Abstract
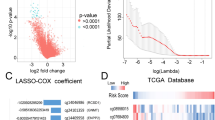
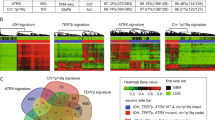
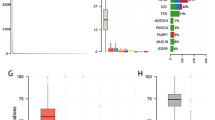
DNA methylation is an important regulator of gene expression, and plays a significant role in carcinogenesis in the brain. Here, we explored specific prognosis-subtypes based on DNA methylation status using 138 Glioblastoma Multiforme (GBM) samples from The Cancer Genome Atlas (TCGA) database. The methylation profiles of 11,637 CpG sites that significantly correlated with survival in the training set were employed for consensus clustering. We identified three GBM molecular subtypes, and their survival curves were distinct from each other. Furthermore, ten feature CpG sites were obtained on conducting a weighted gene co-expression network analysis (WGCNA) of the CpG sites. We were able to classify the samples into high- and low-methylation groups, and classified the prognosis information of the samples after cluster analysis of the training set samples using the hierarchical clustering algorithm. Similar results were obtained in the test set and clinical GBM specimens. Finally, we found that a positive relationship existed between methylation level and sensitivity to temozolomide (or radiotherapy) or anti-migration ability of GBM cells. Taken together, these results suggest that the model constructed in this study could help explain the heterogeneity of previous molecular subgroups in GBM and can provide guidance to clinicians regarding the prognosis of GBM.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Friedmann-Morvinski D. Glioblastoma heterogeneity and cancer cell plasticity. Crit Rev Oncog. 2014;19:327–36.
Lee E, Yong RL, Paddison P, Zhu J. Comparison of glioblastoma (GBM) molecular classification methods. Semin Cancer Biol. 2018;53:201–11.
Crespo I, Vital AL, Gonzalez-Tablas M, Patino Mdel C, Otero A, Lopes MC, et al. Molecular and genomic alterations in glioblastoma multiforme. Am J Pathol. 2015;185:1820–33.
Hill C, Hunter SB, Brat DJ. Genetic markers in glioblastoma: prognostic significance and future therapeutic implications. Adv Anat Pathol. 2003;10:212–7.
Lieberman F. Glioblastoma update: molecular biology, diagnosis, treatment, response assessment, and translational clinical trials. F1000Res 2017;6:1892.
Sasmita AO, Wong YP, Ling APK. Biomarkers and therapeutic advances in glioblastoma multiforme. Asia Pac J Clin Oncol. 2018;14:40–51.
Han TS, Ban HS, Hur K, Cho HS. The epigenetic regulation of HCC metastasis. Int J Mol Sci. 2018;19:E3978.
Porten SP. Epigenetic alterations in bladder cancer. Curr Urol Rep. 2018;19:102.
Romani M, Pistillo MP, Banelli B. Epigenetic targeting of glioblastoma. Front Oncol. 2018;8:448.
Muhammad JS, Khan MR, Ghias K. DNA methylation as an epigenetic regulator of gallbladder cancer: an overview. Int J Surg. 2018;53:178–83.
Yamashita K, Hosoda K, Nishizawa N, Katoh H, Watanabe M. Epigenetic biomarkers of promoter DNA methylation in the new era of cancer treatment. Cancer Sci. 2018;109:3695–706.
Koch A, Joosten SC, Feng Z, de Ruijter TC, Draht MX, Melotte V, et al. Author correction: analysis of DNA methylation in cancer: location revisited. Nat Rev Clin Oncol. 2018;15:467.
Gustafsson JR, Katsioudi G, Degn M, Ejlerskov P, Issazadeh-Navikas S, Kornum BR. DNMT1 regulates expression of MHC class I in post-mitotic neurons. Mol Brain. 2018;11:36.
Li Z, Takenobu H, Setyawati AN, Akita N, Haruta M, Satoh S, et al. EZH2 regulates neuroblastoma cell differentiation via NTRK1 promoter epigenetic modifications. Oncogene. 2018;37:2714–27.
Perez RF, Tejedor JR, Bayon GF, Fernandez AF, Fraga MF. Distinct chromatin signatures of DNA hypomethylation in aging and cancer. Aging Cell. 2018;17:e12744.
Kim DS, Lee WK, Park JY. Promoter methylation of Wrap53alpha, an antisense transcript ofp53, is associated with the poor prognosis of patients with non-small cell lung cancer. Oncol Lett. 2018;16:5823–8.
