Abstract
Zika virus (ZV) infects neural stem cells (NSCs) and causes quiescence in NSCs, reducing the pool of brain cells, leading to microcephaly. Despite conscientious efforts, the molecular mechanisms for ZV-mediated effects on NSCs lack clarity. This study aimed to explore the underlying mechanisms for ZV-mediated induction of quiescence in the primary cultures of human fetal neural stem cells (fNSCs). We demonstrate that expression of ZV envelope (E) protein displays maximum quiescence in human fNSCs by accumulating cells in the G0/G1 phase of the cell cycle as compared to other non-structural proteins, viz. NS2A, NS4A and NS4B. E protein induces immature differentiation by induction of pro-neuronal genes in proliferating fNSCs, induces apoptosis in differentiating fNSCs 3 days post differentiation, and disrupts migration of cells from differentiating neurospheres. In utero electroporation of mouse brain with E protein shows drastic downregulation of proliferating cells in ventricular and subventricular zone regions. Global microRNA sequencing suggests that E protein modulates miRNA circuitry. Among differentially expressed miRNAs, we found 14 upregulated and 11 downregulated miRNAs. Mir-204-3p and mir-1273g-3p directly regulate NOTCH2 and PAX3 expression, respectively, by binding to their 3′UTR. Bioinformatic analysis using GO analysis for the targets of differentially expressed miRNAs revealed enrichment of cell cycle and developmental processes. Furthermore, WNT, CCKR, PDGF, EGF, p53, and NOTCH signaling pathways were among the top enriched pathways. Thus, our study provides evidence for the involvement of ZV E protein and novel insights into the molecular mechanism through identification of miRNA circuitry.

Art work depicting the effect of Zika virus E protein on human fetal neural stem cells.
Similar content being viewed by others
Introduction
Zika virus (ZV) infection has gathered worldwide attention following outbreaks in Brazil during 2015–2016 epidemics, and recently WHO has defined it as a global public health emergency [1,2,3]. Zika virus belongs to the flavivirus family and is spread by Aedes aegypti and Aedes albopictus mosquitoes, though other mosquito species may also contribute to its transmission. It causes a severe in utero clinical condition called microcephaly where fetuses are born with abnormally small brain [4]. TORCHS (toxoplasmosis, rubella, cytomegalovirus, herpes virus, and syphilis) are main congenital infections that hamper in utero brain development [5]. ZV is a new TORCH member that compromises the developing brain in utero [6]. ZV has been detected in the amniotic fluids of pregnant women and in the brain tissues of microcephalic fetuses, suggesting that ZV can potentially cross the placental barrier and blood brain barrier to infect the fetus [7], similar to Epstein–Barr virus, which infects fetal as well as adult brain [8, 9]. ZV has a 10.617-kb long single-stranded, positive sense RNA genome that encodes for a polyprotein with three structural proteins and seven non-structural proteins. The structural proteins are capsid (C), premembrane/membrane (PrM), and envelope (E protein), and non-structural proteins are NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5 [10]. It is largely unknown how ZV develops neurotropism and pathogenicity, though in recent isolates genetic changes caused variations in 10 amino acids of the viral envelope (E) protein near a glycosylation site, whereas this site is lacking in many isolates from the African lineage. Also, N-linked glycosylation of E protein is considered an important determinant of ZV virulence and neuroinvasion [11].
Proliferation and differentiation of NSCs are key processes for maintaining the pool of cells in developing fetal brain [12, 13]. ZV attenuates the growth of neurospheres and brain organoids derived from induced pluripotent stem cells (iPSCs) [4, 14, 15]. Expression of ZV proteins NS4A and NS4B inhibit differentiation of human NSCs into neuronal lineage by inhibiting the mTOR signaling pathway in differentiating neurons, thereby preventing neurogenesis [16]. In vivo studies on mice model systems demonstrate that ZV disrupts brain development and alters the properties of neural stem cells [17,18,19,20]. To date, the detailed molecular basis of ZV-induced alteration in the properties of NSCs is unknown and poses the biggest challenge for development of anti-viral therapy; this lacunae in the ZV biology field necessitated this study.
Small RNAs or microRNAs (miRNAs) are endogenous, small (~18–24 nt), non-coding RNAs that are key regulators of gene expression. They function post-transcriptionally by binding to complementary sequences of the 3′-untranslated region (3′-UTR) of target mRNAs [21] and play an important role in the regulation of proliferation, differentiation, migration, and apoptosis [22]. The role of miRNA in brain development, neurodevelopmental disorders, and viral infections is well established [23,24,25,26,27,28]. Dysregulation of miRNA circuitry is reported in several mosquito-borne flavivirus infections [29]. However, till date, research on whether ZV alters the cellular miRNome in human NSCs and an understanding of the mechanisms at miRNA level are lacking. As miRNAs are a crucial part of cellular transcriptome and are critical for flavivirus infection, we hypothesized that ZV infection modulates fNSC miRNome.
NOTCH molecules are single-pass transmembrane receptors with characteristic epidermal growth factor (EGF)-like repeats on the extracellular portion [30]. There are four paralogs of NOTCH: NOTCH 1–4, the receptors are activated by specific ligands of Jagged and Delta serrate member families. These receptors play a crucial role in cell-fate determination during embryonic development [31]. NOTCH2 is involved in maintaining the proliferative state of NSCs and inhibiting differentiation [32]. It functions as a mitotic and an anti-apoptotic factor in other cell types also [33,34,35,36]. Further paired box (PAX) genes code for transcription factors, which contain a highly conserved DNA-binding domain called the paired domain. The PAX family of proteins is among the principal regulators of gene expression due to their involvement in compartmentalization and body patterning of developing organisms. PAX3 plays key roles in specification, migration, and differentiation of neural crest cells into the various tissue lineages. We provide evidence that miRNAs dysregulated in response to ZV E protein are involved in modulating the properties of fNSCs by regulating important developmental genes. In this report, for the first time we present detailed evidence that ZV E protein alters the miRNome profile of fNSCs, leading to alterations in proliferation and differentiation of fNSCs. Our data also suggest that in utero electroporation of E protein at embryonic day (E) 13.5 in mouse brain reduced proliferation of neural stem cells at E 15.5, hence indicating the crucial role of E protein in ZV pathology.
Materials and methods
Human fetal neural stem cell (fNSC) isolation and differentiation
Handling of human fetal brain tissues was done only after obtaining the mother’s informed consent. The tissues were processed according to the protocol approved by the Institutional Human Ethics and Stem Cell and Research Committee of National Brain Research Centre, India, in compliance with the recommendations of the Indian Council of Medical Research (ICMR), India. The telencephalon of 10–15-week-old human aborted fetuses was used to isolate fNSCs [37]. Isolated fNSCs were cultured in poly-d-lysine (Sigma-Aldrich, St. Louis, MO, USA) coated culture dishes in neurobasal media (Invitrogen, San Diego, CA, USA). The media was supplemented with Neural Survival Factor-1 (Lonza, Charles City, IA, USA), N2 supplement (Invitrogen, San Diego, CA, USA), bovine serum albumin (Sigma-Aldrich, St. Louis, MO, USA), glutamine (Sigma, St. Louis, MO, USA), 25 ng/ml basic fibroblast growth factor (bFGF) (Peprotech, Rocky Hill, NJ, USA), 20 ng/ml EGF (Peprotech, Rocky Hill, NJ, USA), penicillin and streptomycin solution (Invitrogen, San Diego, CA, USA), and gentamycin (Sigma, St. Louis, MO, USA). Cells were passaged at least nine times and characterized before performing experiments. The fNSCs were characterized by assessing the expression of neural stem cell markers SOX2 and Nestin, and lineage specific markers GFAP and MAP2 by western blotting and immunocytochemistry. Almost 99% fNSCs showed immunoreactivity towards SOX2 and Nestin, and were negative for both GFAP and MAP2. Differentiation of fNSCs into neurons was achieved by withdrawing bFGF and EGF from neurobasal media, and supplementing it with 10 ng/ml brain-derived neurotrophic factor (BDNF) (Peprotech, Rocky Hill, NJ, USA) and 10 ng/ml platelet-derived growth factor (PDGF-AB) (Peprotech, Rocky Hill, NJ, USA). For this particular study two human fetal tissues were employed for various assays and results obtained from both tissues were comparable. Cells were maintained in differentiating media for at least 21 days, and were assessed for their immunoreactivity towards neuronal markers TUJ-1 and MAP2; >95% of the cells were positive for TUJ-1 and MAP2 after differentiation.
