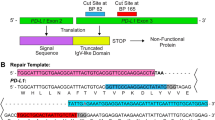
Abstract
Hepatoblastoma remains one of the most difficult childhood tumors to treat and is alarmingly understudied. We previously demonstrated that Proviral Insertion site in Maloney murine leukemia virus (PIM) kinases, specifically PIM3, are overexpressed in human hepatoblastoma cells and function to promote tumorigenesis. We aimed to use CRISPR/Cas9 gene editing with dual gRNAs to introduce large inactivating deletions in the PIM3 gene and achieve stable PIM3 knockout in the human hepatoblastoma cell line, HuH6. PIM3 knockout of hepatoblastoma cells led to significantly decreased proliferation, viability, and motility, inhibited cell-cycle progression, decreased tumor growth in a xenograft murine model, and increased animal survival. Analysis of RNA sequencing data revealed that PIM3 knockout downregulated expression of pro-migratory and pro-invasive genes and upregulated expression of genes involved in apoptosis and differentiation. Furthermore, PIM3 knockout decreased hepatoblastoma cancer cell stemness as evidenced by decreased tumorsphere formation, decreased mRNA abundance of stemness markers, and decreased cell surface expression of CD133, a marker of hepatoblastoma stem cell-like cancer cells. Reintroduction of PIM3 into PIM3 knockout cells rescued the malignant phenotype. Successful CRISPR/Cas9 knockout of PIM3 kinase in human hepatoblastoma cells confirmed the role of PIM3 in promoting hepatoblastoma tumorigenesis and cancer cell stemness.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Feng J, Polychronidis G, Heger U, Frongia G, Mehrabi A, Hoffmann K. Incidence trends and survival prediction of hepatoblastoma in children: a population-based study. Cancer Commun (Lond). 2019;39:62.
Zhang X, Song M, Kundu JK, Lee MH, Liu ZZ. PIM kinase as an executional target in cancer. J Cancer Prev. 2018;23:109–16.
Narlik-Grassow M, Blanco-Aparicio C, Carnero A. The PIM family of serine/threonine kinases in cancer. Med Res Rev. 2014;34:136–59.
Stafman LL, Mruthyunjayappa S, Waters AM, Garner EF, Aye JM, Stewart JE, et al. Targeting PIM kinase as a therapeutic strategy in human hepatoblastoma. Oncotarget. 2018;9:22665–79.
Popivanova BK, Li YY, Zheng H, Omura K, Fujii C, Tsuneyama K, et al. Proto-oncogene, Pim-3 with serine/threonine kinase activity, is aberrantly expressed in human colon cancer cells and can prevent Bad-mediated apoptosis. Cancer Sci. 2007;98:321–8.
Santio NM, Eerola SK, Paatero I, Yli-Kauhaluoma J, Anizon F, Moreau P, et al. Pim Kinases Promote Migration and Metastatic Growth of Prostate Cancer Xenografts. PLoS One. 2015;10:e0130340.
Morishita D, Katayama R, Sekimizu K, Tsuruo T, Fujita N. Pim kinases promote cell cycle progression by phosphorylating and down-regulating p27Kip1 at the transcriptional and posttranscriptional levels. Cancer Res. 2008;68:5076–85.
Stafman LL, Williams AP, Garner EF, Aye JM, Stewart JE, Yoon KJ, et al. Targeting PIM Kinases Affects Maintenance of CD133 Tumor Cell Population in Hepatoblastoma. Transl Oncol. 2019;12:200–8.
Stafman LL, Waldrop MG, Williams AP, Aye JM, Stewart JE, Mroczek-Musulman E, et al. The presence of PIM3 increases hepatoblastoma tumorigenesis and tumor initiating cell phenotype and is associated with decreased patient survival. J Pediatr Surg. 2019;54:1206–13.
Mocellin S, Provenzano M. RNA interference: learning gene knock-down from cell physiology. J Transl Med. 2004;2:39.
Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–78.
Gillory LA, Stewart JE, Megison ML, Nabers HC, Mroczek-Musulman E, Beierle EA. FAK inhibition decreases hepatoblastoma survival both in vitro and in vivo. Transl Oncol. 2013;6:206–15.
Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–308.
Stephenson FH, ScienceDirect. Calculations for molecular biology and biotechnology: a guide to mathematics in the laboratory, Chapter 3 - Cell Growth: Academic Press; 2010.
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21.
Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–78.
Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–5.
Kramer A, Green J, Pollard J Jr, Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics. 2014;30:523–30.
Cheng F, Weidner-Glunde M, Varjosalo M, Rainio EM, Lehtonen A, Schulz TF, et al. KSHV reactivation from latency requires Pim-1 and Pim-3 kinases to inactivate the latency-associated nuclear antigen LANA. PLoS Pathog. 2009;5:e1000324.
Hu Y, Smyth GK. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009;347:70–8.
Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–86.
Winer J, Jung CK, Shackel I, Williams PM. Development and validation of real-time quantitative reverse transcriptase-polymerase chain reaction for monitoring gene expression in cardiac myocytes in vitro. Anal Biochem. 1999;270:41–9.
Graham D, Magee H, Kierce B, Ball R, Dervan P, O’Meara A. Evaluation of Ki-67 reactivity in neuroblastoma using paraffin embedded tissue. Pathol Res Pr. 1995;191:87–91.
Aldinucci D, Borghese C, Casagrande N. The CCL5/CCR5 axis in cancer progression. Cancers (Basel). 2020;12:1765.
Baldin V, Lukas J, Marcote MJ, Pagano M, Draetta G. Cyclin D1 is a nuclear protein required for cell cycle progression in G1. Genes Dev. 1993;7:812–21.
Eichmann A, Yuan L, Breant C, Alitalo K, Koskinen PJ. Developmental expression of pim kinases suggests functions also outside of the hematopoietic system. Oncogene. 2000;19:1215–24.
Yan B, Yau EX, Samanta S, Ong CW, Yong KJ, Ng LK, et al. Clinical and therapeutic relevance of PIM1 kinase in gastric cancer. Gastric Cancer. 2012;15:188–97.
Weirauch U, Beckmann N, Thomas M, Grunweller A, Huber K, Bracher F, et al. Functional role and therapeutic potential of the pim-1 kinase in colon carcinoma. Neoplasia. 2013;15:783–94.
Bellon M, Lu L, Nicot C. Constitutive activation of Pim1 kinase is a therapeutic target for adult T-cell leukemia. Blood. 2016;127:2439–50.
Cohen AM, Grinblat B, Bessler H, Kristt D, Kremer A, Schwartz A, et al. Increased expression of the hPim-2 gene in human chronic lymphocytic leukemia and non-Hodgkin lymphoma. Leuk Lymphoma. 2004;45:951–5.
Li YY, Popivanova BK, Nagai Y, Ishikura H, Fujii C, Mukaida N. Pim-3, a proto-oncogene with serine/threonine kinase activity, is aberrantly expressed in human pancreatic cancer and phosphorylates bad to block bad-mediated apoptosis in human pancreatic cancer cell lines. Cancer Res. 2006;66:6741–7.
Fujii C, Nakamoto Y, Lu P, Tsuneyama K, Popivanova BK, Kaneko S, et al. Aberrant expression of serine/threonine kinase Pim-3 in hepatocellular carcinoma development and its role in the proliferation of human hepatoma cell lines. Int J Cancer. 2005;114:209–18.
Li YY, Mukaida N. Pathophysiological roles of Pim-3 kinase in pancreatic cancer development and progression. World J Gastroenterol. 2014;20:9392–404.
Mikkers H, Nawijn M, Allen J, Brouwers C, Verhoeven E, Jonkers J, et al. Mice deficient for all PIM kinases display reduced body size and impaired responses to hematopoietic growth factors. Mol Cell Biol. 2004;24:6104–15.
van der Lugt NM, Domen J, Verhoeven E, Linders K, van der Gulden H, Allen J, et al. Proviral tagging in E mu-myc transgenic mice lacking the Pim-1 proto-oncogene leads to compensatory activation of Pim-2. EMBO J. 1995;14:2536–44.
