70 (clone N6, a gift from Dr. Robin Andersson, Peter MacCallum Cancer Centre, Melbourne, Australia); the last one used as a loading control. Bound primary antibodies were detected by goat anti-rat IgG or goat anti-mouse IgG antibodies conjugated to HRP (Southern Biotech). The ECL reaction was used for detection, and blots were developed using the ChemiDoc Touch System (Bio-Rad Laboratories). Protein concentrations were determined by Bradford assay using Protein Assay Dye Reagent Concentrate as per the manufacturer’s instructions (product code 5000006).
Generation of rat monoclonal antibodies against caspase-1 and caspase-11
Monoclonal antibodies against caspase-1 and caspase-11 were produced as we have described previously [46, 47]. Briefly, Wistar rats were initially immunised by subcutaneous injection (s.c.) with the p20 fragment of mouse caspase-1, a KLH conjugated mouse caspase-1 peptide (aa 206–220) or the p20 fragment of mouse caspase-11 dissolved in complete Freund’s adjuvant (Difco, Detroit, MI). Two subsequent boosts with each of the immunogens resuspended in incomplete Freund’s adjuvant (Difco) were given 3 and 6 weeks later. A final boost with the immunogen dissolved in phosphate-buffered saline (PBS) was given i.v. and i.p. 4 weeks later. Three days later, spleen cells from each of the immunised rats were fused with the SP2/0 myeloma cell line as previously described [47]. Hybridomas producing antibodies against either caspase-1 or caspase-11 and their isotypes were identified by a screening strategy that we have previously described. Briefly, 293T cells were transiently transfected with an EE-tagged inactive cysteine mutant of either mouse caspase-1 (to screen for antibodies against casapse-1) or caspase-11 (to screen for antibodies against casapse-11), fixed in 1% paraformaldehyde/PBS, permeabilised with 0.3% saponin (Sigma) and stained with hybridoma supernatants. Bound antibodies were revealed with fluorescein isothiocyanate (FITC)-conjugated goat anti-rat Ig antibodies (Southern Biotechnology) and analysed by flow cytometry. A single peak of low immunofluorescence indicated that a particular antibody did not recognise caspase-1 or caspase-11. A single peak of high intensity indicated binding to molecules other than caspase-1 or caspase-11. A double peak histogram of low and high intensity staining, due to the presence of both non-transfected and transfected cells, identified hybridomas producing antibodies specific to either caspase-1 or caspase-11. Antibodies against the EE epitope tag (BabCO) were used as a positive control and hybridomas producing antibodies against KLH were eliminated. Hybridomas producing antibodies of the desired specificity were cloned twice and adapted for growth in low serum medium. The caspase-1 antibody producing hydridoma clone 1H11 and the caspase-11 antibody producing hybridoma clone 4E11 were selected for further use. For production of large amounts of antibodies, hybridomas were cultured for several weeks in the miniPERM classic 12.5-kd production and nutrient modules (Heraeus). Antibodies were purified on protein G-sepharose columns (Pharmacia) according to the manufacturer’s instructions.
In vitro stimulation with LPS and ELISA to detect IL-18
For in vitro stimulation of caspase-1, primary BMDMs were primed with 20 ng/mL ultrapure LPS (LPS from E. coli O111:B4, InvivoGen, catalogue code: tlrl-eblps) for 3 h and subsequently activated using 5 mM ATP (Sigma, product number A 2383) for 1 h, after which the cells and supernatants were harvested individually. For caspase-11 activation, the cells were primed with 500 ng/mL ultrapure LPS for 3 h before activation with 2 μg/mL ultrapure LPS that was transfected into the cells using lipofectamine (LPS: lipofectamine was used at a ratio of 1:1.3). Priming and activation were performed in reduced serum opti-MEM media (Gibco). After 6 h, this medium was replaced with complete medium supplemented with 20% L929 cell conditioned medium (a source of M-CSF), and after an additional 16 h incubation the supernatants as well as cell pellets were harvested. The levels of cytokines and chemokines in the cell supernatants were determined by ELISA (IL-1β and Rantes/CCL5 from R&D, lL-18 from MBL International Corp.).
