Abstract
Oncogene-induced senescence (OIS) is a critical tumor-suppressor mechanism, which prevents hyper-proliferation and transformation of cells. c-Myc promotes OIS through the transcriptional activation of p14ARF followed by p53 activation. Although the oncogene-mediated transcriptional regulation of p14ARF has been well addressed, the post-translational modification of p14ARF regulated by oncogenic stress has yet to be investigated. Here, we found that c-Myc increased p14ARF protein stability by inducing the transcription of ubiquitin-specific protease 10 (USP10). USP10, in turn, mediated the deubiquitination of p14ARF, preventing its proteasome-dependent degradation. USP10-null mouse embryonic fibroblasts and human primary cells depleted of USP10 bypassed c-Myc-induced senescence via the destabilization of p14ARF, and these cells displayed accelerated hyper-proliferation and transformation. Clinically the c-Myc-USP10-p14ARF axis was disrupted in non-small cell lung cancer patients, resulting in significantly worse overall survival. Our studies indicate that USP10 induced by c-Myc has a crucial role in OIS by maintaining the stability of key tumor suppressor p14ARF.
Similar content being viewed by others
Introduction
Activated oncogenes such as RAS and c-Myc promote fail-safe programs to prevent the hyper-proliferation and transformation of cells [1,2,3]. RAS mediates the induction of p16INK4A, which prevents the cyclin-D- and CDK4-mediated hyper-phosphorylation of RB and induces the suppression of E2Fs, leading to cellular senescence [2, 4, 5]. Activated c-Myc induces the transcription of p14ARF, which stabilizes p53 and accelerates cellular senescence and apoptosis [1, 3]. Thus, the failure of RAS or c-Myc to induce a fail-safe program is a pivotal step in the transformation and tumorigenesis of normal cells [1].
p14ARF is an alternative reading frame product of the INK4/ARF locus [4]. The function of p14ARF as a potent tumor suppressor mainly depends on p53 stabilization, which induces cellular senescence and prevents tumor cell growth [6, 7]. One mode of action of p14ARF is its ability to interact with MDM2, a well-known E3-ubiquitin ligase of p53 and to induce nucleolus localization as well as inactivation, preventing MDM2-dependent p53 ubiquitination and degradation [8,9,10]. Alternatively, p14ARF also plays tumor-suppressive functions via interactions with numerous proteins, such as TIP60, HIF1a, XPC, NPM, and others [11,12,13]. The physiological roles of p14ARF have been comprehensively investigated in mouse models and human cancers. The p14ARF-KO mouse develops various cancers, including sarcoma and lymphoma, at an early age [14]. In contrast, the super INK4a/ARF mouse strain, which carries a transgenic copy of the entire INK4a/ARF locus, is highly resistant to carcinogen-induced cancer development [15]. p14ARF has been reported to be down-regulated or deleted in various human cancers, including gastric, breast, colon, pancreas, bladder, and lung cancer [16,17,18,19]. Studies examining the regulation of p14ARF expression have chiefly focused on transcriptional regulation, gene loss or silencing by promoter hyper-methylation and mutation [13, 20,21,22]. However, recent studies have indicated that p14ARF can be ubiquitinated by several E3 ubiquitin ligases, including ULF, MKRN1, and Siva1, which can affect p14ARF protein turnover through post-translational modifications (PTM) [23,24,25]. These results highlight the possible implication of enzymes involved in PTMs affecting tumorigenesis by regulating the stability of p14ARF. How such regulators of p14ARF can be controlled under normal, as well as stressed conditions to modulate tumor formation remains to be addressed.
Ubiquitin-specific protease 10 (USP10) is a deubiquitinating enzyme (DUB) that removes ubiquitins from its substrates. Its substrates include sorting nexin 3 (SNX3), the cystic fibrosis transmembrane conductance regulator (CFTR), H2A.Z., MutS Homologs2 (MSH2), topoisomerase IIα, AMPKα and p53 [26,27,28,29,30,31,32]. Although the stabilization of p53 suggests a possible role of USP10 as a tumor suppressor, USP10 functions in tumor prevention and how it is regulated upon oncogenic stimuli are largely unknown. Here, we showed that c-Myc could increase the stability of p14ARF by transcriptionally inducing USP10, which was shown to be a deubiquitinase of p14ARF. Corroborating these findings, USP10 overexpression induced cellular senescence, while its depletion prevented c-Myc-induced senescence and promoted cellular growth and transformation in normal cells. The disruption of c-Myc-USP10-p14ARF in NSCLC supported the critical roles of this axis in tumorigenesis. These findings reveal that c-Myc instigates another PTM program in addition to transcriptional regulatory pathways to maintain the stability of its target protein and accomplish its fail-safe program.
Results
USP10 transcription induced by c-Myc increases p14ARF protein stability
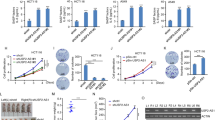
It is well known that c-Myc activates cellular senescence by inducing p14ARF transcription [3]. To determine whether c-Myc could increase p14ARF protein stability as well as transcription, we evaluated its protein stability in human foreskin fibroblasts (HFFs) and human fetal lung cells (IMR90) upon a translation inhibitor, cycloheximide (CHX) treatment after infection with c-Myc-expressing lentivirus. As both cell lines harbor wild type p53, p14ARF dependent senescent pathway could be delivered through the p14ARF-p53 axis, as previously known [6]. Interestingly, p14ARF protein stability was increased by c-Myc overexpression (Fig. 1a and Supplementary Fig. 1a). Accordingly, c-Myc also increased the protein stability of p53, a downstream target of p14ARF, through an indirect or a direct pathway (Fig. 1a and Supplementary Fig. 1a) [6, 7, 33]. Furthermore, treatment with the proteasome inhibitor MG132 increased p14ARF protein levels >2-fold; however, there was a more modest increase in p14ARF protein levels in response to MG132 treatment upon c-Myc overexpression (Fig. 1b and Supplementary Fig. 1b). Based on these observations, we hypothesized that DUB might be affected by c-Myc, which could subsequently increase p14ARF protein stability via a deubiquitination process. To identify a possible DUB induced by c-Myc, three independent microarray analyses were conducted using HFF cell lines stably expressing c-Myc (Supplementary Fig. 2). USP1, USP10, USP13, and USP31, which displayed >1.5-fold induction by c-Myc, were further tested for their correlation with p14ARF. When these DUBs were ablated using siRNAs, only the USP10 knockdown induced p14ARF destabilization without any change of its mRNA level (Fig. 1c, d and Supplementary Fig. 1c). We further confirmed that p14ARF protein stability induced by c-Myc expression was decreased upon the stable ablation of USP10 (Fig. 1e). Notably, the half-life of p53, an alternative known target of USP10, was also decreased (Fig. 1e) [26]. Corroborating these findings, c-Myc overexpression increased USP10 protein levels as well as its mRNA in HFF and IMR90 cells (Fig. 1f; Supplementary Fig. 1d). These observations suggest that the increase in p14ARF by c-Myc appeared to be mediated by post-translational, as well as transcriptional control. Overall, these results imply that c-Myc can stabilize p14ARF proteins through USP10 induction.
