Abstract
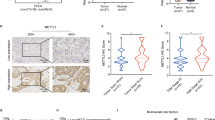
N6-methyladenosine (m6A) RNA methylation and its associated methyltransferase METTL3 play an important role in tumorigenesis of a series of tumors. However, dysregulation of METTL3 in gallbladder cancer (GBC) remains obscure. Here, we showed that upregulated METTL3 level predicted poor prognosis and correlated with increased lymphatic metastasis and high TNM stage. Functionally, we found that METTL3 could promote cell proliferation, invasion, and migration of GBC-SD and NOZ cells. Mechanistically, we revealed the METTL3-mediated m6A-modification profile in GBC cells and identified DUSP5 as the downstream gene of METTL3. METTL3 promoted the degradation of DUSP5 mRNA in a YTHDF2-dependent manner. Rescue assays showed that downregulation of DUSP5 could attenuate the knockdown METTL3-mediated inhibition of cell proliferation, invasion, and migration of GBC-SD and NOZ cells. Thus, our finding shows that elevated METTL3 expression contributes to tumor aggression in GBC, suggesting that METTL3 is a possible prognostic predictor and therapeutic target against GBC.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Misra S, Chaturvedi A, Misra NC, Sharma ID. Carcinoma of the gallbladder. Lancet Oncol. 2003;4:167–76.
Baiu I, Visser B. Gallbladder cancer. JAMA 2018;320:1294.
Lazcano-Ponce EC, Miquel JF, Muñoz N, Herrero R, Ferrecio C, Wistuba II, et al. Epidemiology and molecular pathology of gallbladder cancer. CA Cancer J Clin. 2001;51:349–64.
Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10:93–5.
Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18–29.
Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7:885–7.
Patil DP, Pickering BF, Jaffrey SR. Reading m(6)A in the Transcriptome: m(6)A-Binding Proteins. Trends Cell Biol. 2018;28:113–27.
Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014;505:117–20.
Bartosovic M, Molares HC, Gregorova P, Hrossova D, Kudla G, Vanacova S. N6-methyladenosine demethylase FTO targets pre-mRNAs and regulates alternative splicing and 3’-end processing. Nucleic Acids Res. 2017;45:11356–70.
Wang Y, Li Y, Toth JI, Petroski MD, Zhang Z, Zhao JC. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol. 2014;16:191–8.
Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, et al. 5’ UTR m(6)A promotes cap-independent translation. Cell 2015;163:999–1010.
Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, et al. N(6)-methyladenosine modulates messenger rna translation Efficiency. Cell 2015;161:1388–99.
Chen M, Wei L, Law CT, Tsang FH, Shen J, Cheng CL, et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology. 2018;67:2254–70.
Yang X, Zhang S, He C, Xue P, Zhang L, He Z, et al. METTL14 suppresses proliferation and metastasis of colorectal cancer by down-regulating oncogenic long non-coding RNA XIST. Mol Cancer. 2020;19:46.
Xie A, Hanif S, Ouyang J, Tang Z, Kong N, Kim NY, et al. Stimuli-responsive prodrug-based cancer nanomedicine. EBioMedicine 2020;56:102821.
Chen HD, Huang CS, Xu QC, Li F, Huang XT, Wang JQ, et al. Therapeutic targeting of CDK7 suppresses tumor progression in intrahepatic Cholangiocarcinoma. Int J Biol Sci. 2020;16:1207–17.
He L, Li H, Wu A, Peng Y, Shu G, Yin G. Functions of N6-methyladenosine and its role in cancer. Mol Cancer. 2019;18:176.
Wang Q, Chen C, Ding Q, Zhao Y, Wang Z, Chen J, et al. METTL3-mediated m(6)A modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut 2020;69:1193–205.
Xie H, Li J, Ying Y, Yan H, Jin K, Ma X, et al. METTL3/YTHDF2 m(6) a axis promotes tumorigenesis by degrading SETD7 and KLF4 mRNAs in bladder cancer. J Cell Mol Med. 2020;24:4092–104.
Lin S, Choe J, Du P, Triboulet R, Gregory RI. The m(6)A methyltransferase METTL3 promotes translation in human cancer cells. Mol Cell. 2016;62:335–45.
