Abstract
Herein, Tudor-SN was identified as a DNA damage response (DDR)-related protein that plays important roles in the early stage of DDR. X-ray or laser irradiation could evoke the accumulation of Tudor-SN to DNA damage sites in a poly(ADP-ribosyl)ation-dependent manner via interaction with PARP-1. Additionally, we illustrated that the SN domain of Tudor-SN mediated the association of these two proteins. The accumulated Tudor-SN further recruited SMARCA5 (ATP-dependent chromatin remodeller) and GCN5 (histone acetyltransferase) to DNA damage sites, resulting in chromatin relaxation, and consequently activating the ATM kinase and downstream DNA repair signalling pathways to promote cell survival. Consistently, the loss-of-function of Tudor-SN attenuated the enrichment of SMARCA5, GCN5 and acetylation of histone H3 (acH3) at DNA break sites and abolished chromatin relaxation; as a result, the cells exhibited DNA repair and cell survival deficiency. As Tudor-SN protein is highly expressed in different tumours, it is likely to be involved in the radioresistance of cancer treatment.
Similar content being viewed by others
Introduction
The integrity of genetic information is constantly subjected to exogenous threats, such as genotoxic agents, ionizing radiation (IR), ultraviolet radiation (UV) and endogenous changes [1, 2]. These threats induce an extensive range of DNA damage. Double-strand breaks (DSBs) are potentially the most harmful lesions, as they can eventually cause inflammation or tumourigenesis if the lesions are not promptly and correctly repaired [3, 4].
Upon encountering these threats, cells activate a DNA damage response (DDR), which is a sophisticated and coordinated process consisting of DNA lesion recognition, checkpoint activation, DNA repair and chromatin remodelling and apoptosis [5, 6]. One of the earliest events of DDR is the recruitment and activation of poly(ADP-ribose) polymerase-1 (PARP-1), which acts as a key molecular sensor of DNA breaks [7]. After activation, PARP-1 transfers ADP-ribose units from NAD+ to histones, acceptor proteins and PARP-1 itself, which is essential for initiating various forms of DDR pathways and facilitating DNA repair [7]. Either directly or via the recruitment of proteins, DNA damage-associated poly(ADP-ribosyl)ation (PAR), such as macroH2A1.1, the chromatin remodelling enzyme ALC1 and CHD4, induces the alteration of chromatin structures, resulting in chromatin relaxation and recruitment of other DDR proteins to the sites of DNA break [8,9,10]. Chromatin conformation is an essential event of DDR, in which DNA damage is efficiently accomplished [11, 12], as chromatin needs to undergo relaxation and be accessible to enable the accumulation of repair factors to the lesions. Although ATP-dependent chromatin remodelling and histone modifications are acknowledged for the modulation of chromatin structures in response to DNA damage [13], the underlying molecular mechanisms are sophisticated and remain elusive.
Tudor staphylococcal nuclease (Tudor-SN), which is also known as p100 or SND1 (staphylococcal nuclease domain containing 1), is implicated in a variety of cellular processes [14,15,16,17]. Tudor-SN was recently reported to be involved in apoptotic cell death [18, 19], which supports the idea that Tudor-SN is a likely participant in DDR. In this study, we identified Tudor-SN as a DDR-related protein, which plays important roles in regulating chromatin relaxation and activating the DDR pathway.
Results
PARP-1 interacts with and poly(ADP-ribosyl)ates Tudor-SN in response to DNA damage
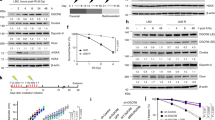
To investigate the biological function of Tudor-SN in DDR, we employed co-immunoprecipitation and mass spectrometry in wild-type mouse embryonic fibroblast (MEF-WT) to interrogate Tudor-SN interactome. The mass spectrometry data indicated that Tudor-SN was associated with multiple proteins (Fig. 1a). Interestingly, PARP-1—a DNA damage sensor—was also identified in the Tudor-SN-containing protein complex (Fig. 1a). We therefore performed a co-immunoprecipitation assay to verify that the endogenous Tudor-SN could physically interact with PARP-1 in an IR treatment-independent manner (Fig. 1b, c).
PARP-1 interacts with and poly(ADP-ribosyl)ates Tudor-SN upon DNA damage. a Mass spectrometry analysis of Tudor-SN-associated proteins visualized by silver staining. b, c Endogenous Tudor-SN associates with PARP-1 in vivo. d Tudor-SN is poly(ADP-ribosyl)ated by PARP-1 in vivo. MEF-WT cells pretreated with vehicle or PJ-34 (10 μm) were untreated or exposed to 10 Gy IR and were immunoprecipitated with anti-Tudor-SN. e GST pulldown assay was performed to identify the interaction domain of Tudor-SN to PARP-1 (middle panel). GST fusion proteins were visualized by Coomassie Blue staining (lower panel). f Mapping the interaction domain of Tudor-SN to PARP-1
Poly(ADP-ribosyl)ation, which is catalysed by poly(ADP-ribose) polymerases (PARPs), has been demonstrated as a pivotal process of DDR to monitor DNA breaks and facilitate DNA repair [20]. To determine whether Tudor-SN is modified by PARP-1, MEF-WT cells were treated with or without PJ-34, which is a PARP-1 inhibitor. As shown in Fig. 1d, a very low level of poly(ADP-ribosyl)ated Tudor-SN was observed in untreated cells; however, it was significantly increased after exposure to IR. As expected, the addition of PJ-34 severely suppressed the DNA damage-induced poly(ADP-ribosyl)ation of Tudor-SN. These data suggest that Tudor-SN is modified by poly(ADP-ribosyl)ation in response to DNA damage.
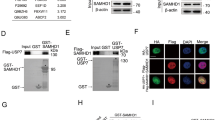
A GST pulldown assay and co-immunoprecipitation assay were performed to map the specific domain of Tudor-SN that interacts with PARP-1. The bacterially produced GST-fusion proteins that contain full-length Tudor-SN (GST-FL), SN domain (GST-SN), TSN domain (GST-TSN) or GST alone were purified using glutathione-Sepharose beads. Equal amounts of fusion proteins were employed to bind assays with PARP-1 protein in MEF-WT cells. As shown in Fig. 1e, the full-length Tudor-SN and SN domain efficiently associated with PARP-1; the TSN domain was not effective. The physical interaction was determined in HeLa cells with ectopic overexpression of Flag-tagged full-length Tudor-SN (Flag-FL), the SN domain (Flag-SN), the TSN domain (Flag-TSN) or the control vector, as indicated. As shown in Fig. 1f, either the FL domain or the SN domain but not the TSN domain or negative ctrl associated with endogenous PARP-1. These data indicate that the SN domain of Tudor-SN mediates the interaction of these two proteins. To investigate whether SN domain can be modified by PARP-1, we ectopically overexpressed different Flag-tagged domains of Tudor-SN in HeLa cells. As shown in Fig. 1f, the SN domain and the full-length Tudor-SN were able to be poly(ADP-ribosyl)ated upon DNA damage. However, no detectable poly(ADP-ribosyl)ation of TSN domain was observed. Collectively, these data strongly support the idea that Tudor-SN is a potential substrate of PARP-1 that can be poly(ADP-ribosyl)ated in response to DNA damage.
