Abstract
p53 mediates the DNA damage response (DDR) by regulating the expression of genes implicated in cell cycle arrest, senescence, programmed cell death (PCD), and metabolism. Herein we demonstrate that human alkaline ceramidase 2 (ACER2) is a novel transcriptional target of p53 and that its transactivation by p53 mediates the DDR. We found that p53 overexpression or its activation by ionizing radiation (IR) upregulated ACER2 in cells. Two putative p53 responsive elements (p53REs) were found in its first intron of the ACER2 gene, and Chromatin Immunoprecipitation (ChIP) assays in combination with promoter activity assays demonstrated that these p53REs are the bona fide p53 binding sites that mediate ACER2 transactivation by p53. As ACER2 catalyzes the hydrolysis of ceramides into sphingosine, which in turn is phosphorylated to form sphingosine-1-phosphate (S1P), ACER2 upregulation increased the levels of both sphingosine and S1P while decreasing the levels of ceramides in cells. A moderate upregulation of ACER2 inhibited cell cycle arrest and cellular senescence in response to low-level expression of p53 or low-dose IR by elevating S1P, a pro-proliferative and pro-survival bioactive lipid, and/or decreasing ceramides whereas its robust upregulation mediated PCD in response to high-level expression of p53 or high-dose IR likely by accumulating cellular sphingosine, a pro-death bioactive lipid. ACER2 is frequently inactivated in various cancers due to its deletion or mutations, and restoring its expression inhibited the growth of tumor xenografts in mice. These results suggest that p53 mediates DDR and exerts its tumor suppressive role in part by regulating the expression of ACER2, which in turn regulates the bioactive sphingolipid lipids.
Similar content being viewed by others
Introduction
Human cells respond to different forms of stress, such as DNA damage, by activating p53, the most frequently inactivated tumor suppressor [1,2,3], and p53 activation in turn regulates cell cycle arrest, DNA repair, senescence, and programmed cell death (PCD) [4]. The abilities of p53 to regulate these stress responses are critical for its role in tumor suppression.
Previous studies demonstrated that p53 regulates enzymes responsible for the metabolism of sphingolipid (SPLs) [5]. Sawada et al. [6] showed that etoposide activated neutral sphingomyelinase catalyzing the hydrolysis of sphingomyelins into ceramides in glioma cells in a p53-dependent manner. Recently, we demonstrated that DNA damage upregulated the alkaline ceramidase ACER2 that catalyzes the hydrolysis of ceramides into sphingosine, the precursor of sphingosine-1-phosphate (S1P), in p53-proficient but not p53-deficient or mutated cell lines [7]. These findings suggest that p53 may regulate the levels of ceramides, SPH, and S1P in cells by controlling the expression of their metabolizing enzymes.
Ceramides, SPH, and S1P, are bioactive lipids that mediate various biological processes. ceramides have been suggested to be anti-proliferative, pro-apoptotic, pro-senescent bioactive lipids [8,9,10]. SPH has also been shown to mediate cell cycle arrest [11, 12], differentiation [12, 13], and PCD [7, 11, 12, 14, 15]. In contrast, S1P mainly promotes cell proliferation and survival and to inhibit senescence [16]. As p53 has a potential role in regulating SPL-metabolizing enzymes, some bioactive SPL metabolites may serve as mediators of p53 in cellular responses such as cell cycle arrest and PCD. However, it is unclear how p53 and bioactive SPLs interplay to regulate biological processes important for the tumor suppressive function of p53.
In this study, we demonstrate that ACER2 is a direct transcriptional target of p53 and a mediator of p53 in DDR. Our study not only provides novel insights into the mechanism by which p53 regulates bioactive SPLs and cellular responses in human cells but also identifies ACER2 as a novel tumor suppressor.
Results
ACER2 a direct transcriptional target of p53
We performed a SPL pathway-specific qPCR array to investigate how p53 regulates SPL-metabolizing enzymes in H1299:p53TRE cells [17]. In these cells, p53 expression is repressed in the presence of tetracycline (TET) in medium but can be induced upon TET withdrawal or its replacement with the vehicle ethanol (ET) (Fig. 1a). p53 expression caused a fourfold increase in ACER2 mRNA levels with minimal or no effect on other SPL genes (Fig. 1b). p53 overexpression also increased ACER2 protein level (Fig. 1c) and enzymatic activity (Fig. 1d). These results suggest that ACER2 is the major SPL-metabolizing enzyme that is regulated by p53.
a, b H1299:p53TRE cells were grown to an 80% confluence in the presence of TET (2 μM) before being switched to fresh medium containing ET or TET (1 μM). At 48 h post the medium change, the cells were collected and subjected to western blot analyzes for p53 expression a or a pathway-specific qPCR array for quantifying mRNA levels of SPL enzymes b according to the manufacturer’s instructions and Clarke et al. [39]. c, d H1299:p53TRE cells were cultured as in b before microsomes were prepared as described in our previous study [14]. The microsomes from H1299:p53TRE cells grown in the presence of ET or TET were subjected to western blot analyzes with anti-ACER2 or βeta-actin antibody c or assays of alkaline ceramidase activity on D-e-C16-ceramide d as described in our previous study [42]. e Two putative p53 responsive elements (RE1 and RE2) were identified by DNA sequence analysis in the first intron of the ACER2 gene. E1–E6 represent Exons 1–6 of the ACER2 gene. f H1299:p53TRE cells were cultured in the presence of ET or TET (1 μM) for 48 h before being subjected to CHIP assays using anti-p53 antibody or naive IgG. Immunoprecipitated DNAs were subjected to qPCR analyzes using the primer pair that amplified an ACER2 gene region encompassing RE1 or RE2 or the primer pair that amplified a region distal to RE1 or RE2 as a negative control. g the full-length ACER2 promoter and its shorter promoter variants were cloned into pGL3-basic and the resulting promoter-reporter constructs were subjected to promoter activity assays as described under Methods section. The promoter activity is expressed as fold change versus pGL3-basic. pGL3-ACER2P, the promoter report construct with the ACER2 core promoter of 193 bp, showed a minimal promoter activity. h the putative p53 responsive element RE1 or RE2 was cloned downstream of the ACER2 core promoter in pGL3-ACER2P to generate the enhancer reporter construct with RE1 (pGL3-ACER2P/RE1) or RE2 (pGL3-ACER2P/RE2). pGL3-ACER2P, pGL3-ACER2P/RE1, or pGL3-ACER2P/RE2 was transfected into H1299:p53TRE cells grown in the presence of TET (2 μM). At 24 h post enhancer construction transfection, the cells were switched to fresh medium containing ET or TET (1 μM). At 48 h medium change, the cells were collected and subjected to promoter activity assays as described under Methods section. The promoter activity is expressed as fold change versus the activity of pGL3-ACER2P in cells in the presence of TET. The data represent mean values ± SD of three independent experiments. Statistical analyzes were performed as described in Methods section. *represents p < 0.001
p53 regulates gene expression by binding a consensus DNA response element(s) (p53RE) [18]. Two putative p53REs, designated RE1 and RE2, were found in the first intron of the ACER2 gene (Fig. 1e). Chromatin immunoprecipitation (ChIP) assays demonstrated that p53 antibody enriched a DNA fragment encompassing RE1 or RE2 in H1299:p53TRE cells treated with ET but not TET (Fig. 1f) but failed to enrich a fragment (non-RE) distal to RE1 and RE2 in the ACER2 gene in H1299:p53TRE cells treated with either ET or TET (Fig. 1f). These results suggest that RE1 and RE2 are p53 binding sites.
