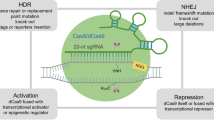
Abstract
Continued advancements in CRISPR-Cas systems have accelerated genome research. Use of CRISPR-Cas in cancer research has been of great interest that is resulting in development of orthogonal methods for drug target validations and discovery of new therapeutic targets through genome-wide screens of cancer cells. CRISPR-based screens have also revealed several new cancer drivers through alterations in tumor suppressor genes (TSGs) and oncogenes inducing resistance to targeted therapies via activation of alternate signaling pathways. Given such dynamic status of cancer, we review the application of CRISPR-Cas in non-small cell lung cancer (NSCLC) for development of mutant models, drug screening, target validation, novel target discoveries, and other emerging potential applications. In addition, CRISPR-based approach for development of novel anticancer combination therapies is also discussed in this review.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Lee W, Lee JH, Jun S, Lee JH, Bang D. Selective targeting of KRAS oncogenic alleles by CRISPR/Cas9 inhibits proliferation of cancer cells. Sci Rep. 2018;8:11879. https://doi.org/10.1038/s41598-018-30205-2
Tong Y, Whitford CM, Robertsen HL, Blin K, Jorgensen TS, Klitgaard AK, et al. Highly efficient DSB-free base editing for streptomycetes with CRISPR-BEST. Proc Natl Acad Sci USA. 2019;116:20366–75. https://doi.org/10.1073/pnas.1913493116
Kim W, Lee S, Kim HS, Song M, Cha YH, Kim YH et al. Targeting mutant KRAS with CRISPR-Cas9 controls tumor growth. Genome Res. 2018. https://doi.org/10.1101/gr.223891.117
Sachdeva M, Sachdeva N, Pal M, Gupta N, Khan IA, Majumdar M, et al. CRISPR/Cas9: molecular tool for gene therapy to target genome and epigenome in the treatment of lung cancer. Cancer Gene Ther. 2015;22:509–17. https://doi.org/10.1038/cgt.2015.54
Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI, et al. Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature. 2017;551:464–71. https://doi.org/10.1038/nature24644
Sharma, S, Petsalaki, E. Application of CRISPR-Cas9 based genome-wide screening approaches to study cellular signalling mechanisms. Int J Mol Sci. 2018;19. https://doi.org/10.3390/ijms19040933
Yin H, Xue W, Anderson DG. CRISPR-Cas: a tool for cancer research and therapeutics. Nat Rev Clin Oncol. 2019;16:281–95. https://doi.org/10.1038/s41571-019-0166-8
Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32:347–55. https://doi.org/10.1038/nbt.2842
Maruyama T, Dougan SK, Truttmann MC, Bilate AM, Ingram JR, Ploegh HL. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat Biotechnol. 2015;33:538–42. https://doi.org/10.1038/nbt.3190
Chu VT, Weber T, Wefers B, Wurst W, Sander S, Rajewsky K, et al. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat Biotechnol. 2015;33:543–8. https://doi.org/10.1038/nbt.3198
Bae T, Kim H, Kim JH, Kim YJ, Lee SH, Ham BJ et al. Specificity assessment of CRISPR genome editing of oncogenic EGFR point mutation with single-base differences. Molecules. 2019;25. https://doi.org/10.3390/molecules25010052
Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553:446–54. https://doi.org/10.1038/nature25183
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. https://doi.org/10.3322/caac.21492
Schiller JH, Harrington D, Belani CP, Langer C, Sandler A, Krook J, et al. Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N. Engl J Med. 2002;346:92–8. https://doi.org/10.1056/NEJMoa011954
Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3:75ra26. https://doi.org/10.1126/scitranslmed.3002003
Yang JC, Wu YL, Schuler M, Sebastian M, Popat S, Yamamoto N, et al. Afatinib versus cisplatin-based chemotherapy for EGFR mutation-positive lung adenocarcinoma (LUX-Lung 3 and LUX-Lung 6): analysis of overall survival data from two randomised, phase 3 trials. Lancet Oncol. 2015;16:141–51. https://doi.org/10.1016/S1470-2045(14)71173-8
Nair J, Nair A, Veerappan S, Sen D. Translatable gene therapy for lung cancer using Crispr CAS9-an exploratory review. Cancer Gene Ther. 2020;27:116–24. https://doi.org/10.1038/s41417-019-0116-8