qRT-PCR analysis
For measuring the levels of mRNA expression, total RNA was isolated from the organs or cultured cells and cDNA was synthesised by using the SuperScript III first-strand synthesis system (ThermoFisher). qRT-PCR was performed in triplicates using TaqMan assay from ThermoFisher Scientific with primers for mouse caspase-1 (Mm00438023_m1), caspase-11 (Mm00432304_m1), caspase-12 (Mm00438038_m1), β-actin (Mm02619580_g1) and hmbs (Mm01143545_m1) (loading control). qRT-PCR was run using Viia7 real-time PCR and an ABI7900 machine. β-actin was chosen as a internal control gene and ΔCt calculation was performed as fallows: ΔCt = (Ct gene of interest−Ct internal control). 2−ΔΔCt values were calculated as described in ref. [48] using the equation: 2−ΔΔCt = ΔCt (treated sample)−ΔCt (untreated sample). The relative difference of gene expression was calculated using β-actin as a reference for comparison. To set a threshold qRT-PCR cycle number for each gene of interest (considered as background noise), we tested Ct values for caspase-1, caspase-11 and caspase-12 in cells from caspase-1/11/12 triple knockout mice (data not shown). For caspase-1 expression, the maximum cycle threshold was set at 35 cycles, anything over 37 cycles in the case of caspase-11 and higher Ct than 39 for caspase-12 were considered as no RNA detected.
Statistical analysis
Mouse survival curves were generated and analysed with GraphPad Prism (GraphPad Software Inc., La Jolla, CA, USA). Mouse survival cohorts were compared using the log-rank Mantel–Cox test. P values of <0.05 were considered significant. In vitro cell survival data were plotted and analysed with GraphPad Prism using one-way or two-way ANOVA statistical test. For ELISA data on cytokine levels in the sera from the mice, graphs were plotted using Prism software and examined by using the ordinary one-way ANOVA test. ELISA data on the levels of cytokines in the supernatants from cultured cells were evaluated by using Prism, applying a two-way ANOVA tests. Data are presented as standard error of the mean (±S.E.M.).
References
Martinon F, Tschopp J. Inflammatory caspases: linking an intracellular innate immune system to autoinflammatory diseases. Cell. 2004;117:561–74.
Man SM, Kanneganti TD. Gasdermin D: the long-awaited executioner of pyroptosis. Cell Res. 2015;25:1183–4.
Ramirez MLG, Salvesen GS. A primer on caspase mechanisms. Semin Cell Dev Biol. 2018 Jan 12. pii: S1084-9521(17)30108-8. https://doi.org/10.1016/j.semcdb.2018.01.002.
Czabotar PE, Lessene G, Strasser A, Adams JM. Control of apoptosis by the BCL-2 protein family: implications for physiology and therapy. Nat Rev Mol Cell Biol. 2014;15:49–63.
Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol. 2008;9:47–59.
Strasser A, Jost PJ, Nagata S. The many roles of FAS receptor signaling in the immune system. Immunity. 2009;30:180–92.
Feltham R, Vince JE, Lawlor KE. Caspase-8: not so silently deadly. Clin Transl Immunol. 2017;6:e124.
Sagulenko V, Lawlor KE, Vince JE. New insights into the regulation of innate immunity by caspase-8. Arthritis Res Ther. 2016;18:4.
Pop C, Salvesen GS. Human caspases: activation, specificity, and regulation. J Biol Chem. 2009;284:21777–81.
Latz E, Xiao TS, Stutz A. Activation and regulation of the inflammasomes. Nat Rev Immunol. 2013;13:397–411.
Man SM, Kanneganti TD. Regulation of inflammasome activation. Immunol Rev. 2015;265:6–21.
Shaw PJ, McDermott MF, Kanneganti TD. Inflammasomes and autoimmunity. Trends Mol Med. 2011;17:57–64.
Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70.
Black RA, Kronheim SR, Merriam JE, March CJ, Hopp TP. A pre-aspartate-specific protease from human leukocytes that cleaves pro-interleukin-1 beta. J Biol Chem. 1989;264:5323–6.
Kostura MJ, Tocci MJ, Limjuco G, Chin J, Cameron P, Hillman AG, et al. Identification of a monocyte specific pre-interleukin 1 beta convertase activity. Proc Natl Acad Sci USA. 1989;86:5227–31.
