Abstract
The progression of localized breast cancer to distant metastasis results in a poor prognosis and a high mortality rate. In this study, the contributions of miRNAs to tumor progression and the regulatory mechanisms leading to their expression alterations were investigated. Using highly lung-metastatic sub-lines from parental breast cancer cells, miRNA expression profiling revealed that the miR-17-92 cluster is significantly downregulated and the miR-18a-5p is the most evidently decreased. Ectopic expression and inhibition of miR-18a-5p demonstrated its capacity in suppressing migration and invasion of breast cancer cells. Further research identified sterol regulatory element binding transcription protein 1 (SREBP1), the master transcription factor that controls lipid metabolism, as a candidate target of miR-18a-5p. SREBP1 is overexpressed and strongly associated with worse clinical outcomes in breast cancer. Functionally SREBP1 promotes growth and metastasis of breast cancer both in vitro and in vivo. To unravel the underlying mechanism of SREBP1-mediated metastasis, mRNA profiling and subsequent gene set enrichment analyses (GSEA) were performed and SREBP1 was demonstrated to be significantly associated with epithelial-mesenchymal transition (EMT). Furthermore, SREBP1-mediated repression of E-cadherin was found to be deacetylation dependent and was augmented by recruiting Snail/HDAC1/2 repressor complex. In the light of these data, we propose that reduced expression of miR-18a-5p and concomitant overexpression of SREBP1 lead to induction of EMT states that in turn, promote breast cancer progression and metastasis. Taken together, our study reveals the crucial role of miR-18a-5p and SREBP1 in the EMT and metastasis, thus providing promising drug targets for tailored therapy in the advanced breast cancer setting.
Similar content being viewed by others
Introduction
Breast cancer is the second leading cause of cancer death among women [1, 2]. The majority of breast cancer patients have operable tumors; however, cancer metastasis rather than the primary tumor imperils patients’ lives by reducing the chance of long-term survival from 90 to 5% [3]. Epithelial-mesenchymal transition (EMT) is regarded as the first and foremost step of cancer metastasis [4]. Although plenty of EMT and metastasis-related genes have been described, the underlying mechanisms of metastatic spread are yet largely unexplored. Therefore, identifying functional molecules involved in cancer metastasis that could effectively be targeted for breast cancer treatment is an important area of investigation.
MicroRNAs (miRNAs) are small non-coding RNA molecules, which regulate multiple central mechanisms of tumorigenesis and metastasis [5, 6]. Amongst the numerous identified miRNAs, the miR-17-92 has been identified to be involved in multiple molecular mechanisms of tumorigenesis and metastasis and is located in the third intron of the open reading frame 13 (C13orf25), encompassing six individual miRNAs (miR-17-5p, miR-18a-5p, miR-19a-3p, miR-19b-3p, miR-20a-5p, and miR-92a-3p) [7]. Recently, miR-17-92 has been reported to represent distinctive expression via different oncogenic molecular mechanisms or pathways in a variety of cancer types [8,9,10]. The miR-17-92 was first designated as oncomir-1 considering its oncogenic role in hematopoietic malignancies [11]. However, increasing evidence suggests its role as a tumor suppressor in solid tumors. For instance, loss of heterozygosity at chromosome 13q31, where human miR-17-92 is mapped, was detected in approximately 25% of human breast tumors [12].
In this study, highly lung-metastatic sub-lines from parental breast cancer cells was utilized. We demonstrated that miR-18a-5p, the relatively understudied member of miR-17-92, was the most decreased miRNAs in the derived cell lines. Furthermore, we verified sterol regulatory element binding transcription protein 1 (SREBP1) as the direct target of miR-18a-5p, which is the main regulator in lipid metabolism [13]. Previous studies have demonstrated that aberrant expression of SREBP1 is not only reported in some metabolic diseases [14], but also is closely related to cancer [15]. In this study, we observed that depletion of SREBP1 in breast cancer cell lines was sufficient to decrease the metastatic ability of cancer cells by interfering with the EMT program. All these data indicate that reduced expression of miR-18a-5p and concomitant overexpression of SREBP1 may be critical components required in the complex process of distant metastasis in breast cancer. Thus, this study further elucidates the potential mechanisms underlying metastasis, and promisingly provides novel targets for therapeutic strategies.
Results
In vivo selection of breast cancer cells with highly metastatic potential
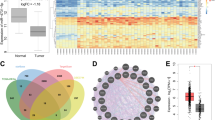
In vivo selection provides an effective way to isolate highly metastatic sub-lines from the original mixture [16, 17]. We generated highly metastatic breast cancer sub-lines using MDA-MB-231EGFP-fLUC cells as a starting point (Fig. 1a). Briefly, MDA-MB-231EGFP-fLUC cells were inoculated into the tail vein of nude mice to form lung metastasis. Eight weeks later, the mice were sacrificed and the lung tissues were subjected to primary tissue culture to obtain the first-round sub-line LM-R1EGFP-fLUC. After expansion and flow sorting, LM-R1EGFP-fLUC cells were re-inoculated into mice to generate the second-round sub-line LM-R2EGFP-fLUC (Fig. 1b). Transwell assay results showed the metastatic ability of LM-R1EGFP-fLUC and LM-R2EGFP-fLUC cells increased dramatically through two rounds of in vivo serial selection when compared with the parental cells (Fig. 1c).
In vivo selection of highly lung-metastatic breast cancer cells identifies miR-18a-5p as candidate miRNA inhibiting breast cancer invasiveness. a Flowchart delineating in vivo selection procedures of highly lung-metastatic breast cancer cells. Parental MDA-MB-231EGFP-Fluc was injected into the Balb/c female mice from tail vein to form lung metastasis. Eight weeks later, the first round sub-line LM-R1EGFP-fLUC was harvested, cultured, sorted by flow-cytometry and reinoculated into mice to obtain the second-round sub-line LM-R2EGFP-fLUC. b Representative fluorescent and phase contrast images of LM-R1EGFP-fLUC. Scale bar: 20 μm. c Transwell assay showed the migration and invasion ability of MDA-MB-231EGFP-Fluc increases in succession during the in vivo selection (LM-R2EGFP-fLUC > LM-R1EGFP-fLUC > MDA-MB-231EGFP-Fluc). Representative areas of invaded cells stained with crystal violet (n = 3). Scale bar: 20 μm. d Expression heatmap of differential miRNAs profile of MDA-MB-231 EGFP-fLUC, LM-R1EGFP-fLUC, and LM-R2EGFP-fLUC. e The expression of six members in miR-17-92 cluster detected by qPCR is coherent to that in microarray (n = 3). f Transwell assay showed that miR-18a-5p impaired the migration and invasion ability of MDA-MB-231 EGFP-fLUC, LM-R1EGFP-fLUC, and LM-R2EGFP-fLUC. (n = 3). Left panel, representative areas of invaded cells stained with crystal violet; right panel, relative quantitation of invaded cells of three random fields. Scale bar: 20 μm. g Transwell assay showed that miR-18a-5p mimics impaired the migration and invasion ability of MDA-MB-231 and MCF-7 while miR-18a-5p inhibitors improved the metastatic effect (n = 3). Scale bar: 20 μm. *P < 0.05, **P < 0.01
MiR-18a-5p was identified as candidate miRNA inhibiting breast cancer invasiveness
We then compared the miRNA expression profile of the two derivative sub-lines with parental cells using a custom miRNA microarray [18]. Amongst the 1105 miRNAs detected, 29 miRNAs in LM-R1EGFP-fLUC cells while 38 miRNAs in LM-R2EGFP-fLUC cells were found to be significantly down-regulated compared to the parental cells. It is noteworthy that the most significantly down-regulated miRNAs in the lung-metastatic sub-lines were members from miR-17-92 clusters (Fig. 1d). In accordance with our previous study [19], three miRNAs (miR-30a-5p, miR-30d-5p, and miR-30e-5p) from the miR-30 family were down-regulated significantly in LM-R2EGFP-fLUC cells when compared to MDA-MB-231 cells, which independently affirmed the reliability of the microarray data.