Transient expression of ZV proteins
Full-length ZV proteins E protein, NS2B, NS4A, and NS4B were cloned in pCAGIG-IRES-GFP expression vector and were a gift from Dr. Shyamala Mani (IISc, Bangalore, India). The vectors were transfected in fNSCs at a confluency of 70–80% using Lipofectamine 2000 (Invitrogen, San Diego, CA, USA) according to the manufacturer’s protocol. Briefly, cells were kept in opti-MEM (Invitrogen, San Diego, CA, USA), plasmid, and Lipofectamine 2000 complex for 5 h, followed by at least 24 h incubation in proliferation or differentiation medium [38]. The transfected cells were processed for different experiments depending upon the requirement of the experiment. The transfection efficiency of all vectors in fNSCs was >25% (Supplementary figure S1) by Lipofectamine 2000 and were comparable.
Experimental animals and in utero electroporation of vectors
All animal experiments were performed according to the protocol sanctioned by the Institute Animal Ethics Committee of Indian Institute of Technology (IIT), Kanpur, registered with CPCSEA (Reg. No. 810/03/ac/CPCSEA, dated 15/10/2003). Wild-type mice (C57BL6/JNCBS; The Jackson Laboratory) were crossed to generate timed pregnant females for the experiments. In utero electroporation was performed with timed pregnant female mice according to established protocols [39].
Immunohistochemistry and quantification of Ki67- and GFP-positive cells
Mouse forebrain sections of the desired stage (E 15.5) were subjected to immunohistochemistry using the established protocol [39] with the primary antibody anti-Ki67 (1:200; Ab16667, Abcam, Cambridge, UK) and secondary antibody anti-rabbit (1:250; Cat. No. 111-545-003, Jackson ImmunoResearch Laboratories Inc., USA). Cryosections of mouse cortex electroporated with the constructs were imaged and analyzed using the multiphoton laser scanning confocal microscope LSM780 (Carl Zeiss Inc. Germany) and ZEN 2011 software. For quantification, each electroporated embryo analyzed was considered as a biological replicate (n) and for each biological replicate, at least three consecutive brain sections were analyzed, which were considered as technical replicates (N). The percentage of GFP-positive cells that were double positive for Ki67 and GFP were calculated from each of the technical replicates corresponding to each biological replicate (n) and the mean was calculated. The mean and standard error mean (SEM) plotted on the graph correspond to the mean of all biological replicates.
Immunocytochemistry
Immunocytochemistry experiments were done in Permanox four-well or eight-well chamber slides (Nunc, Kamstrupvej, Denmark). 20,000 cells/well were seeded in eight-well chamber slides and 50,000 cells/well in four-well chamber slides. After transfection, cells were washed with 1X PBS and fixed with 4% paraformaldehyde for 20 min, then washed thrice with PBS. Cells were blocked and permeabilized with 4% bovine serum albumin (BSA) and 0.3% triton X-100. Cells were probed with following primary antibodies overnight at 4 °C: anti-Ki67 (Novacastra, Wetzlar, Germany; 1:1000), anti-BrdU (Santa Cruz Biotechnolgy, Santa Cruz, CA, USA; 1:250), anti-TUJ-1 (Promega, Madison, WI, USA; 1:2000), and anti-DCX (Abcam, Cambridge, UK; 1:1000). After overnight incubation, cells were washed thrice with PBS and incubated with suitable Alexa Flour tagged 594 or 488 secondary antibodies (1:1000) followed by washing with PBS. Cells were mounted using Hardset mounting media with DAPI (Vector Labs, Burlingame, CA, USA). AxioImager.Z1 microscope (Carl Zeiss, Heidenheim, Germany) was used for visualization of slides using 10× or 20× objectives at room temperature. For analysis, 5–8 images were captured from random fields from each group.
Assessment of apoptosis by TUNEL assay
Experiments were performed in poly-d-lysine-coated Permanox eight-well chamber slides (Nunc, Kamstrupvej, Denmark). Cells were seeded at the density of 20,000 cells/well. Cells were transfected either with empty or with E protein expression vector using lipofection. The transfection medium was replaced with neuronal differentiation media as mentioned above and cells were differentiated for 72 h. Cells were then fixed with 4% paraformaldehyde for 20 min and washed. Blocking and permeabilization of cells was done using 4% BSA containing 0.3% Triton-X 100. TUNEL assay was performed using an in situ cell death detection kit, TMR red (Roche, Mannheim, Germany), as per the manufacturer’s protocol to detect apoptotic cells. For each experimental group, at least five images were captured from random fields. Images were acquired using AxioImager.Z1 microscope (Carl Zeiss, Heidenheim, Germany).
Bromodeoxyuridine (BrdU) labeling
After transfection of vectors, cells were incubated with 10 µM BrdU for 12 h in culturing media and washed with PBS followed by fixation in 4% paraformaldehyde for 20 min at room temperature. After washing, cells were treated with 2 N HCl for 30 min at room temperature for DNA denaturation. Following washing, cells were blocked with 4% BSA and were processed for immunostaining using anti-BrdU antibody.
Western blotting
We performed western blotting of cells transfected either with empty or with E protein expression vector. Briefly, transfected cells were lysed, protein was quantitated, run on 12% SDS-PAGE, and transferred onto nitrocellulose membrane (MDI, Ambala, India) [38]. After blocking, membranes were incubated with primary antibodies anti-caspase3/anti-cleaved caspase3 (Abcam, Cambridge, UK) and beta-tubulin (Sigma–Aldrich, St. Louis, MO, USA). Dilutions used were 1:2000 and 1:4000, respectively, overnight at 4 °C. HRP-conjugated (Vector Labs, USA) secondary antibodies (dilution 1:4000) were used and blots were developed using chemiluminescence reagent (Millipore, Billerica, MA, USA) on ChemiGenius Bio-imaging System (Syngene, Cambridge, UK). Densitometric analysis of the developed blots was carried out using ImageJ software (NIH). Normalization was done using beta-tubulin. Representative blots are depicted in figures.
Neurosphere culture and assay
Transfected fNSCs were cultured in a non-poly-d-lysine (PDL)-coated 25-cm2 flask (Tarsons, West Bengal, India) and kept upright with 3 ml neural stem cell medium (with EGF and FGF) for 72 h at 37 °C and 5% CO2 as described by Mishra et al. [37]. fNSCs formed neurospheres in 30 h and were maintained as neurospheres till termination of the experiments. Images of neurospheres from control and test groups were captured from at least five random fields using a phase-contrast microscope (Leica DM IL; Leica Microsystems, Germany). The size of neurospheres from each area was measured using Image-J. Neurospheres were categorized as small when their size was <8 × 103 mm2. Neurospheres were counted from each group in more than three separate experiments. At least 50 neurospheres were counted in each group and represented as percentage of small neurosphere based on their area.
Neurosphere migration assay
Neurospheres that were cultured from transfected fNSCs were seeded in poly-d-lysine coated eight-well chamber slides and allowed to adhere. Once the neurospheres adhered, the proliferation medium was replaced with neural differentiation medium (with PDGF and BDNF). Three days post differentiation, cells were fixed, blocked, and permeabilized using 4% BSA, 3% normal goat serum, and 0.3% triton-100. Cells were co-labeled with DCX and TUJ-1 antibodies (early markers for neurogenesis and newly formed neurons, respectively). For each experimental group, at least five similar-sized neurospheres were imaged on an AxioImager.Z1 microscope (Carl Zeiss, Heidenheim, Germany). Migration of cells from the periphery of the neurospheres was measured using Image-J [40, 41].