To test whether RE1 or RE2 can enhance ACER2 promoter activity in a p53-dependent manner, we first defined the core promoter of the ACER2 gene, the 190-base pair DNA fragment (−190 bp) upstream of the translational start codon (ATG) of the ACER2 gene (Fig. 1g). We then cloned RE1 and RE2 downstream of the ACER2 core promoter in the promoter-reporter construct (pGL3-ACER2P) to generate the promoter-reporter constructs with the insertion of RE1 (pGL3-ACER2P/RE1) and RE2 (pGL3-ACER2P/RE2), respectively. Compared to transfection with pGL3-ACER2P, transfection with pGL3-ACER2P/RE1 or pGL3-ACER2P/RE2 did not increase luciferase activity in H1299:p53TRE cells when p53 expression was repressed in the presence of TET (Fig. 1h). However, upon p53 expression induction, the luciferase activity in H1299:p53TRE cells transfected with pGL3-ACER2P/RE1 or pGL3-ACER2/RE2 but not pGL3-ACER2P was markedly increased (Fig. 1h). These results suggest that both RE1 and RE2 are bona fide p53REs and that ACER2 is a novel direct transcriptional target of p53.
p53 overexpression increases SPH and S1P by upregulating ACER2
To investigate whether p53 regulates the levels of SPLs through ACER2, we knocked-out ACER2 from H1299:p53TRE cells by the RNA-guided clustered regularly interspaced short palindromic repeats-associated Cas9 (CRISPR-Cas9) genome-editing technology (Supplementary Fig. S2A). p53 overexpression increased ACER2 enzymatic activity in cells expressing wild-type ACER2 (H1299:p53TRE/ACER2+/+ cells) but not ACER2 knockout cells (H1299:p53TRE/ACER2−/− cells) (Fig. 2a). Treatment with nutlin 3a (NUT), which stabilizes p53 by inhibiting the interaction between p53 and MDM2 [19], markedly augmented the increase in p53 levels (Fig. 2b), ACER2 mRNA levels (Fig. 2c), and its enzymatic activity (Fig. 2d) in H1299:p53TRE/ACER2+/+ cells in response to p53 overexpression. p53 overexpression increased the levels of SPH (Fig. 2e) and S1P (Fig. 2f) while decreasing the levels of ceramides in H1299:p53TRE/ACER2+/+ (Fig. 2g), and these effects were markedly inhibited in H1299:p53TRE/ACER2−/− cells (Fig. 2e–g). Treatment with NUT enhanced the increases in the levels of SPH (Fig. 2e) in H1299:p53TRE/ACER2+/+ but not in H1299:p53TRE/ACER2−/− cells in response to p53 overexpression (Fig. 2e). NUT only slightly enhanced an increase in the levels of S1P in H1299:p53TRE/ACER2+/+ cells but not in H1299:p53TRE/ACER2−/− cells (Fig. 2f). NUT significantly enhanced a decrease in the levels of ceramides in H1299:p53TRE/ACER2+/+ cells but not in H1299:p53TRE/ACER2−/− cells in response to p53 overexpression (Fig. 2g). These results suggest that p53 regulates cellular levels of ceramides, SPH, and S1P through ACER2.
a H1299:p53TRE/ACER2+/+ cells or H1299:p53TRE/ACER2−/− cells were grown in the presence of TET or ET for 48 h before ACER2 ceramidase activity was measured. b–d H1299:p53TRE cells were grown in the presence of ET plus DMSO, ET plus NUT (10 μM), TET (1 μM) plus DMSO, or TET plus NUT for 72 h p53 and ACER2 protein levels b, ACER2 mRNA levels c, ACER2 enzymatic activity d, and e–g, H1299:p53TRE/ACER2+/+ cells or H1299:p53TRE/ACER2−/− cells were grown in the presence of ET plus DMSO, ET plus NUT (10 μM), TET (1 μM) plus DMSO, or TET plus NUT for 72 h before the levels of SPH e, S1P f, and ceramides g were determined. The data represent mean values ± SD of three independent experiments. Statistical analyzes were performed as described in Methods section. *represents p < 0.001 and #p < 0.01
ACER2 is a mediator of p53 in cell cycle arrest and cellular senescence
We investigated whether knocking-out ACER2 affects cellular responses to p53 overexpression. ACER2 knockout decreased the viable population of H1299:p53TRE cells in which p53 expression was repressed (Fig. 3a) and arrested these cells in the G1 phase of cell cycle (Fig. 3b), suggesting that basal expression of ACER2 promotes cell proliferation. ACER2 knockout caused a further reduction in the viable cell population (Fig. 3a) while enhancing the cell cycle arrest (Fig. 3b) in response to p53 overexpression. Furthermore, ACER2 knockout significantly augmented cellular senescence induced by p53 overexpression (Fig. 3c). However, these levels of p53 overexpression induced neither DNA fragmentation (Fig. 3d) nor caspase 3/7 activation (Fig. 3e) in either H1299:p53TRE/ACER2+/+ cells or H1299:p53TRE/ACER2−/− cells. These results suggest that a moderate upregulation of ACER2 inhibits both cell cycle arrest and cellular senescence in response to p53 overexpression without affecting PCD.
a–f, H1299:p53TRE/ACER2+/+ cells or H1299:p53TRE/ACER2−/− cells were grown in the presence of ET or TET (1 μM) before being subjected to MTT assays a, cell cycle analysis b, SA-β-gal activity assays c, Cell Death Detection ELISA d, or caspase 3/7 activity assays e, or western blot analysis for p21 protein levels f. g, h H1299:p53TRE cells grown in the presence of ET or TET (1 μg/ml) were treated with PF-543 (200 nM) or the vehicle DMSO for 24 h before LC–MS/MS analysis of S1P g or for 5 days before cellular senescence assays were performed h. i, j H1299:p53TRE cells grown in the presence of ET or TET (1 μg/ml) were treated with bSMase (0.6 unit per ml) or the vehicle buffer for 24 h before LC–MS/MS analysis of ceramides i or for 5 days before cellular senescence assays j. The data represent mean values ± SD of three independent experiments. Statistical analyzes were performed as described in Methods section. *represents p < 0.001. In c, *represents the p-values for the cell populations in the S and G1 phases
The cyclin-dependent kinase inhibitor p21 (CDKN1A), a key regulator for both cell cycle arrest [20] and senescence [21], is regulated by sphingolipids [22,23,24]. We found that knocking-out ACER2 augmented an increase in p21 levels in response to p53 overexpression (Fig. 3f), suggesting that a moderate upregulation of ACER2 promotes cell cycle progression and inhibits cellular senescence by decreasing p21 protein levels.