Next, qPCR was performed to validate the expression of the six members from miR-17-92 clusters. As is shown in Fig. 1e, the expressions of the six miRNAs were all down-regulated in LM-R1EGFP-fLUC or LM-R2EGFP-fLUC cells compared with MDA-MB-231EGFP-fLUC cells. Notably, miR-18a-5p was the most significantly down-regulated miRNAs among the six members, emphasizing its potential role in human breast cancer metastasis. Transwell assays showed the significant suppression of miR-18-5p mimics on the migration and invasion in MDA-MB-231EGFP-fLUC, LM-R1EGFP-fLUC, and LM-R2EGFP-fLUC cells (Fig. 1f). To further confirm the findings, miR-18a-5p mimics and inhibitors were transfected into MDA-MB-231 and MCF-7 cells and the transfection efficiency was verified by qPCR (Supplemental_Fig_S1A). As shown in Fig. 1g, overexpression of miR-18a-5p significantly impaired the migration and invasion in both cell lines. Furthermore, when titrating the amount of miR-18a mimics to more physiological levels, the suppressive effect of miR-18a-5p mimics on migration and invasion of MDA-MB-231 cells was still significant (Supplemental_Fig_S1B). On the contrary, inhibition of miR-18a-5p with the oligonucleotide inhibitor markedly enhanced the migration and invasiveness in both cells. Taken together, these results strongly suggest that miR-18a-5p may play a key role in the metastasis of breast cancer cells.
MiR-18a-5p suppressed migration and invasion by directly targeting SREBP1
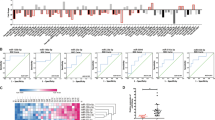
To identify the candidate targets of miR-18a-5p (Fig. 2a), the mRNA microarray and bioinformatic strategy were applied. Assuming a high probability that direct targets of miR-18a-5p were included in the up-regulated gene list, we compared the mRNA expression profile of the two derivative sub-lines with the parental cells. Next, we analyzed the target genes of miR-18a-5p shared by two widely used bioinformatics algorithms: miRWalk and MicroCosm. Finally, four predicted targets of miR-18a-5p meet all these above requirements, including SREBP1, PIK3C2A, RBBP8, and SLC30A7. Promisingly, SREBP1 was markedly down-regulated after miR-18a-5p mimics transfection (Fig. 2b), indicating SREBP1 may be a downstream target of miR-18a-5p. We further found the SREBP1 mRNA level was gradually elevated during the in vivo selection course (Fig. 2c). Then, we found transfection of miR-18a-5p inhibitors could upregulate SREBP1 expression in both cell lines (Supplemental_Fig_S2A, 2B).
MiR-18a-5p targets SREBP1 and suppresses migration and invasion. a Venn diagrams represent the mutual candidate target genes of miR-18a-5p identified by mRNA microarray, miRWalk, and MicroCosm. b The expression of SREBP1 is inhibited most evidently in the four candidate targets (SREBP1, PIK3C2A, RBBP8, and SLC30A7) by qPCR after treatment of miR-18a-5p mimics (n = 3). c The mRNA expression of SREBP1 in MDA-MB-231, LM-R1, and LM-R2 (n = 3). d The alignment of predicted binding sites in SREBP1 3′UTR for miR-18a-5p from eight organisms. The structure of corresponding substitution sequence in the mutant vector. Mut mutated. e Luciferase reporter assays in MDA-MB-231 cells after co-transfection of SREBP1 wild-type or mutated 3′UTR with miR-18a-5p mimics increasing from 0 to 100 nM (n = 3). WT wild-type, MUT mutated. f Luciferase reporter assays after co-transfection of SREBP1 wild-type 3′UTR with miR-18a mimics ± inhibitors in MDA-MB-231 cells (n = 3). g Transwell assay showed restoration of SREBP1 with transient transfection method could partially abrogate miR-18a-5p-induced suppression of migration and invasion in both MDA-MB-231 and MCF-7. Scale bar: 20 μm. h Migration and invasion of the above cells were quantitatively analyzed (n = 3). *P < 0.05, **P < 0.01
The mRNA: miRNA alignment analysis demonstrated the evolutionary conservation of the binding sites for miR-18a-5p (Fig. 2d). We then cloned sequence of SREBP1 3′UTR which contains the putative binding site of miR-18a-5p for the luciferase assay. Meanwhile, a corresponding mutant vector was also constructed. The luciferase activity of wide-type SREBP1 3′UTR vector was significantly decreased when MDA-MB-231 cells were transfected with miR-18a-5p mimics in a dose gradient manner (Fig. 2e). Conversely, the reduction of luciferase activity was abolished when the mutant SREBP1 3′UTR vector was co-transfected with miR-18a-5p mimics. We observed the consistent phenomenon in MCF-7 cells (Supplemental_Fig_S2C). Furthermore, introducing with the miR-18a-5p inhibitors effectively restored the luciferase activity of wide-type SREBP1 3′UTR when it was co-transfected with miR-18a-5p mimics (Fig. 2f). Collectively, these data demonstrate that SREBP1 is a direct target of miR-18a-5p.
Furthermore, we restored SREBP1 expression in the presence of miR-18a-5p mimics and adopted qPCR to detect the SREBP1 mRNA after the cells were manipulated accordingly (Supplemental_Fig_S3A). The ‘rescue’ experiment showed restoration of SREBP1 could partially abrogated miR-18a-5p-induced suppression of migration and invasion in both cells (Fig. 2g, h). Furthermore, the concomitant knockdown of SREBP1 and miR-18a-5p inhibition abrogated the pro-metastatic effect of miR-18a inhibition (Supplemental_Fig_S3B, 3C), hence supporting the hypothesis that SREBP1 is a direct and functional target of miR-18a-5p.
SREBP1 expression is upregulated and associated with survival in breast cancer
To assess the potential clinical relevance of our observations, we detected SREBP1 expression in breast normal tissues and cancer tissues by IHC (Fig. 3a). The staining intensity of SREBP1 was found to be significantly elevated in tumor tissues compared with normal tissues. Interestingly, the staining distribution was also different, with nuclear SREBP1 being negligible and most of the SREBP1 located in cytoplasm in normal tissues, whereas nuclear SREBP1 staining significantly intensified with higher grades of breast tumor tissues, indicating the increased SREBP1 expression and activation (nuclear translocation) may correlate with de-differentiation of breast tumors. Furthermore, analysis of mRNA from breast tumor tissues and matched normal tissues consistently demonstrated that SREBP1 was upregulated in a majority of tumor samples examined (18/29) when compared to paired normal samples (Student’s t-test, P = 0.0471) (Fig. 3b).
SREBP1 expression is upregulated in breast cancer and indicated poor overall survival. a Representative images of immunohistochemistry staining of SREBP1 expression in breast normal tissues and cancer tissues. IDC II Invasive ductal carcinoma, grade II, IDC III Invasive ductal carcinoma, grade III. b The qPCR of SREBP1c mRNA expression in breast tumor tissues and paired normal tissues (n = 30 pairs). c Representative immunohistochemistry images of the intensity of the nuclear and cytoplasmic staining of SREBP1 in 329 human breast tumor tissues on the tissue microarrays. d Kaplan–Meier survival analysis of overall survival and cause specific survival in patients with high or low SREBP1 expression, from the aspect of nucleus, cytoplasm and total scoring of SREBP1.