DDR causes the accumulation of poly(ADP-ribosyl)ated Tudor-SN protein to the DNA damage sites
One of the hallmarks of DDR-related proteins is the local accumulation at DNA damage sites [21]. To investigate whether Tudor-SN is a bona fide protein that is involved in DNA damage, we performed a laser-induced DNA breaks assay to detect the accumulation of Tudor-SN at DNA damage sites. HeLa cells were transfected with GFP alone (ctrl) or ectopically overexpressed with GFP-tagged full-length Tudor-SN (GFP-FL) and then subjected to laser microirradiation. Analysis of GFP signal in the cells (Fig. 2a) indicated that GFP-tagged Tudor-SN was rapidly accumulated at irradiated sites. We further demonstrated that the SN domain (GFP-SN) was recruited to the damaged sites after laser exposure but the TSN domain (GFP-TSN) was not recruited (Fig. 2b). Poly(ADP-ribosyl)ation modification has been reported to play an important role in the recruitment of DDR-related proteins to DNA damage sites [22, 23]. Consistent with this concept, no detectable accumulation of SN domain or Tudor-SN was observed at the DNA lesion region with the addition of PJ-34 (Fig. 2c).
PARP-1 regulates Tudor-SN recruitment to DSBs. HeLa cells transiently transfected with GFP-FL (a), GFP-SN and GFP-TSN (b), were subjected to laser microirradiation. Representative fields were acquired at the indicated time points after microirradiation. Scale bar: 10 μm. c HeLa cells transiently transfected with GFP-FL and GFP-SN were subjected to laser microirradiation in the absence or presence of PJ-34 (10 μm). Scale bar: 10 μm. d ChIP was performed with anti-Tudor-SN or anti-IgG as a negative control in DR-GFP U2OS cells transfected with control vector or I-SceΙ. Error bars indicate the s.d. Statistical analysis was performed using a two-tailed unpaired Student’s ttest; **P < 0.01 (n = 3). e Western blotting of HA-I-SceΙ
To verify that Tudor-SN can be recruited to the DNA damage sites, we took advantage of chromatin immunoprecipitation (ChIP) assay with U2OS-DR-GFP reporter cells [24, 25]. As shown in Fig. 2d, the ChIP assay revealed that Tudor-SN was enriched in the DNA damage sites. The overexpression of the endonuclease I-SceΙ was verified by western blotting (Fig. 2e). These data are consistent with previous results that reveal that DNA damage can enhance poly(ADP-ribosyl)ation modification of Tudor-SN, which is required for the efficient accumulation of Tudor-SN at the DNA damage sites, and demonstrate that Tudor-SN is a novel DDR-related protein.
Tudor-SN plays an important role in cell survival in response to DNA damage
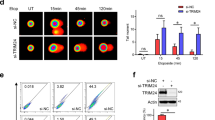
To investigate the physiological function of Tudor-SN on the cellular response to radiation, we performed MTT and clonogenic cell survival assays to assess cell viability. As shown in Fig. 3a, b, the loss-of-function of Tudor-SN in MEF-KO cells exhibited a significant reduction in cell viability and survival in response to IR compared with MEF-WT cells. A statistical analysis (Fig. 3c) verified that the loss of Tudor-SN function sensitized cells to DNA damage. As shown in Fig. 3d, Tudor-SN was efficiently knocked out in MEF-KO cells. Supportively, knockout of Tudor-SN in HeLa (HeLa-KO) cells by CRISPR/Cas9 system strongly impeded colony formation (Fig. 3e). We performed rescue experiments in HeLa-KO cells. The colony formation assays indicate that ectopic overexpression of Tudor-SN, but not the negative vector control, can largely rescue the phenotype evoked by loss-of-function of Tudor-SN, which indicates that Tudor-SN is a key mediator of DNA damage-induced cell viability (Fig. 3e, f). The protein levels of Tudor-SN in different samples were verified by western blotting (Fig. 3g). Moreover, we employed flow cytometry to determine that the apoptosis rate of MEF-KO cells significantly increased with IR treatment in a time-dependent manner compared with MEF-WT cells (Fig. 3h, i). Collectively, these data demonstrate that Tudor-SN plays an important role in the evasion of cell death to promote cell survival.
Downregulation of Tudor-SN sensitizes cells to DNA damage. a Cell viability of MEF cells after IR. MEF-WT and MEF-KO cells were treated with the indicated doses of radiation and analysed using an MTT assay after 3 days in culture. Error bars indicate the s.d. Statistical analysis was performed using a two-tailed unpaired Student’s ttest; *, P < 0.05 (n = 3). b MEF-WT and MEF-KO cells were treated with increasing doses of radiation and evaluated by a colony-forming assay after 10 days of culture. c Quantification of survival rate. Error bars indicate the s.d. Statistical analysis was performed using a two-tailed unpaired Student’s ttest; *P < 0.05 (n = 3); **P < 0.01 (n = 3). d Expression level of Tudor-SN in MEF-WT and MEF-KO cells. e Colony formation assay. HeLa-KO cells were transfected with Flag-Tudor-SN or vector and irradiated with the indicated doses of IR. f Quantification of survival rate. Error bars indicate the s.d. Statistical analysis was performed using a two-tailed unpaired Student’s ttest; *, P < 0.05 (n = 3). g Western blotting of Tudor-SN. h Compared with MEF-WT cells (24 h: 10.83 ± 1.26%, 48 h: 14.37 ± 1.76%, 72 h: 25.67 ± 2.43%), the apoptosis rate of MEF-KO cells was significantly increased (24 h: 20.06 ± 1.54%, 48 h: 29.08 ± 2.44%, 72 h: 41.42 ± 3.08%) with 20 Gy IR treatment. i Quantification of apoptosis percentage. Error bars indicate the s.d. Statistical analysis was performed using a two-tailed unpaired Student’s ttest; *P < 0.05 (n = 3); **P < 0.01 (n = 3)
Tudor-SN accelerates chromatin relaxation in response to DNA damage
An early event in the DDR associated with post-translational poly(ADP-ribosyl)ation modification activity is the induction of chromatin relaxation, which enables DDR-related proteins to access the lesion sites [26]. Consistently, chromatin relaxation analysis showed that, in MEF-WT cells (lanes 6 and 7), but not MEF-KO cells with loss-of-function of Tudor-SN (lanes 9 and 10), the chromatin degraded into fragments with the addition of micrococcal nuclease (MNase) after IR treatment. The protein levels of Tudor-SN in different samples are shown in Fig. 4b. This finding demonstrates that Tudor-SN plays a crucial role in chromatin relaxation in response to DNA damage.