Suresh R, Mosser DM. Pattern recognition receptors in innate immunity, host defense, and immunopathology. Adv Physiol Educ. 2013;37:284–91.
Lamkanfi M, Dixit VM. Mechanisms and functions of inflammasomes. Cell. 2014;157:1013–22.
Lamkanfi M. Emerging inflammasome effector mechanisms. Nat Rev Immunol. 2011;11:213–20.
de Zoete MR, Palm NW, Zhu S, Flavell RA. Inflammasomes. Cold Spring Harb Perspect Biol. 2014;6:a016287.
Vince JE, Silke J. The intersection of cell death and inflammasome activation. Cell Mol Life Sci. 2016;73:2349–67.
Hagar JA, Powell DA, Aachoui Y, Ernst RK, Miao EA. Cytoplasmic LPS activates caspase-11: implications in TLR4-independent endotoxic shock. Science. 2013;341:1250–3.
Kayagaki N, Warming S, Lamkanfi M, Vande Walle L, Louie S, Dong J, et al. Non-canonical inflammasome activation targets caspase-11. Nature. 2011;479:117–21.
Kayagaki N, Wong MT, Stowe IB, Ramani SR, Gonzalez LC, Akashi-Takamura S, et al. Noncanonical inflammasome activation by intracellular LPS independent of TLR4. Science. 2013;341:1246–9.
Ramirez MLG, Poreba M, Snipas SJ, Groborz K, Drag M, Salvesen GS. Extensive peptide and natural protein substrate screens reveal that mouse caspase-11 has much narrower substrate specificity than caspase-1. J Biol Chem. 2018;293:7058–67.
Lamkanfi M, Kalai M, Vandenabeele P. Caspase-12: an overview. Cell Death Differ. 2004;11:365–8.
Hermel E, Klapstein KD. A possible mechanism for maintenance of the deleterious allele of human CASPASE-12. Med Hypotheses. 2011;77:803–6.
Kalai M, Lamkanfi M, Denecker G, Boogmans M, Lippens S, Meeus A, et al. Regulation of the expression and processing of caspase-12. J Cell Biol. 2003;162:457–67.
Nakagawa T, Zhu H, Morishima N, Li E, Xu J, Yankner BA, et al. Caspase-12 mediates endoplasmic-reticulum-specific apoptosis and cytotoxicity by amyloid-beta. Nature. 2000;403:98–103.
Obeng EA, Boise LH. Caspase-12 and caspase-4 are not required for caspase-dependent endoplasmic reticulum stress-induced apoptosis. J Biol Chem. 2005;280:29578–87.
Wang S, Miura M, Jung YK, Zhu H, Li E, Yuan J. Murine caspase-11, an ICE-interacting protease, is essential for the activation of ICE. Cell. 1998;92:501–9.
Ghayur T, Banerjee S, Hugunin M, Butler D, Herzog L, Carter A, et al. Caspase-1 processes IFN-gamma-inducing factor and regulates LPS-induced IFN-gamma production. Nature. 1997;386:619–23.
Nakagawa T, Yuan J. Cross-talk between two cysteine protease families. Activation of caspase-12 by calpain in apoptosis. J Cell Biol. 2000;150:887–94.
Rao RV, Hermel E, Castro-Obregon S, del Rio G, Ellerby LM, Ellerby HM, et al. Coupling endoplasmic reticulum stress to the cell death program. Mechanism of caspase activation. J Biol Chem. 2001;276:33869–74.
Murphy JM, Czabotar PE, Hildebrand JM, Lucet IS, Zhang JG, Alvarez-Diaz S, et al. The pseudokinase MLKL mediates necroptosis via a molecular switch mechanism. Immunity. 2013;39:443–53.
Jimbo A, Fujita E, Kouroku Y, Ohnishi J, Inohara N, Kuida K, et al. ER stress induces caspase-8 activation, stimulating cytochrome c release and caspase-9 activation. Exp Cell Res. 2003;283:156–66.
Wang Y, Gao W, Shi X, Ding J, Liu W, He H, et al. Chemotherapy drugs induce pyroptosis through caspase-3 cleavage of a gasdermin. Nature. 2017;547:99–103.