IHC staining analysis of SREBP1 expression on tissue microarrays (TMA) containing 329 breast cancer samples (Fig. 3c) showed overexpression of SREBP1 was correlated to HER2 status of breast tumors (Supplementary Table 2). Furthermore, patients with high nuclear SREBP1 expression had worse overall survival (OS) and cause specific survival (CSS) (Fig. 3d). However, the cytoplasmic staining of SREBP1 failed to identify the patients with poor OS and CSS. Combination of nuclear and cytoplasmic analysis approximated the result of the nuclear analysis. Univariate Cox survival analysis also indicated that high SREBP1 expression is strongly associated with worse clinical outcomes (Supplementary Table 3). When all the other factors were considered, the multivariate Cox analysis still showed that the survival was significantly lower in patients with high SREBP1 expression (Supplementary Table 4), suggesting that SREBP1 may serve as an independently prognostic factor in breast cancer.
SREBP1 promotes cell growth and invasiveness in vitro and in vivo
To investigate the role of SREBP1 in breast cancer metastatic processes, stable knockdown and overexpression cells were established. Among four shRNAs targeting at SREBP1 mRNA, we adopted sh2 and sh4 in the subsequent experiments (Supplemental_Fig_S4A). The stable transfection efficiency was then validated with qPCR and western blot (Fig. 4a, Supplemental_Fig_S4B, S4C). Subsequently, IF analysis showed that both shRNA constructs significantly reduced the staining of SREBP1 (red), indicating the active form of SREBP1 could be diminished by shRNA silence (Fig. 4b). Further detection of SCD1 expression level showed knockdown of SREBP1 significantly reduced SCD1 expression, supporting that SREBP1 is a prerequisite to its downstream lipogenic gene expression (Supplemental_Fig_S4D, S4E).
SREBP1 promotes cell growth and invasiveness in vitro and in vivo. a Validation of the efficiency in stably knockdown (left panel) and overexpression (right panel) MDA-MB-231 and MCF-7 cell lines by western blot. b Immunofluorescence analyses showed the active form of SREBP1 locating in the nucleus is diminished by shRNA silencing in both MDA-MB-231 and MCF-7 cell lines. Scale bar: 20 μm. c Transwell migration and invasion assays of MDA-MB-231 and MCF -7 cells after stable transfection with SREBP1 knockdown or overexpression vector (n = 3). Representative areas of invaded cells stained with crystal violet were displayed and invaded cells of three random fields were quantitatively analyzed. Scale bar: 20 μm. d Images of the xenograft tumors in nude mice implanted with the MDA-MB-231 control cells and stable SREBP1 knockdown cells (1 × 107) (n = 6 in each group). e Tumor growth curves of SREBP1 knockdown or control MDA-MB-231 cells in nude mice (n = 6 in each group). The volume of tumors was assessed in indicated days. f Tumor weight of nude mice implanted with the control and stable SREBP1 knockdown cells was measured at the completion of the experiment (n = 3). g Representative xenograft tumors stained for Ki67 and TUNEL. Scale bar: 20 μm. h Lung metastasis was diminished with SREBP1 knockdown (n = 7 in each group). Upper panel: whole-lung images of mice after injection of SREBP1 knockdown or control MDA-MB-231 cells; lower panel: representative hematoxylin and eosin staining of lung-metastatic tumors are shown. i Quantification of metastasis nodules at the lung surface after orthotopic injection of SREBP1 knockdown or control MDA-MB-231 cells (n = 7 in each group). j Survival of mice injected with SREBP1 knockdown or control MDA-MB-231 cells (n = 6 in each group). *P < 0.05, **P < 0.01
The impact of SREBP1 on cell proliferation and oncogenic potential was investigated. The depletion of SREBP1 suppressed both MDA-MB-231 and MCF-7 proliferation significantly (Supplemental_Fig_S5A). We then observed an incremental effect on cancer cell proliferation after overexpressing SREBP1. Similarly, a remarkable inhibitory effect on colony formation was observed when SREBP1 was silenced (Supplemental_Fig_S5B). Transwell assays were employed to detect the migration and invasion ability of breast cancer cells. As shown in Fig. 4c, knockdown of SREBP1 could potentially suppress migration and invasion of MDA-MB-231 and MCF-7 cells, while overexpression of SREBP1 could enhance these invasive abilities. All these data support the hypothesis that SREBP1 has an intensive effect on growth and invasion of breast cancer cells.
Investigation of the role of SREBP1 in tumor growth in vivo showed that the knockdown of SREBP1 could significantly inhibited tumor volume, rate of tumor growth, and tumor weight (Fig. 4d–f). Further analysis of these harvested xenografts revealed that knockdown of SREBP1 significantly decreased proliferation and increased apoptosis (Fig. 4g) as indicated with staining of Ki67 and TUNEL, respectively. We also investigated the function of SREBP1 in breast cancer lung metastasis via intravenous injection of MDA-MB-231 cells. Concordant with the in vitro observations, silencing of SREBP1 alleviated the lung colonization of breast cancer cells in nude mice (Fig. 4h, i). In addition, inoculation of MDA-MB-231 cells with SREBP1 knockdown effectively prolonged the survival time (Fig. 4j). Together, these data demonstrated a functional role of SREBP1 in promoting breast cancer growth and metastasis both in vitro and in vivo.
SREBP1 promotes cell metastasis via inducing EMT
Microarray analysis was adopted to further elucidate the underlying mechanisms of SREBP1-mediated metastasis (Fig. 5a). We performed gene set enrichment analysis (GSEA) to evaluate the global transcriptomic variations associated with SREBP1. Using the hallmarks of cancer-related gene signatures, we found that the SREBP1-enriched cells were significantly associated with EMT gene list (Fig. 5b). In both breast cancer cell lines, silencing of SREBP1 aroused a pronounced elevation of E-cadherin while a decrement of N-cadherin and Fibronection (Fig. 5c–e). We also observed down-regulation of EMT-related transcription factors such as Snail, Zeb1, and Twist after silencing of SREBP1.
SREBP1 promotes cell metastasis via inducing EMT. a Microarray-based transcription profiling of differential gene expression in stable SREBP1 knockdown and the control breast cells. b Gene set enrichment analysis showed that SREBP1-enriched cells were significantly associated with EMT gene lists in both MDA-MB-231 and MCF-7 cells. c Immunofluorescence analyses of E-Cadherin, N-Cadherin, and Fibronectin expression in MDA-MB-231 and MCF-7 cells transfected with control vector or shSREBP1 vector. DAPI was used to indicate the nuclear location. Scale bar: 20 μm. d The mRNA level of the EMT-related markers in MDA-MB-231 and MCF-7 cells transfected with the control vector and the shSREBP vector (n = 3). e Western blot results of EMT-related markers in MDA-MB-231 and MCF-7 cells are shown after SREBP1 was knockdown. *P < 0.05, **P < 0.01
Next, we investigated whether SREBP1 expression significantly correlated with EMT markers in human breast tumor tissue (Fig. 6a). The results revealed that high mRNA expression of SREBP1 was significantly correlated with low E-cadherin, high N-cadherin, and Fibronection expression. Furthermore, the expression patterns of these EMT markers in SREBP1 knockdown cells was additionally validated by IHC analysis using xenograft tumors previously mentioned (Fig. 6b). We further examined whether SREBP1 mRNA expression could predict outcome when combined with E-cadherin mRNA expression (Fig. 6c). As anticipated, patients with high E-cadherin mRNA and low SREBP1 mRNA expression experienced better OS and CSS compared with patients with low E-cadherin mRNA and high SREBP1 mRNA expression.