c-Myc increases p14ARF protein stability through the induction of USP10. a HFF cells stably expressing c-Myc by lentivirus injection were treated with CHX (100 ng/ml) as indicated, and cell lysates were detected with p14ARF, p53, c-Myc, and actin antibodies. Graphs indicate the protein levels of p14ARF and p53 quantified using the ImageJ program. b HFF cells were infected with lentiviral c-Myc and treated with 20 μM MG132 for 3 h. Cell lysates were immunoblotted with c-Myc, p14ARF and actin antibodies. The graph indicates the average of the protein levels of p14ARF quantified using the ImageJ program. Error bars indicate 95% confidence intervals. Three independent experiments were performed. c A microarray was conducted using HFF cells stably expressing mock vector and c-Myc. Graphs indicate the fold change in mRNA levels of USPs. This experiment was conducted in triplicate. d H1299 cells were transfected with 20 nM USP1, 10, 13, and 31 siRNAs for 72 h. Cell lysates were immunoblotted with p14ARF and actin antibodies. e HFF cells were injected with lentiviral c-Myc and shUSP10 and treated with CHX as indicated, followed by immunoblotting using specific antibodies. Graphs indicate the protein levels of p14ARF and p53 quantified using the ImageJ program. f HFF cells were injected with lentiviral c-Myc. Cell lysates were immunoblotted with specific antibodies, and mRNA levels were analyzed by Q-RT-PCR using p14ARF- and USP10-specific primers. Three independent experiments were performed
c-Myc functions as a transcriptional activator of USP10 expression
We next investigated whether c-Myc could be a direct transcriptional activator of USP10. There are five E-box sequences upstream of the USP10 transcription start site (Fig. 2a) [34]. To determine whether c-Myc was able to bind to these E-box sequences, ChIP assays were performed in HFF and IMR90 cells using the indicated primers (Fig. 2a, b). The results indicated that c-Myc could bind to the second E-box, named E#2, of the USP10 gene in the tested cell lines (Fig. 2b). To confirm these observations, luciferase assays were conducted using luciferase reporter constructs containing separate endogenous E-box elements of USP10, L#1 and L#2 (Fig. 2c). In accordance with the ChIP analyses, there was a significant increase in luciferase activity with construct L#1 by c-Myc overexpression (Fig. 2c). Furthermore, increasing the levels of the c-Myc expression vector could proportionally augment the luciferase signal. These kinetic data implicitly indicated that the promoter of USP10 could be indeed recognized by c-Myc (Fig. 2c). To identify the essential E-box in construct L#1 required for c-Myc binding, each E-box was tested for its ability to response to c-Myc expression (Fig. 2d). In these experiments, each of wild type E-box construct named W1, W2, W3 or W4 with the rest of E-box sites mutated as indicated in Fig. 2d was employed. The results showed that c-Myc was capable of promoting luciferase activity of W2, the data of which corroborates the results of the ChIP analyses of Fig. 2a, b, suggesting that the second E-box is an essential promoter for c-Myc-mediated transcription of USP10 (Fig. 2d). Overall, these data suggest that USP10 can be transcriptionally induced by c-Myc to function as a transcriptional factor for the USP10 promoter.
USP10 is a transcriptional target of c-Myc. a Diagram of the USP10 promoter. There are five c-Myc binding elements, E-box sequences, upstream of the transcription start site of USP10. b A ChIP assay was performed using c-Myc antibodies in HFF and IMR90 cells stably expressing c-Myc. Relative enrichment was determined by quantifying PCR products obtained from three experiments. c 0.5 μg USP10 luciferase reporter constructs and 0.3–0.5 μg c-Myc-expressing plasmid were transfected into HFF cells as indicated. Luciferase activities of each reporter were analyzed by the luciferase assay. Cell lysates were immunoblotted with specific antibodies. This experiment was conducted in triplicate. d Luciferase activities of 0.5 μg USP10 luciferase reporter constructs, which have E-box sequence mutations of CAATTG, were analyzed using a luciferase assay. This experiment was conducted in triplicate
USP10 functions as a DUB of p14ARF
Our previous data indicated that c-Myc could stabilize the levels of p14ARF protein through the transcriptional induction of USP10, a plausible DUB of p14ARF. Corroborating these findings, exogenously expressed USP10 increased exogenous and endogenous p14ARF protein levels in a dose-dependent manner (Fig. 3a, b). The half-life of exogenous as well as endogenous p14ARF upon treatment with CHX was also increased by the exogenous expression of USP10 (Fig. 3c, d). However, USP10 ablation using both siRNA #1 and #3 of USP10 decreased p14ARF protein level without any change in its mRNA levels, indicating that USP10 could regulate p14ARF in a post-translational level (Fig. 3e). As both siRNAs of USP10 were able to ablate the protein levels of USP10, we employed siRNA #3 for subsequent experiments. Confirming these findings, the half-life of endogenous p14ARF upon CHX treatment was decreased by the ablation of USP10 (Fig. 3f). Employing immunoprecipitation analyses, we found that p14ARF and USP10 either in overexpressed or endogenous states could bind to each other (Fig. 3g–j). Moreover, a GST pulldown assay showed that USP10 could directly interact with p14ARF (Fig. 3k). Domain mapping analyses indicated that the interaction between the two proteins was mediated through the N-terminal regions of each protein (Supplementary Fig. 3a, b). It is known that p19ARF, a mouse orthologue of human p14ARF, displays similar physiological effects as a tumor suppressor [4]. However, because it also exhibits different interacting partners with some functional differences compared with p14ARF [35], we investigated whether mouse USP10 (mUSP10) could regulate p19ARF protein levels as observed in their human counterparts. The results showed that mUSP10 could bind to p19ARF and increase its protein levels (Supplementary Fig. 4a–c). Ablation of mUSP10 by siRNA also decreased p19ARF protein levels in mouse embryonic fibroblasts (MEFs), suggesting a conserved function of USP10 as a DUB for p14ARF or p19ARF (Supplementary Fig. 4 d).