Qu Y, Zhang C, Du E, Wang A, Yang Y, Guo J, et al. Pim-3 is a critical risk factor in development and prognosis of prostate cancer. Med Sci Monit. 2016;22:4254–60.
Zhuang H, Zhao MY, Hei KW, Yang BC, Sun L, Du X, et al. Aberrant expression of pim-3 promotes proliferation and migration of ovarian cancer cells. Asian Pac J Cancer Prev. 2015;16:3325–31.
Liu B, Wang Z, Li HY, Zhang B, Ping B, Li YY. Pim-3 promotes human pancreatic cancer growth by regulating tumor vasculogenesis. Oncol Rep. 2014;31:2625–34.
Leroux AE, Biondi RM. Renaissance of allostery to disrupt protein kinase interactions. Trends Biochem Sci. 2020;45:27–41.
Nair JR, Caserta J, Belko K, Howell T, Fetterly G, Baldino C, et al. Novel inhibition of PIM2 kinase has significant anti-tumor efficacy in multiple myeloma. Leukemia. 2017;31:1715–26.
Narlik-Grassow M, Blanco-Aparicio C, Cecilia Y, Peregrina S, Garcia-Serelde B, Munoz-Galvan S, et al. The essential role of PIM kinases in sarcoma growth and bone invasion. Carcinogenesis. 2012;33:1479–86.
Garcia PD, Langowski JL, Wang Y, Chen M, Castillo J, Fanton C, et al. Pan-PIM kinase inhibition provides a novel therapy for treating hematologic cancers. Clin Cancer Res. 2014;20:1834–45.
Sharpe JJ, Cooper TA. Unexpected consequences: exon skipping caused by CRISPR-generated mutations. Genome Biol. 2017;18:109.
Joberty G, Falth-Savitski M, Paulmann M, Bosche M, Doce C, Cheng AT, et al. A tandem guide RNA-based strategy for efficient CRISPR gene editing of cell populations with low heterogeneity of edited alleles. CRISPR J. 2020;3:123–34.
Zhang XH, Tee LY, Wang XG, Huang QS, Yang SH. Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol Ther Nucleic Acids. 2015;4:e264.
Ma H, Dang Y, Wu Y, Jia G, Anaya E, Zhang J, et al. A CRISPR-based screen identifies genes essential for west-nile-virus-induced cell death. Cell Rep. 2015;12:673–83.
Heck JE, Meyers TJ, Lombardi C, Park AS, Cockburn M, Reynolds P, et al. Case-control study of birth characteristics and the risk of hepatoblastoma. Cancer Epidemiol. 2013;37:390–5.
Turusov VS, Torii M, Sills RC, Willson GA, Herbert RA, Hailey JR, et al. Hepatoblastomas in mice in the US National Toxicology Program (NTP) studies. Toxicol Pathol. 2002;30:580–91.
Horton JD, Lee S, Brown SR, Bader J, Meier DE. Survival trends in children with hepatoblastoma. Pediatr Surg Int. 2009;25:407–12.
Mertens B, Cristina de Araujo Nogueira T, Topalis D, Stranska R, Snoeck R, Andrei G. Investigation of tumor-tumor interactions in a double human cervical carcinoma xenograft model in nude mice. Oncotarget. 2018;9:21978–2000.
Wu Y, Wang YY, Nakamoto Y, Li YY, Baba T, Kaneko S, et al. Accelerated hepatocellular carcinoma development in mice expressing the Pim-3 transgene selectively in the liver. Oncogene. 2010;29:2228–37.
Chang JC. Cancer stem cells: role in tumor growth, recurrence, metastasis, and treatment resistance. Medicine (Baltim). 2016;95:S20–5.
Jensen JB, Parmar M. Strengths and limitations of the neurosphere culture system. Mol Neurobiol. 2006;34:153–61.
Villodre ES, Kipper FC, Pereira MB, Lenz G. Roles of OCT4 in tumorigenesis, cancer therapy resistance and prognosis. Cancer Treat Rev. 2016;51:1–9.