SREBP1 expression is significantly correlated with EMT markers in clinical samples. a The correlation between SREBP1 expression and EMT-related markers (E-Cadherin, N-Cadherin, and Fibronectin) was investigated by qPCR methods (n = 3). The defined “High” and “Low” expression levels of SREBP1 were stratified by the median expression level. b The expression of SREBP1, E-Cadherin, N-Cadherin, and Fibronectin in controls and shSREBP1 xenograft tumors was stained by immunohistochemistry. Scale bar: 20 μm. c Kaplan–Meier survival analysis of overall survival and cause specific survival indicated that SREBP1 mRNA may predict survival outcomes in combination with E-cadherin mRNA in breast cancer patients. *P < 0.05, **P < 0.01
SREBP1 regulates EMT by forming a co-repressor complex with HDAC1/2 and Snail to suppress E-cadherin
Up-regulation of E-cadherin, the gatekeeper of EMT and metastasis, is a hallmark of cells with SREBP1 knockdown. Therefore, we investigated the mechanisms accounted for the regulation of E-cadherin by SREBP1. Epigenetic regulation is an explanation for the alteration of E-cadherin expression. To validate this, we targeted the epigenetic machinery in a series of breast cancer cell lines with either the demethylation agent decitabine or the HDAC inhibitor Trichostatin A (TSA). Decitabine treatment did not result in a significant elevation in E-cadherin expression (Supplemental_Fig_S6A), while treatment with TSA markedly up-regulated E-cadherin mRNA expression in a dose-dependent manner (Supplemental_Fig_S6B). Assessing whether deacetylation machinery contributes to the silencing of E-cadherin by SREBP1 was further evaluated by two experimental designs. Firstly, SREBP1 was overexpressed for 48 h and then followed by the treatment with TSA. As shown in Fig. 7a, overexpression of SREBP1 significantly downregulated E-cadherin expression, while treatment of TSA restored the expression of E-cadherin. Secondly, we reversed the procedure by pretreating cells with TSA for 48 h before introducing SREBP1 overexpression vector (Fig. 7b). These results revealed overexpression of SREBP1 impaired the increment of E-cadherin triggered by TSA treatment. Taken together, these data strongly suggest that silencing of E-cadherin by SREBP1 results from deacetylation machinery.
SREBP1 regulates EMT by forming a co-repressor complex with HDAC1/2 and Snail to suppress E-cadherin. a The mRNA expression of E-cadherin expression was detected by qPCR when MDA-MB-231 was transfected with SREBP1 overexpression vector for 48 h and followed by the treatment with 0, 100, and 300 nM TSA (n = 3). b The mRNA expression of E-cadherin expression was detected by qPCR when MDA-MB-231 was firstly treated with 0, 100, and 300 nM TSA for 48 h and then transfected with SREBP1 overexpression vector later (n = 3). c Luciferase reporter assays was used to determine transcriptionally suppressive effect of SREBP1 on E-cadherin promoter in the presence of Snail or not (n = 3). d The physical interaction of SREBP1 with Snail and HDAC1/2 was observed by the co-IP assay. e Luciferase reporter assays validated that Snail/HDAC1/2 contribute to the suppressive effect of SREBP1 on E-Cadherin expression (n = 3). f ChIP-qPCR analyses confirmed the recruitment of SREBP1/Snail/HDAC1/2 to the E-cadherin promoter. Silencing of SREBP1 did not affect the recruitment of Snail, HDAC1, and HDAC2 to the E-Cadherin promoter (left panel), while silencing of snail significantly dissociated Snail, SREBP1, and HDAC1/2 from E-cadherin promoter (middle panel). Concurrent knockdown of HDAC1 and HDAC2 did not impair the recruitment of Snail as well as SREBP1 to the promoter region (right panel). g Proposed schematic representation depicting the mechanism of miR-18a-5p targeting SREBP1 to suppress SNAIL/HDAC1/2-induced EMT and metastasis of breast cancer. *P < 0.05, **P < 0.01
SREBP1 has been predicted to be one of the transcriptional factors in the promoter region of E-cadherin (Supplemental_Fig_S7A); nevertheless, all published studies suggest that SREBP1 requires additional co-regulatory factors to function properly [20]. Snail, the principal transcriptional repressor of E-cadherin [21], was predicted to interact with SREBP1 along with the deacetylation enzymes HDAC1/2 (Supplemental_Fig_S7B). To validate this, we applied luciferase assay to determine the transcriptional activity of SREBP1 on E-cadherin promoter in the presence of Snail or not (Fig. 7c). The promoter activity of E-cadherin was suppressed by SREBP1 overexpression alone in a dose-dependent pattern, and the suppressive effect was substantially enhanced by co-transfection of Snail vector. Next, we observed the physical interaction of SREBP1 with Snail and HDAC1/2 in the co-immunoprecipitation assay (Fig. 7d, Supplemental_Fig_S8A). Additionally, we involved luciferase assay to verify the requirement of Snail and HDAC1/2 in the suppressive function of SREBP1 toward E-cadherin promoter (Fig. 7e). Consistently, the suppressive effect of SREBP1 on the E-cadherin promoter was significantly reinforced in the presence of Snail, and was further augmented by introducing either HDAC1 or HDAC2. Subsequently, we performed ChIP to confirm the recruitment of SREBP1, Snail and HDAC1/2 to the E-cadherin promoter. As shown in Fig. 7f, the recruitment of Snail and HDAC1/2 to the E-cadherin promoter was not affected by SREBP1 depletion. Nevertheless, silencing of Snail significantly dissociated SREBP1 and HDAC1/2 from E-cadherin promoter, suggesting that Snail may be the essential element in the complex. Additionally, concurrent knockdown of HDAC1 and HDAC2 did not impair the recruitment of Snail as well as SREBP1 to the promoter region. The knockdown efficiency of each siRNA was showed in Supplemental_Fig_S8B. Furthermore, we applied IF staining to detect the E-cadherin expression upon knockdown of SREBP1, Snail and SREBP1 + Snail. We found that concurrent knockdown of SREBP1 and Snail did not exert additive effect on E-cadherin expression when compared with knockdown of SREBP1 or Snail alone (Supplemental_Fig_S9). Overall, these findings demonstrate that a Snail/HDAC1/2 containing repressor complex plays an important role in SREBP1-mediated E-cadherin silencing during EMT process of breast cancer cells (Fig. 7g).
Discussion
Emerging evidence has highlighted the fundamental role of miRNAs in the progression of cancer [22, 23]. The miR-17-92 is one of the best characterized miRNAs, whose aberrant expression is observed in a variety of tumor types [24, 25]. Our results suggested downregulation of miR-17-92 is an acquired trait associated with distant metastasis of breast cancer, which are concordant with previously findings [26, 27]. While many studies argue for the oncogenic role of miR-17-92 cluster in different cancer types [7], this discrepancy may be related to the uniquely polycistronic structure of miR-17-92 [28]. Among the members of miR-17-92, limited studies have been published on the specific functionality of miR-18a-5p in human breast cancer. With microarray and bio-informatic strategies, we reduced the predictive targets of miR-18a-5p to four candidate genes, amongst which SREBP1 was verified to be the target of miR-18a-5p. It is notable that in the rescue experiment SREBP1 expression could be suppressed by miR-18a-5p mimics in the absence of 3′UTR of SREBP1, indicating an alternative regulation of SREBP1 by miR-18a-5p. Reconsidering the sequence complementarity between miR-18a-5p and SREBP1 regardless of the location revealed a 6-mer binding site located in the coding sequence of SREBP1. We speculated that miR-18a-5p might regulate SREBP1 expression via targeting the coding sequence besides 3′UTR as reported in other studies [29]. Nevertheless, whether this interpretation could account for the results needs further study.