USP10 interacts with and stabilizes p14ARF. a H1299 cells were transfected with plasmids expressing p14ARF (0.05 μg) and USP10 (0.5 μg). Cell lysates were evaluated using specific antibodies. b H1299 cells were transfected with plasmids expressing p14ARF (0.05 μg) and USP10 (0.5 μg) and were treated with CHX as indicated. p14ARF and USP10 were detected using specific antibodies. c H1299 cells were transfected with plasmids expressing USP10 (1.9 μg). Cell lysates were detected using specific antibodies. d H1299 cells were transfected with plasmids expressing USP10 (1.9 μg) and treated with CHX as indicated. p14ARF and USP10 were detected using specific antibodies. e H1299 cell were transfected with 30 nM USP10 siRNAs for 72 h. Protein levels of p14ARF and USP10 were detected using specific antibodies, and mRNA levels were analyzed by Q-RT-PCR. Three independent experiments were performed. f H1299 cell were transfected with 30 nM USP10 siRNA #3 for 72 h and treated with CHX as indicated. Cell lysates were immunoblotted with specific antibodies. g, h Cells were transfected with plasmids expressing p14ARF (3 μg) and USP10 (3 μg) as indicated. Cell lysates were immunoprecipitated with HA or FLAG antibodies. p14ARF and USP10 were detected using specific antibodies. i, j Cell lysates were immunoprecipitated with p14ARF or USP10 antibodies. k Purified GST/USP10 and in vitro-translated p14ARF were incubated at 37 °C (input) followed by pulldown (PD) using glutathione-Sepharose. The precipitated samples were detected using USP10 and p14ARF antibodies
Next, we investigated whether USP10 could protect p14ARF protein from the proteasome-dependent degradation pathway. The levels of endogenously or exogenously expressed p14ARF stabilized by USP10 overexpression were similar to those stabilized by MG132 treatment, indicating that the proteasome-dependent pathway might be involved in the interaction of USP10 and p14ARF (Fig. 4a, b). USP10 C424A, a mutant that is defective in DUB activity, was able to bind to p14ARF but could not induce its stabilization like wild-type (WT) USP10, indicating that deubiquitination processes are necessary for the stabilization of p14ARF (Fig. 4c–e) [28]. We have previously reported that E3 ligase MKRN1 induces the ubiquitination of p14ARF followed by degradation [24]. MKRN1-mediated degradation, as well as ubiquitination was blocked by WT USP10 but not by C424A, indicating that USP10 reversed the E3 ligase-mediated ubiquitination of p14ARF (Fig. 4f, g). Finally USP10 ablation induced an increase in the ubiquitinated forms of endogenous p14ARF (Fig. 4h). Overall, these results indicate that USP10 is a DUB of p14ARF, which prevents its ubiquitination and thus subsequent proteasome-mediated degradation.
USP10 functions as a DUB of p14ARF. a, b H1299 cells were transfected with 0.8 μg USP10- and 0.1 μg p14ARF-expressing plasmids and treated with 20 μM MG132 for 3 h. Cell lysates were detected with specific antibodies. c Cells were transfected with plasmids expressing USP10 WT (3 μg), USP10 C424A (3 μg), and p14ARF (3 μg) as indicated, and cell lysates were immunoprecipitated with FLAG antibodies. d, f Cells were transfected with plasmids expressing USP10 WT (0.8 μg), USP10 C424A (0.8 μg), MKRN1 (0.8 μg), and p14ARF (0.1 μg) as indicated. Cell lysates were immunoblotted with specific antibodies. g Cells were transfected with plasmids expressing USP10 (4 μg), MKRN1 (4 μg), p14ARF (2 μg), and Ub (2 μg) as indicated. Cell lysates were immunoprecipitated with HA antibodies under denaturation conditions. h Cells were transfected with 30 nM control siRNA or USP10 siRNA for 72 h followed by transfection with HA/Ub (10 μg). Cell lysates were immunoprecipitated with p14ARF antibodies under denaturation conditions
USP10-mediated p14ARF stabilization is required for c-Myc-induced cellular senescence
As it is well known that activated oncogenic c-Myc turns on oncogene-induced senescence (OIS) through the activation of tumor suppressors to prevent abnormal cell proliferation and tumorigenesis, we investigated whether c-Myc-induced USP10 is required for p14ARF-mediated cellular senescence [1]. HFF cells stably expressing either c-Myc or shUSP10, or both, were established. As expected, while c-Myc overexpression increased p14ARF and USP10 protein levels, c-Myc-induced p14ARF accumulation was blocked upon USP10 ablation (Fig. 5a). This finding indicates that the USP10-mediated deubiquitination process was important for maintaining the stability of c-Myc-induced p14ARF. Furthermore, while c-Myc overexpression facilitated cellular senescence, which was detected by β-galactosidase staining, this phenomenon was subdued following the depletion of USP10 (Fig. 5b). Next, we investigated whether USP10 ablation could induce cell transformation upon c-Myc overexpression. Overexpression of c-Myc alone could not induce the colony formation of normal cells in soft agar as previously reported [36]. Interestingly, USP10 depletion under the condition of c-Myc overexpression induced anchorage-independent colony formation (Fig. 5c). Quantification analyses of cell growth further indicated that USP10 ablation under c-Myc overexpression significantly increased anchorage-independent cell growth compared with c-Myc overexpression alone (Fig. 5c). To verify the tumor-suppressive effects, USP10 was stably overexpressed in HFF and IMR90, and its effects on cellular growth and senescence were examined. USP10 overexpression stabilized p14ARF protein levels in both cell lines (Fig. 5d). In line with these observations, these cells exhibited accelerated cellular senescence with decreased cellular growth (Fig. 5e, f). These phenomena were prevented by p14ARF ablation under USP10 overexpression (Fig. 5e, f). Overall, these results indicated that USP10 could accelerate cellular senescence through p14ARF stabilization.