Li Y, Gong Y, Ning X, Peng D, Liu L, He S, et al. Downregulation of CLDN7 due to promoter hypermethylation is associated with human clear cell renal cell carcinoma progression and poor prognosis. J Exp Clin Cancer Res. 2018;37:276.
Hao X, Luo H, Krawczyk M, Wei W, Wang W, Wang J, et al. DNA methylation markers for diagnosis and prognosis of common cancers. Proc Natl Acad Sci USA. 2017;114:7414–9.
Bell EH, Zhang P, Fisher BJ, Macdonald DR, McElroy JP, Lesser GJ, et al. Association of MGMT promoter methylation status with survival outcomes in patients with high-risk glioma treated with radiotherapy and temozolomide: an analysis from the NRG oncology/RTOG 0424 Trial. JAMA Oncol. 2018;4:1405–9.
Johannessen LE, Brandal P, Myklebust TA, Heim S, Micci F, Panagopoulos I. MGMT gene promoter methylation status—assessment of two pyrosequencing kits and three methylation-specific PCR methods for their predictive capacity in glioblastomas. Cancer Genomics Proteom. 2018;15:437–46.
Kyrochristos ID, Ziogas DE, Roukos DH. Dynamic genome and transcriptional network-based biomarkers and drugs: precision in breast cancer therapy. Med Res Rev. 2019;39:1205–27.
Liu S, Xu C, Zhang Y, Liu J, Yu B, Liu X, et al. Feature selection of gene expression data for Cancer classification using double RBF-kernels. BMC Bioinf. 2018;19:396.
Xiao M, Liu L, Zhang S, Yang X, Wang Y. Cancer stem cell biomarkers for head and neck squamous cell carcinoma: a bioinformatic analysis. Oncol Rep. 2018;40:3843–51.
Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, Ellrott K, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45:1113–20.
Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The SVA package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 2012;28:882–3.
Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics 2010;26:1572–3.
Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–97.
Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 2008;9:559.
Aquilanti E, Miller J, Santagata S, Cahill DP, Brastianos PK. Updates in prognostic markers for gliomas. Neuro Oncol 2018;20:vii17–vii26.
Kumar R, Liu APY, Orr BA, Northcott PA, Robinson GW. Advances in the classification of pediatric brain tumors through DNA methylation profiling: from research tool to frontline diagnostic. Cancer 2018;124:4168–80.
Villanueva A, Portela A, Sayols S, Battiston C, Hoshida Y, Mendez-Gonzalez J, et al. DNA methylation-based prognosis and epidrivers in hepatocellular carcinoma. Hepatology 2015;61:1945–56.
Stefansson OA, Moran S, Gomez A, Sayols S, Arribas-Jorba C, Sandoval J, et al. A DNA methylation-based definition of biologically distinct breast cancer subtypes. Mol Oncol. 2015;9:555–68.
Exner R, Pulverer W, Diem M, Spaller L, Woltering L, Schreiber M, et al. Potential of DNA methylation in rectal cancer as diagnostic and prognostic biomarkers. Br J Cancer 2015;113:1035–45.
Ceccarelli M, Barthel FP, Malta TM, Sabedot TS, Salama SR, Murray BA, et al. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell 2016;164:550–63.
de Souza CF, Sabedot TS, Malta TM, Stetson L, Morozova O, Sokolov A, et al. A distinct DNA methylation shift in a subset of glioma CpG island methylator phenotypes during tumor recurrence. Cell Rep. 2018;23:637–51.
Leonhardt H, Cardoso MC. DNA methylation, nuclear structure, gene expression and cancer. J Cell Biochem Suppl. 2000;Suppl 35:78–83.
Rountree MR, Bachman KE, Herman JG, Baylin SB. DNA methylation, chromatin inheritance, and cancer. Oncogene 2001;20:3156–65.
Kawano H, Saeki H, Kitao H, Tsuda Y, Otsu H, Ando K, et al. Chromosomal instability associated with global DNA hypomethylation is associated with the initiation and progression of esophageal squamous cell carcinoma. Ann Surg Oncol. 2014;21(Suppl 4):S696–702.
El-Osta A. The rise and fall of genomic methylation in cancer. Leukemia. 2004;18:233–7.
Issa JP. DNA methylation as a therapeutic target in cancer. Clin Cancer Res. 2007;13:1634–7.