SREBP1 is a major transcription factor directly activating the key and rate-limiting enzymes in cholesterol and fatty acid [30, 31]. It is noteworthy that reprogramming of cellular metabolism is known to be a hallmark of cancer [32] and lipid metabolism is very often disrupted in cancer cells to fuel the biosynthetic demands for malignant behaviors. Many investigations have reported the increased expression of SREBP1 is associated with a worse prognosis in a variety of solid tumors [15, 33,34,35]. Similarly, we found that patients with high SREBP1 expression had worse survival outcomes and may function as a prognostic indicator in breast cancer, which are in accordance with the previously reported study [36]. Furthermore, the histopathological concentration of SREBP1 in cells during pathological mitosis was intriguing (Supplemental_Fig_S10), suggesting a latent role of SREBP1 in cell mitotic program. Several studies have demonstrated that mature form of SREBP1 is hyperphosphorylated to avoid degradation and maintain stabilized during mitosis [37, 38], thereby providing a stable amount of active transcription factors to support lipogenesis and cell division [39].
Many attentions have been paid on the role of SREBP1 in tumor cell proliferation and apoptosis [15, 34, 40,41,42]. Nevertheless, little is known about the underlying mechanism of SREBP1 in breast cancer metastasis. In this study, we found that SREBP1 prompted breast cancer cell migration and invasion both in vitro and in vivo. Though SREBP1 has been reported to be involved in the processes of invasion and metastasis in human liver cancer [43], ovarian cancer [42], and endometrial cancer [15], the molecular mechanism has not been clearly delineated. Our findings suggest the involvement of EMT in SREBP1-mediated metastatic program and overexpression of E-cadherin has been reported to correlate with downregulation of SREBP1. In our study, the prognostic assessment revealed that patients with high E-cadherin and low SREBP1 expression had favorable survival compared with patients with low E-cadherin and high SREBP1 expression.
Our data demonstrates that the upregulation of E-cadherin by SREBP1 knockdown is deacetylation dependent. As a critical transcriptional regulator of cholesterol and fatty acid metabolism, SREBP1 has been reported to occupy the E-box motifs on the promoters of numerous target genes by ChIP-chip assay [44]. To date, all the studies have shown that SREBP1 itself is a very weak transcriptional regulator and requires co-regulators to maximize the effect [45]. Snail, the key transcriptional factor of EMT, was unveiled to mediate the repression of E-cadherin by recruiting a co-repressor complex containing HDAC1/2 and Sin3A [46]. Other groups have also reported the suppression of E-cadherin by the Snail/HDAC1/2 complex for the regulation of metastasis in nasopharyngeal carcinoma and pancreatic cancer [47, 48]. Likewise, our results verified that Snail was required for the suppression of E-cadherin along with HDAC1/2. Alternatively, YY1 was identified as one of the common binding partners of SREBP1 [49] and has been reported to interact with HDAC3 to exert inhibitory effect on E-cadherin promoter [50]. However, we failed to find any evidence to suggest the binding of SREBP1 at the YY1 binding sites on the E-cadherin promoter region, indicating that Snail, but not YY1, was responsible for the suppressive role of SREBP1 toward E-cadherin. However, our data are limited when interpreting whether these components in the complex interact in a direct or indirect fashion, thus further study is needed.
In summary, we reported that miR-18a-5p could inhibit lung metastasis of breast cancer by directly targeting SREBP1, which could form a co-repressor complex with Snail and HDAC1/2 to modulate EMT. All these findings underscore the significant role of miR-18a-5p as well as SREBP1 in the progression and metastasis of breast cancer, and also provide a specific target for the development of novel therapeutic strategies for invasive and metastatic breast cancer patients in the future.
Methods
Cell lines and cell transfection
Breast cancer cell lines MCF-7, MDA-MB-231 as well as the Human Embryonic Kidney (HEK) 293T were obtained from American Type Culture Collection (ATCC), and were cultured in DMEM/high glucose medium (Hyclone) supplemented with 10% FBS (Clark), 100 U/ml penicillin, and 100 μg/ml streptomycin (Millipore). All the cells were cultured at 37 °C and 5% CO2. The cell lines were characterized by Genetic Testing Biotechnology Corporation (Suzhou, China) using short tandem repeat (STR) markers. Transient transfection was performed with lipofectamine 2000 regent (Invitrogen). To generate stable cell lines, the lenti-viral vector expressing shRNA was introduced into HEK293T cells by transient transfection. After 6 h, the cell culture medium was replaced and cells were allowed to grow for 36 h to produce virus. The supernatant was then collected and filtered through a 0.45-μm filter. Cells were infected at approximately 70% confluence in complete medium supplemented with 8 μg/ml of polybrene (Sigma), followed by puromycin (Sigma) selection of the infected cells. The knockdown efficiency was determined by either western blot or qPCR assays.
Primary culture of sub-lines from lung metastasis
To generate experimental lung metastasis, 5 × 105 MDA-MB-231EGFP-fLUC cells were injected into the lateral tail veins of 4–5 weeks-old BALB/c nu/nu female mice. Eight weeks later, the mice were sacrificed and the lungs were extracted. To establish the lung-metastatic sub-line, lungs were dissected, rinsed in PBS, digested with 1 mg/ml collagenase type I (Sigma) + hyaluronidase (Sigma) at 37 °C for 2 h, lysed in red blood cell lysis buffer (Sigma) at 4 °C for 15 min, washed for three times with PBS and plated in 6 cm disk containing DMEM medium supplemented with 10% FBS, 100 U/ml penicillin, 100 μg/ml streptomycin and 10 mg/ml Amphotericin B (Sigma). Adherent cells were then expanded and sorted based on positive selection with flow cytometer (BD Biosciences). Cells with EGFP positive was designated the first-round LM-R1EGFP-fLUC sub-line. The same experimental procedures were used to generate second-round LM-R2EGFP-fLUC sub-line derived from mice inoculated with LM-R1EGFP-fLUC cells. All animal experiments were performed with the approval of Shandong University Animal Care and Use Committee.
Plasmids, antibodies and reagents
A set of pLKO.1 shRNAs targeting human SREBF1 were a gift from Wang [15]. For 3′UTR reporter assays, a 1.1 kb fragment of SREBF1 3′UTR was amplified from genomic DNA of MDA-MB-231 cells and subcloned into XhoI/XbaI of pmiRGLO vector (Promega). The miR-18a-5p seed sequence was further mutated to 5′-CGAGCUUA-3′. For promoter reporter assays, a 2 kb fragment upstream of the CDH1 gene was synthetized and subcloned into pGL3-Basic vector (GenePharma). All constructs were confirmed by sequencing. The pEnter-SREBF1, pEnter-HDAC1, pEnter-HDAC2 vector (Vigene Bioscience), and pReceiver-SNAIL (GeneCopoeia) were all commercially obtained. The SREBP1 antibody (Proteintech, Cat. 14088-1-AP), E-cadherin antibody (Proteintech, Cat. 20874-1-AP), N-cadherin antibody (Immunoway, Cat. YT2988), FN1 antibody (Proteintech, Cat. 15613-1-AP), Snail antibody (Proteintech, Cat.13099-1-AP), Zeb1 antibody (CST, Cat. 3396S), VIM antibody (CST, Cat. 5741S), Twist antibody (CST, Cat. 46702S), Slug antibody (CST, Cat. 9585S), HDAC1 (Proteintech, Cat. 10197-1-AP), and HDAC2 (Proteintech, Cat. 12922-3-AP) were applied in the study. The miR-18a-5p mimics/inhibitor and the negative control (NC) were purchased from GenePharma. SREBF1 siRNA (5′-CUCCUGCUUGAGUUUCUGGTT-3′), SNAIL siRNA pool (5′-GCAAAUACU GCAACAAGGATT-3′,’-GCUUGGGCCAAGUGCCCAATT-3′), HDAC1 siRNA (5′-GCAGAUGCAGAGAUUCAACTT-3′), HDAC2 siRNA (5′-GCAUCAGGAUU CUGUUACGTT-3′), and NC (5′-UUCUCCGAACGUGUCACGUTT-3′) were synthesized from GenePharma. Decitabine (Cat. S1200) and TSA (Cat.S1045) was obtained from Selleck. All other chemicals and reagents were from Solarbio and Sigma unless otherwise indicated.