Tudor-SN mediates chromatin relaxation in response to DNA damage. a Chromatin relaxation assay was performed with different concentration of the nuclease (0, 0.5, or 0.8 U) to determine the extent of MNase sensitivity in MEF cells with 20 Gy IR. b Expression level of Tudor-SN in MEF-WT and MEF-KO cells. c, d Endogenous Tudor-SN associates with SMARCA5 in vivo. e, f Endogenous Tudor-SN associates with GCN5 in vivo. MEF-WT cells untreated or irradiated with 10 Gy IR. g Knockout of Tudor-SN reduces the acetylation level of H3K9, 14 and 18 upon DNA damage. h ChIP assay was performed with anti-Tudor-SN, anti-γH2AX, anti-acH3, anti-GCN5, anti-SMARCA5 or anti-IgG as a negative control in DR-GFP U2OS cells with or without I-SceΙ transfection. Error bars indicate the s.d. Statistical analysis was performed using a two-tailed unpaired Student’s ttest; **P < 0.01 (n = 3). i−k ChIP of SMARCA5 (i), GCN5 (j) and acH3 (k) in DR-GFP U2OS cells transfected with Tudor-SN siRNA or scramble (Scr) siRNA in the absence or presence of I-SceΙ expression. Error bars indicate the s.d. Statistical analysis was performed using a two-tailed unpaired Student’s ttest; *P < 0.05 (n = 3); **P < 0.01 (n = 3). l Western blotting of Tudor-SN and HA-I-SceΙ
SMARCA5 ATPase, which was also identified in our mass spectrometry data, has been reported to be the catalytic subunit of the SWI/SNF family that contributes to remodel chromatin and facilitates the DNA repair process [25, 27]. Therefore, co-immunoprecipitation assays were performed to show that IR treatment significantly enhanced the association of endogenous Tudor-SN with SMARCA5 (Fig. 4c, d). Additionally, a cooperative activation between ATP-dependent chromatin remodelling complexes and GCN5 is required to facilitate DSB repair [28]. We have recently reported that GCN5, which directly implicates in DSB response, can interact with the Tudor-SN [29]. Thus, we performed co-immunoprecipitation assays to reveal that IR treatment significantly enhanced the interaction of endogenous Tudor-SN and GCN5 compared with the untreated cells (Fig. 4e, f).
GCN5 is a histone acetyltransferase that acetylates histone H3 on lysine 9, 14 and 18 (H3K9ac, H3K14ac and H3K18ac) [30, 31]. The acetylation of these marks is directly involved in the unfolding of high-order chromatin structures in the DDR; thus, we examined the different acetylation status of H3. As shown in Fig. 4g, the acetylation level of H3K9ac, H3K14ac and H3K18ac was significantly increased in an IR dose-dependent manner in MEF-WT cells; however, very low acetylation levels were observed in MEF-KO cells without an observed difference.
Furthermore, we investigate whether the Tudor-SN-dependent chromatin remodelling specifically occurs at DDR foci. We performed a ChIP assay in U2OS-DR-GFP reporter cells and discovered that the effective accumulation of Tudor-SN, γH2AX, acetylation of histone H3 (acH3), GCN5 and SMARCA5 coincided at the I-SceΙ-generated break sites (Fig. 4h). We subsequently performed a ChIP assay with anti-SMARCA5, anti-GCN5 and anti-acH3 in U2OS cells to verify that Tudor-SN is required for chromatin remodelling. As shown in Fig. 4i–k, the loss-of-function of Tudor-SN remarkably attenuates the enrichment of SMARCA5, GCN5 and acH3 at the defined DNA break sites compared with wild-type U2OS cells. The knockdown effect of Tudor-SN and the overexpression of the endonuclease I-SceΙ were verified by western blotting (Fig. 4l). These data indicate that Tudor-SN is involved in the early stage of DDR, which contributes to chromatin relaxation at DNA damage sites.
The activation of ATM signal pathway is remarkably inhibited due to un-sufficient chromatin relaxation impaired by loss-of-function of Tudor-SN protein
DNA damage-induced chromatin relaxation is required and can initiate ATM activation, which is crucial for the initiation of DDR signal pathways [32, 33]. To reveal the essential role of Tudor-SN as a regulator of chromatin remodelling in DDR, we detected the phosphorylation of a subset of ATM substrates, which were consequently activated by triggered ATM activation. As shown in Fig. 5a, IR treatment induced or enhanced the phosphorylation of ATM and downstream targets, including Chk2 and p53 in MEF-WT cells compared with MEF-KO cells.
Tudor-SN facilitates the DNA damage-induced phosphorylation of a subset of ATM substrates. a Total extracts from MEF-WT and MEF-KO cells were immunoblotted against the indicated antibodies following 10 Gy IR. b Expression level of γH2AX in MEF-WT and MEF-KO cells after the indicated doses (0, 2, 6 and 10 Gy) of IR. c The graph represents densitometric units normalized to H2AX. Error bars indicate the s.d. Statistical analysis was performed using a two-tailed unpaired Student’s ttest; **P < 0.01 (n = 3). d γH2AX expression was detected by flow cytometry after the indicated doses of IR. e Quantification of γH2AX percentage. Error bars indicate the s.d. Statistical analysis was performed using a two-tailed unpaired Student’s ttest; *P < 0.05 (n = 3); **P < 0.01 (n = 3). f Immunofluorescence assay shows significantly reduced γH2AX foci formation in MEF-KO cells than MEF-WT cells after 10 Gy IR. Scale bar: 10 μm. g The quantification of γH2AX foci number per nucleus. Fifty nuclei were counted in each group. Error bars indicate the s.d. Statistical analysis was performed using a two-tailed unpaired Student’s ttest; **P < 0.01 (n = 3). h Co-localization of endogenous Tudor-SN with γH2AX upon DNA damage. The distribution of endogenous Tudor-SN and γH2AX was detected using immunofluorescence with anti-Tudor-SN and anti-γH2AX by confocal microscopy analysis. Scale bar: 10 μm
Phosphorylation of histone variant H2AX (γH2AX) is a hallmark of DDR activation mediated by ATM and is essential for the accumulation of repair factors and amplification of the DNA damage signal [34]. Therefore, we employed different methodologies to examine the γH2AX level in MEF cells with loss-of-function of Tudor-SN. As shown in Fig. 5b, c, the protein level of γH2AX was significantly increased in MEF-WT cells in a dose-dependent manner with IR treatment but was rarely detectable in MEF-KO cells. Flow cytometry demonstrated an increased number of MEF-WT cells with γH2AX in a dose-dependent manner after IR treatment, but this was not observed in MEF-KO cells with impaired γH2AX levels (Fig. 5d, e). In addition, the immunofluorescence assay illustrated that MEF-WT cells displayed a significantly large number of γH2AX foci after IR, whereas less and weaker γH2AX foci formation was detected in MEF-KO cells for the same condition (Fig. 5f, g), which suggests a prominent role of Tudor-SN in the formation of γH2AX foci. Additionally, immunofluorescence assay demonstrated the co-localization of Tudor-SN and γH2AX in the foci of the cells with IR treatment (Fig. 5h). Collectively, these findings support the idea that less phosphorylation of ATM signal pathway in MEF-KO cells is not attributed to less DNA damage but rather is due to inefficient chromatin relaxation at DSBs, which is consistent with the observation that loss-of-function of Tudor-SN causes inefficient chromatin relaxation.