USP10 is required for c-Myc-induced cellular senescence. a HFF cells were infected with lentiviral c-Myc and shUSP10. Cell lysates were immunoblotted with specific antibodies. b Stable cells were stained for β-galactosidase activity, and β-gal-positive cells from three independent experiments were counted. c Stable cells were seeded in a soft agar matrix and cultured for 8 days. Anchorage-independent cell growth was analyzed using the MTT assay. d HFF and IMR90 cells were injected with lentiviral USP10 and shp14ARF. Cell lysates were immunoblotted with specific antibodies. e Cells were stained for β-galactosidase activity. β-gal-positive cells from three independent experiments were counted. f The same number of cells was seeded and counted for 7 days. Error bars indicate 95% confidence intervals. Three independent experiments were performed
USP10−null MEFs are resistant to cellular senescence
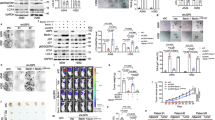
To further investigate the role of USP10 in vivo, USP10-knockout (KO) mice were generated using the CRISPR/Cas9 system. We designed 4 sgRNAs targeting USP10 exon 2 sequences (Supplementary Fig. 5a) and tested their ability to cleave the USP10 gene in vitro. Among 4 sgRNAs, #3 efficiently cleaved the USP10 gene (Supplementary Fig. 5b). The sgRNA #3 was confirmed to effectively cleave genomic DNA of USP10 but not cDNA because one ‘g’ of the PAM domain was present in an intron (Supplementary Fig. 5b). This sgRNA and cas9 were injected into an embryo. We acquired 38 newborn mice, and after genotyping and high-resolution melt (HRM) analysis, we found that ~90% of the mice had a mutation in the USP10 gene, including a deletion, substitution and insertion (Supplementary Fig. 5c and data not shown). Among them, we selected #34, which had an early stop codon in the USP10 gene, and obtained USP10-KO MEFs.
USP10-KO MEFs showed decreased p19ARF protein levels, as well as p53 (Fig. 6a). Accordingly, they displayed decreased cellular senescence with increased cellular growth compared with WT MEFs (Fig. 6b, c). c-Myc overexpression in two different lines of USP10-KO MEFs did not lead to p19ARF accumulation as proficiently as in WT MEFs (Fig. 6d). Notably, c-Myc overexpression in WT MEFs displayed increased levels of USP10 as well as p19ARF, which indicates that c-Myc could similarly enforce the expression of USP10 in mouse cells (Fig. 6d). As expected, USP10-KO MEFs did not display c-Myc-induced cellular senescence as detected by β-galactosidase staining but rather showed rapid cellular growth with a transformed morphology (Fig. 6e, f). Furthermore, USP10-KO MEFs increased anchorage-independent colony formation in soft agar by c-Myc overexpression, which could not be accomplished by WT MEFs (Fig. 6g). Based on these results, we suggest that c-Myc-induced USP10 could increase the stability of p19ARF and thus p53, which reinforces the fail-safe program in response to oncogenic stimuli.
USP10-KO MEFs escape from c-Myc-induced cellular senescence. a WT and USP10-KO MEFs were immunoblotted with specific antibodies. b MEFs were stained for β-galactosidase activity, and β-gal-positive cells from three independent experiments were counted. c The same number of cells was seeded and counted for 5 days. Error bars indicate 95% confidence intervals. Three independent experiments were performed. d MEFs were infected with retroviral c-Myc. Cell lysates were immunoblotted with specific antibodies. e Cells were stained for β-galactosidase activity, and β-gal-positive cells from three independent experiments were counted. f The same number of cells was seeded and counted for 5 days. Error bars indicate 95% confidence intervals. Three independent experiments were performed. g Cells were seeded in a soft agar matrix and cultured for 8 days. Colonized cells from three independent experiments were counted
c-Myc/p14ARF/USP10 expression in tumor cells correlates with a poor prognosis in human non-small cell lung cancer (NSCLC)
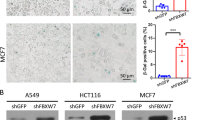
The fail-safe program induced by c-Myc is largely disrupted in various cancers [37]. This phenomenon led us to examine whether the positive correlation between c-Myc and USP10 is maintained in various cancer cell lines, including breast, pancreatic and lung. When c-Myc was knocked down in these cell lines, the protein and mRNA levels of USP10 were diminished in two out of four breast cancer cell lines we tested. These levels were not reduced in two breast cancer cell lines: MCF7 and SKBR3. On the other hand, all five pancreatic cell lines we tested displayed decreased levels of USP10 mRNA upon c-Myc ablation (Supplementary Fig. 6a, b). Interestingly, all four non-small cell lung cancer (NSCLC) cell lines we analyzed, A549, PC-9, H23, and H460, did not display any decrease in USP10 levels upon knockdown of c-Myc. (Supplementary Fig. 7). Having clarified the molecular mechanism by which c-Myc increased p14ARF protein stability by activating USP10 transcription and observing a substantial disruption of the correlation between c-Myc and USP10 among NSCLC cancer cell lines, we aimed to determine the clinical relevance of the c-Myc, p14ARF, and USP10 axis in human NSCLC, a cancer exhibiting c-Myc overexpression (Fig. 7a) [38, 39]. The clinicopathological characteristics of the study are summarized in Supplementary Table 2. c-Myc immunoreactivity was found more frequently in males (p = 0.01) and squamous cell carcinomas (p < 0.001). In contrast, p14ARF expression was more prominent in adenocarcinomas (p = 0.045). USP10 expression was associated with an earlier stage (p = 0.01) (Supplementary Table 2). As expected, USP10 expression showed a positive correlation with that of p14ARF in NSCLC (Spearman’s rho = 0.315, p = 0.001) (Supplementary Fig. 8a). Interestingly, c-Myc expression negatively correlated with that of USP10 (Spearman’s rho = −0.561, p < 0.001) and p14ARF (Spearman’s rho = −0.388, p < 0.001) (Supplementary Fig. S8A). These observations indicate that unlike normal cells, the induction of USP10 by c-Myc was disrupted in NSCLC by unknown mechanism, corroborating the data shown in lung cancer cell lines (Supplementary Fig. 7).