RNA isolation and qPCR assay
Total RNA was extracted from cultured cells with TRIzol reagents (Invitrogen) as per the manufacturer’s protocol. Total RNA was reverse-transcribed to cDNA using PrimeScript RT reagent Kit (Takara), which was reverse transcribed to miRNA with Mir-X miRNA First-Strand Synthesis Kit (Takara). Real-time PCR was performed with the SYBR Premix Ex Taq (Tli RNaseH Plus) (Takara) with Light Cycler 480 II Real-Time PCR System (Roche). GAPDH was used as the endogenous control for detection of mRNA expression level, while U6 were used as endogenous control for miRNA expression analysis. Relative quantification analysis was performed using the comparative C (T) (2(−ΔΔCT)) method. Primer information used in the study can be found in Supplementary Table 1.
Western blotting and immunoprecipitation (IP) assay
Western blotting analysis was performed as described previously [51]. Cells were collected and lysed with Western and IP lysis buffer (Beyotime) with protease inhibitors. Equal amounts of proteins was loaded on SDS-PAGE gels and then transferred to a PVDF membrane (Millipore). After blocking with 5% non-fat milk, the membrane was incubated overnight at 4 °C with the primary antibody and then with horseradish peroxydase-coupled secondary antibody (Millipore). Signal was detected with enhanced chemiluminescence (Millipore). For IP, cells were harvested at 48 h post-transfection and lysed in NP40 lysis buffer supplemented with protease inhibitor cocktail (Roche) for 30 min on ice. Indicated primary antibody and control IgG were added to the lysate separately and incubated on a rotator at 4 °C for 1 h. Afterwards, 20 μl protein A/G-agarose beads (Santa Cruz) were added and incubated at 4 °C overnight on a rotator. Immunoprecipitates were examined using the indicated primary antibodies in the same way as the immunoblotting assay.
Immunofluorescence (IF), immunohistochemistry (IHC), and TUNEL
For IF, cells cultivated on glass cover slides were fixed in 4% paraformaldehyde for 10 min, permeabilized in 0.2% Triton X-100 for 20 min, and blocked in 100% FBS for 1 h. Chromatin was stained by DAPI (Invitrogen). Specimens were covered with ProLong Gold Antifade (Invitrogen). As a NC, all staining were performed without primary antibody. The IHC method to study altered protein expression was performed as previously described [52]. In summary, paraffin-embedded tissue slides were de-paraffinized with xylene, rehydrated through a graded alcohol series and retrieved in 10 mM sodium citrate buffer (pH 6.0). After cooling down to room temperature, the slides were incubated with 3% hydrogen peroxide, followed by incubation with normal serum to block nonspecific binding. The slides were then incubated with the primary antibody overnight at 4 °C in a humidified chamber. The secondary antibody was from PV9000 IHC reagent kit (Zhongshan Biotechnology). Sections were stained with diaminobenzidine (DAB) and counterstained with hematoxylin. All of staining was assessed by two pathologists blinded to the origination of the samples using a semi-quantitative method. Each specimen was assigned a score according to the intensity and percentage of the nucleic and cytoplasmic staining. TUNEL assay was conducted using the in situ Cell Death Detection Kit (Roche).
Cell viability and clonogenic assay
Cell proliferation was assessed by the 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) assay. Breast cancer cells were plated in 96-well plates (2 × 103 per well). After incubation at 37 °C for 1–5 days, 20 μl of MTT (5 mg/ml in PBS) was added to each well, and cells were incubated for another 4 h at 37 °C. The supernatants were carefully removed and 100 μl of dimethyl sulfoxide (DMSO) was added to each well. The plates were gently shaken and the absorbance values were measured at 550 nm with a Microplate Reader (Perkin Elmer). To determine clonogenic ability, cells were plated in triplicate at a density of 8 × 102 per 6 cm disk and were allowed to grow for 14 days to form colonies, which were then stained with 2% crystal violet (Sigma).
Migration and invasion assay
Migration and invasion assays were performed as described previously [53] using transwell system (BD Biosciences). The invasion assay was performed using the same protocol as the migration assay, except that the membrane was coated with Matrigel (Corning). Briefly, the polycarbonate membranes were coated with 40 μL matrigel at 37 °C for 2 h to form a reconstituted basement membrane. Seven hundred microliters of the medium with 20% FBS was added to the lower well of each chamber and 1 × 105 of cells resuspended in the serum-free medium were added to the upper inserts. The total number of cells adhering to the lower surface of the membrane was acquired in six representative fields.
In vivo tumorigenesis and metastasis assay
The in vivo tumorigenesis and metastasis assay was performed as described previously [19]. Briefly, breast cancer cells (1 × 107 cells in 100 µl of PBS: Matrigel (1:3, v/v)) were injected subcutaneously into each flank of 4–5 weeks-old BALB/c nu/nu female mice. Tumor growth rate was monitored by measuring tumor diameters every 4 days and the tumor growth curve was recorded accordingly. Both maximum (L) and minimum (W) length of the tumor were measured using a slide caliper, and the tumor volume was calculated as ½ LW2. Once the mice were sacrificed, tumors were collected and weighed. To produce experimental lung metastasis, 5 × 105 cells were injected into the lateral tail veins of 4–5 weeks-old BALB/c nu/nu female mice. All the mice were killed under anesthesia. The lungs were collected and fixed in 10% formalin. For tissue morphology evaluation, hematoxylin and eosin (HE) staining was performed on sections from embedded samples.
Dual luciferase reporter assay
The 3′UTR of SREBP1 containing one putative miR-18a-5p binding sites was amplified and cloned into pmirGLO vector separately (Promega). The miR-18a-5p complementary sites with the sequence GCACCUUA in SREBP1 3′UTR were mutated singly to remove complementarity to miR-18a-5p as described previously [54]. Cells cultured in 96-well plates were co-transfected with 100 ng of wide-type or mutated SREBP1 3′UTR constructs, and NC or miR-18a-5p mimics. The human CDH1 gene promoter segment covering from ‒2000 bp to +58 bp was cloned into the pGL3-basic vector (Promega), which was designated as pGL3 -2000 vector. The vector of pEnter-SREBP1 with or without pReceiver-SNAIL, pEnter-HDAC1, and pEnter-HDAC2 vector was co-transfected with pGL3 -2000 and pRL-TK vector. Luciferase activity was measured with the dual-luciferase reporter assay system (Promega).
Chromatin immunoprecipitation (ChIP) assay
ChIP assay was performed with EZ-ChIP Kit (Millipore) according to the manufacturer’s protocol. To be brief, MCF-7 cells were transfected with pEnter-SREBP1 or pEnter vector before ChIP assay. Forty-eight hours after transfection, cells were cross-linked with 1% formaldehyde and terminated after 5 min by addition of glycine. Cells were harvested with SDS lysis buffer and were sheared by sonication cycles to generate DNA fragments with an average size of 200–1000 bp for qChIP. Preclearing and incubation with antibody or IgG control overnight was performed, followed by incubation with protein-A/G beads for 2 h at 4 °C. Washing and reversal of crosslinking was performed. The immunoprecipitated DNA was extracted with Phenol-chloroform followed by ethanol precipitation. Purified DNA was analyzed by quantitative real-time PCR (qPCR) and the enrichment was expressed as fold enrichment compared with IgG.