DNA repair deficiency occurs in the cells with loss-of-function of Tudor-SN
The functional consequences of ATM signal pathway phosphorylation include the activation of cell cycle checkpoints, including the G2/M checkpoint in response to IR [35]. To address the possible involvement of Tudor-SN in cell cycle arrest after DNA damage, we performed cell cycle analyses. As shown in Fig. 6a, b, IR induced the accumulation of MEF-WT cells in the G2/M phases of the cell cycle. This accumulation was not observed in the MEF-KO cells, which suggests that Tudor-SN is important for the regulation of the cell cycle arrest in the G2/M phases after DNA damage, which provides time for repair of the DNA lesion.
Tudor-SN depletion impairs G2/M arrest and DNA repair. a The cell cycle distribution was detected by flow cytometry after IR treatment. b The percentage of cells in each phase are shown in the histogram. Error bars indicate the s.d. Statistical analysis was performed using a two-tailed unpaired Student’s ttest; *P < 0.05 (n = 3); **P < 0.01 (n = 3). c Assessment of DNA repair by comet assay. The comet tails of MEF-KO cells (52.2 ± 4.07 μm) were longer than the comet tails of MEF-WT cells (23.5 ± 3.51 μm) after 10 Gy IR treatment. d Comet tail length was determined for 50 cells using Casplab-Comet Assay Software. Error bars indicate the s.d. Statistical analysis was performed using a two-tailed unpaired Student’s ttest; **P < 0.01 (n = 3). e Expression level of Tudor-SN in MEF-WT and MEF-KO cells. f In the HR assay, with ectopic overexpression of I-SceI in the cells, approximately 4.42±0.55% of GFP-positive cells were observed in the control U2OS-Scr cells, whereas fewer GFP-positive cells (2.65±0.31%) appeared in the U2OS-siRNA cells. g Quantification of GFP-positive cell percentage in the HR assay. Error bars indicate the s.d. Statistical analysis was performed using a two-tailed unpaired Student’s ttest; *P < 0.05 (n = 3). h In the NHEJ assay, with ectopic overexpression of I-SceI, the percentage of GFP-positive cells (3.66±0.49%) in HEK 293-siRNA cells was lower than the percentage of GFP-positive cells (6.32±0.71%) in the HEK 293-Scr control group. i Quantification of GFP-positive cell percentage in the NHEJ assay. Error bars indicate the s.d. Statistical analysis was performed using a two-tailed unpaired Student’s ttest; *P < 0.05 (n = 3). j U2OS and HEK 293 cells were transfected with Tudor-SN siRNA or Scr siRNA, and the transfection efficiency was detected by western blotting
Next, we investigate the impact of the loss-of-function of Tudor-SN on DNA repair activity to address the essential function of Tudor-SN in the early events of DDR. We performed a comet assay, which is an extremely sensitive assay, to evaluate the occurrence of DNA repair in MEF cells. As shown in Fig. 6c, d, the comet tails of MEF-KO cells were, on average, approximately 2.2-fold longer than the comet tails of MEF-WT cells after IR treatment, which indicated that the loss-of-function of Tudor-SN in MEF-KO cells impaired the DNA repair ability. The protein level of Tudor-SN in different samples is shown in Fig. 6e.
We also performed HR and NHEJ assays, which are two major methods for repairing DSBs, to demonstrate the importance of Tudor-SN in activating the early stage of DDR. DSBs were induced by the transient expression of I-SceΙ in different cells. Once the DSB is repaired by HR or NHEJ, a functional GFP gene is generated. Analysis of HR and NHEJ assay (Fig. 6f–i) indicated that depletion of Tudor-SN expression was associated with a reduced percentage of GFP-positive cells to nearly 40% of that of control cells, suggesting that Tudor-SN is required for efficient DNA repair. The knockdown effect of Tudor-SN and the overexpression of the endonuclease I-SceΙ were verified by western blotting (Fig. 6j). These data are consistent with our findings that Tudor-SN plays an important role in regulating chromatin relaxation to enable the accumulation of repair factors to the lesions, efficiently accomplishing DNA damage repair to promote cell survival under genotoxic stress.
Discussion
Cells utilize a coordinated series of events to counteract DNA damage. Herein, we demonstrate that the recruitment of Tudor-SN to the DNA damage sites is dependent on poly(ADP-ribosyl)ation modification. We first observed the physical interaction between PARP-1 and Tudor-SN and illustrated that Tudor-SN, as a novel substrate of PARP-1, can be efficiently poly(ADP-ribosyl)ated after initiation of the DDR. Most important, the laser microirradiation data indicate that poly(ADP-ribosyl)ation modification is required for the accumulation of Tudor-SN at DSB sites after DNA damage. Although the exact function of poly(ADP-ribosyl)ation formation at DSBs and its impact on DDR remains ambiguous, an increasing number of chromatin remodellers are recruited to DNA break sites in a poly(ADP-ribosyl)ation dependent-manner to assist the chromatin conformation change, for example, the early recruitment of BRCA1-BARD complex [36] and scaffold attachment factor B1 (SAFB1) [37].
Chromatin conformation is an essential component of the initiation of DDR signal and DNA repair [13]. Our data suggest that Tudor-SN is a critical factor that is capable of remodelling chromatin at the early stage in DDR. The recruitment of chromatin modifiers at DNA damage sites involves many different proteins with highly specific functions. For example, RBMX [38], hnRNPUL1/2 [39] and SAFB1 [37] can be highly diverse. Unknown factors may determine the formation of DNA-damage-induced chromatin structural components. Thus, Tudor-SN is likely to be an uncharacterized regulator of architectural chromatin conformation, which relocalizes in response to DNA damage and plays an important role in chromatin remodelling.