Quantitative real-time PCR
Total RNA was isolated using TRIZOL (Sigma-Aldrich, St. Louis, MO, USA) following the manufacturer’s protocol. cDNA preparation from RNA was done using a high-capacity cDNA reverse transcription kit (Applied Biosystems, USA) in accordance with the manufacturer’s instructions. SYBER Green master mix (Applied Biosystems, USA) was used for real-time detection of transcripts. 18S ribosomal RNA was used as internal control for normalization. Specific primers for PTN, DCX, SPOCK1, ROBO2, NOTCH2, PAX3, and 18S are mentioned in Supplementary document S3. The cycling conditions used for qPCR were 95 °C for 10 min (1 cycle), 95 °C for 20 s, 62 °C for 30 s, and 72 °C for 40 s (35 cycles).
miRNAs from fNSCs were isolated using miRNAeasy mini kit (Qiagen, Hilden, Germany). cDNA from miRNA was synthesized using miScript II RT kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. miRNAs were detected using specific primers for miRNAs (Qiagen, Hilden, Germany). U6 small nuclear RNA was used as internal control for normalization. qPCR was performed for detection of levels of specific miRNAs using miScript SYBER Green PCR kit (Qiagen, Hilden, Germany). Quantification of total RNA, miRNA, and cDNA was done using the Nanodrop1000 Spectrophotometer (Applied Biosystems, USA). All qPCR experiments were performed using the ViiA 7 Real-Time PCR System (Applied Biosystems, USA). Specificity of primers was confirmed with a single-product peak in the melt curve analysis.
miRNA mimic transfection
fNSCs were seeded at ~70% confluency in 35 mm PDL-coated plates. For mimic studies, 50 pmol of Syn-hsa-miR-1273g-3p or Syn-hsa-miR-204-3p miScript miRNA mimic (Qiagen, Hilden, Germany) was used to transfect the cells. All stars negative control siRNA (Qiagen, Hilden, Germany) was used as a mimic control.
miRNA target prediction
TargetScan database (http://www.targetscan.org/) was used for prediction of potential miRNA binding sites on 3′UTR of transcripts.
Cloning of 3′UTR and luciferase assay
3′UTR of PAX3 was amplified using specific primers mentioned in Supplementary document S3. Restriction sites used were NaeI and HindIII for cloning in pMIR-Reporter (Ambion) downstream of the luciferase coding region. The 1970-bp-long amplified product contained two predicted hsa-mir-1273g-3p binding sites. The 1253-bp-long 3′UTR of NOTCH2 was amplified using specific primers mentioned in Supplementary document S3. Restriction sites used for NOTCH2 3′UTR cloning in pMIR-Reporter were SacI and NaeI. The amplified product contained one mir-204-3p binding site. Then, 1 μg of the cloned product and 50 pmol of the mimic (mir-1273g-3p or mir-204-3p) were used to transfect the cells (HeLa cell line) using Lipofectamine 2000 in 35 mm plates as per the manufacturer′s protocol. Samples were harvested for luciferase detection post 24 h using passive lysis buffer (Promega, Madison, WI, USA). Luciferase activity was measured with Luciferase detection kit (Promega, Madison, WI, USA) according to the manufacturer’s protocol. Detection was performed using a Sirius single tube luminometer (Berthold Detection Systems, Pforzheim, Germany) and luciferase units were normalized with the total protein.
Global miRNA analysis using next-generation sequencing
Four samples consisting two each of empty vector and E protein were outsourced for RNA isolation and small RNA next-generation sequencing to BioServe Biotechnologies, Hyderabad, India. Briefly, the workflow involved polyadenylation, cDNA synthesis, and PCR amplification steps, resulting in an indexed cDNA library, followed by library purification, validation, gel-free bead-based size selection, and post-size selection library validation. The resulting validated cDNA indexed libraries were sequenced on Illumina NextSeq 500 for paired end (2 × 75 bp) to generate ≥20 million reads per sample. A minimum of 70% of the read pairs are of Q30 value. Sequenced data were processed to generate FASTQ files. The quality of FASTQ reads was checked using fastQC tool set in CLC bio Genomic Workbench ver 9.0 (CGWB). Raw data for each sample were aligned to human microRNAnome database. Pair-wise comparison of the alignment results was done for identification of miRNAs that are differentially expressed at a significant level, i.e. upregulated or downregulated.
Gene ontology (GO) analysis and pathway enrichment
Prediction of targets of upregulated and downregulated miRNAs was done using Genecards database. All targets were further considered for protein classification using PantherdB (http://www.pantherdb.org). GO derived from Gene ontology consortium (www.geneontology.org) was applied to check the roles of targets of differentially expressed miRNAs. Biological processes were derived from GO according to their annotations. PANTHER platform was used for enrichment of pathways of targets of differentially expressed miRNAs.
Statistical analysis
Results are represented as mean ± SEM. At least three independent experiments (n = 3) were carried out for each data set; the exact number of experiments is specified in the legends. Results were analyzed using Student’s t-test in order to determine the significance of the means. A value of p < 0.05 was taken as the level of significance.
Results
Viability of fNSCs was unaffected post ZV proteins transfection
ZV targets neural stem cells and causes death and quiescence of NSCs [14]; however, the effects of its envelope (E) protein and other non-structural protein (NS2A, NS4A, NS4B) components on fNSCs were not much explored. Hence, we examined the impact of ZV proteins on fNSCs. The viability of highly proliferative human fNSCs was assayed 24 h after transfection of E protein, NS2A, NS4A, NS4B, or empty vector, followed by TUNEL (terminal deoxynucleotidyl transferase dUTP nick end labeling) assay [42]. TUNEL-positive cells were 2.33 ± 0.42%, 2.44 ± 0.32%, 2.62 ± 0.59%, 1.70 ± 0.31%, and 1.90 ± 0.37% in empty vector, E protein, NS2A, NS4A, and NS4B, respectively (Fig. 1a–f). Significant cell death was not observed in E protein, NS2A, NS4A, or NS4B proteins; hence the viability of fNSCs remained unaffected.
ZV proteins reduced proliferation of human fNSCs without affecting the viability. a–e Immunocytochemistry images showing TUNEL-positive cells (red) and DAPI (blue) for nucleus in indicated groups (Empty vector, E protein, NS2B, NS4A, and NS4B) 24 h post transfection. Scale bar: 50 µm. f Bar graph shows percentage of TUNEL-positive cells over DAPI in the indicated groups. g–k Immunocytochemistry images showing Ki67-positive, proliferation marker (red) and DAPI (blue) for nucleus 24 h post transfection in indicated groups. Scale bar: 50 µm. l Bar graph shows relative percentage of Ki67-positive cells over DAPI in the indicated groups. The values represent the mean ± standard error of three independent experiments (n = 3); *p < 0.05, **p < 0.005, #statistically insignificant when compared to vector control
ZV E protein impairs proliferation of fNSCs and neurospheres
We then investigated the effect of these proteins on proliferation of fNSCs and immunostained the cells with Ki67 antibody, a well-accepted proliferation marker [43]. All experimental groups transfected with ZV proteins, except NS2A, showed significantly reduced number of Ki67-positive cells after 24 h of transfection. However, E protein demonstrated the maximum and most significant reduction in Ki67-positive fNSCs. The percentage of Ki67-positive cells in E protein, NS2A, NS4A, and NS4B experimental groups were 74.99 ± 3.31%, 83.16 ± 9.88%, 81.50 ± 7.06%, and 86.41 ± 3.71%, respectively, relative to empty vector-transfected group (p < 0.005 for E protein and p < 0.05 for NS4A and NS4B) (Fig. 1g–l). Since E protein was most potent in inducing quiescence in fNSCs, we further explored the effect of E protein on properties of fNSCs. To confirm the effect of ZV E protein on proliferation of fNSC, we performed BrdU incorporation assay [43] as BrdU labels cells in S phase. Number of cells actively involved in DNA synthesis (S phase) was significantly reduced post E protein transfection as depicted in Fig. 2a–f. Percentage of BrdU-positive cells in empty vector-transfected groups were 15.65 ± 2.12%, whereas it reduced to 5.95 ± 1.72% in E protein-transfected group (p < 0.05) (Fig. 2g). To further investigate that E protein induced quiescence in primary cultures of fNSCs, we relied upon the ability of fNSCs to form neurospheres on non-adherent surface, as neurosphere formation is an important property of neural stem cells and reflects the proliferative ability of fNSCs. In E protein group, number of small neurospheres increased (indicating decrease in proliferation) as revealed by quantitative assessment of the size of cultured neurospheres (Fig. 2h–j). Percentage of small neurospheres in E protein-transfected group was 174.68 ± 4.29% in relative to empty vector- transfected groups (p < 0.05) (Fig. 2j). This further substantiated our finding that E protein impairs fNSC proliferation.