Microarray hybridization and data analysis
Total RNA from MDA-MB-231EGFP-fLuc, LM-R1EGFP-fLUC, and LM-R2EGFP-fLUC cells was subjected to hybridization to Affymetrix U133 plus 2.0 arrays, performed by Shanghai Gene Company. The total RNA from MDA-MB-231 and MCF-7 cells with SREBP1 shRNA or control vector was subjected to hybridization to Affymetrix GeneChip Array performed by Shanghai Genminix Company. As for the miRNA microarray for MDA-MB-231EGFP-fLuc, LM-R1EGFP-fLUC, and LM-R2EGFP-fLUC cells, all of the human mature miRNAs in the miRBase database (Release 13.0) were used for designing probes for constructing the in-house miRNA microarray and a total of 1105 probes have been successfully designed according to the principle proposed by Wang [55]. The microarray was fabricated and hybridized as described previously [56]. All microarray data were normalized according to the median intensity of each sample. The microarray data have been deposited in Gene Expression Omnibus of the National Center for Biotechnology Information (GSE106144).
Patients and tissue samples
A paraffin-embedded, archived breast cancer tissue microarray with 329 patients had been clinically and histopathologically diagnosed at Qilu hospital of Shandong University was subjected to IHC of SREBP1. Furthermore, the infiltrating carcinoma tissues were collected from January 2007 to December 2016 in Qilu hospital at the time of surgery and cDNA of these tissues were stored in −80 °C until use. A total of 74 patients were involved to study the association between SREBP1 and E-cadherin/N-cadherin/Fibronectin mRNA expression as well as the survival for SREBP1 and E-cadherin combined analysis.
Statistics
Unless stated otherwise, data are presented as mean ± SD in the figures. The software SPSS V22.0 was used for statistical analysis. Student’s t-test and one-way ANOVA analysis were used to determine the significance of two groups and multiple groups respectively. Clinicalpathological characteristics were compared between groups using Pearson’s Chi-square test. Survival curves were generated using the Kaplan–Meier method, and differences between curves were analyzed with log-rank test. Univariate and multivariate Cox proportional hazard model was applied to estimate the association of covariates with OS and disease-specific survival (DSS). Differences with P < 0.05 were considered significant.
Study approval
For all participants in this study, written informed consent was obtained from participants prior to inclusion in the study as delineated by the protocol that was approved by the Ethics Committee on Scientific Research of Shandong University Qilu Hospital. All animal experiments were performed with the approval of the Ethics Committee on Scientific Research of Shandong University Qilu Hospital.
References
DeSantis CE, Fedewa SA, Goding Sauer A, Kramer JL, Smith RA, Jemal A. Breast cancer statistics, 2015: convergence of incidence rates between black and white women. CA Cancer J Clin. 2016;66:31–42.
Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7–30.
Greenberg PA, Hortobagyi GN, Smith TL, Ziegler LD, Frye DK, Buzdar AU. Long-term follow-up of patients with complete remission following combination chemotherapy for metastatic breast cancer. J Clin Oncol. 1996;14:2197–205.
Gupta GP, Massague J. Cancer metastasis: building a framework. Cell . 2006;127:679–95.
Jafri MA, Al-Qahtani MH, Shay JW. Role of miRNAs in human cancer metastasis: implications for therapeutic intervention. Semin Cancer Biol. 2017;44:117–31.
Valinezhad Orang A, Safaralizadeh R, Kazemzadeh-Bavili M. Mechanisms of miRNA-mediated gene regulation from common downregulation to mRNA-specific upregulation. Int J Genom. 2014;2014:970607.
Mogilyansky E, Rigoutsos I. The miR-17/92 cluster: a comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 2013;20:1603–14.
Sand M, Hessam S, Amur S, Skrygan M, Bromba M, Stockfleth E, et al. Expression of oncogenic miR-17-92 and tumor suppressive miR-143-145 clusters in basal cell carcinoma and cutaneous squamous cell carcinoma. J Dermatol Sci. 2017;86:142–8.
Liu XS, Chopp M, Wang XL, Zhang L, Hozeska-Solgot A, Tang T, et al. MicroRNA-17-92 cluster mediates the proliferation and survival of neural progenitor cells after stroke. J Biol Chem. 2013;288:12478–88.
Hsu TI, Hsu CH, Lee KH, Lin JT, Chen CS, Chang KC, et al. MicroRNA-18a is elevated in prostate cancer and promotes tumorigenesis through suppressing STK4 in vitro and in vivo. Oncogenesis. 2014;3:e99.
He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–33.
Eiriksdottir G, Johannesdottir G, Ingvarsson S, Bjornsdottir IB, Jonasson JG, Agnarsson BA, et al. Mapping loss of heterozygosity at chromosome 13q: loss at 13q12-q13 is associated with breast tumour progression and poor prognosis. Eur J Cancer. 1998;34:2076–81.
Shimano H, Sato R. SREBP-regulated lipid metabolism: convergent physiology - divergent pathophysiology. Nat Rev Endocrinol. 2017;13:710–30.
Ruiz R, Jideonwo V, Ahn M, Surendran S, Tagliabracci VS, Hou Y, et al. Sterol regulatory element-binding protein-1 (SREBP-1) is required to regulate glycogen synthesis and gluconeogenic gene expression in mouse liver. J Biol Chem. 2014;289:5510–7.
Li W, Tai Y, Zhou J, Gu W, Bai Z, Zhou T, et al. Repression of endometrial tumor growth by targeting SREBP1 and lipogenesis. Cell Cycle. 2012;11:2348–58.
Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordon-Cardo C, et al. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3:537–49.
Clark EA, Golub TR, Lander ES, Hynes RO. Genomic analysis of metastasis reveals an essential role for RhoC. Nature. 2000;406:532–5.
Wei R, Huang GL, Zhang MY, Li BK, Zhang HZ, Shi M, et al. Clinical significance and prognostic value of microRNA expression signatures in hepatocellular carcinoma. Clin Cancer Res. 2013;19:4780–91.
Zhang N, Wang X, Huo Q, Sun M, Cai C, Liu Z, et al. MicroRNA-30a suppresses breast tumor growth and metastasis by targeting metadherin. Oncogene. 2014;33:3119–28.
Dooley KA, Millinder S, Osborne TF. Sterol regulation of 3-hydroxy-3-methylglutaryl-coenzyme A synthase gene through a direct interaction between sterol regulatory element binding protein and the trimeric CCAAT-binding factor/nuclear factor Y. J Biol Chem. 1998;273:1349–56.
Goossens S, Vandamme N, Van Vlierberghe P, Berx G. EMT transcription factors in cancer development re-evaluated: beyond EMT and MET. Biochim Biophys Acta. 2017;1868:584–91.
Geiger TR, Peeper DS. Metastasis mechanisms. Biochim Biophys Acta. 2009;1796:293–308.
Bertoli G, Cava C, Castiglioni I. MicroRNAs: new biomarkers for diagnosis, prognosis, therapy prediction and therapeutic tools for breast cancer. Theranostics. 2015;5:1122–43.
Olive V, Li Q, He L. mir-17-92: a polycistronic oncomir with pleiotropic functions. Immunol Rev. 2013;253:158–66.
Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133:217–22.
Krutilina R, Sun W, Sethuraman A, Brown M, Seagroves TN, Pfeffer LM, et al. MicroRNA-18a inhibits hypoxia-inducible factor 1alpha activity and lung metastasis in basal breast cancers. Breast Cancer Res. 2014;16:R78.
Yu Z, Willmarth NE, Zhou J, Katiyar S, Wang M, Liu Y, et al. microRNA 17/20 inhibits cellular invasion and tumor metastasis in breast cancer by heterotypic signaling. Proc Natl Acad Sci USA. 2010;107:8231–6.
Olive V, Jiang I, He L. mir-17-92, a cluster of miRNAs in the midst of the cancer network. Int J Biochem Cell Biol. 2010;42:1348–54.