Ng SR, Rideout WM 3rd, Akama-Garren EH, Bhutkar A, Mercer KL, Schenkel JM, et al. CRISPR-mediated modeling and functional validation of candidate tumor suppressor genes in small cell lung cancer. Proc Natl Acad Sci USA. 2020;117:513–21. https://doi.org/10.1073/pnas.1821893117
Fellmann C, Gowen BG, Lin PC, Doudna JA, Corn JE. Cornerstones of CRISPR-Cas in drug discovery and therapy. Nat Rev Drug Disco. 2017;16:89–100. https://doi.org/10.1038/nrd.2016.238
Smurnyy Y, Cai M, Wu H, McWhinnie E, Tallarico JA, Yang Y, et al. DNA sequencing and CRISPR-Cas9 gene editing for target validation in mammalian cells. Nat Chem Biol. 2014;10:623–5. https://doi.org/10.1038/nchembio.1550
Kasap C, Elemento O, Kapoor TM. DrugTargetSeqR: a genomics- and CRISPR-Cas9-based method to analyze drug targets. Nat Chem Biol. 2014;10:626–8. https://doi.org/10.1038/nchembio.1551
Aregger M, Chandrashekhar M, Tong AHY, Chan K, Moffat J. Pooled lentiviral CRISPR-Cas9 screens for functional genomics in mammalian cells. Methods Mol Biol. 2019;1869:169–88.https://doi.org/10.1007/978-1-4939-8805-1_15
Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–4. https://doi.org/10.1126/science.1246981
Koike-Yusa H, Li Y, Tan EP, Velasco-Herrera Mdel C, Yusa K. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat Biotechnol. 2014;32:267–73. https://doi.org/10.1038/nbt.2800
Zhou Y, Zhu S, Cai C, Yuan P, Li C, Huang Y, et al. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature. 2014;509:487–91. https://doi.org/10.1038/nature13166
Manguso RT, Pope HW, Zimmer MD, Brown FD, Yates KB, Miller BC, et al. In vivo CRISPR screening identifies Ptpn2 as a cancer immunotherapy target. Nature. 2017;547:413–8. https://doi.org/10.1038/nature23270
Shi J, Wang E, Milazzo JP, Wang Z, Kinney JB, Vakoc CR. Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat Biotechnol. 2015;33:661–7. https://doi.org/10.1038/nbt.3235
Pikor LA, Ramnarine VR, Lam S, Lam WL. Genetic alterations defining NSCLC subtypes and their therapeutic implications. Lung Cancer. 2013;82:179–89. https://doi.org/10.1016/j.lungcan.2013.07.025
Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, Maki RG, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693–703. https://doi.org/10.1056/NEJMoa1006448
Pao W, Chmielecki J. Rational, biologically based treatment of EGFR-mutant non-small-cell lung cancer. Nat Rev Cancer. 2010;10:760–74. https://doi.org/10.1038/nrc2947
Burgess AW. EGFR family: structure physiology signalling and therapeutic targets. Growth Factors. 2008;26:263–74. https://doi.org/10.1080/08977190802312844
Ferguson KM. Structure-based view of epidermal growth factor receptor regulation. Annu Rev Biophys. 2008;37:353–73. https://doi.org/10.1146/annurev.biophys.37.032807.125829
Oda K, Matsuoka Y, Funahashi A, Kitano H. A comprehensive pathway map of epidermal growth factor receptor signaling. Mol Syst Biol. 2005;1:2005 0010. https://doi.org/10.1038/msb4100014
Gazdar AF. Activating and resistance mutations of EGFR in non-small-cell lung cancer: role in clinical response to EGFR tyrosine kinase inhibitors. Oncogene. 2009;28:S24–31. https://doi.org/10.1038/onc.2009.198
Yun CH, Mengwasser KE, Toms AV, Woo MS, Greulich H, Wong KK, et al. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc Natl Acad Sci USA. 2008;105:2070–5. https://doi.org/10.1073/pnas.0709662105
Eck MJ, Yun CH. Structural and mechanistic underpinnings of the differential drug sensitivity of EGFR mutations in non-small cell lung cancer. Biochim Biophys Acta. 2010;1804:559–66. https://doi.org/10.1016/j.bbapap.2009.12.010
Suda K, Onozato R, Yatabe Y, Mitsudomi T. EGFR T790M mutation: a double role in lung cancer cell survival? J Thorac Oncol. 2009;4:1–4. https://doi.org/10.1097/JTO.0b013e3181913c9f
Zhou W, Ercan D, Chen L, Yun CH, Li D, Capelletti M, et al. Novel mutant-selective EGFR kinase inhibitors against EGFR T790M. Nature. 2009;462:1070–4. https://doi.org/10.1038/nature08622
Balak MN, Gong Y, Riely GJ, Somwar R, Li AR, Zakowski MF, et al. Novel D761Y and common secondary T790M mutations in epidermal growth factor receptor-mutant lung adenocarcinomas with acquired resistance to kinase inhibitors. Clin Cancer Res. 2006;12:6494–501. https://doi.org/10.1158/1078-0432.CCR-06-1570