Tumourigenesis is accompanied by various stresses that tumour cells have to conquer, including oncogene-induced DNA replication or DNA damage stress [40]. DDR is activated by oncogenic stress and constitutes barriers to tumourigenesis. The interaction of Tudor-SN with MTDH has been reported to promote tumourigenesis by regulating tumour-initiating cells [40]. As a novel DDR-related protein, how does Tudor-SN function in tumourigenesis? One possibility is that Tudor-SN acts as a pro-survival protein and prevents cell death in various stress conditions. Derangements of DDR in cells trigger tumourigenesis by allowing evasion of anti-proliferative and cell death, which may have otherwise occurred secondary to DNA damage [41]. According to recent reports, Tudor-SN is a bona fide novel component of stress granules to protect cells from a variety of environmental stress stimuli [15, 16]. Our data consistently suggest the dual role of Tudor-SN in pro-survival: it accelerates chromatin relaxation, which enables DDR-related proteins to access DNA damage sites, to robustly promote cell survival in response to DNA damage in both normal cells and cancer cells.
Ionizing radiation represents the most effective therapy for a variety of tumours with the aim of inducing cell death. However, DNA damage activates the DDR to alleviate this damage and promote cell survival [3]. Enhancement of DDR activation in cancer cells contributes to radioresistance through preferential activation of the DNA-damage checkpoint and increased efficiency of DNA repair [42]. The increased expression of Ku80, which is a key regulator of NHEJ DNA repair, is reported to be involved in radioresistance, with worsened locoregional recurrence and total survival [43]. Thyroid hormone receptor interactor 13 (TRIP13), which facilitates NHEJ DNA repair, has also been identified as a key contributor to radioresistance [44]. In addition, overexpression of the Rad51 has also been implicated in radioresistance due to its role in HR DNA repair [45]. Due to the frequently observed and increased expression of Tudor-SN in various human tumours [49, 50, 51], and most importantly, we observed that the cells with loss-of-function of Tudor-SN cannot efficiently accomplish either NHEJ or HR DNA repair; it suggests that Tudor-SN is likely to be involved in the radioresistance of cancer treatment, which is consistent with the effects of Tudor-SN on DNA repair and cell survival after IR. Although DDR and the alternative reading frame (ARF) are commonly considered to function independently, it was reported recently that inhibition of ATM correlated with increased ARF levels and stimulated the tumour-suppressive effects [46]. As Tudor-SN is highly expressed in tumours and facilitates chromatin relaxation and activates the ATM signal pathway, it is highly possible that there is a potential link between increased expression of Tudor-SN in human tumours with aberrant expression of DDR and possible suppression of ARF via ATM conferring to radioresistance. However, further studies are needed to determine the hypothesis.
In summary, our study provides evidence that Tudor-SN is a novel regulator in DDR (Fig. 7). After DNA damage, Tudor-SN is accumulated at DSB sites in a poly(ADP-ribosyl)ation modification dependent-manner mediated by PARP-1, in which Tudor-SN acts as a novel regulator that facilitates chromatin relaxation and activates the ATM signal pathway to initiate the DDR. Defects in the capacity to precisely respond to the DDR from DNA damage signalling to cell cycle checkpoints or DNA repair are associated with the initiation and progression of many forms of cancer [47, 48]. Tudor-SN has been reported to be highly expressed in various tumours; our findings may provide new insight into potential therapeutic targets that can sensitize cancer cells to radiotherapy or chemotherapy.
Materials and methods
Cell culture and transfections
MEF and HeLa cells were obtained from the American Type Culture Collection (Manassas, VA) and cultured according to the manufacturer’s instructions. The Tudor-SN knockout mouse was generated in the C57BL/6N background in the Silvennoinen laboratory. MEF-KO cells were generated from at least six generations of backcrossing to Tudor-SN knockout C57BL/6N at the Turku Center for Disease Modelling (TCDM) by standard procedures and generously sent to us as a gift. DR-GFP-U2OS and EJ5-GFP-HEK 293 cell lines were provided by Dr. Lei Shi (Tianjin Medical University, Tianjin). The cells were maintained in Dulbecco’s modified Eagle’s medium, which was supplemented with 10% fetal bovine serum. Transfection of U2OS and HEK 293 cells was performed using the Lipofectamine 2000 transfection reagent (11668-019, Invitrogen) according to the manufacturer’s protocol. Tudor-SN siRNA was generated as previously reported [52]. A HeLa stable cell line with Tudor-SN-Knockout (HeLa-KO) was generated via a modified CRISPR/Cas9 double-nicking gene editing system [16]. All of the cells were authenticated by examination of morphology and growth characteristics, and were confirmed to be mycoplasma-free.
Western blotting and antibodies
Total cell lysates or the immunoprecipitated proteins were finally analysed by SDS-PAGE. Antibodies against PAR (ab14459, 1:500 for WB), γH2AX (phosphor S139) (ab2893, 1:200 for IF and 1:1000 for WB), Histone H3 (ab10799, 1:1000 for WB), Histone H3 (acetyl k9) (ab10812, 1:500 for WB) from Abcam; ATM (NB100-309, 1:1000 for WB) from Novus Biologicals; PARP-1 (sc-8007, IP and 1:1000 for WB) from Santa Cruz Biotechnology, SMARCA5 (A2000, IP and 1:1500 for WB) from abclonal; GCN5 (3305, IP and 1:1000 for WB), p53 (2524, 1:1000 for WB) from CST; acety-Histone H3 (06-599, IP and 1:20,000 for WB) from Millipore. DNA Damage Antibody Sampler Kit (9947) was purchased from CST. The mouse monoclonal anti-Tudor-SN antibody was generated against the SN4 domain (amino acids 507–674) of Tudor-SN at the Institute of Medical Technology, University of Tampere, Finland.
Co-immunoprecipitation
Total cell lysates were collected with Nonidet P-40 lysis buffer (50 mm Tris-HCl, pH 7.6, 150 mm NaCl, 0.1 mm EDTA, 0.5% Nonidet P-40, 20% glycerol, 0.1 mm sodium orthovanadate, 1 mm sodium butyrate) supplemented with PMSF and protease inhibitor mixture (04693124001, Roche Applied Science). Protein concentrations of the lysates were measured using the Pierce BCA Protein Assay Kit (number 23227, Thermo Scientific). Total cell lysates were incubated with the interesting antibodies, mouse polyclonal anti-IgG antibody (Santa Cruz Biotechnology) as a negative control, followed by incubation with Pierce Protein A/G agarose (20422, Thermo Pierce) overnight at 4 °C in a roller. Ten percent of the total cell lysates were used as input. The bound proteins were subjected to SDS-PAGE and blotted with the corresponding antibodies.