We hypothesized that ACER2 inhibits cellular senescence probably by increasing S1P levels and/or decreasing ceramide levels. As the formation of S1P is catalyzed by the sphingosine kinases SPHK1 and 2, we determined whether inhibiting SPHK1 or SPHK2 affected cellular senescence in response to p53 overexpression. Treatment with PF-543, a specific and potent inhibitor of SPHK1 [25], nearly blocked the increase in the levels of S1P (Fig. 3g) without affecting cellular senescence (Fig. 3h) in response to p53 overexpression in H1299-p53TRE cells. We then tested whether treatment with bacterial sphingomyelinase (bSMase), which catalyzes the hydrolysis of sphingomyelins into ceramides [14], affects cellular senescence in response to p53 overexpression. We confirmed that treatment with bSMase inhibited a decrease in the levels of ceramides in H1299:p53TRE cells in response to p53 overexpression (Fig. 3i). Treatment with bSMase enhanced cellular senescence of H1299:p53TRE cells in response to p53 overexpression (Fig. 3j). Taken together, these results suggest that a moderate upregulation of ACER2 inhibits cellular senescence in response to p53 overexpression likely by decreasing the cellular levels of ceramides.
ACER2 is a mediator of p53 in PCD
High but not low levels of p53 induce PCD in H1299 cells [17], and we previously demonstrated that similar to p53, ACER2 at high but not low levels markedly induces PCD [7, 14], suggesting that ACER2 may mediate PCD in response to a marked increase in p53 levels. p53 overexpression along with NUT treatment induced DNA fragmentation (Fig. 4a) and an increase in caspase 3/7 activity (Fig. 4b) in H1299:p53TRE/ACER2+/+ cells, and these effects were significantly inhibited in H1299:p53TRE/ACER2−/− cells (Fig. 4a, b). Expectedly, treatment with NUT did not induce DNA fragmentation (Fig. 4a) or caspase 3/7 activation (Fig. 4b) in H1299:p53TRE/ACER2+/+ or H1299:p53TRE/ACER2−/− cells when p53 expression was repressed. These results suggest that a robust upregulation of ACER2 mediates PCD in response to a marked increase in p53 levels.
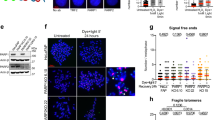
a, b H1299:p53TRE/ACER2+/+ cells or H1299:p53TRE/ACER2−/− cells were grown in the presence of ET plus DMSO, ET plus NUT (10 μM), TET (1 μM) plus DMSO, or TET plus NUT for 72 h before DNA fragmentation a and caspase 3/7 activity b were determined. c, d HeLa:ACER2TO cells were grown in the presence of ET or TET (100 ng/ml) for 48 h before mitochondria were imaged under the confocal microscope C and the percentages of cells with MF were computed d. e, f HeLa:ACER2TO cells were grown in the presence of ET or TET (100 ng/ml) for 48 h before being subjected to MMP assays by confocal microscopy e or fluorometry f as described in Methods section. The data represent mean values ± SD of three independent experiments. Statistical analyzes were performed as described in Methods section. *represents p < 0.001
We recently demonstrated that overexpression of the yeast alkaline ceramidase YPC1, a yeast ortholog of ACER2, induces mitochondrial fragmentation (MF) and ROS production by accumulating phytosphingosine (yeast sphingosine) in mitochondria [26]. This prompted us to test whether overexpression of ACER2 also induces MF. We previously generated a HeLa stable cell line, HeLa:ACER2TO where ACER2 overexpression can be induced by TET but not ET [14]. By labeling cells with MitoTracker green, a mitochondrion-specific fluorescent dye, we revealed that HeLa:ACER2TO cells grown in the presence of ET had interconnected elongated and tubular mitochondria whereas those grown in the presence of TET had individual circular mitochondria (Fig. 4c, d), suggesting that overexpression of ACER2 induces MF. MF has been shown to cause loss of mitochondrial membrane potential (MMP) [27, 28], and we found that ACER2 overexpression significantly induced loss of MMP (Fig. 4e, f). These studies suggest that ACER2 is a key regulator of mitochondrial dynamics and that its marked upregulation induces MF and loss of MMP.
The p53/ACER2 pathway mediates DNA damage response to ionizing radiation
As DNA damage markedly upregulates p53, we determined whether DNA damage upregulates ACER2 by activating p53. Indeed, treatment with ionizing radiation (IR) increased ACER2 mRNA levels (Fig. 5a) and its enzymatic activity (Fig. 5b) in a dose-dependent manner in HCT116 cells (HCT116:p53+/+) expressing WT p53 but not in isogenic p53-deficient cells (HCT116:p53−/−). IR also increased ACER2 protein levels in HCT116:p53+/+ cells but not HCT116:p53−/− cells (Fig. 5c).
a–c HCT116:p53+/+ or HCT116:p53−/− were treated with IR at different doses (2–10 Gy) or sham (0 Gy), and 48 h later the cells were subjected to qPCR analyzes for ACER2 mRNA levels a, ACER2 enzymatic activity assays b, or western blot analyzes with anti-ACER2 or calnexin antibody c. d–h HCT116:ACER2+/+ cells and HCT116:ACER2−/− cells were treated with 4 Gy (low-dose IR; IR-low), 8 Gy (high-dose IR; IR-high), or sham, and 48 h later the cells were subjected to ACER2 enzymatic activity assays d, MTT assays e, cell cycle analysis f, cellular senescence assays g, Cell Death Detection ELISA h, caspase 3/7 activity assays i. The data represent mean values ± SD of three independent experiments. Statistical analyzes were performed as described in Methods section. *represents p < 0.001. In f, *represents the p-values for the cell populations in the S and G2 phases
We then tested if knocking-out ACER2 inhibited IR-induced cellular effects in HCT116:TP3+/+ cells. HCT116:p53+/+ cells. HCT116:p53+/+ cells were transduced with lentiviruses expressing Cas9 and sgACER2 or sgCON to generate ACER2-deficient (HCT116:ACER2−/−) and ACER2-proficeint (HCT116:ACER2+/+) cells, respectively. The ACER2 gene inactivation in HCT116:ACER2−/− was confirmed by DNA sequencing (Supplementary Fig. S2B). We treated HCT116:ACER2+/+ and HCT116:ACER2−/− cells with IR at 0 Gy (sham), 4 Gy (IR-low), and 10 Gy (IR-high), and cell cycle progression, PCD, or senescence were assessed at different time points. Either IR-low or IR-high increased ACER2 enzymatic activity in HCT116:ACER2+/+ but not in HCT116:ACER2−/− cells (Fig. 5d). IR-low induced growth inhibition (Fig. 5e), cell cycle arrest (Fig. 5f), or cellular senescence (Fig. 5g) to a greater extent in HCT116:ACER2−/− cells than in HCT116:ACER2+/+ cells. IR-high caused DNA fragmentation (Fig. 5h) and caspase 3/7 activation (Fig. 5i) to a greater extent in HCT116:ACER2+/+ cells than in HCT116:ACER2−/− cells. These results suggest that low-level upregulation of ACER2 protects cells from cell cycle arrest and cellular senescence in response to IR-low whereas its high-level upregulation mediates PCD in response to IR-high.