Chu J, Galicia-Vazquez G, Cencic R, Mills JR, Katigbak A, Porco JA Jr., et al. CRISPR-mediated drug-target validation reveals selective pharmacological inhibition of the RNA helicase, eIF4A. Cell Rep. 2016;15:2340–7. https://doi.org/10.1016/j.celrep.2016.05.005
Wanzel M, Vischedyk JB, Gittler MP, Gremke N, Seiz JR, Hefter M, et al. CRISPR-Cas9-based target validation for p53-reactivating model compounds. Nat Chem Biol. 2016;12:22–8. https://doi.org/10.1038/nchembio.1965
Kurata M, Yamamoto K, Moriarity BS, Kitagawa M, Largaespada DA. CRISPR/Cas9 library screening for drug target discovery. J Hum Genet. 2018;63:179–86. https://doi.org/10.1038/s10038-017-0376-9
Ma P, Fu Y, Chen M, Jing Y, Wu J, Li K, et al. Adaptive and acquired resistance to EGFR inhibitors converge on the mapk pathway. Theranostics. 2016;6:1232–43. https://doi.org/10.7150/thno.14409
Cheung AH, Chow C, Zhang J, Zhou Y, Huang T, Ng KC, et al. Specific targeting of point mutations in EGFR L858R-positive lung cancer by CRISPR/Cas9. Lab Invest. 2018;98:968–76. https://doi.org/10.1038/s41374-018-0056-1
Park MY, Jung MH, Eo EY, Kim S, Lee SH, Lee YJ, et al. Generation of lung cancer cell lines harboring EGFR T790M mutation by CRISPR/Cas9-mediated genome editing. Oncotarget. 2017;8:36331–8. https://doi.org/10.18632/oncotarget.16752
Floc’h N, Martin MJ, Riess JW, Orme JP, Staniszewska AD, Menard L, et al. Antitumor activity of osimertinib, an irreversible mutant-selective EGFR tyrosine kinase inhibitor, in NSCLC harboring EGFR exon 20 insertions. Mol Cancer Ther. 2018;17:885–96. https://doi.org/10.1158/1535-7163.MCT-17-0758
Arcila ME, Nafa K, Chaft JE, Rekhtman N, Lau C, Reva BA, et al. EGFR exon 20 insertion mutations in lung adenocarcinomas: prevalence, molecular heterogeneity, and clinicopathologic characteristics. Mol Cancer Ther. 2013;12:220–9. https://doi.org/10.1158/1535-7163.MCT-12-0620
Yasuda H, Park E, Yun CH, Sng NJ, Lucena-Araujo AR, Yeo WL, et al. Structural, biochemical, and clinical characterization of epidermal growth factor receptor (EGFR) exon 20 insertion mutations in lung cancer. Sci Transl Med. 2013;5:216ra177. https://doi.org/10.1126/scitranslmed.3007205
Vyse S, Huang PH. Targeting EGFR exon 20 insertion mutations in non-small cell lung cancer. Signal Transduct Target Ther. 2019;4:5. https://doi.org/10.1038/s41392-019-0038-9
Yasuda H, Kobayashi S, Costa DB. EGFR exon 20 insertion mutations in non-small-cell lung cancer: preclinical data and clinical implications. Lancet Oncol. 2012;13:e23–31. https://doi.org/10.1016/S1470-2045(11)70129-2
Fang W, Huang Y, Hong S, Zhang Z, Wang M, Gan J, et al. EGFR exon 20 insertion mutations and response to osimertinib in non-small-cell lung cancer. BMC Cancer. 2019;19:595. https://doi.org/10.1186/s12885-019-5820-0
Sisdelli L, Cordioli M, Vaisman F, Moraes L, Colozza-Gama GA, Alves PAG Jr., et al. AGK-BRAF is associated with distant metastasis and younger age in pediatric papillary thyroid carcinoma. Pediatr Blood Cancer. 2019;66:e27707 https://doi.org/10.1002/pbc.27707
Ross JS, Wang K, Chmielecki J, Gay L, Johnson A, Chudnovsky J, et al. The distribution of BRAF gene fusions in solid tumors and response to targeted therapy. Int J Cancer. 2016;138:881–90. https://doi.org/10.1002/ijc.29825
Farago AF, Azzoli CG. Beyond ALK and ROS1: RET, NTRK, EGFR and BRAF gene rearrangements in non-small cell lung cancer. Transl Lung Cancer Res. 2017;6:550–9. https://doi.org/10.21037/tlcr.2017.08.02
Vad-Nielsen J, Gammelgaard KR, Daugaard TF, Nielsen AL. Cause-and-Effect relationship between FGFR1 expression and epithelial-mesenchymal transition in EGFR-mutated non-small cell lung cancer cells. Lung Cancer. 2019;132:132–40. https://doi.org/10.1016/j.lungcan.2019.04.023
Gammelgaard KR, Vad-Nielsen J, Clement MS, Weiss S, Daugaard TF, Dagnaes-Hansen F, et al. Up-regulated FGFR1 expression as a mediator of intrinsic tki resistance in EGFR-mutated NSCLC. Transl Oncol. 2019;12:432–40. https://doi.org/10.1016/j.tranon.2018.11.017
Schrock AB, Zhu VW, Hsieh WS, Madison R, Creelan B, Silberberg J, et al. Receptor tyrosine kinase fusions and BRAF kinase fusions are rare but actionable resistance mechanisms to EGFR tyrosine kinase inhibitors. J Thorac Oncol. 2018;13:1312–23. https://doi.org/10.1016/j.jtho.2018.05.027