Iv Santaliz-Ruiz LE, Xie X, Old M, Teknos TN, Pan Q. Emerging role of nanog in tumorigenesis and cancer stem cells. Int J Cancer. 2014;135:2741–8.
Mamun MA, Mannoor K, Cao J, Qadri F, Song X. SOX2 in cancer stemness: tumor malignancy and therapeutic potentials. J Mol Cell Biol. 2020;12:85–98.
Neradil J, Veselska R. Nestin as a marker of cancer stem cells. Cancer Sci. 2015;106:803–11.
Bahnassy AA, Fawzy M, El-Wakil M, Zekri AR, Abdel-Sayed A, Sheta M. Aberrant expression of cancer stem cell markers (CD44, CD90, and CD133) contributes to disease progression and reduced survival in hepatoblastoma patients: 4-year survival data. Transl Res. 2015;165:396–406.
Rikhi RR, Spady KK, Hoffman RI, Bateman MS, Bateman M, Howard LE. Hepatoblastoma: a need for cell lines and tissue banks to develop targeted drug therapies. Front Pediatr. 2016;4:22.
Costantini S, Di Bernardo G, Cammarota M, Castello G, Colonna G. Gene expression signature of human HepG2 cell line. Gene. 2013;518:335–45.
Acknowledgements
We wish to thank Vidya Sagar Hanumanthu and the UAB Comprehensive Flow Cytometry Core (supported by NIH P30 AR048311 and NIH P30 AI27667), Dr. Michael Crowley and the UAB Genomics Core (supported by NIH CA013148), and Dr. Anita Hjemeland’s laboratory for their assistance with the qPCR.
Funding
This project was made possible by funding from the National Cancer Institute of the National Institutes of Health under award numbers T32 CA229102 (RM and LVB), T32 CA091078 (LLS), T32 CA183926 (APW), 5T32GM008361 (CHQ), P30 AR048311, and P30 AI027767 to the Flow Cytometry Core, and CA013148 to the UAB Genomics Core. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Other funding sources include Cannonball Kids cancer, Sid Strong Foundation, Elaine Roberts Foundation, Open Hands Overflowing Hearts, and Starr Fund-Vince Lombardi Cancer Foundation (EAB), and the Society of University Surgeons (RM).
Author information
Authors and Affiliations
Contributions
R Marayati and LL Stafman were involved in study concept and design, development of methodology, data collection, data analysis, and manuscript preparation. AP Williams, LV Bownes, CH Quinn, HR Markert, and JE Stewart contributed with data collection and analysis. JL Easlick was involved in the development of the CRISPR/Cas9 PIM3 knockout. DK Crossman performed bioinformatics analyses of the RNA sequencing data. E Mroczek-Musulman was involved in the immunohistochemistry evaluation and analysis. EA Beierle provided senior guidance with study concept and design, data analysis, and manuscript preparation.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Marayati, R., Stafman, L.L., Williams, A.P. et al. CRISPR/Cas9-mediated knockout of PIM3 suppresses tumorigenesis and cancer cell stemness in human hepatoblastoma cells. Cancer Gene Ther 29, 558–572 (2022). https://doi.org/10.1038/s41417-021-00334-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41417-021-00334-4
This article is cited by
-
BEX1 supports the stemness of hepatoblastoma by facilitating Warburg effect in a PPARγ/PDK1 dependent manner
British Journal of Cancer (2023)
-
A review on CRISPR/Cas: a versatile tool for cancer screening, diagnosis, and clinic treatment
Functional & Integrative Genomics (2023)
-
LncRNA SNHG1 Accelerates Cell Proliferation, Migration, and Invasion of Hepatoblastoma Through Mediating miR-6838-5p/PIM3/RhoA Axis
Biochemical Genetics (2023)
-
Mechanism of cancer stemness maintenance in human liver cancer
Cell Death & Disease (2022)
-
PIM3 kinase promotes tumor metastasis in hepatoblastoma by upregulating cell surface expression of chemokine receptor cxcr4
Clinical & Experimental Metastasis (2022)