IR regulates bioactive SPLs through the p53/ACER2 pathway
To determine whether IR regulates ceramides, SPH, and S1P through the p53/ACER2 pathway, we treated HCT116:p53+/+, HCT116:p53−/−, HCT116:ACER2+/+, and HCT116:ACER2−/− cells with sham, IR-low (4 Gy), or IR-high (10 Gy) before SPLs were analyzed. IR-high induced an increase in the levels of SPH to a greater extent than IR-low in HCT116:p53+/+ or HCT116:ACER2+/+ (Fig. 6a) whereas IR-low and IR-high increased the levels of S1P to a similar extent in these cells (Fig. 6b). Knocking-out either p53 or ACER2 robustly inhibited the increases in the levels of both SPH (Fig. 6a) and S1P (Fig. 6b) in response to either IR-low or IR-high. Interestingly, neither IR-low nor IR-high increased the levels of ceramides in HCT116:ACER2+/+ cells and IR-high but not IR-low increased the levels of ceramides in HCT116:p53−/− or HCT116:ACER2−/− cells (Fig. 6c). These results suggest that IR upregulates SPH in a dose-dependent manner and S1P in a dose-independent manner by activating the p53/ACER2 pathway and that the activation of the p53/ACER2 pathway inhibits IR-induced upregulation of ceramides in cells.
a, b HCT116:p53+/+, HCT116:p53−/− cells, HCT116:ACER2+/+, or HCT116:ACER2+/+ cells were treated with IR-low, IR-high, or sham, and 24 h later, the cells were subjected to LC/MS/MS analyzes for SPH, S1P, or ceramides. The data represent mean values ± SD of three independent experiments. Statistical analyzes were performed as described in Methods section. *represents p < 0.001 and #p < 0.01
ACER2 overexpression inhibits tumor growth
To determine the role of ACER2 tumor suppression, we tested whether increasing the expression of ACER2 in tumor cells is sufficient to inhibit the growth of tumor xenografts in immunodeficient nude mice. For this experiment, we used the cervical cancer cell line HeLa:ACER2TO in which expression of ACER2 can be induced by doxycycline (DOX), a water-soluble analog of TET [14]. HeLa:ACER2TO cells were subcutaneously inoculated into SCID mice that were fed a regular diet (control group) or a diet supplied with DOX (DOX group). As shown in Fig. 7a, tumor growth was inhibited in the DOX group compared to that in the control group, and the tumors from the DOX group were significantly smaller than those from the control group. It is worth to note that DOX did not affect the growth of tumor xenografts derived from HeLa T-REx cells that were stably transfected with the empty vector pcDNA4/TO (Fig. 7b). ACER2 enzymatic activity was higher in the tumors from the DOX group than in the tumors from the control group (Fig. 7c), suggesting that DOX indeed induced the expression of ACER2 in tumors. As expected, DOX-induced expression increased the levels of SPH (Fig. 7d) and S1P (Fig. 7e) while decreasing the levels of ceramides in tumors (Fig. 7f). DOX-induced expression also increased caspase 3/7 activity in tumors (Fig. 7g). These results suggest that increasing ACER2 expression indeed mediates tumor growth inhibition in response to RT.
a, b HeLa:ACER2TO cells A or HeLa T-REx cells (HeLa:pcDNA4TO) stably transfected with the empty vector pcDNA4/TO b were subcutaneously inoculated into immunodeficient nude mice (n = 6 and 5 per group for HeLa:ACER2TO and HeLa: pcDNA4TO cells, respectively), which were fed regular or doxycycline (DOX)-containing chow. Tumor volumes were determined as described under Methods section (upper panels). Tumors were collected and imaged on Day 20 (lower panels). c–f the collected tumors were homogenized and tissue homogenates were subjected to ACER2 enzymatic activity assays c, LC–MS/MS for the levels of SPH d, S1P e, and ceramides f, or caspase 3/7 activity assays g. The data represent mean values ± SD. Statistical analyzes were performed as described in Methods section. *represents p < 0.001 and #p < 0.01
Discussion
In this study, we identify ACER2 as a novel transcriptional target of p53. Depending its expression levels, upregulation of ACER2 either inhibits cell cycle arrest and senescence in response to a low-grade activation of p53 or mediates PCD in response to a hyperactivation of p53. Our compelling data strongly suggest that ACER2 is a novel tumor suppressor that serves as an important mediator of p53 in the DDR.
p53 has been shown to regulate several SPL-metabolizing enzymes, such as neutral sphingomyelinase [6], ceramide synthase 5 (Cers5) [29], and ceramide synthase 6 (CERS6) [30]. However, in this study, we uncovered that in the SPL metabolic network, ACER2 is the only enzyme that is transcriptionally regulated by p53 in H1299 cells. Previous studies showed that p53 activation contributes to an increase in the cellular levels of ceramides in response to genotoxic stress [6, 31]. However, in this study, we demonstrated that DNA damage induces the generation of ceramides in HCT116 cells through a p53-independent mechanism and that p53 activation attenuates the ceramide rise in response to DNA damage mainly through ACER2 upregulation. Therefore, the p53/ ACER2 pathway is a negative regulator for ceramides and a positive regulator for both SPH and S1P.
In this study, we provide strong evidence that ACER2, as an important downstream effector of p53, plays an important role in dictating a cell’s fate in response to p53 activation. ACER2 expression levels correlate with p53 levels, so drastic upregulation of ACER2 mediates PCD in response to a hyperactivation of p53, whereas its moderate upregulation inhibits cell cycle arrest and cellular senescence in response to a mild activation of p53. The inhibition of cell cycle arrest and senescence by low ACER2 expression may allow cells that are temporarily arrested to re-enter cell cycle after damaged DNA is repaired. In contrast, the pro-apoptotic effect conveyed by high ACER2 expression ensures that cells with irreparable damaged DNA be eliminated by PCD. Therefore, ACER2, like p53, has an important role in maintaining the integrity of the genome by coordinating cell cycle progression, senescence, and PCD.