Wegert J, Vokuhl C, Collord G, Del Castillo Velasco-Herrera M, Farndon SJ, Guzzo C, et al. Recurrent intragenic rearrangements of EGFR and BRAF in soft tissue tumors of infants. Nat Commun. 2018;9:2378. https://doi.org/10.1038/s41467-018-04650-6
Vojnic M, Kubota D, Kurzatkowski C, Offin M, Suzawa K, Benayed R, et al. Acquired BRAF rearrangements induce secondary resistance to EGFR therapy in EGFR-mutated lung cancers. J Thorac Oncol. 2019;14:802–15. https://doi.org/10.1016/j.jtho.2018.12.038
Ware KE, Hinz TK, Kleczko E, Singleton KR, Marek LA, Helfrich BA, et al. A mechanism of resistance to gefitinib mediated by cellular reprogramming and the acquisition of an FGF2-FGFR1 autocrine growth loop. Oncogenesis. 2013;2:e39. https://doi.org/10.1038/oncsis.2013.4
Zhu X, Chen L, Liu L, Niu X. EMT-mediated acquired EGFR-TKI resistance in NSCLC: mechanisms and strategies. Front Oncol. 2019;9:1044. https://doi.org/10.3389/fonc.2019.01044
Kubo T, Yamamoto H, Lockwood WW, Valencia I, Soh J, Peyton M, et al. MET gene amplification or EGFR mutation activate MET in lung cancers untreated with EGFR tyrosine kinase inhibitors. Int J Cancer. 2009;124:1778–84. https://doi.org/10.1002/ijc.24150
Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–43. https://doi.org/10.1126/science.1141478
Zhang Z, Yang S, Wang Q. Impact of MET alterations on targeted therapy with EGFR-tyrosine kinase inhibitors for EGFR-mutant lung cancer. Biomark Res. 2019;7:27 https://doi.org/10.1186/s40364-019-0179-6
Hussmann D, Madsen AT, Jakobsen KR, Luo Y, Sorensen BS, Nielsen AL. IGF1R depletion facilitates MET-amplification as mechanism of acquired resistance to erlotinib in HCC827 NSCLC cells. Oncotarget. 2017;8:33300–15. https://doi.org/10.18632/oncotarget.16350
Togashi Y, Mizuuchi H, Tomida S, Terashima M, Hayashi H, Nishio K, et al. MET gene exon 14 deletion created using the CRISPR/Cas9 system enhances cellular growth and sensitivity to a MET inhibitor. Lung Cancer. 2015;90:590–7. https://doi.org/10.1016/j.lungcan.2015.10.020
Al-Saad S, Richardsen E, Kilvaer TK, Donnem T, Andersen S, Khanehkenari M, et al. The impact of MET, IGF-1, IGF1R expression and EGFR mutations on survival of patients with non-small-cell lung cancer. PLoS ONE. 2017;12:e0181527 https://doi.org/10.1371/journal.pone.0181527
Li B, Ren S, Li X, Wang Y, Garfield D, Zhou S, et al. MiR-21 overexpression is associated with acquired resistance of EGFR-TKI in non-small cell lung cancer. Lung Cancer. 2014;83:146–53. https://doi.org/10.1016/j.lungcan.2013.11.003
Wang YS, Wang YH, Xia HP, Zhou SW, Schmid-Bindert G, Zhou CC. MicroRNA-214 regulates the acquired resistance to gefitinib via the PTEN/AKT pathway in EGFR-mutant cell lines. Asian Pac J Cancer Prev. 2012;13:255–60. https://doi.org/10.7314/apjcp.2012.13.1.255
Liu WB, Jiang X, Han F, Li YH, Chen HQ, Liu Y, et al. LHX6 acts as a novel potential tumour suppressor with epigenetic inactivation in lung cancer. Cell Death Dis. 2013;4:e882. https://doi.org/10.1038/cddis.2013.366
Liao J, Lin J, Lin D, Zou C, Kurata J, Lin R, et al. Down-regulation of miR-214 reverses erlotinib resistance in non-small-cell lung cancer through up-regulating LHX6 expression. Sci Rep. 2017;7:781. https://doi.org/10.1038/s41598-017-00901-6
Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. https://doi.org/10.1126/science.1247005
Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–78. https://doi.org/10.1016/j.cell.2014.05.010
Kiessling MK, Schuierer S, Stertz S, Beibel M, Bergling S, Knehr J, et al. Identification of oncogenic driver mutations by genome-wide CRISPR-Cas9 dropout screening. BMC Genomics. 2016;17:723. https://doi.org/10.1186/s12864-016-3042-2
Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108–12. https://doi.org/10.1038/nature08460
Ou YH, Torres M, Ram R, Formstecher E, Roland C, Cheng T, et al. TBK1 directly engages Akt/PKB survival signaling to support oncogenic transformation. Mol Cell. 2011;41:458–70. https://doi.org/10.1016/j.molcel.2011.01.019
