Abstract
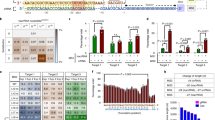
Genome editing tools targeting high-risk human papillomavirus (HPV) oncogene could be a promising therapeutic strategy for the treatment of HPV-related cervical cancer. We aimed to improve the editing efficiency and detect off-target effects concurrently for the clinical translation strategy by using CRISPR-Cas9 system co-transfected with 34nt non-homologous double-stranded oligodeoxynucleotide (dsODN). We firstly tested this strategy on targeting the Green Fluorescent Protein (GFP) gene, of which the expression is easily observed. Our results showed that the GFP+ cells were significantly decreased when using GFP-sgRNAs with dsODN, compared to using GFP-sgRNAs without donors. By PCR and Sanger sequencing, we verified the dsODN integration into the break sites of the GFP gene. And by amplicon sequencing, we observed that the indels% of the targeted site on the GFP gene was increased by using GFP-sgRNAs with dsODN. Next, we went on to target the HPV18 E7 oncogene by using single E7-sgRNA and multiplexed E7-sgRNAs respectively. Whenever using single sgRNA or multiplexed sgRNAs, the mRNA expression of HPV18 E7 oncogene was significantly decreased when adding E7-sgRNAs with dsODN, compared to E7-sgRNAs without donor. And the indels% of the targeted sites on the HPV18 E7 gene was markedly increased by adding dsODN with E7-sgRNAs. Finally, we performed GUIDE-Seq to verify that the integrated dsODN could serve as the marker to detect off-target effects in using single or multiplexed two sgRNAs. And we detected fewer on-target reads and off-target sites in multiplexes compared to the single sgRNAs when targeting the GFP and the HPV18 E7 genes. Together, CRISPR-Cas9 system co-transfected with 34nt dsODN concurrently improved the editing efficiency and monitored off-target effects, which might provide new insights in the treatment of HPV infections and related cervical cancer.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Cohen PA, Jhingran A, Oaknin A, Denny L. Cervical cancer. Lancet. 2019;393:169–82.
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424.
Crosbie EJ, Einstein MH, Franceschi S, Kitchener HC. Human papillomavirus and cervical cancer. Lancet. 2013;382:889–99.
Munoz N, Bosch FX, de Sanjose S, Herrero R, Castellsague X, Shah KV, et al. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N. Engl J Med. 2003;348:518–27.
Hu Z, Zhu D, Wang W, Li W, Jia W, Zeng X, et al. Genome-wide profiling of HPV integration in cervical cancer identifies clustered genomic hot spots and a potential microhomology-mediated integration mechanism. Nat Genet. 2015;47:158–63.
Adey A, Burton JN, Kitzman JO, Hiatt JB, Lewis AP, Martin BK, et al. The haplotype-resolved genome and epigenome of the aneuploid HeLa cancer cell line. Nature. 2013;500:207–11.
Nakamura Y, Migita T, Hosoda F, Okada N, Gotoh M, Arai Y, et al. Kruppel-like factor 12 plays a significant role in poorly differentiated gastric cancer progression. Int J Cancer. 2009;125:1859–67.
Dong Z, Yang L, Lai D. KLF5 strengthens drug resistance of ovarian cancer stem-like cells by regulating survivin expression. Cell Prolif. 2013;46:425–35.
Narisawa-Saito M, Kiyono T. Basic mechanisms of high-risk human papillomavirus-induced carcinogenesis: roles of E6 and E7 proteins. Cancer Sci. 2007;98:1505–11.
Halbert CL, Demers GW, Galloway DA. The E7 gene of human papillomavirus type 16 is sufficient for immortalization of human epithelial cells. J Virol. 1991;65:473–8.
Barbosa MS, Vass WC, Lowy DR, Schiller JT. In vitro biological activities of the E6 and E7 genes vary among human papillomaviruses of different oncogenic potential. J Virol. 1991;65:292–8.
Haurwitz RE, Jinek M, Wiedenheft B, Zhou K, Doudna JA. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science. 2010;329:1355–8.
Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–6.
Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–78.
Zhen S, Hua L, Takahashi Y, Narita S, Liu YH, Li Y. In vitro and in vivo growth suppression of human papillomavirus 16-positive cervical cancer cells by CRISPR/Cas9. Biochem Biophys Res Commun. 2014;450:1422–6.
Zhen S, Hua L, Liu YH, Gao LC, Fu J, Wan DY, et al. Harnessing the clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated Cas9 system to disrupt the hepatitis B virus. Gene Ther. 2015;22:404–12.
Ye L, Wang J, Beyer AI, Teque F, Cradick TJ, Qi Z, et al. Seamless modification of wild-type induced pluripotent stem cells to the natural CCR5Delta32 mutation confers resistance to HIV infection. Proc Natl Acad Sci USA. 2014;111:9591–6.
Hu Z, Yu L, Zhu D, Ding W, Wang X, Zhang C, et al. Disruption of HPV16-E7 by CRISPR/Cas system induces apoptosis and growth inhibition in HPV16 positive human cervical cancer cells. Biomed Res Int. 2014;2014:612823.
Yoshiba T, Saga Y, Urabe M, Uchibori R, Matsubara S, Fujiwara H, et al. CRISPR/Cas9-mediated cervical cancer treatment targeting human papillomavirus E6. Oncol Lett. 2019;17:2197–206.