Flavahan WA, Drier Y, Liau BB, Gillespie SM, Venteicher AS, Stemmer-Rachamimov AO, et al. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature. 2016;529:110–4.
Hashimoto Y, Zumwalt TJ, Goel A. DNA methylation patterns as noninvasive biomarkers and targets of epigenetic therapies in colorectal cancer. Epigenomics. 2016;8:685–703.
Kaminska K, Nalejska E, Kubiak M, Wojtysiak J, Zolna L, Kowalewski J, et al. Prognostic and predictive epigenetic biomarkers in oncology. Mol Diagn Ther. 2019;23:83–95.
Zhu X, Wang Y, Tan L, Fu X. The pivotal role of DNA methylation in the radio-sensitivity of tumor radiotherapy. Cancer Med. 2018;7:3812–9.
Paul Y, Mondal B, Patil V, Somasundaram K. DNA methylation signatures for 2016 WHO classification subtypes of diffuse gliomas. Clin Epigenetics. 2017;9:32.
Yin AA, Lu N, Etcheverry A, Aubry M, Barnholtz-Sloan J, Zhang LH, et al. A novel prognostic six-CpG signature in glioblastomas. CNS Neurosci Ther. 2018;24:167–77.
Kloosterhof NK, de Rooi JJ, Kros M, Eilers PH, Sillevis Smitt PA, van den Bent MJ, et al. Molecular subtypes of glioma identified by genome-wide methylation profiling. Genes Chromosomes Cancer. 2013;52:665–74.
Wu X, Rauch TA, Zhong X, Bennett WP, Latif F, Krex D, et al. CpG island hypermethylation in human astrocytomas. Cancer Res. 2010;70:2718–27.
Yu ZQ, Zhang BL, Ren QX, Wang JC, Yu RT, Qu DW, et al. Changes in transcriptional factor binding capacity resulting from promoter region methylation induce aberrantly high GDNF expression in human glioma. Mol Neurobiol. 2013;48:571–80.
Pangeni RP, Zhang Z, Alvarez AA, Wan X, Sastry N, Lu S, et al. Genome-wide methylomic and transcriptomic analyses identify subtype-specific epigenetic signatures commonly dysregulated in glioma stem cells and glioblastoma. Epigenetics. 2018;13:432–48.
Wenger A, Ferreyra Vega S, Kling T, Bontell TO, Jakola AS, Caren H. Intratumor DNA methylation heterogeneity in glioblastoma: implications for DNA methylation-based classification. Neuro Oncol. 2019;21:616–27.
Acknowledgements
This research was supported by the National Natural Science Foundation of China (81872066, 31571433, 81773131, and 81972635), Innovative Program of Development Foundation of Hefei Center for Physical Science and Technology (2018CXFX004 and 2017FXCX008), the CASHIPS Director’s Fund (YZJJ201704) and the Youth Innovation Promotion Association of the Chinese Academy of Sciences (2018487).
Author information
Authors and Affiliations
Contributions
XRC and LJW conceived and designed the experiments. HHM, CGZ, ZYZ, and LZH collected the data. HHM, CGZ, and FY performed the analysis. XRC, LJW, HZW, and ZYF participated in the discussion of the algorithm. HHM and XRC prepared and edited the paper. All authors have read and approved the final paper.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics statement
The protocol for this article was approved by the Institutional Review Board of Hefei Institutes of Physical Science, CAS.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ma, H., Zhao, C., Zhao, Z. et al. Specific glioblastoma multiforme prognostic-subtype distinctions based on DNA methylation patterns. Cancer Gene Ther 27, 702–714 (2020). https://doi.org/10.1038/s41417-019-0142-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41417-019-0142-6
This article is cited by
-
DNA methylation-based subtypes of acute myeloid leukemia with distinct prognosis and clinical features
Clinical and Experimental Medicine (2023)
-
Systematic transcriptome profiling of pyroptosis related signature for predicting prognosis and immune landscape in lower grade glioma
BMC Cancer (2022)
-
Bisulfite profiling of the MGMT promoter and comparison with routine testing in glioblastoma diagnostics
Clinical Epigenetics (2022)
-
Genome-wide profiling of alternative splicing in glioblastoma and their clinical value
BMC Cancer (2021)
-
Perspective of mesenchymal transformation in glioblastoma
Acta Neuropathologica Communications (2021)