Muvaffak A, Pan Q, Yan H, Fernandez R, Lim J, Dolinski B, et al. Evaluating TBK1 as a therapeutic target in cancers with activated IRF3. Mol Cancer Res. 2014;12:1055–66. https://doi.org/10.1158/1541-7786.MCR-13-0642
Raoof S, Mulford IJ, Frisco-Cabanos H, Nangia V, Timonina D, Labrot E, et al. Targeting FGFR overcomes EMT-mediated resistance in EGFR mutant non-small cell lung cancer. Oncogene. 2019;38:6399–413. https://doi.org/10.1038/s41388-019-0887-2
Liao S, Davoli T, Leng Y, Li MZ, Xu Q, Elledge SJ. A genetic interaction analysis identifies cancer drivers that modify EGFR dependency. Genes Dev. 2017;31:184–96. https://doi.org/10.1101/gad.291948.116
Krall EB, Wang B, Munoz DM, Ilic N, Raghavan S, Niederst MJ et al. KEAP1 loss modulates sensitivity to kinase targeted therapy in lung cancer. Elife 2017;6. https://doi.org/10.7554/eLife.18970
Kansanen E, Kuosmanen SM, Leinonen H, Levonen AL. The Keap1-Nrf2 pathway: Mechanisms of activation and dysregulation in cancer. Redox Biol. 2013;1:45–49. https://doi.org/10.1016/j.redox.2012.10.001
Mitsuishi Y, Taguchi K, Kawatani Y, Shibata T, Nukiwa T, Aburatani H, et al. Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell. 2012;22:66–79. https://doi.org/10.1016/j.ccr.2012.05.016
Baker NM, Der CJ. Cancer: drug for an ‘undruggable’ protein. Nature. 2013;497:577–8. https://doi.org/10.1038/nature12248
Srikar R, Suresh D, Zambre A, Taylor K, Chapman S, Leevy M, et al. Targeted nanoconjugate co-delivering siRNA and tyrosine kinase inhibitor to KRAS mutant NSCLC dissociates GAB1-SHP2 post oncogene knockdown. Sci Rep. 2016;6:30245. https://doi.org/10.1038/srep30245
Hallin J, Engstrom LD, Hargis L, Calinisan A, Aranda R, Briere DM, et al. The KRAS(G12C) inhibitor MRTX849 provides insight toward therapeutic susceptibility of KRAS-mutant cancers in mouse models and patients. Cancer Discov. 2020;10:54–71. https://doi.org/10.1158/2159-8290.CD-19-1167
Canon J, Rex K, Saiki AY, Mohr C, Cooke K, Bagal D, et al. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature. 2019;575:217–23. https://doi.org/10.1038/s41586-019-1694-1
Chan RJ, Feng GS. PTPN11 is the first identified proto-oncogene that encodes a tyrosine phosphatase. Blood. 2007;109:862–7. https://doi.org/10.1182/blood-2006-07-028829
Mainardi S, Mulero-Sanchez A, Prahallad A, Germano G, Bosma A, Krimpenfort P, et al. SHP2 is required for growth of KRAS-mutant non-small-cell lung cancer in vivo. Nat Med. 2018;24:961–7. https://doi.org/10.1038/s41591-018-0023-9
Gavine PR, Wang M, Yu D, Hu E, Huang C, Xia J, et al. Identification and validation of dysregulated MAPK7 (ERK5) as a novel oncogenic target in squamous cell lung and esophageal carcinoma. BMC Cancer. 2015;15:454. https://doi.org/10.1186/s12885-015-1455-y
Hayashi M, Kim SW, Imanaka-Yoshida K, Yoshida T, Abel ED, Eliceiri B, et al. Targeted deletion of BMK1/ERK5 in adult mice perturbs vascular integrity and leads to endothelial failure. J Clin Invest. 2004;113:1138–48. https://doi.org/10.1172/JCI19890
Rovida E, Spinelli E, Sdelci S, Barbetti V, Morandi A, Giuntoli S, et al. ERK5/BMK1 is indispensable for optimal colony-stimulating factor 1 (CSF-1)-induced proliferation in macrophages in a Src-dependent fashion. J Immunol. 2008;180:4166–72. https://doi.org/10.4049/jimmunol.180.6.4166
Dompe N, Klijn C, Watson SA, Leng K, Port J, Cuellar T, et al. A CRISPR screen identifies MAPK7 as a target for combination with MEK inhibition in KRAS mutant NSCLC. PLoS ONE. 2018;13:e0199264. https://doi.org/10.1371/journal.pone.0199264
Lanman BA, Allen JR, Allen JG, Amegadzie AK, Ashton KS, Booker SK, et al. Discovery of a covalent inhibitor of KRAS(G12C) (AMG 510) for the treatment of solid tumors. J Med Chem. 2020;63:52–65. https://doi.org/10.1021/acs.jmedchem.9b01180
Hong DS, Kuo J, Sacher AG, Barlesi F, Besse B, Kuboki Y, et al. CodeBreak 100: Phase I study of AMG 510, a novel KRASG12C inhibitor, in patients (pts) with advanced solid tumors other than non-small cell lung cancer (NSCLC) and colorectal cancer (CRC). J Clin Oncol. 2020;38:3511–3511. https://doi.org/10.1200/JCO.2020.38.15_suppl.3511
Wang Y, Li N, Jiang W, Deng W, Ye R, Xu C, et al. Mutant LKB1 confers enhanced radiosensitization in combination with trametinib in KRAS-mutant non-small cell lung cancer. Clin Cancer Res. 2018;24:5744–56. https://doi.org/10.1158/1078-0432.CCR-18-1489