E protein reduced cells in S phase of cell cycle in monolayer cultures and hampered growth of neurospheres. a–f Monolayer fNSCs were assessed for BrdU (red) incorporation 24 h post transfection and mounted with DAPI (blue) for nucleus. Scale bar: 20 µm. g Bar graph depicts percentage of cells positive for BrdU over DAPI in indicated groups. The values represent the mean ± standard error of three independent experiments (n = 3). h, i Bright field images of neurospheres 72 h post transfection from indicated groups. Scale bar: 500 µm. j Bar graph shows percentage of small neurospheres in indicated groups. The values represent the mean ± standard error of five independent experiments (n = 5); *p < 0.05
In utero electroporation of E protein hampers cell proliferation in developing mouse cortex
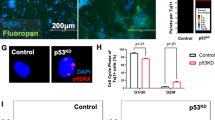
Various in vivo studies have demonstrated that ZV hampers growth of NSCs in developing brain [17,18,19,20]. To see the effect of E protein expression on the developing mouse cortex, we performed in utero electroporation in embryonic mouse cortex at E13.5 followed by harvesting at E15.5. Figure 3c depicts schematic representation of plan for in utero electroporation experiments. Lower panel of Fig. 3c shows electroporated whole mice brains showing GFP expression in empty vector and E protein groups. We carried out immunohistochemistry with anti-Ki67 antibody to assess the effect on cell proliferation. Indeed, we observed a significant decrease in percentage of GFP-positive cells that also expressed Ki67 (Fig. 3a, b, white arrowheads) in VZ and SVZ regions of cortex electroporated with pCAGIG-PrE-IRES-GFP (19.51 ± 1.96%) as compared with control construct pCAGIG-IRES-GFP (79.41 ± 2.22) (p < 0.0005) (Fig. 3d). This indicated that, on expression of E protein, there is dramatic effect on cell proliferation in VZ and SVZ of developing mouse cortex.
E protein reduced proliferating cells in developing mouse cortex. a, b E15.5 mouse cerebral cortex overexpressing control and E protein constructs. Ki67 is stained red, green is EGFP and DAPI is blue. White arrowheads showing Ki67-EGFP double-positive cells in indicated groups. Scale 50 µm. c Schematic showing plan for in utero electroporation of mice brain. Lower panel shows electroporated brains showing GFP expression in indicated groups. d Bar graph represents percentage of double-positive cells (EGFP and Ki67). The values represent the mean ± standard error of three independent experiments (n = 3); ***p < 0.0005
ZV E protein alters cell cycle dynamics of fNSCs and induces pro-neural genes
As number of cells in S-phase reduced after E protein transfection, we next determined potential impact of ZV E protein on cell cycle of fNSCs. Twenty-four hours post transfection, cells were stained and analyzed with propidium iodide for DNA content analysis using FACS [44]. E protein caused the accumulation of fNSCs in G0 phase of cell cycle with concomitant reduction in percentage of cells in S and G2 phase (Fig. 4a, b) (p < 0.05). Percentage of cells in G0/G1, S, and G2/M phase of cell cycle in E protein-expressing fNSCs were 65.48 ± 2.08%, 15.20 ± 3.8%, and 5.53 ± 1.01%, respectively, whereas in empty vector-transfected groups percentage of cells in G0/G1, S, and G2/M phase were 42.71 ± 5.81%, 24.52 ± 0.83%, and 11.84 ± 1.19%, respectively (Fig. 4c). Cell cycle analysis post transfection of other proteins viz. NS2A, NS4A, and NS4B was also performed, data shown in Supplementary Figure S2 A–F.
ZV E protein modulates cell cycle dynamics of fNSCs and induces expression of pro-neural genes. a, b Representative images of DNA content analysis by propidium iodide (PI) staining using FACS 24 h post transfection of either empty vector or E protein in human fNSCs. c Bar graph shows quantitative analysis of data obtained from PI staining depicting percentage of cells in G0/G1, S phase and G2/M phase of cell cycle in indicated groups. d Bar graph represents relative mRNA levels of pro-neural genes (PTN, ROBO2, SPOCK1, and DCX) normalized with 18S mRNA levels, 24 h post transfection of empty vector and E protein in indicated groups. The values represent the mean ± standard error of three independent experiments (n = 3); *p < 0.05
A delay in cell cycle or proliferation arrest is associated with NSC differentiation [45, 46]. Further, it is also established that ZV induces immature differentiation of NSCs in organoids [14]. Therefore, we asked whether E protein has any role in immature differentiation of fNSCs. Cells were transfected either with empty or with E protein expression vector, and were cultured in proliferation medium (with EGF and FGF). Transcript levels of pro-neural genes pleotrophin (PTN), SPOCK1, doublecortin (DCX), and ROBO2 were analyzed in fNSCs 24 h post transfection [47,48,49,50,51]. Robust increase in mRNA levels of these pro-neural genes was observed in response to E protein. Expression of PTN, ROBO2, SPOCK1, and DCX significantly increased to 195.31 ± 19.14%, 218.44 ± 25.23%, 180.40 ± 31.95%, and 158.39 ± 26.39%, respectively, with respect to empty vector-transfected group (p < 0.05) (Fig. 4d).
ZV E protein alters differentiation of fNSCs and neurospheres
During differentiation of NSCs into astrocytes and neurons, programmed cell death occur due to lineage selection process [52, 53]. As our previous result indicated premature expression of pro-neural genes in E protein-expressing fNSCs, we asked whether E protein hampers viability of differentiating fNSCs. Therefore, transfected cells were differentiated by withdrawal of mitotic factors (EGF and FGF) and addition of neuronal differentiation factors PDGF and BDNF to the media for 3 days. Percentage of TUNEL-positive cells significantly increased in E protein-expressing group as compared to empty vector-transfected group, suggesting E protein perturbed neurogenesis and induced programmed cell death in differentiating neurons. Percentage of TUNEL-positive cells over DAPI in E protein and empty vector-transfected cells post 3 days of differentiation were 22.08 ± 0.80% and 12.42 ± 1.11% (p < 0.0005) (Fig. 5a–g). To confirm this, activation of executioner caspase-3 was assessed in transfected fNSCs by western blot. As shown in Fig. 5h, i, levels of cleaved form of caspase-3 was robustly increased in E protein-expressing cells as compared to empty vector. Relative cleaved caspase3 level was 1319.56 ± 256% with respect to empty vector control (p < 0.05).