Hamdorf M, Idica A, Zisoulis DG, Gamelin L, Martin C, Sanders KJ, et al. miR-128 represses L1 retrotransposition by binding directly to L1 RNA. Nat Struct Mol Biol. 2015;22:824–31.
Menendez JA, Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat Rev Cancer. 2007;7:763–77.
Shao W, Espenshade PJ. Expanding roles for SREBP in metabolism. Cell Metab. 2012;16:414–9.
Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell . 2011;144:646–74.
Huang WC, Li X, Liu J, Lin J, Chung LW. Activation of androgen receptor, lipogenesis, and oxidative stress converged by SREBP-1 is responsible for regulating growth and progression of prostate cancer cells. Mol Cancer Res. 2012;10:133–42.
Lee JH, Jeon YG, Lee KH, Lee HW, Park J, Jang H, et al. RNF20 suppresses tumorigenesis by inhibiting SREBP1c-PTTG1 axis in kidney cancer. Mol Cell Biol. 2017;37:e00265–17.
Li N, Zhou ZS, Shen Y, Xu J, Miao HH, Xiong Y, et al. Inhibition of the sterol regulatory element-binding protein pathway suppresses hepatocellular carcinoma by repressing inflammation in mice. Hepatology. 2017;65:1936–47.
Bao J, Zhu L, Zhu Q, Su J, Liu M, Huang W. SREBP-1 is an independent prognostic marker and promotes invasion and migration in breast cancer. Oncol Lett. 2016;12:2409–16.
Bengoechea-Alonso M, Ericsson J. The phosphorylation-dependent regulation of nuclear SREBP1 during mitosis links lipid metabolism and cell growth. Cell Cycle. 2016;15:2753–65.
Bengoechea-Alonso MT, Punga T, Ericsson J. Hyperphosphorylation regulates the activity of SREBP1 during mitosis. Proc Natl Acad Sci USA. 2005;102:11681–6.
Bengoechea-Alonso MT, Ericsson J. Cdk1/cyclin B-mediated phosphorylation stabilizes SREBP1 during mitosis. Cell Cycle. 2006;5:1708–18.
Siqingaowa Sekar S, Gopalakrishnan V, Taghibiglou C. Sterol regulatory element-binding protein 1 inhibitors decrease pancreatic cancer cell viability and proliferation. Biochem Biophys Res Commun. 2017;488:136–40.
Guo D, Reinitz F, Youssef M, Hong C, Nathanson D, Akhavan D, et al. An LXR agonist promotes glioblastoma cell death through inhibition of an EGFR/AKT/SREBP-1/LDLR-dependent pathway. Cancer Discov. 2011;1:442–56.
Nie LY, Lu QT, Li WH, Yang N, Dongol S, Zhang X, et al. Sterol regulatory element-binding protein 1 is required for ovarian tumor growth. Oncol Rep. 2013;30:1346–54.
Li C, Yang W, Zhang J, Zheng X, Yao Y, Tu K, et al. SREBP-1 has a prognostic role and contributes to invasion and metastasis in human hepatocellular carcinoma. Int J Mol Sci. 2014;15:7124–38.
Reed BD, Charos AE, Szekely AM, Weissman SM, Snyder M. Genome-wide occupancy of SREBP1 and its partners NFY and SP1 reveals novel functional roles and combinatorial regulation of distinct classes of genes. PLoS Genet. 2008;4:e1000133.
Magana MM, Koo SH, Towle HC, Osborne TF. Different sterol regulatory element-binding protein-1 isoforms utilize distinct co-regulatory factors to activate the promoter for fatty acid synthase. J Biol Chem. 2000;275:4726–33.
Peinado H, Ballestar E, Esteller M, Cano A. Snail mediates E-cadherin repression by the recruitment of the Sin3A/histone deacetylase 1 (HDAC1)/HDAC2 complex. Mol Cell Biol. 2004;24:306–19.
Tong ZT, Cai MY, Wang XG, Kong LL, Mai SJ, Liu YH, et al. EZH2 supports nasopharyngeal carcinoma cell aggressiveness by forming a co-repressor complex with HDAC1/HDAC2 and Snail to inhibit E-cadherin. Oncogene. 2012;31:583–94.
von Burstin J, Eser S, Paul MC, Seidler B, Brandl M, Messer M, et al. E-cadherin regulates metastasis of pancreatic cancer in vivo and is suppressed by a SNAIL/HDAC1/HDAC2 repressor complex. Gastroenterology. 2009;137:361–71.
Ai W, Liu Y, Wang TC. Yin yang 1 (YY1) represses histidine decarboxylase gene expression with SREBP-1a in part through an upstream Sp1 site. Am J Physiol Gastrointest Liver Physiol. 2006;290:G1096–104.
Yao R, Jiang H, Ma Y, Wang L, Wang L, Du J, et al. PRMT7 induces epithelial-to-mesenchymal transition and promotes metastasis in breast cancer. Cancer Res. 2014;74:5656–67.
Zhang N, Wang X, Huo Q, Li X, Wang H, Schneider P, et al. The oncogene metadherin modulates the apoptotic pathway based on the tumor necrosis factor superfamily member TRAIL (Tumor Necrosis Factor-related Apoptosis-inducing Ligand) in breast cancer. J Biol Chem. 2013;288:9396–407.
Su P, Zhang Q, Yang Q. Immunohistochemical analysis of Metadherin in proliferative and cancerous breast tissue. Diagn Pathol. 2010;5:38.
Kong X, Ding X, Li X, Gao S, Yang Q. 53BP1 suppresses epithelial-mesenchymal transition by downregulating ZEB1 through microRNA-200b/429 in breast cancer. Cancer Sci. 2015;106:982–9.
Heckman KL, Pease LR. Gene splicing and mutagenesis by PCR-driven overlap extension. Nat Protoc. 2007;2:924–32.
Wang H, Ach RA, Curry B. Direct and sensitive miRNA profiling from low-input total RNA. RNA. 2007;13:151–9.
Wang HY, Luo M. Tereshchenko IV, Frikker DM, Cui X, Li JY, et al. A genotyping system capable of simultaneously analyzing >1000 single nucleotide polymorphisms in a haploid genome. Genome Res. 2005;15:276–83.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 81502285, No.81672613, No.81272903), Key Research and Development Program of Shandong Province (No. 2016GSF201119), Shandong Science and Technology Development Plan (2016CYJS01A02), Shandong Provincial Natural Science Foundation of China (No. ZR2017BH050), Special Support Plan for National High Level Talents (“Ten Thousand Talents Program”) to QY and China Postdoctoral Science Foundation to NZ (No. 2018M630787).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Edited by A. Villunger
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Zhang, N., Zhang, H., Liu, Y. et al. SREBP1, targeted by miR-18a-5p, modulates epithelial-mesenchymal transition in breast cancer via forming a co-repressor complex with Snail and HDAC1/2. Cell Death Differ 26, 843–859 (2019). https://doi.org/10.1038/s41418-018-0158-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-018-0158-8
This article is cited by
-
USP30 promotes the progression of breast cancer by stabilising Snail
Cancer Gene Therapy (2024)
-
Targeting KK-LC-1 inhibits malignant biological behaviors of triple-negative breast cancer
Journal of Translational Medicine (2023)
-
Prognostic significance of serum miR-18a-5p in severe COVID-19 Egyptian patients
Journal of Genetic Engineering and Biotechnology (2023)
-
Gene profiles and mutations in the development of cataracts in the ICR rat model of hereditary cataracts
Scientific Reports (2023)
-
Targeting SLP2-mediated lipid metabolism reprograming restricts proliferation and metastasis of hepatocellular carcinoma and promotes sensitivity to Lenvatinib
Oncogene (2023)