Zhou X, Padanad MS, Evers BM, Smith B, Novaresi N, Suresh S, et al. Modulation of mutant Kras(G12D)-driven lung tumorigenesis in vivo by gain or loss of PCDH7 function. Mol Cancer Res. 2019;17:594–603. https://doi.org/10.1158/1541-7786.MCR-18-0739
Li F, Huang Q, Luster TA, Hu H, Zhang H, Ng WL, et al. In vivo epigenetic CRISPR screen identifies Asf1a as an immunotherapeutic target in Kras-mutant lung adenocarcinoma. Cancer Disco. 2020;10:270–87. https://doi.org/10.1158/2159-8290.CD-19-0780
Liu B, Song S, Setroikromo R, Chen S, Hu W, Chen D, et al. CX chemokine receptor 7 contributes to survival of KRAS-mutant non-small cell lung cancer upon loss of epidermal growth factor receptor. Cancers (Basel) 2019;11. https://doi.org/10.3390/cancers11040455
Caiola E, Falcetta F, Giordano S, Marabese M, Garassino MC, Broggini M, et al. Co-occurring KRAS mutation/LKB1 loss in non-small cell lung cancer cells results in enhanced metabolic activity susceptible to caloric restriction: an in vitro integrated multilevel approach. J Exp Clin Cancer Res. 2018;37:302. https://doi.org/10.1186/s13046-018-0954-5
Galan-Cobo A, Sitthideatphaiboon P, Qu X, Poteete A, Pisegna MA, Tong P, et al. LKB1 and KEAP1/NRF2 pathways cooperatively promote metabolic reprogramming with enhanced glutamine dependence in KRAS-mutant lung adenocarcinoma. Cancer Res. 2019;79:3251–67. https://doi.org/10.1158/0008-5472.CAN-18-3527
Romero R, Sayin VI, Davidson SM, Bauer MR, Singh SX, LeBoeuf SE, et al. Keap1 loss promotes Kras-driven lung cancer and results in dependence on glutaminolysis. Nat Med. 2017;23:1362–8. https://doi.org/10.1038/nm.4407
Bialk P, Wang Y, Banas K, Kmiec EB. Functional gene knockout of NRF2 increases chemosensitivity of human lung cancer A549 cells in vitro and in a xenograft mouse model. Mol Ther Oncolytics. 2018;11:75–89. https://doi.org/10.1016/j.omto.2018.10.002
Tang KJ, Constanzo JD, Venkateswaran N, Melegari M, Ilcheva M, Morales JC, et al. Focal adhesion kinase regulates the DNA damage response and its inhibition radiosensitizes mutant KRAS lung cancer. Clin Cancer Res. 2016;22:5851–63. https://doi.org/10.1158/1078-0432.CCR-15-2603
Anderson GR, Winter PS, Lin KH, Nussbaum DP, Cakir M, Stein EM, et al. A landscape of therapeutic cooperativity in KRAS mutant cancers reveals principles for controlling tumor evolution. Cell Rep. 2017;20:999–1015. https://doi.org/10.1016/j.celrep.2017.07.006
Mou H, Moore J, Malonia SK, Li Y, Ozata DM, Hough S, et al. Genetic disruption of oncogenic Kras sensitizes lung cancer cells to Fas receptor-mediated apoptosis. Proc Natl Acad Sci USA. 2017;114:3648–53. https://doi.org/10.1073/pnas.1620861114
Timmer T, de Vries EG, de Jong S. Fas receptor-mediated apoptosis: a clinical application. J Pathol. 2002;196:125–34. https://doi.org/10.1002/path.1028
Gulbins E, Coggeshall KM, Brenner B, Schlottmann K, Linderkamp O, Lang F. Fas-induced apoptosis is mediated by activation of a Ras and Rac protein-regulated signaling pathway. J Biol Chem. 1996;271:26389–94. https://doi.org/10.1074/jbc.271.42.26389
Wu Q, Tian Y, Zhang J, Tong X, Huang H, Li S, et al. In vivo CRISPR screening unveils histone demethylase UTX as an important epigenetic regulator in lung tumorigenesis. Proc Natl Acad Sci USA. 2018;115:E3978–E3986. https://doi.org/10.1073/pnas.1716589115
Jiang C, Lin X, Zhao Z. Applications of CRISPR/Cas9 technology in the treatment of lung cancer. Trends Mol Med. 2019;25:1039–49. https://doi.org/10.1016/j.molmed.2019.07.007
Lee AY, Cho MH, Kim S. Recent advances in aerosol gene delivery systems using non-viral vectors for lung cancer therapy. Expert Opin Drug Deliv. 2019;16:757–72. https://doi.org/10.1080/17425247.2019.1641083
Mari-Alexandre J, Diaz-Lagares A, Villalba M, Juan O, Crujeiras AB, Calvo A, et al. Translating cancer epigenomics into the clinic: focus on lung cancer. Transl Res. 2017;189:76–92. https://doi.org/10.1016/j.trsl.2017.05.008
Zuckermann M, Kawauchi D, Gronych J. Applications of the CRISPR/Cas9 system in murine cancer modeling. Brief Funct Genomics. 2017;16:25–33. https://doi.org/10.1093/bfgp/elw021