LC-MS/MS analysis and data interpretation
The proteins recovered by immunoaffinity purification were resolved on SDS-PAGE and visualized by silver staining. The corresponding bands were further excised and subjected to in-gel digestion as described previously [53]. Resulting peptides were separated by reverse phase liquid chromatography on an easy-nLC 1000 system (Thermo Fisher Scientific) and directly sprayed into a Q-Exactive mass spectrometer (Thermo Fisher Scientific). All MS/MS spectra were searched against the Uniport-Human protein sequence database using the PD search engine (version 2.1.0, Thermo Fisher Scientific) with an overall false discovery rate for peptides of less than 1%. The identified MS/MS spectra were manually verified.
X-ray irradiation and Laser microirradiation
IR was delivered by an X-ray generator (RS2000 PRO, 160 kV, 25 mA; Radsource Corporation). For laser irradiation, DSBs were induced by exposure of cells to a UV-A laser. Cells cultured in 35 mm coverglass dishes were treated with 10 mm 5-bromo-2-deoxyuridine (BrdU) for 24 h. Microirradiation was performed with a Olympus FluoView microscope equipped with a 37 °C heating stage and a 405-nm laser diode. The maximum power of laser setting was chosen to scan lines across nuclei to induce DNA damage.
ChIP assay
ChIP assay was performed in U2OS-DR-GFP reporter cells, in which a defective GFP cassette that contains the I-SceI recognition site is stably incorporated into genome and DNA damage may be caused by a unique DNA cut induced by transient transfection of the I-SceI endonuclease [24, 25]. Formaldehyde was added to the DR-GFP U2OS cells transfected with control or Tudor-SN siRNA in the absence or presence of I-SceΙ expression at a final concentration of 1%. Cross-linking was quenched by the addition of glycine to a final concentration of 100 mm for 10 min. The fixed cells were resuspended in SDS lysis buffer containing 1% SDS, 5 mm EDTA and 50 mm Tris-HCl (pH 8.1). The soluble chromatin was prepared with sonication using a Vibra Cell 500 watt sonicator (Sonics & Materials, Inc., Newtown, CT). After centrifugation, the diluted chromatin was incubated with control or specific antibodies for 12 h at 4 °C, and the immune complexes were collected onto protein G magnetic beads. Beads were washed three times with buffer containing 10 mm Tris-HCl (pH 8.0), 140 mm NaCl, 1 mm EDTA, 0.1% Triton X-100, and once with TE buffer (10 mm Tris-HCl, pH 8.0, 1 mm EDTA). The chromatin fragments were eluted from the beads with elution buffer containing 62.5 mm Tris-HCl (pH 6.8), 200 mm NaCl, 2% SDS, and 10 mm dithiothreitol, and cross-links were reverted by heating at 65 °C overnight. Eluted DNA was purified with PCR purification kit (Qiagen) and analysed by quantitative PCR (qPCR) using primer DR-GFP Set9: forward, ATCACATGGTCCTGCTGGAGTT, reverse, TGGCTGATTATGATCTAGAGTCGCGG.
GST pulldown assays
GST, GST-Tudor-SN, GST-SN or GST-TSN fusion proteins were produced in BL21 Escherichia coli and purified using glutathione-Sepharose 4B beads (Amersham Biosciences) according to the manufacturer’s instructions. The bead-bound GST fusion proteins were incubated with total cell lysates on a rotator for a further 8 h at 4 °C. Beads were then washed five times with binding buffer containing 75 mm NaCl. The bound proteins were separated by SDS-PAGE and analysed by immunoblotting.
Immunofluorescence
MEF cells were grown on glass coverslips, then fixed for 10 min in 4% paraformaldehyde, permeabilized with 0.5% Triton X-100 in PBS for 5 min, and blocked with 0.1% BSA in PBS for 60 min. The cells were then incubated with specific primary antibodies. After washing, cells were incubated with fluorescent dye-conjugated secondary antibodies at 4 °C overnight. The images were collected using an Olympus FV1000 confocal microscope (Japan).
Chromatin relaxation assay
The chromatin relaxation assay, in which MNase preferentially cuts the DNA in the linker regions between nucleosomes and releases chromatin into fragments, was carried out as previously described [54]. Briefly, MEF cells were harvested after IR and lysed in hypotonic cell lysis buffer (10 mm Tris-HCl, pH 8.0, 10 mm MgCl2, 25% glycerol, 0.2% NP-40, 1 mm DTT, and complete protease inhibitor cocktail Roche) for 30 min. Nuclei were isolated by centrifugation, resuspended in MNase buffer (15 mm Tris-HCl, pH 7.4, 60 mm KCl, 15 mm NaCl, 0.25 m sucrose, 1 mm CaCl2, and 0.5 mm DTT), and digested with MNase (M0247S, New England) for 5 min at 37 °C. The reaction was terminated with stop solution (1% SDS, 20 mm EDTA). DNA was extracted with phenol/chloroform, and the fragments were separated on a 1.2% agarose gel.
Comet assay
DNA damage was analysed with a comet assay. Briefly, MEF cells were exposed to IR and subsequently incubated in normal medium for 2 h before processing. Harvested cells (1×105) were mixed with 0.7% low-melting-point agarose and layered onto agarose-coated slides. Slides were then submerged in lysis buffer (10 mm Tris-HCl, pH 7.5, 2.5 m NaCl, 100 mm EDTA, and 1% Triton X-100) for 2 h at 4 °C. After lysis, the slides were incubated for 40 min in electrophoresis buffer (300 mm sodium acetate, 100 mm Tris-HCl, pH 8.3). After electrophoresis, the slides were washed with water and then air dried and stained with ethidium bromide (20 mg/ml). Comet tail length was measured by fluorescence microscopy and then analysed using Casplab-Comet Assay Software.
HR and NHEJ analysis
HR and NHEJ analysis were performed as previously reported [55, 56]. To measure the HR or NHEJ repair of I-SceΙ-generated DSBs, an I-SceΙ expression vector and Tudor-SN-siRNA were transfected to U2OS cells or HEK 293 cells using Lipofectamine 2000. To determine the amount of HR or NHEJ repair, the percentage of GFP-positive cells was quantified by flow cytometer at 3 days after transfection (Guava EasyCyte, Millipore).