Different expression levels of ACER2 exert distinct roles in regulating cell proliferation, senescence, and PCD in response to p53 activation likely by orchestrating a delicate balance among bioactive SPLs that it regulates. We showed that low-level expression of ACER2 in response to low ectopic expression of p53 or IR-low caused a marked increase in S1P levels and a moderate increase in SPH levels while decreasing the levels of ceramides whereas its high-level expression in response to high-level ectopic expression of p53 or IR-high further increased the levels of SPH but not S1P. In cells expressing low levels of p53 and ACER2, the proliferative effect of S1P might counterbalance the anti-proliferative effect of low levels of SPH, thus promoting cell proliferation. In cells expressing high levels of p53 and ACER2, the apoptotic effect of the high levels of SPH might override the anti-apoptotic effects of S1P, thus resulting in PCD (Fig. 8). These speculations are supported by our previous findings that low ectopic expression of ACER2 promotes cell proliferation and survival through S1P whereas high ectopic expression induces PCD through SPH [7, 14]. In addition, we found that a moderate upregulation of ACER2 inhibits cellular senescence in response to p53 overexpression likely by decreasing cellular levels of ceramides but not by increasing S1P. Taken together, our data suggest that high-level expression of ACER2 mediates PCD by accumulating SPH in response to high-level of p53 or high-degree DNA damage and that low-level expression of ACER2 inhibits cell cycle arrest by increasing S1P and cellular senescence by decreasing ceramides in response to low-level of p53 or low-grade DNA damage (Fig. 8).
High-grade DNA damage (red) causes a hyperactivation of the p53/ACER2 pathway, resulting in the dominance of the pro-death signal of SPH over the pro-proliferative and pro-survival of S1P and thereby PCD. Low-grade DNA damage (blue) triggers a moderate activation of the p53/ACER2 pathway, resulting in a balance of the pro-death signal of SPH and pro-senescence signal of ceramides with the pro-proliferative and pro-survival signals of S1P and thereby cell cycle arrest and senescence
Although the role of ACER2 in proliferation inhibition has been firmly established but how it exerts this role remains unclear. In this study, we demonstrated that knocking-out ACER2 enhanced an increase in the levels of p21. As p21 has been implicated in cell cycle arrest in both G1 [20] and G2 phases [32, 33], we postulate that a moderate activation of ACER2 mediates a G1 arrest in cell cycle in response to p53 overexpression or a G2 arrest in response to low-grade DNA damage likely through p21. In addition to cell cycle arrest, p21 has been shown to play an important role in cellular senescence [21, 34]. This may suggest that a moderate activation of the ACER2 pathway inhibits cellular senescence in response to p53 overexpression by inhibiting the p21 pathway. How ACER2 regulates p21 levels is currently under investigation.
Although the role of the ACER2/SPH pathway in PCD has been firmly established, the underlying mechanisms by which SPH mediates PCD remain largely unclear. We previously demonstrated that ACER2 overexpression induces ROS production, which is partially responsible for SPH-induced PCD [7]. In this study, we found that ACER2 overexpression induces MF and loss of MMP. Interestingly, we recently demonstrated that overexpression of YPC1p, a yeast ortholog of ACER2, also induces MF, loss of MMP, and production of ROS likely by increasing the generation of phytosphingosine (the yeast sphingosine), which in turn inhibits mitochondrial fusion [26]. Previous studies by other group suggest that MF may be associated with both loss of MMP and production of ROS in mammalian cells [35,36,37]. These observations suggest that the hyperactivation of the ACER2/SPH pathway induces PCD likely through MF, which in turn resulting in ROS production and loss of MMP.
According to TCGA, ACER2 is frequently deleted or mutated in certain cancers (Supplementary Fig. S3) and numerous cancer cell lines (Supplementary Table S1). In cell culture models, we demonstrated that knocking-out ACER2 renders tumor cells resistant to PCD in response to DNA damage. Our animal model studies demonstrated that restoring ACER2 expression alone could inhibit tumor growth by inducing tumor cell PCD. These results suggest that ACER2 inactivation may promote tumorigenesis by allowing transformed and malignant cells to survive DNA damage stress. Therefore, ACER2 is a novel tumor suppressor whose inactivation may lead to cancer.
p53 is inactivated in over 50% cancers, and its inactivation renders cancers resistant to treatment with either genotoxic chemotherapeutics (CT) or radiotherapy (RT) due to a lack of activation of downstream targets of p53 that mediate cell cycle arrest, cellular senescence, and PCD. As ACER2 is an important mediator of p53 in PCD in response to either CT or RT, a failure in ACER2 upregulation in p53-deficient or mutated cancer cells may contribute to the resistance of p53-deficient or mutated cancer cells to PCD in response to RT or CT. Indeed, knocking-out ACER2 protected tumor cells from PCD in response to IR. This finding provides a strong rationale that restoring ACER2 expression in p53-deficient or mutated cancer cells should improve the anti-cancer efficacy of RT or CT.
In conclusion, our current study identifies ACER2 as a direct gene target of p53 and as a novel tumor suppressor that mediates the anti-cancer actions of p53 by regulating ceramides, SPH, and S1P, respectively (Fig. 8). Moreover, our compelling data strongly suggest that p53-inactivated cancers are resistant to RT or CT at least in part due to the inactivation of the ACER2/SPH pathway and that rectifying the ACER2/SPH pathway would improve the efficacy of CT or RT against p53-deficient or mutated cancers.
Materials and Methods
Reagents
The lentiCRISPR v2 plasmid was a gift from Dr. Feng Zhang (Addgene plasmid #52961). Other reagents used in this study have been listed in our previous study [7].
Mice
Athymic nude-Foxn1nu mice were purchased from Charles River (Wilmington, MA) and were housed under pathogen-free laboratory conditions with constant room temperature (22 °C), humidity level (55%), a 12-h light:12-h dark cycle, and food (W.F. Fisher and Son; Somerville, NJ) and water available ad libitum. Animal studies were approved by the Institutional Animal Care and Use Committee at Stony Brook University, Stony Brook, NY, USA.
Cell lines
The cervical cancer cell line HeLa:ACER2TO (also known as ACER2-Tet-ON) in which expression of ACER2 is under the control of a TET-ON (TO) system was created in our previous study [14]. The HCT116:p53+/+ and HCT116:p53−/− cell lines [38], kind gifts of Dr. B. Vogelstein at Johns Hopkins University, were maintained in DMEM medium with 10% FBS. The non-small cell lung carcinoma cell line H1299 and its derivative H1299:p53TRE expressing p53 under the control of a TET responsive element, the TET-OFF system [17], kind gifts from Dr. Carol Prives at Columbia University, New York City, New York, were maintained in DMEM medium in the presence of TET as described [17].
RT² Profiler PCR Array
qPCR-based gene expression profiling for genes involved in the metabolism of SPLs was performed as described [39].
