Abstract
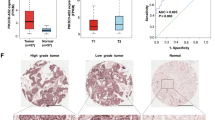
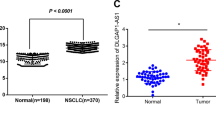
Emerging evidence indicates that the incidence of nasopharyngeal carcinoma (NPC) remains high in endemic regions despite changing environmental factors, suggesting that genetic traits contribute to its development. Recently, long non-coding RNA-microRNA-messenger RNA (lncRNA-miRNA-mRNA) axis has been reported to be implicated in the pathophysiological processes of malignancies. Moreover, initial bioinformatic analysis revealed a highly expressed lncRNA Forkhead box D3 antisense RNA1 (FOXD3-AS1) for mechanistic network underlying NPC in this present study. Therefore, this study aims to delineate the ability of lncRNA FOXD3-AS1 to influence the NPC progression. The relationship among lncRNA FOXD3-AS1, miR-185-3p, and FOXD3 was identified with bioinformatics prediction, dual-luciferase reporter gene assays, RNA-binding protein immunoprecipitation, and RNA pull-down assays. Furthermore, effects of lncRNA FOXD3-AS1 on malignant phenotypes in vitro, alongside tumor formation in vivo, of transfected NPC stem-like cells were examined with gain- and loss-of-function experiments. Our findings revealed that lncRNA FOXD3-AS1 and FOXD3 exhibited increased expression levels, while miR-185-3p exhibited diminished levels in NPC. The levels of lncRNA FOXD3-AS1 and FOXD3 were further correlated with tumor node metastasis stage and pathological type of patients with NPC. LncRNA FOXD3-AS1 was also confirmed to negatively regulate the miR-185-3p expression, which further targeted the downstream gene FOXD3. In addition, lncRNA FOXD3-AS1 knockdown repressed cell stemness, colony formation, viability, invasion, migration, and in vivo tumor growth, and accelerated cell apoptosis. Moreover, FOXD3 silencing or miR-185-3p overexpression reversed the effects of lncRNA FOXD3-AS1. Our findings provide evidence indicating that lncRNA FOXD3-AS1 could bind to miR-185-3p to upregulate the FOXD3 expression, thereby promoting the development of NPC.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout











Similar content being viewed by others
References
Gong Z, Yang Q, Zeng Z, Zhang W, Li X, Zu X, et al. An integrative transcriptomic analysis reveals p53 regulated miRNA, mRNA, and lncRNA networks in nasopharyngeal carcinoma. Tumour Biol. 2016;37:3683–95.
Razak AR, Siu LL, Liu FF, Ito E, O’Sullivan B, Chan K. Nasopharyngeal carcinoma: the next challenges. Eur J Cancer. 2010;46:1967–78.
Bei JX, Li Y, Jia WH, Feng BJ, Zhou G, Chen LZ, et al. A genome-wide association study of nasopharyngeal carcinoma identifies three new susceptibility loci. Nat Genet. 2010;42:599–603.
Su J, Xu XH, Huang Q, Lu MQ, Li DJ, Xue F, et al. Identification of cancer stem-like CD44+ cells in human nasopharyngeal carcinoma cell line. Arch Med Res. 2011;42:15–21.
Guo Y, Chen JX, Yang S, Fu XP, Zhang Z, Chen KH, et al. Selection of reliable reference genes for gene expression study in nasopharyngeal carcinoma. Acta Pharmacol Sin. 2010;31:1487–94.
Nie Y, Liu X, Qu S, Song E, Zou H, Gong C. Long non-coding RNA HOTAIR is an independent prognostic marker for nasopharyngeal carcinoma progression and survival. Cancer Sci. 2013;104:458–64.
Feng XP, Yi H, Li MY, Li XH, Yi B, Zhang PF, et al. Identification of biomarkers for predicting nasopharyngeal carcinoma response to radiotherapy by proteomics. Cancer Res. 2010;70:3450–62.
Qi X, Zhang DH, Wu N, Xiao JH, Wang X, Ma W. ceRNA in cancer: possible functions and clinical implications. J Med Genet. 2015;52:710–8.
Yang X, Song JH, Cheng Y, Wu W, Bhagat T, Yu Y, et al. Long non-coding RNA HNF1A-AS1 regulates proliferation and migration in oesophageal adenocarcinoma cells. Gut. 2014;63:881–90.
Wang A, Meng M, Zhao X, Kong L. Long non-coding RNA ENST00462717 suppresses the proliferation, survival, and migration by inhibiting MDM2/MAPK pathway in glioma. Biochem Biophys Res Commun. 2017;485:513–21.
Sun J, Chen X, Wang Z, Guo M, Shi H, Wang X, et al. A potential prognostic long non-coding RNA signature to predict metastasis-free survival of breast cancer patients. Sci Rep. 2015;5:16553.
Xie X, Xiong G, Chen W, Fu H, Li M, Cui X. FOXD3 inhibits cell proliferation, migration and invasion in nasopharyngeal carcinoma through regulation of the PI3K/Akt pathway. Biochem Cell Biol 2020;27:1–8.
Lu J, He ML, Wang L, Chen Y, Liu X, Dong Q, et al. MiR-26a inhibits cell growth and tumorigenesis of nasopharyngeal carcinoma through repression of EZH2. Cancer Res. 2011;71:225–33.