The functional axis of c-Myc-USP10-p14ARF in human non-small cell lung cancer specimens is disrupted. a Representative images of immunohistochemical staining of c-Myc, USP10, and p14ARF in non-small cell lung cancer tissues. High magnification images are shown in the inset. The scale bar is 100 µm. b Overall survival curves for non-small cell lung cancer patients according to combined markers groups. Patients with combined c-Myc+/USP10-, c-Myc+/p14ARF-, USP10-/p14ARF-, and c-Myc+/USP10-/p14ARF- expression showed significantly worse overall survival (p = 0.008, p = 0.021, p = 0.035 and p = 0.021, respectively) than patients with combined c-Myc-/USP10+, c-Myc-/p14ARF+, USP10+/p14ARF+, and c-Myc-/USP10+/p14ARF+ expression. c Proposed model. Oncogenic c-Myc induces USP10 transcription to strengthen the fail-safe program by deubiquitination-dependent stabilization of p14ARF and p53, which prevents immortalization and tumorigenesis in normal cells
Next, we examined the relationship of each protein expression level with patient survival outcomes. Kaplan-Meier plots demonstrated that patients with high p14ARF expression displayed significantly higher overall survival than patients with low p14ARF expression (72.5% vs. 35.3%, p = 0.027) (Supplementary Fig. 8b). Furthermore, the patients with combined c-Myc+/USP10−, c-Myc+/p14ARF−, USP10−/p14ARF−, and c-Myc+/USP10−/p14ARF− expression showed significantly worse overall survival (38.0% vs. 69.4%, p = 0.008, 30.2% vs. 74.1%, p = 0.021, 32.4% vs. 71.3%, p = 0.035, and 27.8% vs. 73.1%, p = 0.021, respectively) than patients with c-Myc−/USP10+, c-Myc−/p14ARF+, USP10+/p14ARF+, and c-Myc−/USP10+/p14ARF+ expression (Fig. 7b). A combination of c-Myc+/USP10− (hazard ratio = 6.11 (95% CI, 1.54–24.3), p = 0.01), USP10+/p14ARF+ (hazard ratio = 0.30 (95% CI, 0.10–0.88), p = 0.029), and c-Myc+/USP10−/p14ARF− (hazard ratio = 5.12 (95% CI, 1.13–23.27), p = 0.034) expression was found to be independent prognostic factors with respect to overall survival using a Cox proportional hazards model (Supplementary Table 1). Overall these data indicated that c−Myc, USP10, and p14ARF expression levels serve as important prognostic factors in human NSCLC.
Discussion
One way for cancer to avoid the deleterious effects of p14ARF expression is to suppress its activities through PTMs. In support of this hypothesis, p14ARF, a lysine-less protein, was ubiquitinated at a methionine residue located at the N-terminus and degraded by proteasomes [40]. Since this observation was reported, several E3-ligases, including ULF, MKRN1, and Siva1, have been identified; these ligases can mediate p14ARF ubiquitination at the N-terminus and proteasome-dependent degradation, preventing p14ARF-dependent senescence, but its deubiquitinating enzymes have not yet been revealed [23,24,25]. Interestingly, c-Myc is known to interfere with the interaction between ULF and 14ARF, stabilizing p14ARF [23]. However, the lack of a link between oncogenic signaling pathways and the function of most PTM enzymes regulating the stabilization of p14ARF that such interactions are isolated phenomena and are not involved in the circuit of oncogenic stresses.
In this study, we report the association between oncogenic stimulus, c-Myc, and PTM of its target protein through USP10, the first deubiquitinating enzyme of p14ARF. c-Myc mediated OIS is a processes that relies heavily on p14ARF expression followed by p53 stabilization [3]. Because p14ARF is a rather unstable protein that is rapidly degraded in cells, it raises the question as to whether there is any oncogene-dependent signaling feedback that could stimulate p14ARF protein stabilization to reinforce a fail-safe program [24]. Interestingly, the observation that c-Myc overexpression significantly increases the protein stability of p14ARF, as well as its transcription provide a possible clue concerning PTM pathways triggered by c-Myc (Fig. 1a). The microarray and biochemical data showed that to increase the effects of p14ARF following oncogenic stimulus, c-Myc induces the transcription of USP10; this induces the deubiquitination of p14ARF, preventing its proteasome-dependent degradation (Figs. 1–4). Of note, USP10 is also known to be a DUB of p53 [26]. It appears that USP10 could boost p53 activity more potently by simultaneously deubiquitinating p53 and p14ARF (Fig. 7c). The USP10-mediated stabilization of p14ARF suppresses the function of MDM2, a true E3 ligase of p53 and protects p53 from degradation [9, 10]. Moreover, concurrent deubiquitination of p53 by USP10 ensures the protection of p53 from other E3 ligases [26] (Fig. 7c). Thus, USP10 might function as a c-Myc-induced tumor suppressor, enforcing the potent p14ARF-p53 tumor-suppressive axis.
The importance of USP10 induction by c-Myc has clearly been shown by observations that USP10 ablation prevents c-Myc-induced cellular senescence by destabilizing p14ARF (Figs. 5 and 6). A positive correlation between c-Myc and USP10 was observed in various cell lines, including human fibroblasts, MEFs, pancreatic cancer cells, and several breast cancer cell lines. Notably, this correlation between c-Myc and USP10 expression is disrupted in several breast cancer and NSCLC cell lines and tumor tissues of NSCLC, indicating that the disruption of this pathway might contribute to tumorigenesis (Supplementary Figs. 6 and 7). In support of this hypothesis, patients displaying c-Myc+/USP10−, c-Myc+/p14ARF−, USP10−/p14ARF−, and c−Myc+/USP10-/p14ARF− expression exhibited significantly worse overall survival than patients with c−Myc−/USP10+, c-Myc−/p14ARF+, USP10+/p14ARF+, and c-Myc−/USP10+/p14ARF+ expression (Fig. 7a, b). Disrupting the transcriptional effect of c-Myc on USP10 could be a crucial factor in the suppression of OIS in NSCLC, which will instigate escape from the fail-safe program (Fig. 7c). These data underscore the importance of the c-Myc/p14ARF/USP10 axis in human cancer, particularly in human NSCLC, and demonstrate that assessment of the c-Myc/p14ARF/USP10 expressional level in tumor cells could serve as a valuable clinical index. Moreover, further studies about the regulatory mechanisms by which c-Myc mediated USP10 is dysregulated in several cancers will provide clues for identifying pro-senescence therapeutic targets.