Quantitative polymerase chain reaction (qPCR)
Total RNA was isolated from cells using RNeasy kits (Qiagen) according to the manufacturer’s instructions and qPCR analyzes were performed with primer pairs specific for ACER2 (5′-AGTGTCCTGTCTGCGGTTACG-3′/5′-TGTTGTTGATGGCAGGCTTGAC-3′, SPHK2 (5′-CTGTCTGCTCCGAGGACTGC-3′ and 5′-CAAAGGGATTGACCAATAGAAGC-3′), or ACTB (β-actin; 5′-TTGCCGACAGGATGCAGAAG-3′/5′-ACATCTGCTGGAAGGTGGAC-3′) as described in our previous studies [40]. qPCR results were analyzed using Q-Gene software which expresses data as mean normalized expression (MNE) [41]. MNE is directly proportional to the amount of mRNA of a target gene relative to the amount of mRNA of the reference gene (β-actin).
Protein concentration determination
Protein concentrations were determined with BSA as a standard using a BCA protein determination kit (Pierce) according to the manufacturer’s instructions.
Western blot analysis
Western blot analyzes were performed as described in our previous studies [7].
ACER2 ceramidase activity assays
Microsomes were isolated from cells and alkaline ceramidase activity was assayed using D-e-C16-ceramide as a substrate at pH 8.0 as described in our previous study [42, 43]. The ceramidase activity detected under these conditions is encoded specifically by ACER2 because the other alkaline ceramidases ACER1 [13] and ACER3 [44] do not use D-e-C16-ceramide as a substrate.
CHIP assays
CHIP assays were performed using a Magna CHIP assay kit (Millipore EMD) essentially according to the manufacturer’s instructions. DNA fragments were immunoprecipitated with p53 antibody or naïve control IgG and were subjected to qPCR analyzes using the primer pairs P1 (5′-GGGTCTGTGTCAGCCCATTC-3′/5′-GCACTCCGAGAGCAGGTATT-3′), P2 (5′-CACTCCCACCAGGCAGACTT-3′/5′-AGTTGAATTAGCGGCTTGCAC-3′), P3 (5′-AGGTTCAAGGTGGTGGTCAGT-3′/5′-TGCAGTGCAAGGAACTCCCA-3′). P1, P2, and P3 amplified a DNA segment encompassing the p53 putative binding site RE1 or RE2 in the first intron of the ACER2 gene and a distal DNA segment (non-RE) in the fourth exon of the ACER2 gene (a negative control), respectively.
Promoter and transcriptional enhancer activity assays
The 1882 base pair DNA sequence upstream of the ACER2 translation initiation site (ATG), designated as the full-length ACER2 promoter, was cloned into a promoter-reporter vector pGL3-basic (Promega) as described in our previous study [40]. To map the core promoter region, progressive 5′-deletion fragments of the full-length promoter were constructed by PCR and cloned into pGL3-basic. Each of the resulting promoter constructs or pGL3-basic was co-transfected with β-galactosidase-expressing plasmid pcDNA4-β-gal (Promega) as a normalization control into cells using the transfection reagent, X-tremeGene (Roche), according to the manufacturer’s instructions. Luciferase activity was determined using a Luciferase Reporter Assay System (Promega), and β-galactosidase activity a High Sensitivity β-galactosidase assay kit (Agilent Technologies) according to the manufacturer’s instructions. Promoter activity was expressed as the ratio of the firefly luciferase activity (average relative light units (RLU)) to β-galactosidase activity (absorbance at 570 nm (A570)-absorbance at 595 nm (A595)). To assay the enhancer activity of the putative p53 binding sites (RE1 and RE2) on the first intron of the ACER2 gene, RE1 or RE2 was cloned downstream of the luciferase gene of the promoter-reporter construct (pGL3-ACER2P) carrying the ACER2 core promoter. The resulting enhancer reporter construct pGL3-ACER2P/RE1 or pGL3-ACER2/RE2 and pGL3-ACER2P were respectively subjected to promoter activity assays as described above.
Gene disruption by CRISPR/Cas9
A 20-mer CRISPR RNA targeting the third exon of ACER2 (ACER2-crRNA; 5′-UGAUGUGUGCUUUGGCCAUG-3′) was designed using a web tool CHOP-CHOP (http://chopchop.cbu.uib.no), and a control crRNA (Con-crRNA; 5′-UGGCGAUGUGUGCCUUUAUG-3′) was designed by randomly scrambling the sequence of ACER2-crRNA. The double stranded oligonucleotide corresponding to ACER2-crRNA or Con-crRNA was synthesized and cloned into the single-guide RNA (sgRNA) scaffold of the CRISPR/Cas9 vector, lentiCRISPR v2 as described [45]. The resulting DNA construct that directs the expression of Cas9 and sgRNA specific for ACER2 (sgACER2) or control sgRNA (sgCON) was packaged into lentiviral particles using HEK293T cells as host cells, and lentiviral particles were transduced into H1299:p53RTE or HCT116:p53+/+ cells as described in our previous study [7]. Individual clones that stably expressed Cas9 and sgACER2 or sgCON were selected in the presence of puromycin and ACER2 targeting in each clone was verified by DNA sequencing (Supplementary Fig. S2). Three individual clones with ACER2 (ACER2−/−) being mutated or those with the WT ACER2 (ACER2+/+) were combined for further studies. The resulting ACER2-deficent cell lines HCT116:ACER2−/−, H1299:p53TRE/ACER2−/− and their respective isogenic WT cell lines HCT116:ACER2+/+ and H1299:p53TRE/ACER2+/+ were maintained and cultured as described earlier.
Ionizing radiation
Tumor cells grown in 6 or 12-well plates or 10-cm culture plates were exposed to γ-irradiation from a 137Cs source in a Gamma 40 Exactor (Atomic Energy of Canada LTD. Commercial products) with a dose rate of 0.8 Gy/min for a total of 8 Gy.
LC–MS/MS analysis for SPLs
SPLs were analyzed by liquid chromatography tandem mass spectrometry (LC–MS/MS) as described (24) in the Lipidomics Core Facility at Stony Brook University. Briefly, cells were collected after being washed with ice-cold 25 mM Tris-HCl buffer (pH 7.4) containing 150 mM NaCl. Volume of 50 μl of an internal SPL standard mixture (IS) at a concentration of 1 nmol/ml was added to each cell pellet. The IS contains C17SPH, C17SPH-1-phosphate, C17dihydro-SPH, 13C16-ceramide (d13/16), 17C16-ceramide (d17:1/16), C17-ceramide (d18:1/17:0), and 17C24:1-ceramide (d17:1/24:1). Lipids were extracted with 4 ml of the ethyl acetate/iso-propanol/water (60/28/12%;v/v) solvent system. After centrifugation, one ml aliquot of the lipid extract from each sample was used for determination of total phospholipids, and the remaining was used for LC–MS/MS. The lipid extracts from each sample were dried under a stream of nitrogen gas, dissolved in 150 μl of mobile phase (Methanol with 0.2% Formic acid and 1 mM Ammonium formate), and injected on the TSQ Quantum Ultra LC/MS/MS system. The lipids were gradient-eluted from the Spectra C8SR, 150 × 3.0 mm, 3-μm particle size column, with a mobile phase system consisting of methanol acidified with 0.2% formic acid and 1.0 mM ammonium formate. Peaks corresponding to the target analytes and IS were identified and processed using the LC Quan software. Quantitative analyzes of endogenous SPLs were based on calibration curves generated by standards purchased from Avanti polar lipids. For the analytes for which there is not commercially available standard, the closest by mass and structure standard was used for quantification. The target analyte/IS peak area ratios were compared to the calibration curves using a linear regression model. Levels of SPLs were normalized to total protein, and expressed as SPLs/protein (pmol/mg).