Papillon-Cavanagh S, Doshi P, Dobrin R, Szustakowski J, Walsh AM. STK11 and KEAP1 mutations as prognostic biomarkers in an observational real-world lung adenocarcinoma cohort. ESMO Open 2020;5. https://doi.org/10.1136/esmoopen-2020-000706
Bhateja P, Chiu M, Wildey G, Lipka MB, Fu P, Yang MCL, et al. Retinoblastoma mutation predicts poor outcomes in advanced non small cell lung cancer. Cancer Med. 2019;8:1459–66. https://doi.org/10.1002/cam4.2023
Hollstein PE, Eichner LJ, Brun SN, Kamireddy A, Svensson RU, Vera LI, et al. The AMPK-related kinases SIK1 and SIK3 mediate key tumor-suppressive effects of LKB1 in NSCLC. Cancer Disco. 2019;9:1606–27. https://doi.org/10.1158/2159-8290.CD-18-1261
Li F, Ng WL, Luster TA, Hu H, Sviderskiy VO, Dowling CM, et al. Epigenetic CRISPR screens identify npm1 as a therapeutic vulnerability in non-small cell lung cancer. Cancer Res. 2020;80:3556–67. https://doi.org/10.1158/0008-5472.CAN-19-3782
Gao L, Hu Y, Tian Y, Fan Z, Wang K, Li H, et al. Lung cancer deficient in the tumor suppressor GATA4 is sensitive to TGFBR1 inhibition. Nat Commun. 2019;10:1665. https://doi.org/10.1038/s41467-019-09295-7
Yan H, Chen X, Li Y, Fan L, Tai Y, Zhou Y, et al. MiR-1205 functions as a tumor suppressor by disconnecting the synergy between KRAS and MDM4/E2F1 in non-small cell lung cancer. Am J Cancer Res. 2019;9:312.
Foggetti G, Li C, Cai H, Hellyer JA, Lin W-Y, Ayeni D, et al. Genetic determinants of EGFR-driven lung cancer growth and therapeutic response in vivo. bioRxiv, 2020.2004.2013.036921, https://doi.org/10.1101/2020.04.13.036921 (2020).
Sportoletti P, Grisendi S, Majid SM, Cheng K, Clohessy JG, Viale A, et al. Npm1 is a haploinsufficient suppressor of myeloid and lymphoid malignancies in the mouse. Blood. 2008;111:3859–62. https://doi.org/10.1182/blood-2007-06-098251
Maiguel DA, Jones L, Chakravarty D, Yang C, Carrier F. Nucleophosmin sets a threshold for p53 response to UV radiation. Mol Cell Biol. 2004;24:3703–11. https://doi.org/10.1128/mcb.24.9.3703-3711.2004
Gaj T, Perez-Pinera P. The continuously evolving CRISPR barcoding toolbox. Genome Biol. 2018;19:143 https://doi.org/10.1186/s13059-018-1541-y
Kalhor R, Mali P, Church GM. Rapidly evolving homing CRISPR barcodes. Nat Methods. 2017;14:195–200. https://doi.org/10.1038/nmeth.4108
Wong AS, Choi GC, Cui CH, Pregernig G, Milani P, Adam M, et al. Multiplexed barcoded CRISPR-Cas9 screening enabled by CombiGEM. Proc Natl Acad Sci USA. 2016;113:2544–9. https://doi.org/10.1073/pnas.1517883113
Guernet A, Mungamuri SK, Cartier D, Sachidanandam R, Jayaprakash A, Adriouch S, et al. CRISPR-barcoding for intratumor genetic heterogeneity modeling and functional analysis of oncogenic driver mutations. Mol Cell. 2016;63:526–38. https://doi.org/10.1016/j.molcel.2016.06.017
Meyers RM, Bryan JG, McFarland JM, Weir BA, Sizemore AE, Xu H, et al. Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat Genet. 2017;49:1779–84. https://doi.org/10.1038/ng.3984
Han HS, Eom DW, Kim JH, Kim KH, Shin HM, An JY, et al. EGFR mutation status in primary lung adenocarcinomas and corresponding metastatic lesions: discordance in pleural metastases. Clin Lung Cancer. 2011;12:380–6. https://doi.org/10.1016/j.cllc.2011.02.006
Chang YL, Wu CT, Shih JY, Lee YC. Comparison of p53 and epidermal growth factor receptor gene status between primary tumors and lymph node metastases in non-small cell lung cancers. Ann Surg Oncol. 2011;18:543–50. https://doi.org/10.1245/s10434-010-1295-6
Marchetti A, Grammastro Del, Felicioni M, Malatesta L, Filice S, Centi G. et al. Assessment of EGFR mutations in circulating tumor cell preparations from NSCLC patients by next generation sequencing: toward a real-time liquid biopsy for treatment. PLoS ONE. 2014;9:e103883. https://doi.org/10.1371/journal.pone.0103883
Maheswaran S, Sequist LV, Nagrath S, Ulkus L, Brannigan B, Collura CV, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N. Engl J Med. 2008;359:366–77. https://doi.org/10.1056/NEJMoa0800668
De Luca F, Rotunno G, Salvianti F, Galardi F, Pestrin M, Gabellini S, et al. Mutational analysis of single circulating tumor cells by next generation sequencing in metastatic breast cancer. Oncotarget. 2016;7:26107–19. https://doi.org/10.18632/oncotarget.8431