Overall, our findings suggest that oncogenic c-Myc induces USP10 transcription to strengthen the fail-safe program by deubiquitination-dependent stabilization of p14ARF and p53, which prevents immortalization and tumorigenesis in normal cells. Given that USP10 depletion induced c-Myc-mediated transformation of normal cells and that the c-Myc/USP10/p14ARF axis is disrupted in NSCLC, defects in this axis can accelerate tumorigenesis without the intervention of oncogene-induced senescence, which partly explains the bypass of OIS in human cancers.
Methods
Cell culture
Lung fibroblast (IMR90), human foreskin fibroblasts (HFFs), an embryonic kidney cell line expressing SV40 large T antigen (293 T) and human non-small cell lung carcinoma (H1299) were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). Breast cancer cell lines including MDA-MB231, T47D, MCF7, and SKBR3 and non-small cell lung cancer cell lines including A549, H460 and H23 were provided from the U.S. National Cancer Institute (NCI; MTA no. 2702-09). PC-9 cells were purchased in 2015 from Sigma-Aldrich Corporation (St. Louis, MO). All cell lines were grown in MEM (Gibco), DMEM (HyClone) and RPMI (Gibco) supplemented with 10% fetal bovine serum. All human pancreatic cancer cell lines were purchased from ATCC and grown in RPMI, IMDM or Mccoy supplemented with 10% fetal bovine serum. All lung cancer cell lines except PC-9 and H1299 were tested and authenticated annually by STR test in National Cancer Center Core Facility, Korea. STR profile was analyzed using Gene Mapper v 5.0 software. All the provided cell lines were aliquoted and frozen within passage 10, and experiments were performed using thawed cell lines from passage 7 to 18. Cell lines were tested for infection of mycoplasma as soon as receipt using e-Myco™ plus Mycoplasma PCR Detection Kit (Intron, Seongnam, Korea). Whenever subculture was performed, cells were treated with plasmosin (Invivogen, San Diego, CA) in order to prevent infection of mycoplasma.
Generation of USP10 KO mice and MEFs using CRISPR/Cas9
Four candidate sgRNAs targeting USP10 were designed in proximity to exon 2. For the experiments, the optimal sgRNA was experimentally determined by measuring the target DNA cleavage efficiency using an in vitro cleavage assay for PCR products containing the target region of genomic DNA and cDNA of USP10. The genomic DNA and cDNA for USP10 gene were incubated at 37 °C for 1 h with purified Cas9 protein and each sgRNA and then resolved by agarose gel electrophoresis.
To generate USP10-deficient mice, microinjection of fertilized zygotes and embryo transfer into pseudopregnant foster mothers were performed, as previously described [41]. Briefly, the SpCas9 mRNA and sgRNA (candidate #3F) were microinjected into the cytoplasm of one-cell embryos from FVB/NTac mice, followed by transfer into ICR pseudopregnant recipients to produce live animals. To screen founder mice with a mutated USP10 locus, genomic DNA were prepared from tail biopsies of 38 newborn mice. The genomic region contacting the sgRNA target site was PCR-amplified and resolved by agarose gel electrophoresis. The PCR reactions were performed using primers 5′–cctgccctaggaaccttagc–3′ and 5′–ggcgtgaagaagaaagcaac–3′ and analyzed by sequencing analysis.
USP KO MEFs were obtained from 13.5-day-old embryos by washing them with PBS (Welgene) followed by mincing. After mincing, the MEFs were incubated with trypsin/EDTA (Gibco) at 37 °C. Trypsinized MEFs were transferred to culture dishes, followed by addition of DMEM containing 10% fetal bovine serum (Gibco).
Microarrays
HFF cells were infected with lentivirus expressing c-Myc, and RNAs were purified and analyzed by microarray using Human gene 2.0 ST array chips (DNA Link, Korea). Three independent experiments were performed. The microarray data are deposited under GEO number GSE101941
Human tumor samples
A total of 112 NSCLC cases were selected from the pathology case archive of Asan Medical Center based on the diagnosis and quality of the available tissue on the paraffin blocks. None of the patients received neoadjuvant treatments, and all underwent complete resections between 1993 and 2004. The tumors were staged according to the International Union against Cancer tumor-node-metastasis classification, and histology was defined and graded according to 2004 WHO guidelines. A total of 62 patients (58.5%) had adenocarcinomas, and 44 (41.5%) squamous cell carcinomas. In total 72 (64.3%) were stage I, and 40 (35.7%) were stage II or higher. Of the 61 cases with a known smoking history, 31 (50.8%) patients had a smoking history. Tissue samples were collected from patients who had signed informed consent forms, which was approved by the Institutional Review Boards of Asan Medical Center (approval no.S2017-0068-0001; Seoul, South Korea). This study was additionally approved by the Office of Human Subjects Research at the National Institutes of Health.
Immunohistochemistry and scoring
Tissue microarrays (TMAs) were constructed from 112 formalin-fixed, paraffin-embedded (FFPE) tissue specimens. For each case, areas with the most representative histology were selected from review of hematoxylin and eosin (H&E)-stained slides. Three 0.6-mm-diameter tissue cores were retrieved from FFPE tissue blocks and arrayed on a recipient paraffin block using a manual tissue arrayer MTA-1 (Beecher Instruments Inc., Silver Spring, MD, USA). Multiple 5-μm-thick sections were cut with a microtome, and H&E-stained TMA slides were examined every 50th section for the presence of tumor cells.