ZV E protein induces apoptosis in differentiating fNSCs and disrupts migration from neurospheres. a–f Human fNSCs were transfected either with empty vector or with E protein and were cultured in neuronal differentiation medium for 3 days; immunocytochemistry images showing TUNEL-positive cells (red) and DAPI (blue) for nucleus in indicated groups. Scale bar: 50 µm. g Bar graph shows percentage of TUNEL-positive cells over DAPI in indicated groups. The values represent the mean ± standard error of five independent experiments (n = 5). h The human fNSCs were transfected either with empty vector or with E protein and were cultured in neural differentiation medium. Western blot shows levels of caspase3 and cleaved caspase3 in differentiating fNSCs three day post transfection in indicated groups. i Bar graph shows relative levels of cleaved caspase3 in indicated groups. The values represent the mean ± standard error of three independent experiments (n = 3). j, k Neurospheres transfected either with empty vector or with E protein were cultured on adherent surface and progenitor media was replaced with neural differentiation medium. Distance migrated by differentiating human fNSCs is shown in immunocytochemistry images from indicated groups. DAPI (blue) for nucleus, tuj-1 (green) for sprouting neurons and DCX (red) for fNSCs committed to form neurons. Scale bar: 100 µm. l Bar graph represents relative percentage migration of cells from periphery of neurospheres. The values represent the mean ± standard error of four independent experiments (n = 4). *p < 0.05, **p < 0.005 and ***p < 0.0005
New neurons or astrocytes “sprout” from neurospheres, when differentiated on adherent surface [41]. We employed this in vitro model system for studying neurogenesis. The neurospheres that were cultured from fNSCs transfected either with empty vector or with E protein were seeded onto adherent surface and neuronal differentiation media was added. Three days post differentiation, migration of cells from neurospheres was assessed by staining with DCX and TUJ1 (early markers for neurogenesis and immature neurons, respectively). Migration of neurons from neurospheres was disrupted in E protein transfected neurospheres as compared to vector control (Fig. 5j–l). Quantitative measurement of distance migrated by cells showed that in E protein transfected group distance migrated by cells significantly reduced to 64.81 ± 8.49% with respect to empty vector-transfected cells (p < 0.005) (Fig. 5l). These results further suggest that ZV E protein impairs neuronal differentiation.
ZV E protein modulates miRNA profile of fNSC
To investigate the impact of E protein on fNSCs at molecular level, we employed global small RNA sequencing (small RNA-seq) (Fig. 6a). Our global miRNA analysis revealed a number of differentially expressed miRNAs in response to ZV E protein. Among differentially expressed miRNAs, we found 14 upregulated and 11 downregulated miRNAs (Fig. 6b). Expression of miRNAs is represented as log2 of transcripts per million (TPM) in heat map. Four miRNAs; hsa-mir-4745-3p, hsa-mir-765, hsa-mir676-3p, and hsa-mir-7112-3p could not be included in heat map due to zero TPM in at least one of the samples, fold change for these miRNAs are 12.5, −8.5, −12.33, and −31, respectively. We validated differentially expressed miRNAs hsa-miR-1273g-3p, hsa-miR-204-3p, hsa-miR-1306-5p, hsa-miR-6087, hsa-miR-23c, and hsa-miR-3128 using qPCR (Fig. 7a), these miRNAs have been reported in other viral infections and hence were highly relevant for the study. Expression of mir-1273g-3p, mir-204-3p, miR-1306-5p, mir-6087, mir-23c, and mir-3128 was 127.36 ± 9.53%, 208.23 ± 11.69%, 193.29 ± 21.59%, 168.29 ± 16.08%, 56.00 ± 7.89%, and 34.86 ± 14.74%, respectively (p < 0.05) with respect to empty vector-transfected group.
ZV E protein alters miRNome of fNSCs. a Schematic representation of experimental plan for global miRNA analysis. b Heatmap shows the comparison of miRNA in indicated groups. Expression of miRNAs is represented as log2 of transcripts per million (TPM). Four miRNAs; hsa-mir-4745-3p, hsa-mir-765, hsa-mir676-3p, and hsa-mir-7112-3p could not be included in the heat map due to zero TPM in at least one of the samples, fold change for these miRNAs are 12.5, −8.5, −12.33, and −31, respectively
Targets of differentially expressed miRNAs affect important biological processes. a Bar graph represents relative expression of hsa-miR-1273g-3p, hsa-miR-204-3p, hsa-miR-1306-5p, hsa-miR-6087, hsa-miR-23c, and hsa-miR-3128 normalized with U6 levels, 24 h post transfection of empty vector and E protein in indicated groups. The values represent the mean ± standard error of three independent experiments (n = 3). b Schematic representation of plan for GO analysis after obtaining differentially expressed miRNAs. c, d Targets of differentially expressed miRNAs were subjected to GO analysis. The 15 most significant and relevent terms are shown for (b) downregulated miRNA targets and (d) upregulated miRNAs targets. The −log(p) values are indicated in bar plots
Targets of differentially expressed miRNAs are involved in developmental pathways, cell cycle, and metabolic processes
GO enrichment analysis is an important tool for finding patterns in high volume data sets; it provides comprehensive understanding of high volume data sets. Figure 7b is schematic representation of plan for GO analysis of huge list of genes obtained as targets of differentially expressed miRNAs. GO enrichment analysis using targets of differentially expressed miRNAs revealed that upregulated miRNAs targets are enriched in metabolic process (−log(p) = 15.40), regulation of gene expression (−log(p) = 11.82), cellular response to stress (−log(p) = 4.44), viral transcription (−log(p) = 2.11), and viral gene expression (−log(p)= 1.69), whereas downregulated miRNAs regulate cell cycle (−log(p) = 19.75), development (−log(p) = 5.02), cell cycle regulation (−log(p) = 10.34), DNA replication (−log(p) = 5.79), neurogenesis (−log(p) = 2.55) related processes (Fig. 7c, d and Document S1). Further, Molecular pathway analysis using PANTHER pathway analysis highlighted developmental pathways as highly enriched pathways including WNT, CCKR, PDGF, EGF, p53, FGF, and NOTCH signaling pathways (Table 1 and Document S2). Thus, our study provides novel insights into the involvement of potential miRNAs in ZV induced microcephaly, and is a valuable resource to neuroscience field for further understanding the molecular mechanism of ZV induced complications in human fetal brain.
Mir-1273g-3p and mir-204-3p targets PAX3 and NOTCH2, respectively
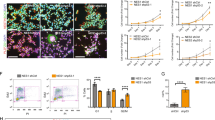
Bioinformatic (http://www.targetscan.org/) analysis shows that mir-1273g-3p and mir-204-3p posses potential binding sites on 3′UTR of PAX3 and NOTCH2, respectively (Fig. 8a, b). When we checked transcript levels of PAX3 and NOTCH2, we found significant attenuation of these genes in response to E protein (p < 0.005 for PAX3 and p < 0.05 for NOTCH2) (Fig. 8c, d). To experimentally prove that mir-1273g-3p and mir-204-3p target PAX3 and NOTCH2, respectively, we employed microRNA mimics and luciferase assays. Mimics of mir-1273g-3p and mir-204-3p were transfected in fNSCs for 24 h. Efficacy analysis through qPCR showed mir-1273g-3p and mir-204-3p levels being enhanced upto 5.02 ± 0.25 folds (p < 0.0005) and 14394.26 ± 2773.49 folds (p < 0.05), respectively, with mimics as compared with scrambled RNA transfection (Fig. 8e, f). When transcript levels of PAX3 and NOTCH2 were analyzed post mir-1273g-3p and mir-204-3p transfection, a significant decrease to 49.1 ± 6.45% (p < 0.005) and 51.6 ± 2.5% (p < 0.0005) (Fig. 8g, h), respectively, was observed as compared with scrambled transfection group.