MTT assay
Cell viability was determined using an in vitro toxicology assay kit based on 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) (Sigma, Inc.) according to the manufacturer’s instructions.
FACS analysis
Cell cycle profiles were analyzed by fluorescence activated cell sorting (FACS) as described in our previous study [7].
PCD assessment
PCD was determined by the release of cytoplasmic histone-associated DNA fragments (mono- and oligonucleosomes) from cells using Cell Death Detection ELISAPLUS photometric enzyme immunoassay kits (Roche) according to the manufacturer’s instructions and/or caspase 3/7 activity assays as described in our previous study [7].
Senescence assays
Cellular senescence was assessed by SA-β-gal activity assays using fluorometric senescence assay kits (Cell Biolabs, Inc.) according to the manufacturer’s instructions.
Mitochondrial fragmentation assessment
HeLa:ACER2TO cells grown in the presence of ET or TET (10 ng/ml) were incubated with MitoTracker Green FM (200 μM) in Hanks Balanced Salt Solution (HBSS) for 45 min before mitochondria were imaged at the excitation wavelength of 488 nm and emission wavelength range from 500 to 550 nm under a Leica TCS SP8 confocal laser scanning microscope (Leica Microsystems Inc.).
Mitochondrial membrane potential assays
HeLa:ACER2TO cells grown in the presence of ET or TET (10 ng/ml) were incubated with JC-1 (10 μM) in Hanks Balanced Salt Solution (HBSS) with 2% FBS for 30 min before mitochondria were imaged at the excitation wavelength of 488 nm (Ex488) and emission wavelength ranges from 515 to 545 nm (Em530 ± 15) and from 575 to 615 nm (Em596 ± 20), respectively, under the confocal laser scanning microscope or fluorescence intensity (FI) was measured under the multi-mode microplate reader SpectraMax M5 (Molecular Devices) at Ex488 and Em530 or Em595. The ratio of FI at Em595 to Em530, which positively correlates with mitochondrial membrane potential (MMP), was computed.
Tumor xenograft mouse model
HeLa:ACER2TO cells (5 × 106 cells in 0.2 ml per site) were subcutaneously inoculated into the right flank of 6-week-old female athymic nude (nu/nu) mice. When tumors became palpable, the mice were randomly divided into two groups, which were fed regular diet (the control group) or diet containing doxycycline (DOX). Tumor dimensions were measured twice weekly with a digital caliper and tumor volumes computed using the formula V = ½ (L×W2), where L and represent length and width, respectively. At the endpoint of the experiment, the tumors were resected, imaged, and subjected to LC–MS/MS analyzes for SPLs, alkaline ceramidase activity or caspase 3/7 activity assays.
Statistical analysis
The data are presented as mean values ± SD and were compared by two-tailed t-tests using Graphpad Prism (La Jolla, CA). A difference between two groups or treatments was considered significant if p < 0.05.
References
Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8:275–83.
Kruiswijk F, Labuschagne CF, Vousden KH. p53 in survival, death and metabolic health: a lifeguard with a licence to kill. Nat Rev Mol Cell Biol. 2015;16:393–405.
Yeo CQ, Alexander I, Lin Z, Lim S, Aning OA, Kumar R, et al p53 maintains genomic stability by preventing interference between transcription and replication. Cell Rep. 2016;15:132–46.
Chen J. The cell-cycle arrest and apoptotic functions of p53 in tumor initiation and progression. Cold Spring Harb Perspect Med. 2016;6:a026104.
Carroll B, Donaldson JC, Obeid L. Sphingolipids in the DNA damage response. Adv Biol Regul. 2015;58:38–52.
Sawada M, Nakashima S, Kiyono T, Nakagawa M, Yamada J, Yamakawa H, et al p53 regulates ceramide formation by neutral sphingomyelinase through reactive oxygen species in human glioma cells. Oncogene. 2001;20:1368–78.
Xu R, Wang K, Mileva I, Hannun YA, Obeid LM, Mao C. Alkaline ceramidase 2 and its bioactive product sphingosine are novel regulators of the DNA damage response. Oncotarget. 2016;7:18440–57.
Futerman AH, Hannun YA. The complex life of simple sphingolipids. EMBO Rep. 2004;5:777–82.
Hannun YA, Obeid LM. Principles of bioactive lipid signalling: lessons from sphingolipids. Nat Rev Mol Cell Biol. 2008;9:139–50.
Hannun YA, Obeid LM. Many ceramides. J Biol Chem. 2011;286:27855–62.
Ahn EH, Schroeder JJ. Sphingoid bases and ceramide induce apoptosis in HT-29 and HCT-116 human colon cancer cells. Exp Biol Med. 2002;227:345–53.
Ahn EH, Chang CC, Schroeder JJ. Evaluation of sphinganine and sphingosine as human breast cancer chemotherapeutic and chemopreventive agents. Exp Biol Med. 2006;231:1664–72.
Sun W, Xu R, Hu W, Jin J, Crellin HA, Bielawski J, et al Upregulation of the human alkaline ceramidase 1 and acid ceramidase mediates calcium-induced differentiation of epidermal keratinocytes. J Invest Dermatol. 2008;128:389–97.
Xu R, Jin J, Hu W, Sun W, Bielawski J, Szulc Z, et al Golgi alkaline ceramidase regulates cell proliferation and survival by controlling levels of sphingosine and S1P. Faseb J. 2006;20:1813–25.
Lepine S, Lakatos B, Maziere P, Courageot MP, Sulpice JC, Giraud F. Involvement of sphingosine in dexamethasone-induced thymocyte apoptosis. Ann N Y Acad Sci. 2002;973:190–3.
Alvarez SE, Milstien S, Spiegel S. Autocrine and paracrine roles of sphingosine-1-phosphate. Trends Endocrinol Metab. 2007;18:300–7.
Chen X, Ko LJ, Jayaraman L, Prives C. p53 levels, functional domains, and DNA damage determine the extent of the apoptotic response of tumor cells. Genes Dev. 1996;10:2438–51.
Menendez D, Inga A, Resnick MA. The expanding universe of p53 targets. Nat Rev Cancer. 2009;9:724–37.
Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, et al In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–8.