Cai X, Wang X, Cao C, Gao Y, Zhang S, Yang Z, et al. HBXIP-elevated methyltransferase METTL3 promotes the progression of breast cancer via inhibiting tumor suppressor let-7g. Cancer Lett. 2018;415:11–9.
Li F, Yi Y, Miao Y, Long W, Long T, Chen S, et al. N(6)-Methyladenosine modulates nonsense-mediated mRNA decay in human glioblastoma. Cancer Res. 2019;79:5785–98.
Vu LP, Pickering BF, Cheng Y, Zaccara S, Nguyen D, Minuesa G, et al. The N(6)-methyladenosine (m(6)A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat Med. 2017;23:1369–76.
Yankova E, Blackaby W, Albertella M, Rak J, De Braekeleer E, Tsagkogeorga G, et al. Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature 2021;593:597–601.
Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20:285–95.
Ding J, Li J, Wang H, Tian Y, Xie M, He X, et al. Long noncoding RNA CRNDE promotes colorectal cancer cell proliferation via epigenetically silencing DUSP5/CDKN1A expression. Cell Death Dis. 2017;8:e2997.
Min H, He W. Long non-coding RNA ARAP1-AS1 promotes the proliferation and migration in cervical cancer through epigenetic regulation of DUSP5. Cancer Biol Ther. 2020;21:907–14.
Du M, Zhuang Y, Tan P, Yu Z, Zhang X, Wang A. microRNA-95 knockdown inhibits epithelial-mesenchymal transition and cancer stem cell phenotype in gastric cancer cells through MAPK pathway by upregulating DUSP5. J Cell Physiol. 2020;235:944–56.
Wang R, Bao HB, Du WZ, Chen XF, Liu HL, Han DY, et al. P68 RNA helicase promotes invasion of glioma cells through negatively regulating DUSP5. Cancer Sci. 2019;110:107–17.
Wang L, Hu J, Qiu D, Gao H, Zhao W, Huang Y, et al. Dual-specificity phosphatase 5 suppresses ovarian cancer progression by inhibiting IL-33 signaling. Am J Transl Res. 2019;11:844–54.
Liu T, Sun H, Liu S, Yang Z, Li L, Yao N, et al. The suppression of DUSP5 expression correlates with paclitaxel resistance and poor prognosis in basal-like breast cancer. Int J Med Sci. 2018;15:738–47.
Zhang Q, Xing Y, Jiang S, Xu C, Zhou X, Zhang R, et al. Integrated analysis identifies DUSP5 as a novel prognostic indicator for thyroid follicular carcinoma. Thorac Cancer. 2020;11:336–45.
Author information
Authors and Affiliations
Contributions
H.C., F. L., and S. C. performed the experiments and conducted bioinformatics analysis of the sequencing data. S.C. prepared plasmids and lentiviral vectors. Z.Z. and P.G. interpreted the clinical data and revised the paper. H.C. and W.G. designed the experiments, interpreted the data, wrote the paper, and supervised the study.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Chen, HD., Li, F., Chen, S. et al. METTL3-mediated N6-methyladenosine modification of DUSP5 mRNA promotes gallbladder-cancer progression. Cancer Gene Ther 29, 1012–1020 (2022). https://doi.org/10.1038/s41417-021-00406-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41417-021-00406-5
This article is cited by
-
Exosomal long non-coding RNA TRPM2-AS promotes angiogenesis in gallbladder cancer through interacting with PABPC1 to activate NOTCH1 signaling pathway
Molecular Cancer (2024)
-
M6A RNA methylation in biliary tract cancer: the function roles and potential therapeutic implications
Cell Death Discovery (2024)
-
Methyltransferase-like proteins in cancer biology and potential therapeutic targeting
Journal of Hematology & Oncology (2023)
-
N4-acetylcytidine-dependent GLMP mRNA stabilization by NAT10 promotes head and neck squamous cell carcinoma metastasis and remodels tumor microenvironment through MAPK/ERK signaling pathway
Cell Death & Disease (2023)
-
Cancer metastasis under the magnifying glass of epigenetics and epitranscriptomics
Cancer and Metastasis Reviews (2023)