Mir-1273g-3p and mir-204-3p are upregulated in response to E protein and target PAX3 and NOTCH2. a, b Putative binding sites of (a) mir-1273g-3p on 3′UTR of PAX3 mRNA and (b) mir-204-3p on 3′UTR of NOTCH2 mRNA. Numbers written aside PAX3 and NOTCH2 indicate starting nucleotide along the 3′UTR of mRNA. c fNSCs were transfected with E protein and empty vector for 24 h, transcript levels of PAX3 were assessed using qPCR. d fNSCs were transfected with E protein and empty vector for 24 h, transcript levels of NOTCH2 were assessed using qPCR. 18S rRNA was used as normalization internal control. Bar graph represents relative expression of NOTCH2 in indicated groups. e Bar graph showing relative levels of mir-1273g-3p representing efficacy of mir-1273g-3p mimic as compared with control mimic. f Bar graph showing relative levels of mir-204-3p representing efficacy of mir-204-3p mimic as compared with control mimic. U6 was used as normalization internal control. g Bar graph represents PAX3 transcript levels with mir-1273g-3p mimic transfection as compared with control. h Bar graph represents NOTCH2 transcript levels with mir-204-3p mimic transfection as compared with control. i, j 3′UTR of PAX3 and NOTCH2 were cloned downstream of luciferase gene in pMIR vector, and HeLa cells were co-transfected with (i) control mimic or mir-1273g-3p mimic along with pMIR vector containing 3′UTR of PAX3; (j) HeLa cells were co-transfected with control mimic or mir-204-3p mimic along with pMIR vector containing 3′UTR of NOCTH2. Bar graph shows normalized relative luciferase activity in indicated groups. Data sets have been compiled from three independent experiments (mean ± SEM); *p < 0.05, **p < 0.005 and ***p < 0.0005 with respect to control
To further validate that mir-1273g-3p and mir-204-3p are molecular targets of PAX3 and NOTCH2, respectively, 3′UTR of PAX3 and NOTCH2 were cloned downstream of luciferase gene in pMIR-Reporter vector, and luciferase activity was assessed either cotransfecting with mimic control, mir-1273g-3p or mir-204-3p mimic. Transfection was carried out for 24 h in HeLa cell line. A significant downregulation in luciferase activity was observed in case of mir-1273g-3p and mir-204-3p mimics as compared to mimic control. Luciferase activity reduced to 27.01 ± 12.41% (p < 0.05) and 47.17 ± 8.83% (p < 0.005) (Fig. 8i, j) in mir-1273g-3p or mir-204-3p mimic transfected groups, respectively, as compared to scrambled RNA transfected cells. These experiments confirmed that mir-1273g-3p and mir-204-3p target PAX3 and NOTCH2, and hence play a role in ZV pathology.
Discussion
Variations in 10 amino acids of the viral envelope (E) protein near a glycosylation site appear to be the cause of enhanced pathogenicity; however, the exact mechanism has not been explored. Here, we present experimental evidence for the virulent nature of ZV E protein with regard to the properties of fNSCs and demonstrate that E protein causes cell cycle arrest and immature differentiation of fNSCs. In quest of the mechanisms, we employed next-generation sequencing to identify miRNAs that may be dysregulated in response to E protein. The gene expression profiles have been determined by several groups following ZV infection in human and mouse neuronal cells [15, 54, 55]. Our findings suggest that E protein induces differential expression of miRNAs that are highly relevant for the alterations observed in fNSCs. Using targets of these differentially expressed miRNAs and GO analysis; cell cycle and development-related processes were found to be highly enriched biological processes by targets of DEM miRNAs. Besides this, several pathways related to growth and proliferation, including WNT, CCKR, PDGF, EGF, p53, and NOTCH, were found to be top enriched pathways. We also demonstrated that mir-1273 and mir-204 mediate their effects on the properties of fNSCs by disrupting the expression of PAX3 and NOTCH2. Moreover, our in utero electroporation experiments provide substantial evidence that E protein perturbs the proliferation of cells in the VZ and SVZ of developing mouse brain, hence providing strong evidence that E protein plays a crucial role in ZV pathology.
Flavivirus envelope (E) protein is responsible for attachment and entry into host cell. It also represents a major target for neutralizing antibodies [56]. In fact, mutations in dengue virus E protein strongly influence the pathogenicity of virus [57]. We screened four ZV proteins (three non-structural and the E protein) and demonstrated that, among other proteins, ZV E protein induces maximum quiescence in fNSCs and hampers the growth of neurospheres. Furthermore, for the first time, in this study we provide evidence that miRNA circuitry is perturbed in response to ZV envelope protein. We observed that E protein reduced the viability of differentiating fNSCs; however, E protein does not affect the viability of proliferating fNSCs. ZV E protein induced pro-neural genes in quiescent fNSCs, suggesting immature differentiation into neurons, which is consistent with recent findings that ZV induces immature differentiation of NSCs in organoids [14]. E protein has been found to be an important determinant of ZV virulence and neuroinvasion due to gain of N-linked glycosylation property [11]. Our results of cell cycle arrest, pre-mature differentiation and reduced viability during neurogenesis of fNSCs in response to E protein are in line with another report that suggested that cell cycle might be halted in ZV Mexican strain (MEX1-7) infected fNSCs [58]; the study with the Mexican strain however fell short of pinpointing the E protein. In vivo evidence further suggests that ZV infection in pregnant mice leads to cell cycle arrest and inhibits neural progenitor proliferation in mouse embryos [19, 20]. Further, ZV induces apoptosis in iPSC-derived NSCs and disrupts growth of neurospheres and brain organoids [14, 15]. In our study, defects in migration from neurospheres could have been due to excessive cell death as observed in monolayer cultures, or because of inhibition of mTOR pathway as demonstrated by Liang et al. [16]. These studies with ZV infection are important discoveries; however, to date, none of the reports on ZV could (1) identify the viral protein responsible for attenuation in proliferation in fNSCs, and (2) attribute ZV effects to specific miRNAs regulating the stemness of fNSCs, making this study timely and important for understanding the pathobiology of ZV and a major advancement in the field.
miRNAs are considered to be fine regulators of gene expression and play an important role in flavivirus pathogenesis [25, 59, 60]. For example, in Japanese encephalitis virus, miR-33a-5p modulates virus replication by targeting host eukaryotic translation elongation factor 1A1 [60]. miRNA may also act as a serum biomarker, as in the case of dengue virus infection [61]. Knowledge of the impact of ZV infection on miRNome is crucial to further advance our understanding of ZV pathogenesis. Till date, only one study has demonstrated alterations in miRNome during the course of ZV infection, which again is not in fNCSs but in astrocytes [55]. In our study, we found 25 miRNAs significantly altered in response to E protein in fNSCs. Coincidentally, some of these miRNAs have also been associated with other viral infections. For example, mir-1273g-3p modulates activation and apoptosis of hepatic stellate cells by directly targeting PTEN in hepatitis C virus-related liver fibrosis [62]. Further, miR-204 reduces hepatitis B virus (HBV) pregenomic RNA encapsidation and capsid assembly [63]. Our data showed a novel involvement of mir-1273g-3p and mir-204-3p in ZV pathology, as these miRNAs are upregulated in response to ZV E protein and directly target important developmental genes, i.e. PAX3 and NOTCH2, respectively.
Bioinformatic analysis using targets of DEM suggest enrichment of cell cycle and development-related processes. Besides them, WNT, CCKR, PDGF, EGF, p53, FGF, and notch signaling pathways were observed as highly rated pathways affected by these miRNAs. This study provides a molecular mechanism for ZV causing neurodevelopmental defects in fNSCs. ZV upon infection might dysregulate crucial miRNAs, which regulate genes involved in development processes. On cessation of proliferation fNSCs tend to prematurely differentiate into neurons, which are usually accompanied by enhanced apoptosis, culminating in disrupted cell homeostasis and perhaps leading to microcephaly, as shown in some animal studies with ZV. While our study provides novel insights into the involvement of potential miRNAs in ZV-induced pathogenesis in primary cultures of human fNSCs, further studies to probe the consequences of dysregulation of specific miRNAs reported by us, using autopsy samples of ZV-infected human fetal samples and more in utero electroporation studies in animal models, will be helpful.
Accession number
GEO accession is GSE108791.
References
Heukelbach J, Werneck GL. Surveillance of Zika virus infection and microcephaly in Brazil. Lancet. 2016;388:846–7.
Cugola FR, Fernandes IR, Russo FB, Freitas BC, Dias JLM, Guimarães KP, et al. The Brazilian Zika virus strain causes birth defects in experimental models. Nature. 2016;534:267–71.
Organization (WHO) World Health. WHO | Zika situation report. http://www.who.int/emergencies/zika-virus/situation-report/10-march-2017/en/; 2017.