Statistical analysis
Data from biological triplicate experiments are presented with error bar as mean ± s.d. The data were analysed by a two-tailed unpaired Student’s ttest. All of the statistical testing results were determined by Prism 5 programme (GraphPad Software, San Diego, CA, USA). A value of P < 0.05 was considered to be statistically significant. Before statistical analysis, variation within each group of data and the assumptions of the tests were checked.
References
Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204.
Chapman JR, Taylor MR, Boulton SJ. Playing the end game: DNA double-strand break repair pathway choice. Mol Cell. 2012;47:497–510.
Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–8.
Lord CJ, Ashworth A. The DNA damage response and cancer therapy. Nature. 2012;481:287–94.
Downs JA, Nussenzweig MC, Nussenzweig A. Chromatin dynamics and the preservation of genetic information. Nature. 2007;447:951–8.
Giunta S, Belotserkovskaya R, Jackson SP. DNA damage signaling in response to double-strand breaks during mitosis. J Cell Biol. 2010;190:197–207.
Kleine H, Luscher B. Learning how to read ADP-ribosylation. Cell. 2009;139:17–9.
Ahel D, Horejsi Z, Wiechens N, Polo SE, Garcia-Wilson E, Ahel I, et al. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science. 2009;325:1240–3.
Pines A, Vrouwe MG, Marteijn JA, Typas D, Luijsterburg MS, Cansoy M, et al. PARP1 promotes nucleotide excision repair through DDB2 stabilization and recruitment of ALC1. J Cell Biol. 2012;199:235–49.
Li M, Yu X. The role of poly(ADP-ribosyl)ation in DNA damage response and cancer chemotherapy. Oncogene. 2015;34:3349–56.
Wiest NE, Houghtaling S, Sanchez JC, Tomkinson AE, Osley MA. The SWI/SNF ATP-dependent nucleosome remodeler promotes resection initiation at a DNA double-strand break in yeast. Nucleic Acids Res. 2017;45:5887–900.
Ayrapetov MK, Gursoy-Yuzugullu O, Xu C, Xu Y, Price BD. DNA double-strand breaks promote methylation of histone H3 on lysine 9 and transient formation of repressive chromatin. Proc Natl Acad Sci USA. 2014;111:9169–74.
Price BD, D’Andrea AD. Chromatin remodeling at DNA double-strand breaks. Cell. 2013;152:1344–54.
Duan Z, Zhao X, Fu X, Su C, Xin L, Saarikettu J, et al. Tudor-SN, a novel coactivator of peroxisome proliferator-activated receptor gamma protein, is essential for adipogenesis. J Biol Chem. 2014;289:8364–74.
Gao X, Fu X, Song J, Zhang Y, Cui X, Su C, et al. Poly(A)(+) mRNA-binding protein Tudor-SN regulates stress granules aggregation dynamics. FEBS J. 2015;282:874–90.
Su C, Gao X, Yang W, Zhao Y, Fu X, Cui X, et al. Phosphorylation of Tudor-SN, a novel substrate of JNK, is involved in the efficient recruitment of Tudor-SN into stress granules. Biochim Biophys Acta. 2017;1864:562–71.
Gao X, Zhao X, Zhu Y, He J, Shao J, Su C, et al. Tudor staphylococcal nuclease (Tudor-SN) participates in small ribonucleoprotein (snRNP) assembly via interacting with symmetrically dimethylated Sm proteins. J Biol Chem. 2012;287:18130–41.
Sundstrom JF, Vaculova A, Smertenko AP, Savenkov EI, Golovko A, Minina E, et al. Tudor staphylococcal nuclease is an evolutionarily conserved component of the programmed cell death degradome. Nat Cell Biol. 2009;11:1347–54.
Zagryazhskaya A, Surova O, Akbar NS, Allavena G, Gyuraszova K, Zborovskaya IB, et al. Tudor staphylococcal nuclease drives chemoresistance of non-small cell lung carcinoma cells by regulating S100A11. Oncotarget. 2015;6:12156–73.
Kraus WL. PARPs and ADP-ribosylation: 50 years… and counting. Mol Cell. 2015;58:902–10.
Coster G, Goldberg M. The cellular response to DNA damage: a focus on MDC1 and its interacting proteins. Nucleus. 2010;1:166–78.
Khoury-Haddad H, Guttmann-Raviv N, Ipenberg I, Huggins D, Jeyasekharan AD, Ayoub N. PARP1-dependent recruitment of KDM4D histone demethylase to DNA damage sites promotes double-strand break repair. Proc Natl Acad Sci USA. 2014;111:E728–37.
Rulten SL, Rotheray A, Green RL, Grundy GJ, Moore DA, Gomez-Herreros F, et al. PARP-1 dependent recruitment of the amyotrophic lateral sclerosis-associated protein FUS/TLS to sites of oxidative DNA damage. Nucleic Acids Res. 2014;42:307–14.
Li X, Liu L, Yang S, Song N, Zhou X, Gao J, et al. Histone demethylase KDM5B is a key regulator of genome stability. Proc Natl Acad Sci USA. 2014;111:7096–101.
Toiber D, Erdel F, Bouazoune K, Silberman DM, Zhong L, Mulligan P, et al. SIRT6 recruits SNF2H to DNA break sites, preventing genomic instability through chromatin remodeling. Mol Cell. 2013;51:454–68.
Garcia K, Blank JL, Bouck DC, Liu XJ, Sappal DS, Hather G, et al. Nedd8-activating enzyme inhibitor MLN4924 provides synergy with mitomycin C through interactions with ATR, BRCA1/BRCA2, and chromatin dynamics pathways. Mol Cancer Ther. 2014;13:1625–35.
Vidi PA, Liu J, Salles D, Jayaraman S, Dorfman G, Gray M, et al. NuMA promotes homologous recombination repair by regulating the accumulation of the ISWI ATPase SNF2h at DNA breaks. Nucleic Acids Res. 2014;42:6365–79.
Lee HS, Park JH, Kim SJ, Kwon SJ, Kwon J. A cooperative activation loop among SWI/SNF, gamma-H2AX and H3 acetylation for DNA double-strand break repair. EMBO J. 2010;29:1434–45.
Yu L, Di Y, Xin L, Ren Y, Liu X, Sun X, et al. SND1 acts as a novel gene transcription activator recognizing the conserved Motif domains of Smad promoters, inducing TGFbeta1 response and breast cancer metastasis. Oncogene. 2017;36:3903–14.
Guo R, Chen J, Mitchell DL, Johnson DG. GCN5 and E2F1 stimulate nucleotide excision repair by promoting H3K9 acetylation at sites of damage. Nucleic Acids Res. 2011;39:1390–7.
Luijsterburg MS, van Attikum H. Chromatin and the DNA damage response: the cancer connection. Mol Oncol. 2011;5:349–67.
Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506.
Ortega-Atienza S, Wong VC, DeLoughery Z, Luczak MW, Zhitkovich A. ATM and KAT5 safeguard replicating chromatin against formaldehyde damage. Nucleic Acids Res. 2015;44:198–209.
van Attikum H, Gasser SM. Crosstalk between histone modifications during the DNA damage response. Trends Cell Biol. 2009;19:207–17.
Kamer I, Sarig R, Zaltsman Y, Niv H, Oberkovitz G, Regev L, et al. Proapoptotic BID is an ATM effector in the DNA-damage response. Cell. 2005;122:593–603.
Li M, Yu X. Function of BRCA1 in the DNA damage response is mediated by ADP-ribosylation. Cancer Cell. 2013;23:693–704.
Altmeyer M, Toledo L, Gudjonsson T, Grofte M, Rask MB, Lukas C, et al. The chromatin scaffold protein SAFB1 renders chromatin permissive for DNA damage signaling. Mol Cell. 2013;52:206–20.
Adamson B, Smogorzewska A, Sigoillot FD, King RW, Elledge SJ. A genome-wide homologous recombination screen identifies the RNA-binding protein RBMX as a component of the DNA-damage response. Nat Cell Biol. 2012;14:318–28.
Polo SE, Blackford AN, Chapman JR, Baskcomb L, Gravel S, Rusch A, et al. Regulation of DNA-end resection by hnRNPU-like proteins promotes DNA double-strand break signaling and repair. Mol Cell. 2012;45:505–16.
Wan L, Lu X, Yuan S, Wei Y, Guo F, Shen M, et al. MTDH-SND1 interaction is crucial for expansion and activity of tumor-initiating cells in diverse oncogene- and carcinogen-induced mammary tumors. Cancer Cell. 2014;26:92–105.
Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–70.
Yamamoto VN, Thylur DS, Bauschard M, Schmale I, Sinha UK. Overcoming radioresistance in head and neck squamous cell carcinoma. Oral Oncol. 2016;63:44–51.
Moeller BJ, Yordy JS, Williams MD, Giri U, Raju U, Molkentine DP, et al. DNA repair biomarker profiling of head and neck cancer: Ku80 expression predicts locoregional failure and death following radiotherapy. Clin Cancer Res: Off J Am Assoc Cancer Res. 2011;17:2035–43.
Banerjee R, Russo N, Liu M, Basrur V, Bellile E, Palanisamy N, et al. TRIP13 promotes error-prone nonhomologous end joining and induces chemoresistance in head and neck cancer. Nat Commun. 2014;5:4527.
Scheckenbach K, Baldus SE, Balz V, Freund M, Pakropa P, Sproll C, et al. RAD51C--a new human cancer susceptibility gene for sporadic squamous cell carcinoma of the head and neck (HNSCC). Oral Oncol. 2014;50:196–9.
Velimezi G, Liontos M, Vougas K, Roumeliotis T, Bartkova J, Sideridou M, et al. Functional interplay between the DNA-damage-response kinase ATM and ARF tumour suppressor protein in human cancer. Nat Cell Biol. 2013;15:967–77.
Brady CA, Jiang D, Mello SS, Johnson TM, Jarvis LA, Kozak MM, et al. Distinct p53 transcriptional programs dictate acute DNA-damage responses and tumor suppression. Cell. 2011;145:571–83.
Gupta GP, Vanness K, Barlas A, Manova-Todorova KO, Wen YH, Petrini JH. The Mre11 complex suppresses oncogene-driven breast tumorigenesis and metastasis. Mol Cell. 2013;52:353–65.
Yu L, Liu X, Cui K, Di Y, Xin L, Sun X, et al. SND1 acts downstream of TGFbeta1 and upstream of Smurf1 to promote breast cancer metastasis. Cancer Res. 2015;75:1275–86.
Wang N, Du X, Zang L, Song N, Yang T, Dong R, et al. Prognostic impact of Metadherin−SND1 interaction in colon cancer. Mol Biol Rep. 2012;39:10497–504.
Yoo BK, Santhekadur PK, Gredler R, Chen D, Emdad L, Bhutia S, et al. Increased RNA-induced silencing complex (RISC) activity contributes to hepatocellular carcinoma. Hepatology. 2011;53:1538–48.
Valineva T, Yang J, Palovuori R, Silvennoinen O. The transcriptional co-activator protein p100 recruits histone acetyltransferase activity to STAT6 and mediates interaction between the CREB-binding protein and STAT6. J Biol Chem. 2005;280:14989–96.
Chen Y, Kwon SW, Kim SC, Zhao Y. Integrated approach for manual evaluation of peptides identified by searching protein sequence databases with tandem mass spectra. J Proteome Res. 2005;4:998–1005.
Ziv Y, Bielopolski D, Galanty Y, Lukas C, Taya Y, Schultz DC, et al. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat Cell Biol. 2006;8:870–6.
Sakamoto S, Iijima K, Mochizuki D, Nakamura K, Teshigawara K, Kobayashi J, et al. Homologous recombination repair is regulated by domains at the N- and C-terminus of NBS1 and is dissociated with ATM functions. Oncogene. 2007;26:6002–9.
Shibata A, Moiani D, Arvai AS, Perry J, Harding SM, Genois MM, et al. DNA double-strand break repair pathway choice is directed by distinct MRE11 nuclease activities. Mol Cell. 2014;53:7–18.
Acknowledgements
This work was supported by the National Science Foundation for Distinguished Young Scholars of China (31125012); Innovation Team Development Plan of the Ministry of Education (IRT13085); National Science Foundation of China (31670759, 31370749 to JY, 31501056 to XF, 81572882 to ZY); Natural Science Foundation of Tianjin (16JCQNJC09000); and Canadian Institutes of Health Research (MOP-130423).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Edited by M. Oren
Rights and permissions
About this article
Cite this article
Fu, X., Zhang, C., Meng, H. et al. Oncoprotein Tudor-SN is a key determinant providing survival advantage under DNA damaging stress. Cell Death Differ 25, 1625–1637 (2018). https://doi.org/10.1038/s41418-018-0068-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-018-0068-9
This article is cited by
-
Vascular injury activates the ELK1/SND1/SRF pathway to promote vascular smooth muscle cell proliferative phenotype and neointimal hyperplasia
Cellular and Molecular Life Sciences (2024)
-
ELFN1-AS1 promotes GDF15-mediated immune escape of colorectal cancer from NK cells by facilitating GCN5 and SND1 association
Discover Oncology (2023)
-
SND1 confers chemoresistance to cisplatin-induced apoptosis by targeting GAS6-AKT in SKOV3 ovarian cancer cells
Medical Oncology (2022)