Macias M, Alegre E, Diaz-Lagares A, Patino A, Perez-Gracia JL, Sanmamed M, et al. Liquid biopsy: from basic research to clinical practice. Adv Clin Chem. 2018;83:73–119. https://doi.org/10.1016/bs.acc.2017.10.003
Hennigan ST, Trostel SY, Terrigino NT, Voznesensky OS, Schaefer RJ, Whitlock NC, et al. Low abundance of circulating tumor DNA in localized prostate cancer. JCO Precis Oncol. 2019.;3. https://doi.org/10.1200/PO.19.00176
Gu J, Zang W, Liu B, Li L, Huang L, Li S, et al. Evaluation of digital PCR for detecting low-level EGFR mutations in advanced lung adenocarcinoma patients: a cross-platform comparison study. Oncotarget. 2017;8:67810–20. https://doi.org/10.18632/oncotarget.18866
Lee SH, Yu J, Hwang GH, Kim S, Kim HS, Ye S, et al. CUT-PCR: CRISPR-mediated, ultrasensitive detection of target DNA using PCR. Oncogene. 2017;36:6823–9. https://doi.org/10.1038/onc.2017.281
Tsou JH, Leng Q, Jiang F. A CRISPR test for rapidly and sensitively detecting circulating EGFR mutations. i 2020.;10. https://doi.org/10.3390/diagnostics10020114
Aalipour A, Dudley JC, Park SM, Murty S, Chabon JJ, Boyle EA, et al. Deactivated CRISPR Associated Protein 9 for Minor-Allele Enrichment in Cell-Free DNA. Clin Chem. 2018;64:307–16. https://doi.org/10.1373/clinchem.2017.278911
Kim SM, Shin SC, Kim EE, Kim SH, Park K, Oh SJ, et al. Simple in vivo gene editing via direct self-assembly of Cas9 ribonucleoprotein complexes for cancer treatment. ACS Nano. 2018;12:7750–60. https://doi.org/10.1021/acsnano.8b01670
Maddalo D, Manchado E, Concepcion CP, Bonetti C, Vidigal JA, Han YC, et al. In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature. 2014;516:423–7. https://doi.org/10.1038/nature13902
Platt RJ, Chen S, Zhou Y, Yim MJ, Swiech L, Kempton HR, et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159:440–55. https://doi.org/10.1016/j.cell.2014.09.014
Sanchez-Rivera FJ, Papagiannakopoulos T, Romero R, Tammela T, Bauer MR, Bhutkar A, et al. Rapid modelling of cooperating genetic events in cancer through somatic genome editing. Nature. 2014;516:428–31. https://doi.org/10.1038/nature13906
Blasco RB, Karaca E, Ambrogio C, Cheong TC, Karayol E, Minero VG, et al. Simple and rapid in vivo generation of chromosomal rearrangements using CRISPR/Cas9 technology. Cell Rep. 2014;9:1219–27. https://doi.org/10.1016/j.celrep.2014.10.051
Wilbie D, Walther J, Mastrobattista E. Delivery aspects of CRISPR/Cas for in vivo genome editing. Acc Chem Res. 2019;52:1555–64. https://doi.org/10.1021/acs.accounts.9b00106
Mout R, Ray M, Lee YW, Scaletti F, Rotello VM. In vivo delivery of CRISPR/Cas9 for therapeutic gene editing: progress and challenges. Bioconjug Chem. 2017;28:880–4. https://doi.org/10.1021/acs.bioconjchem.7b00057
El Achi H, Khoury JD, Loghavi S. Liquid biopsy by next-generation sequencing: a multimodality test for management of cancer. Curr Hematol Malig Rep. 2019;14:358–67. https://doi.org/10.1007/s11899-019-00532-w
Nambiar TS, Billon P, Diedenhofen G, Hayward SB, Taglialatela A, Cai K, et al. Stimulation of CRISPR-mediated homology-directed repair by an engineered RAD18 variant. Nat Commun. 2019;10:3395. https://doi.org/10.1038/s41467-019-11105-z
Acknowledgements
We thank Jatin Vimal for his insightful suggestions. We are grateful to Levim Biotech LLP for its support in fostering industry-academia collaborations and furthering fundamental research in the field of genetic engineering.
Author information
Authors and Affiliations
Contributions
All authors researched data, performed extensive literature survey, and provided substantial contribution to the content of this article equally. All authors reviewed and edited the manuscript prior to submission.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Sreedurgalakshmi, K., Srikar, R. & Rajkumari, R. CRISPR-Cas deployment in non-small cell lung cancer for target screening, validations, and discoveries. Cancer Gene Ther 28, 566–580 (2021). https://doi.org/10.1038/s41417-020-00256-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41417-020-00256-7
This article is cited by
-
Modern therapies of nonsmall cell lung cancer
Journal of Applied Genetics (2023)