Chimelli L, Melo ASO, Avvad-Portari E, Wiley CA, Camacho AHS, Lopes VS, et al. The spectrum of neuropathological changes associated with congenital Zika virus infection. Acta Neuropathol. 2017;133:983–99.
Stegmann BJ, Carey JC. TORCH infections. toxoplasmosis, other (syphilis, varicella-zoster, parvovirus B19), rubella, cytomegalovirus (CMV), and herpes infections. Curr Women’s Health Rep. 2002;2:253–8.
Relich RF, Loeffelholz M. Zika virus: an explosive pandemic and a new TORCH agent. Clin Lab Med. 2017;37:253–67.
Calvet G, Aguiar RS, Melo ASO, Sampaio SA, de Filippis I, Fabri A, et al. Detection and sequencing of Zika virus from amniotic fluid of fetuses with microcephaly in Brazil: a case study. Lancet Infect Dis. 2016;16:653–60.
Veroni C, Marnetto F, Granieri L, Bertolotto A, Ballerini C, Repice AM, et al. Immune and Epstein-Barr virus gene expression in cerebrospinal fluid and peripheral blood mononuclear cells from patients with relapsing-remitting multiple sclerosis. J Neuroinflamm. 2015;12. https://doi.org/s12985-016-0541-3/s12974-015-0353-1.
Eskild A, Bruu A-L, Stray-Pedersen B, Jenum P. Epstein-Barr virus infection during pregnancy and the risk of adverse pregnancy outcome. BJOG. 2005;112:1620–4.
Kuno G, Chang GJJ. Full-length sequencing and genomic characterization of Bagaza, Kedougou, and Zika viruses. Arch Virol. 2007;152:687–96.
Annamalai AS, Pattnaik A, Sahoo BR, Muthukrishnan E, Natarajan SK, Steffen D, Vu HLX, Delhon G, Osorio FA, Petro TM, Xiang S-H, Pattnaik AK. 2017. Zika virus encoding nonglycosylated envelope protein is attenuated and defective in neuroinvasion. J Virol 91:e01348-17. https://doi.org/10.1128/JVI.01348-17.
De Filippis L, Binda E. Concise review: self-renewal in the central nervous system: neural stem cells from embryo to adult. Stem Cells Transl Med. 2012;1:298–308.
Farkas LM, Huttner WB. The cell biology of neural stem and progenitor cells and its significance for their proliferation versus differentiation during mammalian brain development. Curr Opin Cell Biol. 2008;20:707–15.
Dang J, Tiwari SK, Lichinchi G, Qin Y, Patil VS, Eroshkin AM, et al. Zika virus depletes neural progenitors in human cerebral organoids through activation of the innate immune receptor TLR3. Cell Stem Cell. 2016;19:258–65.
Tang H, Hammack C, Ogden SC, Wen Z, Qian X, Li Y, et al. Zika virus infects human cortical neural progenitors and attenuates their growth. Cell Stem Cell. 2016;18:587–90.
Liang Q, Luo Z, Zeng J, Chen W, Foo SS, Lee SA, et al. Zika virus NS4A and NS4B proteins deregulate Akt-mTOR signaling in human fetal neural stem cells to inhibit neurogenesis and induce autophagy. Cell Stem Cell. 2016;19:663–71.
Lazear HM, Govero J, Smith AM, Platt DJ, Fernandez E, Miner JJ, et al. A mouse model of zika virus pathogenesis. Cell Host Microbe. 2016;19:1–11.
Shao Q, Herrlinger S, Zhu Y-N, Yang M, Goodfellow F, Stice SL, et al. The African Zika virus MR-766 is more virulent and causes more severe brain damage than current Asian lineage and dengue virus. Development. 2017;144:4114–24.
Li C, Xu D, Ye Q, Shi L, Qin C-F, Xu Correspondence Z, et al. Zika virus disrupts neural progenitor development and leads to microcephaly in mice cell stem cell Zika virus disrupts neural progenitor development and leads to microcephaly in mice. Cell Stem Cell. 2016;19:120–6.
Wu KY, Zuo GL, Li XF, Ye Q, Deng YQ, Huang XY, et al. Vertical transmission of Zika virus targeting the radial glial cells affects cortex development of offspring mice. Cell Res. 2016;26:645–54.
Lai EC. Micro RNAs are complementary to 3′UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet. 2002;30:363–4.
Adams BD, Anastasiadou E, Esteller M, He L, Slack FJ. The inescapable influence of noncoding RNAs in cancer. Cancer Res. 2015;75:5206–10.
Somel M, Liu X, Tang L, Yan Z, Hu H, Guo S et al.MicroRNA-driven developmental remodeling in the brain distinguishes humans from other primates. PLoS Biol. 2011; 9: e1001214. https://doi.org/10.1371/journal.pbio.1001214.
Fineberg SK, Kosik KS, Davidson BL. MicroRNAs potentiate neural development. Neuron. 2009;64:303–9.
Aloia AL, Abraham AM, Bonder CS, Pitson SM, Carr JM. Dengue virus induced inflammation of the endothelium and the potential roles of sphingosine kinase-1 and MicroRNAs. Mediat Inflamm. 2015;2015. https://doi.org/10.1155/2015/509306.
Kloosterman WP, Plasterk RHa. The Diverse Functions of MicroRNAs in Animal Development and Disease. Dev Cell. 2006;11:441–50.
Kim AH, Reimers M, Maher B, Williamson V, McMichael O, McClay JL, et al. MicroRNA expression profiling in the prefrontal cortex of individuals affected with schizophrenia and bipolar disorders. Schizophr Res. 2010;124:183–91.
Carrington JC, Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301:336–8.
Bavia L, Mosimann ALP, Aoki MN, Duarte Dos Santos CN. A glance at subgenomic flavivirus RNAs and microRNAs in flavivirus infections. Virol J. 2016;13:84. https://doi.org/10.1186/s12985-016-0541-3.
Rebay I, Fleming RJ, Fehon RG, Cherbas L, Cherbas P, Artavanis-Tsakonas S. Specific EGF repeats of Notch mediate interactions with Delta and serrate: Implications for notch as a multifunctional receptor. Cell. 1991;67:687–99.
Artavanis-Tsakonas S. Notch Signaling: Cell Fate Control and Signal Integration in Development. Science. 1999;284:770–6.
Solecki DJ, Liu XL, Tomoda T, Fang Y, Hatten ME. Activated Notch2 signaling inhibits differentiation of cerebellar granule neuron precursors by maintaining proliferation. Neuron. 2001;31:557–68.
Zou Y, Fang F, Ding Y-J, Dai M-Y, Yi X, Chen C, et al. Notch 2 signaling contributes to cell growth, anti-apoptosis and metastasis in laryngeal squamous cell carcinoma. Mol Med Rep. 2016;14:3517–24.
Tchorz JS, Tome M, Cloëtta D, et al. Constitutive Notch2 signaling in neural stem cells promotes tumorigenic features and astroglial lineage entry. Cell Death & Disease. 2012;3:e325. doi:10.1038/cddis.2012.65.
Riccio O, van Gijn ME, Bezdek AC, Pellegrinet L, van Es JH, Zimber-Strobl U, et al. Loss of intestinal crypt progenitor cells owing to inactivation of both Notch1 and Notch2 is accompanied by derepression of CDK inhibitors p27Kip1 and p57Kip2. EMBO Rep. 2008;9:377–83.
Sakata-Yanagimoto M., Chiba S. (2015) Notch2 Signaling in Mast Cell Development and Distribution in the Intestine. In: Hughes M., McNagny K. (eds) Mast Cells. Methods in Molecular Biology (Methods and Protocols), vol 1220. Humana Press, New York, NY. https://doi.org/10.1007/978-1-4939-1568-2_6.
Mishra M, Taneja M, Malik S, Khalique H, Seth P. Human immunodeficiency virus type 1 Tat modulates proliferation and differentiation of human neural precursor cells: implication in NeuroAIDS. J Neurovirol. 2010;16:355–67.