Ling K, Yang L, Yang N, Chen M, Wang Y, Liang S, et al. Gene Targeting of HPV18 E6 and E7 Synchronously by Nonviral Transfection of CRISPR/Cas9 System in Cervical Cancer. Hum Gene Ther. 2020;31:297–308.
Zhen S, Lu JJ, Wang LJ, Sun XM, Zhang JQ, Li X, et al. In vitro and in vivo synergistic therapeutic effect of cisplatin with human papillomavirus16 E6/E7 CRISPR/Cas9 on cervical cancer cell line. Transl Oncol. 2016;9:498–504.
Tsai SQ, Zheng Z, Nguyen NT, Liebers M, Topkar VV, Thapar V, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol. 2015;33:187–97.
Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–23.
Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890.
Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60.
You Q, Zhong Z, Ren Q, Hassan F, Zhang Y, Zhang T. CRISPRMatch: an automatic calculation and visualization tool for high-throughput CRISPR genome-editing data analysis. Int J Biol Sci. 2018;14:858–62.
Chakravarthy A, Henderson S, Thirdborough SM, Ottensmeier CH, Su X, Lechner M, et al. Human papillomavirus drives tumor development throughout the head and neck: improved prognosis is associated with an immune response largely restricted to the oropharynx. J Clin Oncol. 2016;34:4132–41.
Mirabello L, Yeager M, Yu K, Clifford GM, Xiao Y, Zhu B, et al. HPV16 E7 genetic conservation is critical to carcinogenesis. Cell. 2017;170:1164–74. e1166.
Ding W, Hu Z, Zhu D, Jiang X, Yu L, Wang X, et al. Zinc finger nucleases targeting the human papillomavirus E7 oncogene induce E7 disruption and a transformed phenotype in HPV16/18-positive cervical cancer cells. Clin Cancer Res. 2014;20:6495–503.
Hu Z, Ding W, Zhu D, Yu L, Jiang X, Wang X, et al. TALEN-mediated targeting of HPV oncogenes ameliorates HPV-related cervical malignancy. J Clin Invest. 2015;125:425–36.
Gao X, Jin Z, Tan X, Zhang C, Zou C, Zhang W, et al. Hyperbranched poly(beta-amino ester) based polyplex nanopaticles for delivery of CRISPR/Cas9 system and treatment of HPV infection associated cervical cancer. J Control Release. 2020;321:654–68.
Ceccaldi R, Rondinelli B, D’Andrea AD. Repair pathway choices and consequences at the double-strand break. Trends Cell Biol. 2016;26:52–64.
Jasin M, Haber JE. The democratization of gene editing: Insights from site-specific cleavage and double-strand break repair. DNA Repair. 2016;44:6–16.
Pannunzio NR, Watanabe G, Lieber MR. Nonhomologous DNA end-joining for repair of DNA double-strand breaks. J Biol Chem. 2018;293:10512–23.
Xie A, Kwok A, Scully R. Role of mammalian Mre11 in classical and alternative nonhomologous end joining. Nat Struct Mol Biol. 2009;16:814–8.
Xie A, Hartlerode A, Stucki M, Odate S, Puget N, Kwok A, et al. Distinct roles of chromatin-associated proteins MDC1 and 53BP1 in mammalian double-strand break repair. Mol Cell. 2007;28:1045–57.
Guirouilh-Barbat J, Huck S, Bertrand P, Pirzio L, Desmaze C, Sabatier L, et al. Impact of the KU80 pathway on NHEJ-induced genome rearrangements in mammalian cells. Mol Cell. 2004;14:611–23.
Guirouilh-Barbat J, Rass E, Plo I, Bertrand P, Lopez BS. Defects in XRCC4 and KU80 differentially affect the joining of distal nonhomologous ends. Proc Natl Acad Sci USA. 2007;104:20902–7.
Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–5.
Acknowledgements
This work was supported by the National Science and Technology Major Project of the Ministry of science and technology of China (No. 2018ZX10301402); National Ten Thousand Plan-Young Top Talents of China; General Program of Natural Science Foundation of Guangdong Province of China (No. 2021A1515012438); the National Postdoctoral Program for Innovative Talent (No. BX20200398); the China Postdoctoral Science Foundation (No. 2020M672995); Guangdong Basic and Applied Basic Research Foundation (No.2020A1515110170); Characteristic Innovation Research Project of University Teachers (No. 2020SWYY07).
Author information
Authors and Affiliations
Contributions
Conceptualization: WF, HX, ZH; Data curation: WF, MY; Formal Analysis: WF, WX, MY, XW; Funding acquisition: ZH; Investigation: BD, KS, IH, and PD; Resources: ZH; Validation: RT, ZJ, HX; Software: ZC; Visualization: XT, BL; Writing original draft: WF; Writing – review & editing: WF, HX, ZH, JT.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Fan, W., Yu, M., Wang, X. et al. Non-homologous dsODN increases the mutagenic effects of CRISPR-Cas9 to disrupt oncogene E7 in HPV positive cells. Cancer Gene Ther 29, 758–769 (2022). https://doi.org/10.1038/s41417-021-00355-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41417-021-00355-z