Mariathasan S. ASC, Ipaf and Cryopyrin/Nalp3: bona fide intracellular adapters of the caspase-1 inflammasome. Microbes Infect. 2007;9:664–71.
Mariathasan S, Weiss DS, Newton K, McBride J, O’Rourke K, Roose-Girma M, et al. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature. 2006;440:228–32.
Wang S, Miura M, Jung Y, Zhu H, Gagliardini V, Shi L, et al. Identification and characterization of Ich-3, a member of the interleukin-1beta converting enzyme (ICE)/Ced-3 family and an upstream regulator of ICE. J Biol Chem. 1996;271:20580–7.
Druilhe A, Srinivasula SM, Razmara M, Ahmad M, Alnemri ES. Regulation of IL-1beta generation by Pseudo-ICE and ICEBERG, two dominant negative caspase recruitment domain proteins. Cell Death Differ. 2001;8:649–57.
Humke EW, Shriver SK, Starovasnik MA, Fairbrother WJ, Dixit VM. ICEBERG: a novel inhibitor of interleukin-1beta generation. Cell. 2000;103:99–111.
Li P, Allen H, Banerjee S, Franklin S, Herzog L, Johnston C, et al. Mice deficient in IL-1 beta-converting enzyme are defective in production of mature IL-1 beta and resistant to endotoxic shock. Cell. 1995;80:401–11.
Schott WH, Haskell BD, Tse HM, Milton MJ, Piganelli JD, Choisy-Rossi CM, et al. Caspase-1 is not required for type 1 diabetes in the NOD mouse. Diabetes. 2004;53:99–104.
Jost PJ, Grabow S, Gray D, McKenzie MD, Nachbur U, Huang DC, et al. XIAP discriminates between type I and type II FAS-induced apoptosis. Nature. 2009;460:1035–9.
Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–82.
O’Reilly LA, Cullen L, Visvader J, Lindeman GJ, Print C, Bath ML, et al. The proapoptotic BH3-only protein bim is expressed in hematopoietic, epithelial, neuronal, and germ cells. Am J Pathol. 2000;157:449–61.
O’Reilly LA, Cullen L, Moriishi K, O’Connor L, Huang DCS, Strasser A. Rapid hybridoma screening method for the identification of monoclonal antibodies to low abundance cytoplasmic proteins. Biotechniques. 1998;25:824–30.
Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–8.
Acknowledgements
We thank T Ballinger, S Russo, G Siciliano, C Stivala. and S Stoev for their help with experiments with mice; S Monard and his team for their help with flow cytometry. Our work is supported by the Australian National Health and Medical Research Council (Project Grant 1145728 to MJH and 1143105 to MJH and AS; Programme Grant 1016701 to AS and Fellowship 1020363 to AS), the Leukaemia and Lymphoma Society of America (LLS SCOR 7001-13 to AS and MJH), the Cancer Council of Victoria (1052309 to AS and Venture Grant MJH and AS) and by operational infrastructure grants through the Australian Government Independent Research Institute Infrastructure Support Scheme (9000220) and the Victorian State Government Operational Infrastructure Support Program.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Edited by G. Melino
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Salvamoser, R., Brinkmann, K., O’Reilly, L.A. et al. Characterisation of mice lacking the inflammatory caspases-1/11/12 reveals no contribution of caspase-12 to cell death and sepsis. Cell Death Differ 26, 1124–1137 (2019). https://doi.org/10.1038/s41418-018-0188-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-018-0188-2
This article is cited by
-
Deletion of Gpatch2 does not alter Tnf expression in mice
Cell Death & Disease (2023)
-
Sepsis-induced immunosuppression: mechanisms, diagnosis and current treatment options
Military Medical Research (2022)
-
Silencing of FTX suppresses pancreatic cancer cell proliferation and invasion by upregulating miR-513b-5p
BMC Cancer (2021)
-
Reciprocal regulation between ER stress and autophagy in renal tubular fibrosis and apoptosis
Cell Death & Disease (2021)
-
Emerging connectivity of programmed cell death pathways and its physiological implications
Nature Reviews Molecular Cell Biology (2020)