TMA sections were deparaffinized with xylene and dehydrated through a graded alcohol series, and heat-induced antigen retrieval was performed for 20 min in antigen retrieval buffer pH 9.0 (for c-Myc and p14ARF) or pH 6.0 (for USP10) (Dako, Carpinteria, CA, USA) using a pressure cooker (Pascal, Dako). The endogenous peroxidase activity was quenched with 3% H2O2 in water for 10 min. The TMA sections were incubated with rabbit monoclonal anti-c-Myc antibodies (Abcam, Cambridge, MA, USA; Clone #Y69; dilution 1:100) overnight at 4 °C, mouse monoclonal anti-p14ARF antibodies (Cell Signaling Technology, Denver, MA, USA; Clone #4C6/4; dilution 1:1000) for 1 h at room temperature, or rabbit polyclonal anti-USP10 antibodies (Abcam; Cat. #ab72486; dilution 1:1000) for 1 h at room temperature. The antigen-antibody reaction was detected with EnVision+Dual Link System-HRP (Dako) and visualized with DAB+(3,3′-diaminobenzidine; Dako). The stained slides were lightly counterstained with hematoxylin and then imaged using NanoZoomer 2.0 HT (Hamamatsu Photonics K.K., Japan) at a ×20 objective magnification. Appropriate negative controls were concurrently assessed, and the TMAs included suitable positive control tissues. Images were analyzed using Visiopharm software (Ver. 4.5.1.324; Visiopharm, Hørsholm, Denmark), as previously reported. Briefly, brown-colored (3,3′-diaminobenzidine, DAB) and blue-colored (hematoxylin) cells were separated spectrally. The brown staining intensity (0-negative, 1-weak, 2-moderate, and 3-strong) was determined using a predefined algorithm and optimized settings. The overall immunohistochemical score (histoscore) was expressed as the percentage of positive cells multiplied by their staining intensity (possible range, 0–300).
Statistical analyses
The median values of the histoscores were selected for cut-off for discriminating low and high protein expression. The association between the protein expression and clinicopathological characteristics was determined using a Chi-square test. Overall survival according to the protein expression level was analyzed using Kaplan–Meier methodology, and the survival curves were compared with the log-rank test. Subsequently, we used a multivariate proportional Cox model and adjusted for the following clinical/pathological variables: cell type, pN factor, stage, and vascular invasion. Statistical analyses were performed using SPSS version 21.0 (SPSS, Chicago, IL, USA) and the R statistical package (Version 3.1.2).
References
Schmitt CA. Senescence, apoptosis and therapy–cutting the lifelines of cancer. Nat Rev Cancer. 2003;3:286–95.
Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602.
Zindy F, Eischen CM, Randle DH, Kamijo T, Cleveland JL, Sherr CJ, et al. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 1998;12:2424–33.
Gil J, Peters G. Regulation of the INK4b-ARF-INK4a tumour suppressor locus: all for one or one for all. Nat Rev Mol Cell Biol. 2006;7:667–77.
Lin AW, Barradas M, Stone JC, van Aelst L, Serrano M, Lowe SW. Premature senescence involving p53 and p16 is activated in response to constitutive MEK/MAPK mitogenic signaling. Genes Dev. 1998;12:3008–19.
Zhang Y, Xiong Y, Yarbrough WG. ARF promotes MDM2 degradation and stabilizesp53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell. 1998;92:725–34.
Honda R, Yasuda H. Association ofp19(ARF) with Mdm2 inhibits ubiquitin ligase activity of Mdm2 for tumor suppressor p53. EMBO J. 1999;18:22–27.
Weber JD, Taylor LJ, Roussel MF, Sherr CJ, Bar-Sagi D. Nucleolar Arf sequesters Mdm2 and activates p53. Nat Cell Biol. 1999;1:20–26.
Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303.
Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–9.
Eymin B, Claverie P, Salon C, Leduc C, Col E, Brambilla E, et al. p14ARF activates a Tip60-dependent and p53-independent ATM/ATR/CHK pathway in response to genotoxic stress. Mol Cell Biol. 2006;26:4339–50.
Fatyol K, Szalay AA. The p14ARF tumor suppressor protein facilitates nucleolar sequestration of hypoxia-inducible factor-1alpha (HIF-1alpha) and inhibits HIF-1-mediated transcription. J Biol Chem. 2001;276:28421–9.
Ko A, Han SY, Song J. Dynamics of ARF regulation that control senescence and cancer. BMB Rep. 2016;49:598–606.
Kamijo T, Bodner S, van de Kamp E, Randle DH, Sherr CJ. Tumor spectrum in ARF-deficient mice. Cancer Res. 1999;59:2217–22.
Matheu A, Pantoja C, Efeyan A, Criado LM, Martin-Caballero J, Flores JM, et al. Increased gene dosage of Ink4a/Arf results in cancer resistance and normal aging. Genes Dev. 2004;18:2736–46.
Iida S, Akiyama Y, Nakajima T, Ichikawa W, Nihei Z, Sugihara K, et al. Alterations and hypermethylation of thep14(ARF) gene in gastric cancer. Int J Cancer. 2000;87:654–8.
Hsu HS, Wang YC, Tseng RC, Chang JW, Chen JT, Shih CM, et al. 5′ cytosine-phospho-guanine island methylation is responsible for p14ARF inactivation and inversely correlates with p53 overexpression in resected non-small cell lung cancer. Clin Cancer Res: Off J Am Assoc Cancer Res. 2004;10:4734–41.
Silva J, Dominguez G, Silva JM, Garcia JM, Gallego I, Corbacho C, et al. Analysis of genetic and epigenetic processes that influence p14ARF expression in breast cancer. Oncogene. 2001;20:4586–90.
Lee M, Sup Han W, Kyoung Kim O, Hee Sung S, Sun Cho M, Lee SN, et al. Prognostic value of p16INK4a and p14ARF gene hypermethylation in human colon cancer. Pathol Res Pract. 2006;202:415–24.
Komori H, Enomoto M, Nakamura M, Iwanaga R, Ohtani K. Distinct E2F-mediated transcriptional program regulates p14ARF gene expression. EMBO J. 2005;24:3724–36.