Dulic V, Kaufmann WK, Wilson SJ, Tlsty TD, Lees E, Harper JW, et al p53-dependent inhibition of cyclin-dependent kinase activities in human fibroblasts during radiation-induced G1 arrest. Cell. 1994;76:1013–23.
Flores JM, Martin-Caballero J, Garcia-Fernandez RA. p21 and p27 a shared senescence history. Cell Cycle. 2014;13:1655–6.
Pewzner-Jung Y, Brenner O, Braun S, Laviad EL, Ben-Dor S, Feldmesser E, et al A critical role for ceramide synthase 2 in liver homeostasis: II. insights into molecular changes leading to hepatopathy. J Biol Chem. 2010;285:10911–23.
Sankala HM, Hait NC, Paugh SW, Shida D, Lepine S, Elmore LW, et al Involvement of sphingosine kinase 2 in p53-independent induction of p21 by the chemotherapeutic drug doxorubicin. Cancer Res. 2007;67:10466–74.
Lee YS, Choi KM, Choi MH, Ji SY, Lee S, Sin DM, et al Serine palmitoyltransferase inhibitor myriocin induces growth inhibition of B16F10 melanoma cells through G(2) /M phase arrest. Cell Prolif. 2011;44:320–9.
Schnute ME, McReynolds MD, Kasten T, Yates M, Jerome G, Rains JW, et al Modulation of cellular S1P levels with a novel, potent and specific inhibitor of sphingosine kinase-1. Biochem J. 2012;444:79–88.
Yi JK, Xu R, Jeong E, Mileva I, Truman JP, Lin CL, et al Aging-related elevation of sphingoid bases shortens yeast chronological life span by compromising mitochondrial function. Oncotarget. 2016;7:21124–44.
de Arriba G, Calvino M, Benito S, Parra T. Cyclosporine A-induced apoptosis in renal tubular cells is related to oxidative damage and mitochondrial fission. Toxicol Lett. 2013;218:30–38.
Hoppins S, Lackner L, Nunnari J. The machines that divide and fuse mitochondria. Annu Rev Biochem. 2007;76:751–80.
Panjarian S, Kozhaya L, Arayssi S, Yehia M, Bielawski J, Bielawska A, et al De novo N-palmitoylsphingosine synthesis is the major biochemical mechanism of ceramide accumulation following p53 up-regulation. Prostaglandins Other Lipid Mediat. 2008;86:41–8.
Fekry B, Jeffries KA, Esmaeilneakooshkghazi A, Ogretmen B, Krupenko SA, Krupenko NI. CerS6 is a novel transcriptional target of p53 activated by non-genotoxic stress. J Biol Chem. 2016;291:16586–96.
Dbaibo GS, Pushkareva MY, Rachid RA, Alter N, Smyth MJ, Obeid LM, et al p53-dependent ceramide response to genotoxic stress. J Clin Invest. 1998;102:329–39.
Agarwal ML, Agarwal A, Taylor WR, Stark GR. p53 controls both the G2/M and the G1 cell cycle checkpoints and mediates reversible growth arrest in human fibroblasts. Proc Natl Acad Sci USA. 1995;92:8493–97.
Dulic V, Stein GH, Far DF, Reed SI. Nuclear accumulation of p21Cip1 at the onset of mitosis: a role at the G2/M-phase transition. Mol Cell Biol. 1998;18:546–57.
Campisi J, d’Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol. 2007;8:729–40.
Chen H, Chan DC. Emerging functions of mammalian mitochondrial fusion and fission. Hum Mol Genet 2005;14 Spec No. 2:R283–89.
Wu S, Zhou F, Zhang Z, Xing D. Mitochondrial oxidative stress causes mitochondrial fragmentation via differential modulation of mitochondrial fission-fusion proteins. FEBS J. 2011;278:941–54.
Suen DF, Norris KL, Youle RJ. Mitochondrial dynamics and apoptosis. Genes Dev. 2008;22:1577–90.
Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP, et al Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–501.
Clarke CJ, Mediwala K, Jenkins RW, Sutton CA, Tholanikunnel BG, Hannun YA. Neutral sphingomyelinase-2 mediates growth arrest by retinoic acid through modulation of ribosomal S6 kinase. J Biol Chem. 2011;286:21565–76.
Mao Z, Sun W, Xu R, Novgorodov S, Szulc ZM, Bielawski J, et al Alkaline ceramidase 2 (ACER2) and its product dihydrosphingosine mediate the cytotoxicity of N-(4-hydroxyphenyl)retinamide in tumor cells. J Biol Chem. 2010;285:29078–90.
Muller P, Janovjak H, Miserez A, Dobbie Z. Processing of gene expression data generated by quantitative real time RT-PCR. BioTechniques. 2002;32:1372–9.
Xu R, Sun W, Jin J, Obeid LM, Mao C. Role of alkaline ceramidases in the generation of sphingosine and its phosphate in erythrocytes. FASEB J. 2010;24:2507–5.
Sun W, Jin J, Xu R, Hu W, Szulc ZM, Bielawski J, et al Substrate specificity, membrane topology, and activity regulation of human alkaline ceramidase 2 (ACER2). J Biol Chem. 2010;285:8995–9007.
Hu W, Xu R, Sun W, Szulc ZM, Bielawski J, Obeid LM, et al Alkaline ceramidase 3 (ACER3) hydrolyzes unsaturated long-chain ceramides, and its down-regulation inhibits both cell proliferation and apoptosis. J Biol Chem. 2010;285:7964–76.
Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–4.
Acknowledgments
We thank Izolda Mileva in the Lipidomics Core Facility at Stony Brook University for performing sphingolipidomic analyzes. This work was supported, in whole or in part, by the National Institutes of Health Grants R01CA104834 (to C.M.), R01CA163825 (to C.M.), and P01CA097132 (to Y.A.H. and C.M.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interests
The authors declare that they have no competing interests.
Additional information
Edited by G Del Sal
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made.
The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
To view a copy of this licence, visit https://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Xu, R., Garcia-Barros, M., Wen, S. et al. Tumor suppressor p53 links ceramide metabolism to DNA damage response through alkaline ceramidase 2. Cell Death Differ (2017). https://doi.org/10.1038/s41418-017-0018-y
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41418-017-0018-y
This article is cited by
-
Exosomal miR-125b-5p derived from adipose-derived mesenchymal stem cells enhance diabetic hindlimb ischemia repair via targeting alkaline ceramidase 2
Journal of Nanobiotechnology (2023)
-
miR-196a-5p Correlates with Chronic Atrophic Gastritis Progression to Gastric Cancer and Induces Malignant Biological Behaviors of Gastric Cancer Cells by Targeting ACER2
Molecular Biotechnology (2023)
-
Substituting meat for mycoprotein reduces genotoxicity and increases the abundance of beneficial microbes in the gut: Mycomeat, a randomised crossover control trial
European Journal of Nutrition (2023)