Liu N, Chen NY, Cui RX, Li WF, Li Y, Wei RR, et al. Prognostic value of a microRNA signature in nasopharyngeal carcinoma: a microRNA expression analysis. Lancet Oncol. 2012;13:633–41.
Li G, Wang Y, Liu Y, Su Z, Liu C, Ren S, et al. miR-185-3p regulates nasopharyngeal carcinoma radioresistance by targeting WNT2B in vitro. Cancer Sci. 2014;105:1560–8.
Li J, Jiang Z, Han F, Liu S, Yuan X, Tong J. FOXO4 and FOXD3 are predictive of prognosis in gastric carcinoma patients. Oncotarget. 2016;7:25585–92.
Abel EV, Basile KJ, Kugel CH 3rd, Witkiewicz AK, Le K, Amaravadi RK, et al. Melanoma adapts to RAF/MEK inhibitors through FOXD3-mediated upregulation of ERBB3. J Clin Invest. 2013;123:2155–68.
Liu LL, Lu SX, Li M, Li LZ, Fu J, Hu W, et al. FoxD3-regulated microRNA-137 suppresses tumour growth and metastasis in human hepatocellular carcinoma by targeting AKT2. Oncotarget. 2014;5:5113–24.
Fujita A, Sato JR, Rodrigues Lde O, Ferreira CE, Sogayar MC. Evaluating different methods of microarray data normalization. BMC Bioinform. 2006;7:469.
Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3.
Luo W, Li S, Peng B, Ye Y, Deng X, Yao K. Embryonic stem cells markers SOX2, OCT4 and Nanog expression and their correlations with epithelial-mesenchymal transition in nasopharyngeal carcinoma. PLoS ONE. 2013;8:e56324.
Wei P, Niu M, Pan S, Zhou Y, Shuai C, Wang J, et al. Cancer stem-like cell: a novel target for nasopharyngeal carcinoma therapy. Stem Cell Res Ther. 2014;5:44.
Pellegrino G, Trubert C, Terrien J, Pifferi F, Leroy D, Loyens A, et al. A comparative study of the neural stem cell niche in the adult hypothalamus of human, mouse, rat and gray mouse lemur (Microcebus murinus). J Comp Neurol. 2018;526:1419–43.
Luo W, Yao K. Molecular characterization and clinical implications of spindle cells in nasopharyngeal carcinoma: a novel molecule-morphology model of tumor progression proposed. PLoS ONE. 2013;8:e83135.
Jia R, Chai P, Wang S, Sun B, Xu Y, Yang Y, et al. m(6)A modification suppresses ocular melanoma through modulating HINT2 mRNA translation. Mol Cancer. 2019;18:161.
Li B, Chen P, Qu J, Shi L, Zhuang W, Fu J, et al. Activation of LTBP3 gene by a long noncoding RNA (lncRNA) MALAT1 transcript in mesenchymal stem cells from multiple myeloma. J Biol Chem. 2014;289:29365–75.
Lu Y, Li T, Wei G, Liu L, Chen Q, Xu L, et al. The long non-coding RNA NEAT1 regulates epithelial to mesenchymal transition and radioresistance in through miR-204/ZEB1 axis in nasopharyngeal carcinoma. Tumour Biol. 2016;37:11733–41.
Liu XH, Sun M, Nie FQ, Ge YB, Zhang EB, Yin DD, et al. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer. 2014;13:92.
Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF, et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666–81.
Yang H, Liu P, Zhang J, Peng X, Lu Z, Yu S, et al. Long noncoding RNA MIR31HG exhibits oncogenic property in pancreatic ductal adenocarcinoma and is negatively regulated by miR-193b. Oncogene. 2016;35:3647–57.
Zhang D, Lee H, Haspel JA, Jin Y. Long noncoding RNA FOXD3-AS1 regulates oxidative stress-induced apoptosis via sponging microRNA-150. FASEB J. 2017;31:4472–81.
Chen ZH, Hu HK, Zhang CR, Lu CY, Bao Y, Cai Z, et al. Down-regulation of long non-coding RNA FOXD3 antisense RNA 1 (FOXD3-AS1) inhibits cell proliferation, migration, and invasion in malignant glioma cells. Am J Transl Res. 2016;8:4106–19.
Acknowledgements
The authors would like to acknowledge the helpful suggestions concerning this study received from their colleagues. This work was supported by Key Projects of Natural Science Foundation of Hubei Province in 2015 (No. 2015CFA076).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Hu, J., Pan, J., Luo, Z. et al. Long non-coding RNA FOXD3-AS1 silencing exerts tumor suppressive effects in nasopharyngeal carcinoma by downregulating FOXD3 expression via microRNA-185-3p upregulation. Cancer Gene Ther 28, 602–618 (2021). https://doi.org/10.1038/s41417-020-00242-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41417-020-00242-z
This article is cited by
-
Precision medicine in nasopharyngeal carcinoma: comprehensive review of past, present, and future prospect
Journal of Translational Medicine (2023)
-
LncRNA FOXD3-AS1 promotes breast cancer progression by mediating ARF6
Breast Cancer (2022)
-
Long non-coding RNAs in nasopharyngeal carcinoma: biological functions and clinical applications
Molecular and Cellular Biochemistry (2021)