Fatima M, Kumari R, Schwamborn JC, Mahadevan A, Shankar SK, Raja R, et al. Tripartite containing motif 32 modulates proliferation of human neural precursor cells in HIV-1 neurodegeneration. Cell Death Differ. 2016;23:776–86.
Saxena M, Agnihotri N, Sen J. Perturbation of canonical and non-canonical BMP signaling affects migration, polarity and dendritogenesis of mouse cortical neurons. Development. 2018;145:dev147157.
Jacques TS, Relvas JB, Nishimura S, Pytela R, Edwards GM, Streuli CH, et al. Neural precursor cell chain migration and division are regulated through different beta1 integrins. Development. 1998;125:3167–77.
Ishido M, Suzuki J. Quantitative Analyses of Inhibitory Effects of Bisphenol A on Neural Stem-cell Migration Using a Neurosphere Assay in vitro. J Heal Sci. 2010;56:175–81.
Mishra M, Vetrivel S, Siddappa NB, Ranga U, Seth P. Clade-specific differences in neurotoxicity of human immunodeficiency virus-1 B and C Tat of human neurons: Significance of dicysteine C30C31 motif. Ann Neurol. 2008;63:366–76.
Kee N, Sivalingam S, Boonstra R, Wojtowicz JM. The utility of Ki-67 and BrdU as proliferative markers of adult neurogenesis. J Neurosci Methods. 2002;115:97–105.
Malik S, Saha R, Seth P. Involvement of extracellular signal-regulated kinase (ERK1/2)-p53-p21 axis in mediating neural stem/progenitor cell cycle arrest in co-morbid HIV-drug abuse exposure. J Neuroimmune Pharmacol. 2014;9:340–53.
Ohnuma SI, Harris WA. Neurogenesis and the cell cycle. Neuron. 2003;40:199–208.
Pitto L, Cremisi F. Timing neurogenesis by cell cycle? Cell Cycle. 2010;9:434–5.
Jung C-G, Hida H, Nakahira K, Ikenaka K, Kim H-J, Nishino H. Pleiotrophin mRNA is highly expressed in neural stem (progenitor) cells of mouse ventral mesencephalon and the product promotes production of dopaminergic neurons from embryonic stem cell-derived nestin-positive cells. FASEB J. 2004;18:1237–9.
Garcia-Gutierrez P, Juarez-Vicente F, Wolgemuth DJ, Garcia-Dominguez M. Pleiotrophin antagonizes Brd2 during neuronal differentiation. Journal of cell science 2014;127:2554-64.
Pei Y, Brun SN, Markant SL, Lento W, Gibson P, Taketo MM, et al. WNT signaling increases proliferation and impairs differentiation of stem cells in the developing cerebellum. Development. 2012;139:1724–33.
Blockus H, Chédotal A, Ackerman KG, Greer JJ, Alther TA, Domanitskaya E, et al. Slit-Robo signaling. Development. 2016;143:3037–44.
Gleeson JG, Peter TL, Flanagan LA, Walsh CA. Doublecortin is a microtubule-associated protein and is expressed widely by migrating neurons. Neuron. 1999;23:257–71.
Yamaguchi Y, Miura M. Programmed Cell Death and Caspase Functions During Neural Development. Current topics in developmental biology 2015;114:159-84.
Gross CG. Neurogenesis in the adult brain: death of a dogma. Nat Rev Neurosci. 2000;1:67–73.
Nowakowski TJ, Pollen AA, Di Lullo E, Sandoval-Espinosa C, Bershteyn M, Kriegstein AR. Expression analysis highlights AXL as a candidate zika virus entry receptor in neural stem cells. Cell Stem Cell. 2016;18:591–6.
Kozak RA, Majer A, Biondi MJ, Medina SJ, Goneau LW, Sajesh BV et al. MicroRNA and mRNA dysregulation in astrocytes infected with Zika virus.Viruses 2017; 9:297. https://doi.org/10.3390/v9100297.
Oliphant T, Engle M, Nybakken GE, Doane C, Johnson S, Huang L, et al. Development of a humanized monoclonal antibody with therapeutic potential against West Nile virus. Nat Med. 2005;11:522–30.
dos Santos CND, Frenkiel MP, Courageot MP, Rocha CFS, Vazeille-Falcoz MC, Wien MW, et al. Determinants in the envelope E protein and viral RNA helicase NS3 that influence the induction of apoptosis in response to infection with dengue type 1 virus. Virology. 2000;274:292–308.
McGrath EL, Rossi SL, Gao J, Widen SG, Grant AC, Dunn TJ, et al. Differential Responses of Human Fetal Brain Neural Stem Cells to Zika Virus Infection. Stem Cell Rep. 2017;8:715–27.
Thounaojam MC, Kundu K, Kaushik DK, Swaroop S, Mahadevan A, Shankar SK, et al. MicroRNA 155 regulates Japanese encephalitis virus-induced inflammatory response by targeting Src homology 2-containing inositol phosphatase 1. J Virol. 2014;88:4798–810.
Chen Z, Ye J, Ashraf U, Li Y, Wei S, Wan S, et al. miR-33a-5p modulates Japanese Encephalitis Virus Replication by targeting Eukaryotic Translation Elongation Factor 1A1. J Virol. 2016;90:73722-34
Ouyang X, Jiang X, Gu D, Zhang Y, Kong SK, Jiang C, et al. Dysregulated serum miRNA profile and promising biomarkers in dengue-infected patients. Int J Med Sci. 2016;13:195–205.
Niu X, Fu N, Du J, Wang R, Wang Y, Zhao S, et al. miR-1273g-3p modulates activation and apoptosis of hepatic stellate cells by directly targeting PTEN in HCV-related liver fibrosis. FEBS Lett. 2016;590:2709–24.
Huang JY, Chen HL, Shih C. MicroRNA MIR-204 and MIR-1236 inhibit hepatitis B virus replication via two different mechanisms. Sci Rep. 2016;6. https://doi.org/10.1038/srep34740.
Acknowledgements
Technical assistance from Mr. DL Meena and Mr. Naushad Alam of NBRC, India, is acknowledged. Ms. Bindu Singh and Ms. Priyanka S Kshtriya helped in initial assays. Junior and Senior Research Fellowship to Ms. Reshma Bhagat from CSIR, New Delhi, India, is greatly acknowledged. Expression vectors for the four ZV proteins were kind gift from Dr. Shyamala Mani, IISc, India (These were possible due to financial support from CEFIPRA (4903-02) awarded to P. Gressens (INSERM) and S. Mani). Financial support for the study from NBRC core funds to PS is greatly acknowledged. The authors also wish to acknowledge the support of the facilities provided under the Biotechnology Information System Network (BTISNET) grant, DBT, India, and Distributed Information Centre at NBRC, Manesar, India.
Author contributions
RB designed and performed the experiments, analyzed the data, and wrote the manuscript. BP did the bioinformatics analysis of small RNA sequencing data. SN did qPCR experiments for revised version. NA performed and analyzed the in utero electroporation experiments. JS designed, analyzed and interpreted the in utero electroporation experiments. YKA provided inputs and helped in writing the manuscript. SM designed the vectors, provided the initial inputs, PS designed the study, helped in analyzing and interpretation of data and approved the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Bhagat, R., Prajapati, B., Narwal, S. et al. Zika virus E protein alters the properties of human fetal neural stem cells by modulating microRNA circuitry. Cell Death Differ 25, 1837–1854 (2018). https://doi.org/10.1038/s41418-018-0163-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-018-0163-y
This article is cited by
-
Hypoxia Induces Early Neurogenesis in Human Fetal Neural Stem Cells by Activating the WNT Pathway
Molecular Neurobiology (2023)
-
Interplay Between Zika Virus-Induced Autophagy and Neural Stem Cell Fate Determination
Molecular Neurobiology (2023)
-
Identification of novel lncRNA by reanalysis of RNA-seq data in Zika Virus Infected hiNPCs
VirusDisease (2022)
-
Involvement of host microRNAs in flavivirus-induced neuropathology: An update
Journal of Biosciences (2022)