Pollice A, Nasti V, Ronca R, Vivo M, Lo Iacono M, Calogero R, et al. Functional and physical interaction of the human ARF tumor suppressor with Tat-binding protein-1. J Biol Chem. 2004;279:6345–53.
Parisi T, Pollice A, Di Cristofano A, Calabro V, La Mantia G. Transcriptional regulation of the human tumor suppressorp14(ARF) by E2F1, E2F2, E2F3, and Sp1-like factors. Biochem Biophys Res Commun. 2002;291:1138–45.
Chen DL, Shan J, Zhu WG, Qin J, Gu W. Transcription-independent ARF regulation in oncogenic stress-mediated p53 responses. Nature. 2010;464:624–U193.
Ko A, Shin JY, Seo J, Lee KD, Lee EW, Lee MS, et al. Acceleration of gastric tumorigenesis through MKRN1-mediated posttranslational regulation of p14ARF. J Natl Cancer. 2012;104:1660–72.
Wang XW, Zha M, Zhao XC, Jiang P, Du WJ, Tam AYH, et al. Siva1 inhibits p53 function by acting as an ARF E3 ubiquitin ligase. Nat Commun. 2013;4:1551.
Yuan J, Luo KT, Zhang LZ, Cheville JC, Lou ZK. USP10 regulates p53 localization and stability by deubiquitinating p53. Cell. 2010;140:384–U121.
Boulkroun S, Ruffieux-Daidie D, Vitagliano JJ, Poirot O, Charles RP, Lagnaz D, et al. Vasopressin-inducible ubiquitin-specific protease 10 increases ENaC cell surface expression by deubiquitylating and stabilizing sorting nexin 3. Am J Physiol-Ren. 2008;295:F889–F900.
Bomberger JM, Barnaby RL, Stanton BA. The deubiquitinating enzyme USP10 regulates the post-endocytic sorting of cystic fibrosis transmembrane conductance regulator in airway epithelial cells. J Biol Chem. 2009;284:18778–89.
Deng M, Yang X, Qin B, Liu TZ, Zhang HX, Guo W, et al. Deubiquitination and activation of AMPK by USP10. Mol Cell. 2016;61:614–24.
Draker R, Sarcinella E, Cheung P. USP10 deubiquitylates the histone variant H2A.Z and both are required for androgen receptor-mediated gene activation. Nucleic Acids Res. 2011;39:3529–42.
Guturi KKN, Bohgaki M, Bohgaki T, Srikumar T, Ng D, Kumareswaran R, et al. RNF168 and USP10 regulate topoisomerase II alpha function via opposing effects on its ubiquitylation. Nat Commun. 2016;7:12638.
Zhang M, Hu C, Tong D, Xiang SY, Williams K, Bai WL, et al. Ubiquitin-specific peptidase 10 (USP10) deubiquitinates and stabilizes MutS Homolog 2 (MSH2) to regulate cellular sensitivity to DNA damage. J Biol Chem. 2016;291:10783–91.
Pomerantz J, Schreiber-Agus N, Liegeois NJ, Silverman A, Alland L, Chin L, et al. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2’s inhibition of p53. Cell. 1998;92:713–23.
Adhikary S, Eilers M. Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol. 2005;6:635–45.
Wadhwa R, Sugihara T, Hasan MK, Taira K, Reddel RR, Kaul SC. A major functional difference between the mouse and human ARF tumor suppressor proteins. J Biol Chem. 2002;277:36665–70.
Jacobs JJ, Scheijen B, Voncken JW, Kieboom K, Berns A, van Lohuizen M. Bmi-1 collaborates with c-Myc in tumorigenesis by inhibiting c-Myc-induced apoptosis via INK4a/ARF. Genes Dev. 1999;13:2678–90.
Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35.
Ozkara HA, Ozkara S, Topcu S, Criss WE. Amplification of the c-myc oncogene in non-small cell lung cancer. Tumori. 1999;85:508–11.
Mitani S, Kamata H, Fujiwara M, Aoki N, Tango T, Fukuchi K, et al. Analysis of c-myc DNA amplification in non-small cell lung carcinoma in comparison with small cell lung carcinoma using polymerase chain reaction. Clin Exp Med. 2001;1:105–11.
Kuo ML, den Besten W, Bertwistle D, Roussel MF, Sherr CJ. N-terminal polyubiquitination and degradation of the Arf tumor suppressor. Genes Dev. 2004;18:1862–74.
Sung YH, Jin Y, Kim S, Lee HW. Generation of knockout mice using engineered nucleases. Methods. 2014;69:85–93.
Acknowledgements
This work was supported by grants from the Basic Science Research Program of the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT and Future Planning (2014R1A1A1002589) and Creative Research Initiative Program of NRF (2015R1A3A2066581) funded by the Ministry of Science, ICT.
Author contributions
JS and AK conceived and designed the projects and wrote the manuscript. AK and SYH performed the experiments. MSL, SYK and HWL provided technical and material support. CHC, HC, JSS, KH, SMH, and JYC performed the clinical analysis.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Edited by R.A. Knight
These authors contributed equally: Aram Ko, Su Yeon Han.
Rights and permissions
About this article
Cite this article
Ko, A., Han, S.Y., Choi, C.H. et al. Oncogene-induced senescence mediated by c-Myc requires USP10 dependent deubiquitination and stabilization of p14ARF. Cell Death Differ 25, 1050–1062 (2018). https://doi.org/10.1038/s41418-018-0072-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-018-0072-0
This article is cited by
-
LINC00240 in the 6p22.1 risk locus promotes gastric cancer progression through USP10-mediated DDX21 stabilization
Journal of Experimental & Clinical Cancer Research (2023)
-
A microprotein N1DARP encoded by LINC00261 promotes Notch1 intracellular domain (N1ICD) degradation via disrupting USP10-N1ICD interaction to inhibit chemoresistance in Notch1-hyperactivated pancreatic cancer
Cell Discovery (2023)
-
CRL2-KLHDC3 E3 ubiquitin ligase complex suppresses ferroptosis through promoting p14ARF degradation
Cell Death & Differentiation (2022)
-
Of the many cellular responses activated by TP53, which ones are critical for tumour suppression?
Cell Death & Differentiation (2022)
-
Genetic, parental and lifestyle factors influence telomere length
Communications Biology (2022)